Abstract
Genotyping of cytomegalovirus (CMV) is useful to examine potential differences in the pathogenicity of strains and to demonstrate coinfection with multiple strains involved in CMV disease in adults and congenitally infected newborns. Studies on genotyping of CMV in dried blood spots (DBS) are rare and have been hampered by the small amount of dried blood available. In this study, two multiplex real-time PCR assays for rapid gB and gH genotyping of CMV in DBS were developed. Validation of the assays with 39 CMV-positive plasma samples of transplant recipients and 21 urine specimens of congenitally infected newborns was successful in genotyping 100% of the samples, with gB1 and gB3 being the most prevalent genotypes. Multiple gB and gH genotypes were detected in 36% and 33% of the plasma samples, respectively. One urine sample from a newborn with symptomatic congenital CMV was positive for gB1 and gB2. DBS of congenitally infected newborns (n = 41) were tested using 9 μl of dried blood, and genotypes were detected in 81% (gB) and 73% (gH) of the samples, with gB3 being the most prevalent genotype. No clear association of specific genotypes with clinical outcome was observed. In conclusion, the CMV gB and gH PCR assays were found to be rapid, sensitive for detecting mixed infections, and suitable for direct usage on DBS. These assays are efficient tools for genotyping of CMV in DBS of congenitally infected newborns.
INTRODUCTION
Cytomegalovirus (CMV) is the most common cause of congenital infection worldwide and an important viral pathogen affecting immunocompromised patients (13, 21). Both in congenitally infected newborns and in immunocompromised patients, genotyping of CMV has been used to study potential differences in pathogenicity of specific strains. However, few authors describe a correlation between specific CMV genotypes and severity of disease (29, 32, 34, 40). More important, genotyping of CMV has enabled the discrimination of reactivation of latent virus from reinfection with new CMV strains in transplant patients, allowing a better definition of donor-to-recipient transmission patterns (24). Congenital CMV infections mainly result from recurrent infections among pregnant women (37), comprising both reactivation and reinfection. The discrimination of reactivation from reinfection may give insight into the mother-to-fetus transmission pattern and the possible associations with the outcome of congenital CMV infections.
Genotyping of CMV has mainly focused on envelope glycoproteins gB (UL55) and gH (UL75), which play a role in virus entry and are major targets for neutralizing antibody response. The most frequently used methods for genotyping of CMV are nucleotide sequence analysis (27) and restriction fragment length polymorphism of PCR products (25, 28). Recently, real-time PCR-based assays have been used for rapid detection and quantification of CMV gB and gH genotypes (12, 15, 24, 26). However, they have mainly been applied to plasma or other high-volume samples. Also, deep-sequencing-based methods, sensitive in the detection of genotype mixtures with very low ratios, required a large input of CMV genomes (14). Studies on genotyping of CMV in dried blood spots (DBS) are rare (4, 7) and are hampered by the small amount of dried blood (50 μl per spot) available. In this study, two multiplex real-time PCR assays for rapid gB and gH genotyping of CMV were developed and applied to DBS obtained from congenitally infected neonates.
MATERIALS AND METHODS
Plasma samples of immunocompromised patients.
A total of 39 CMV DNA-positive plasma samples (loads, ≥1,000 copies/ml) were randomly selected from the database of the Department of Medical Microbiology of the Leiden University Medical Center (time period, 2009 to 2011). The samples were from immunocompromised patients (median age, 50 years; range, 7 to 78 years): 26 stem cell transplant patients (of whom 22 were allogeneic), 11 kidney transplant patients, and 2 liver transplant patients (median CMV DNA load of 25,000 copies/ml; range, 1,000 to 25,000,000 copies/ml). The pretransplant donor/recipient (D/R) CMV serostatus was distributed as follows: stem cell transplant patients, D+/R+ (n = 18), D−/R+ (n = 3), D+/R− (n = 1), and D−/R− (n = 1); kidney transplant patients, D+/R+ (n = 10), D−/R+ (n = 3), and D+/R− (n = 5); and liver transplant patient, D−/R+ (n = 1) (D/R serostatus was not available for 5 patients).
Urine samples from newborns with congenital CMV.
Urine samples with control gB1 to gB4 strains (determined by means of restriction fragment length polymorphism [4]) were kindly provided by Maria Barbi, Department of Public Health-Microbiology-Virology, University of Milan, Milan, Italy.
Furthermore, 21 CMV culture-positive urine samples from congenitally infected newborns (sampled within 3 weeks after birth) were derived from the database of the Department of Virology, Erasmus Medical Center, Rotterdam, the Netherlands (n = 19; time period, 2000 to 2011), and the Department of Medical Microbiology of the Leiden University Medical Center (n = 2; time period, 2009 to 2011), irrespective of clinical characteristics at birth (median CMV DNA load, 100,000 copies/ml; range, 4,000 to 20,000,000 copies/ml). No clinical data were available for the congenitally infected newborns tested and the CMV serostatus of the mother.
Dried blood spots (DBS) from newborns with congenital CMV.
A total of 41 DBS from newborns with congenital CMV infection were obtained from earlier studies (median CMV DNA load of 5,000 copies/ml whole blood; range, <1,000 to 800,000 copies/ml). Nine of the 41 newborns participated in the previously described DECIBEL study, which included infants with permanent bilateral hearing impairment (≥40 dB in the better ear) at the age of 3 to 5 years (median CMV DNA load of 32,000 copies/ml whole blood) (22). Clinical data included symptoms at birth, developmental score, and severity of hearing loss. The remaining 32 CMV-positive DBS were derived from a prevalence study in which a random selection of DBS from the Netherlands (2007) was tested for CMV DNA (median load of 5,000 copies/ml whole blood) (11). Due to the anonymization of the samples, no clinical data were available from these 32 newborns.
DNA extraction from plasma and urine samples.
Nucleic acids from plasma samples were extracted using the Cobas AmpliPrep total nucleic acid kit. Nucleic acids from urine samples were extracted on the MagNA Pure LC using the total nucleic acid isolation kit and high-performance kit (both from Roche Diagnostics, Almere, the Netherlands). The input volumes were 350 μl plasma and 200 μl urine, and output volumes were 100 μl.
DNA extraction from DBS.
DNA was extracted from DBS using the QIAamp DNA minikit according to the protocol for isolation of total DNA from FTA and Guthrie cards (Qiagen, Hilden, Germany). Sample input per well was 3 punches each measuring 3.2 mm in diameter, corresponding with approximately 9 μl dried blood per well. DBS were punched using an automated plate punch type 1296-071 (Perkin Elmer-Wallac, Zaventem, Belgium), with a negative-control punch between each sample. Output volume was 100 μl. DNA extraction was followed by CMV amplification in duplicate (DECIBEL DBS samples) or triplicate (DBS from prevalence study).
CMV gB- and gH-specific primers and probes.
For the selection of primers and probes, an alignment of CMV gB and gH gene sequences available in GenBank was made using the AlignX program (Vector NTI Advance 11; Invitrogen). The accession numbers of gB and gH sequences that were used were as follows: CMV gB genotype 1, M60929, EF999921, GQ466044, GQ221974, AY446894, U66425, GQ121041, and FJ616285; gB genotype 2, GQ221975, X17403, FJ527563, BK000394, X04606, M60931, and M60932; gB genotype 3, M60934, M85228, and M60933; gB genotype 4, M60926 and M60924; CMV gH genotype 1, AB275152, AB275255, AJ239007, BK000394, EF999921, FJ527563, GQ396663, GQ466044, GU179290, and X17403; gH genotype 2, AB275156, AY446894, FJ616285, GQ121041, GQ221973, GQ396662, GU179291, and M94233. Subsequently, specific primers and probes were designed for efficient amplification of multiple genotypes in one reaction, supported by the software package Beacon Designer 7.91 (Premier Biosoft International, Palo Alto, CA). The sequences of primers and probes are summarized in Tables 1 (gB) and 2 (gH). The gB3-specific probe was published by Gorzer et al. (15).
Table 1.
Sequences of primers and probes for cytomegalovirus gB genotyping
| Primer/probe name | Sequence (5′–3′)b | Product size (bp) |
|---|---|---|
| gB1 forward | TCA CCA TTC CTC TCR TAC GAC | 93 |
| gB1 reverse | CAC CAT GGC TGA CCG TTT GG | |
| gB1 TaqMan probe | FAM-TCT GCT GCT CAY TCT CGA TCC GGT TC–BHQ-1 | |
| gB2 forward | CTT TAA GGT ACG GGT CTA CCA A | 152 |
| gB2 reverse | GAA CTG TAG CAT TGG GCA AAC T | |
| gB2 TaqMan probe | YAK-CTA CGC TTA CAT CYA CAC CAC TTA TCT GC–BHQ-1 | |
| gB3 forward | CCG GTG TGA ACT CCA CGC G | 73 |
| gB3 reverse | GAT TCG CTT TCA RGY GAC AGG | |
| gB3 XS probe (15)a | TXR-TCG TAT TGC CCG TAC T–BHQ-2 | |
| gB4 forward | TCG TGC AAC TTC TAC TCA TAA TG | 85 |
| gB4 reverse | CGT TAC GCG TTG AGA GGA GAT | |
| gB4 TaqMan probe A | Q705-AAA CCA TAC TTC TCA TAC GAC GTC TGC TC–BHQ-2 | |
| gB4 TaqMan probe B | Q705-AAG CCA TAT TTC TCG TAC AAC GTC TGC TC–BHQ-2 |
XS probe, minor groove binding replacement probe.
Abbreviations: FAM, 6-carboxyfluorescein; BHQ, black hole quencher; YAK, Yakima Yellow; TXR, Texas Red.
Quantitative CMV real-time PCR.
In the sensitivity analysis of the newly developed assays, our diagnostic real-time PCR was used to determine the CMV DNA load of the samples. Amplification of a 126-bp fragment from the CMV immediate-early antigen region was performed using an internally controlled quantitative real-time PCR as described previously (9, 20). Quantification was performed using a dilution series of titrated CMV (strain AD169; Advanced Biotechnologies Inc., Columbia, MD) as an external standard.
Multiplex CMV gB1 to gB4 and gH1 and gH2 real-time PCR assays.
CMV gB1- to gB4- and gH1- and gH2-specific DNA amplification was performed using two multiplex real-time PCR assays. Each multiplex assay contained 10 μl of DNA extract, 25 μl HotStar Master mix (Qiagen, Hilden, Germany), and final concentrations of 0.3 μM (all) specific forward and reverse primers (gB1 to gB4 or gH1 and gH2) (Tables 1 and 2), 0.2 μM (all) specific probes (gB1, gB2, gB3, gB4A, and gB4B or gH1 and gH2) (Tables 1 and 2), and 4.5 mM MgCl2. Template denaturation and activation of HotStar Taq DNA polymerase for 15 min at 95°C were followed by 45 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s. The PCR assays were carried out in a CFX96 real-time PCR detection system (Bio-Rad, Veenendaal, the Netherlands).
Table 2.
Sequences of primers and probes for cytomegalovirus gH genotyping
| Primer/probe name | Sequence (5′–3′)a | Product size (bp) |
|---|---|---|
| gH1 forward | GAG ACT TAA CAC CTA CGC AT | 181 |
| gH1 reverse | CGA TCC CTT CCA GTC G | |
| gH1 TaqMan probe | FAM-GGG TCA GCA GCC CAC CAC C–BHQ-1 | |
| gH2 forward | TGG ACA CGA TCT ACT ATT CA | 134 |
| gH2 reverse | TGT CGT CGT CTA TGG AC | |
| gH2 TaqMan probe | YAK-CAC CGT CAC ACC TTG TTT GCA CC–BHQ-1 |
Abbreviations: FAM, 6-carboxyfluorescein; BHQ, black hole quencher; YAK, Yakima Yellow.
RESULTS
Analytical sensitivity.
The analytical sensitivities of the multiplex real-time CMV gB and gH PCR assays were determined using 10-fold dilution series of CMV-positive plasma samples with single genotypes gB1, gB2, gB3, and gB4 and gH1 and gH2, respectively. Comparison of the multiplex CMV gB and gH PCR assays with the diagnostic CMV PCR (20) resulted in equal detection limits of approximately 250 copies/ml.
Comparison of the results of the multiplex gB and gH PCR assays with the monoplex gB and gH PCR assays, testing the above-mentioned plasma samples, revealed comparable cycle threshold values (<1.5 cycle threshold difference) (data not shown).
Furthermore, plasma mixtures of CMV gB1-gB2, gB1-gB3, gB1-gB4, gB2-gB3, gB2-gB4, gB3-gB4, and gH1-gH2 were prepared, each combination in different ratios. The detection limit of the minor variant in these mixtures was approximately 250 CMV DNA copies/ml, which could be detected in mixtures with a proportion of the minor variant down to about 0.2% (data not shown).
The analytical sensitivity of the multiplex real-time CMV gB and gH PCR assays for DBS was determined using DBS with a broad range of CMV DNA loads (range, 50 to 20,000 copies/ml whole blood) (the Quality Control for Molecular Diagnostics [QCMD] CMV DBS 2011 panel and DBS samples prepared in-house with CMV-positive blood from transplant recipients [20]). Comparison of the multiplex CMV gB and gH PCR assays with the diagnostic CMV PCR (20) using DBS resulted in equal detection limits of approximately 1,000 to 2,500 copies/ml (data not shown).
Good precision was observed in the multiplex real-time CMV gB and gH PCR assays. Replicates of DNA from each genotype (gB1 to gB4 and gH1 and gH2) run on different days resulted in a mean difference of cycle threshold values of 0.6 ± 0.4 (standard deviation [SD]) (range, 0 to 1.5) (data not shown).
Analytical specificity.
The multiplex real-time CMV gB and gH PCR assays were negative for plasma and urine samples with noncorresponding gB1 to gB4 and gH1 and gH2 genotypes; no cross-reactions were observed. Furthermore, the assays tested negative for plasma samples with the genomes of Epstein-Barr virus, herpes simplex virus, and varicella-zoster virus.
Detection of CMV gB and gH genotypes in plasma samples of immunocompromised patients.
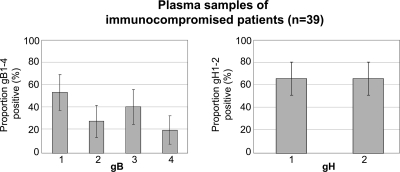
A random selection of CMV DNA-positive plasma samples of 39 transplant patients was tested using the multiplex CMV gB and gH PCR assays (Fig. 1). All 39 samples could be assigned to gB and gH genotypes (median cycle threshold value, 32; range, 24 to 42). The most prevalent genotypes were gB1 (54%; 21/39) and gB3 (41%; 16/39).
Fig. 1.
Distribution of CMV gB and gH genotypes (%) detected in CMV DNA-positive plasma samples of immunocompromised patients (n = 39). Note that the percentages add up to more than 100% due to the detection of multiple gB and gH genotypes in 36% (14/39) and 33% (13/39), respectively, of the CMV-positive plasma samples, with double and triple gB types in 28% (11/39) and 8%, respectively, of the samples. Error bars represent the 95% confidence interval. CMV, cytomegalovirus.
Multiple CMV gB and gH genotypes were detected in 36% (14/39) and 33% (13/39), respectively, of the CMV-positive plasma samples (median CMV DNA load of 40,000 copies/ml versus 25,000 copies/ml in single infections). Of these mixed gB infections, 28% (11/39) were double and 8% (3/39) were triple infections. Double gB genotype infections included gB1-gB2 (n = 4), gB1-gB3 (n = 4), gB2-gB3 (n = 2), and gB1-gB4 (n = 1). Triple gB/gH genotype infections included gB1-gB3-gB4/gH1-gH2 (n = 1 stem cell transplant, D+/R+), gB2-gB3-gB4/gH1-gH2 (n = 1 kidney transplant, D−/R+), and gB1-gB2-gB3/gH1-gH2 (n = 1 kidney transplant, D−/R+). The pretransplant CMV serostatuses of single compared to mixed gB infections were not significantly different (single gB: D+/R+, n = 12; D−/R+, n = 5; D+/R−, n = 5; mixed gB: D+/R+, n = 8; D−/R+, n = 2; D+/R−, n = 1; D−/R−, n = 1).
Detection of CMV gB and gH genotypes in urine samples from newborns with congenital CMV.
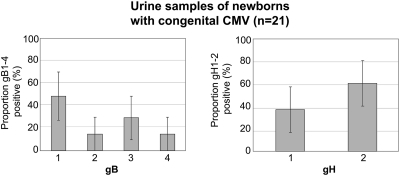
Urine samples obtained from 21 newborns with congenital CMV infection were tested using the multiplex real-time gB and gH assays (Fig. 2). A genotype could be assigned to all urine samples, with CMV gB1 (48%, 10/21), gB3 (29%, 6/21), and gH2 (62% 13/21) being the most prevalent genotypes (median cycle threshold value, 27; range, 21 to 36). One urine sample of a newborn with symptomatic congenital CMV was positive for both gB1 and gB2 (and gH2), indicating a mixed congenital infection. Clinical data revealed that this newborn was severely symptomatic at birth, with microcephaly, hyperbilirubinemia, thrombocytopenia, petechiae, and hepatosplenomegaly, and at a later age was diagnosed with mental retardation and hearing impairment. No clinical data were available from the other congenitally infected newborns tested.
Fig. 2.
Distribution of CMV gB and gH genotypes (%) detected in CMV-positive urine samples (<3 weeks of age) of congenitally infected newborns (n = 21). Error bars represent the 95% confidence interval.
Detection of CMV gB and gH genotypes in dried blood spots.
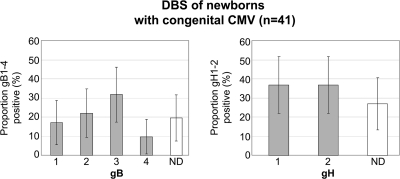
Dried blood spots (DBS) from 41 newborns with congenital CMV infection were tested in the multiplex real-time CMV gB and gH assays (Fig. 3). In total, 33 (81%) and 30 (73%) of the 41 DBS could be assigned a gB and gH genotype, respectively (median cycle threshold value, 36; range, 29 to 39). The most prevalent genotype was gB3 (32%, 13/41). The gH genotypes were distributed evenly.
Fig. 3.
Distribution of CMV gB and gH genotypes (%) detected in CMV-positive dried blood spots (DBS) of newborns with congenital CMV (n = 41). Error bars represent the 95% confidence interval. ND, not detected.
Clinical data were known for 9 of the 41 newborns and are shown in Table 3. These children had permanent bilateral hearing impairment at the age of 3 to 5 years, since that was an inclusion criterion for participation in the DECIBEL study from which they were recruited (22). A genotype could be assigned to 7 of the 9 (78%) DBS from children with hearing impairment. Genotype gB1 was not detected in DBS of these infants with hearing impairment. All 3 newborns with symptoms at birth had CMV loads of 200,000 copies/ml or higher and were genotyped as gB3/gH2, gB2/gH1, and gB4/gH1, respectively. No clear differences were seen in the gH and gB genotype distributions between the CMV-positive DBS from the children with hearing impairment and the DBS from the prevalence study.
Table 3.
Detection of CMV gB and gH genotypes in DBS from children with hearing impairment at the age of 3 to 5 years (22)b
| DBS from newborn | gB genotype | gH genotype | CMV IE load in DBS (log10 copies/ml whole blood) | Symptom(s) at birth | Developmental scorea | Severity of hearing loss |
|---|---|---|---|---|---|---|
| 1 | 3 | 2 | 4.1 | None | 59 | 60–90 dB |
| 2 | ND | ND | 3.3 | None | 84 | 40–60 dB |
| 3 | 4 | 1 | 4.9 | None | 72 | >90 dB (CI) |
| 4 | 2 | 2 | 3.6 | None | 68 | 60–90 dB |
| 5 | 3 | 2 | 5.9 | IUGR, microcephaly, seizures | 30 | >90 dB (CI) |
| 6 | 2 | 1 | 5.3 | IUGR, petechiae, hepatosplenomegaly, jaundice, thrombocytopenia | 50 | >90 dB (CI) |
| 7 | 4 | 1 | 5.3 | IUGR, jaundice, microcephaly | Unknown | >90 dB |
| 8 | 2 | 1 | <3.0 | None | 82 | >90 dB (CI) |
| 9 | ND | ND | 3.6 | Cataract | 56 | >90 dB (CI) |
The general development score is a summary score that provides an overall index of development by including 10 of the most age-discriminating items from each scale of the Child Development Inventory. The Child Development Inventory is a standardized instrument (parent questionnaire) designed to assess the social development, language development, and motor development of young children. Higher scores indicate better development.
Abbreviations: IE, immediate-early antigen; DBS, dried blood spots; gB, glycoprotein B; gH, glycoprotein H; CI, cochlear implant; IUGR, intrauterine growth retardation; ND, not detected.
DISCUSSION
In this study, two multiplex real-time PCR assays were used for rapid CMV gB and gH genotyping on DBS. Validation of these gB1- to gB4- and gH1- and gH2-specific PCR assays showed excellent sensitivity for genotyping plasma samples of immunocompromised patients and urine samples of congenitally infected newborns. Furthermore, the assays were able to detect a high number of mixed infections (>30%) in CMV-positive plasma samples and in one urine sample of a severely symptomatic newborn. In DBS of congenitally infected newborns, using only 9 μl of dried blood, a CMV genotype could be determined in 81% (gB) and 73% (gH), respectively, of the cases.
Our finding that genotypes gB1 and gB3 were the most prevalent genotypes in immunocompromised patients and congenitally infected infants is in agreement with previous studies assessing the genotype distribution of CMV (4, 8, 15, 18, 19, 23, 26, 38, 41). Potential significant variances in genotype distribution found in different studies are potentially based on geographical distribution, the population of patients tested, and/or CMV tissue tropism. In our study, no significant differences were found between the genotype distributions as detected in the urine samples, which were taken from mainly hospitalized newborns, and the DBS, which were from a different group of newborns, including a selection of children with hearing loss at the age of 3 to 5 years. It must be noted that the number of congenitally infected newborns tested in our study is small, and therefore, we cannot exclude differences in genotype distribution. Potential differences might be based on the population of newborns tested (symptomatology/hearing impairment), and also a slight difference in sensitivity between the gB1 and gB3 assays cannot be excluded (due to the lack of genotype-specific standards).
We could detect mixed-genotype infections in >30% of plasma samples from immunocompromised patients. It must be mentioned that potential mixed infections with viral loads below the detection limit could be missed and, therefore, the actual proportion of mixed infections might even be higher. The high proportion of mixed infections detected in our study is comparable to or exceeds the proportion found in previous studies, with a mixed genotype detected in 15 to 21% of the (solid organ) transplant recipients (12, 15, 16, 24, 26). This would suggest that our assays were at least as sensitive. The risk of competitive amplification of multiple genotypes by generic primers has been reduced by using genotype-specific primers and probes. This method was found to be more sensitive for detecting mixed infections (data not shown). Furthermore, the high proportion of D+/R+ and stem cell transplant recipients in our study might also contribute to the high proportion of mixed infections detected, since the highest genotype diversity has been found in these populations (14). It has been demonstrated that the CMV load after transplantation reflects the sum of relative levels of individual genotypes in time (15). Mixed CMV genotypes could be detected significantly more often in patients with higher CMV loads than in patients with lower loads (26), though the interpretation might be biased by underdetection of mixed infections with low viral loads. In this way, an association of mixed infections (and corresponding higher CMV loads) with clinical outcome has been demonstrated in transplant patients (1, 8, 30).
The occurrence of mixed congenital CMV infections in live newborns has rarely been described before. Though coinfection with multiple gB genotypes has been reported in two postpartum mothers of congenitally infected infants (38), one report casually noted the detection of mixed gB types in urine specimens from two congenitally infected newborns (33), and another report suggested the presence of multiple US28 and UL144 genotypes in 8 of 10 autopsy tissues from fatal cases of congenital CMV infection (2). The association of mixed infections with severe disease found in immunocompromised patients, combined with the single report on frequent mixed congenital CMV infections in fetal deaths, may lead to the speculation that congenital CMV infections by multiple strains (correlated with higher viral loads) could be associated with severe symptomatology and possibly fetal death. Interestingly, very recent genome-wide next-generation sequencing of CMV present in urine of three congenitally infected newborns revealed mixed gB and gN genotypes and offered strong evidence that CMV exists as a complex mixture of genome variants, with intrahost variability (0.2%) comparable to that of many RNA viruses (31).
In agreement with earlier studies that attempted genotyping CMV on DBS, a genotype could not be assigned to all strains from positive DBS (4, 7). Detection of CMV DNA in DBS has been shown to be a challenge (3, 5, 6, 9, 10, 35, 36, 39) due to the small amount of dried blood (50 μl per spot) available. Optimizing the DNA extraction step from the DBS has been shown to result in significantly increased sensitivity of CMV DNA detection in DBS. The DNA extraction method used in this study has been optimized previously, and despite a limited input of only 9 μl dried blood per well, a genotype could be determined in approximately 75 to 80% of the DBS samples. Recently developed deep-sequencing methods (14) have been reported to be highly sensitive because of their ability to detect genotype mixtures in low ratios, but they require a large input of approximately 280 μl of whole blood (30 times more than that used in our assay) containing 50,000 to 500,000 CMV DNA copies/ml plasma (14). For comparison, the median CMV DNA whole-blood load in congenitally CMV-infected newborns (symptomatic and asymptomatic newborns) has been reported to be 2,300 copies/ml blood (17).
The association of specific CMV genotypes with congenital CMV disease has previously been addressed with controversial results and is limited to the association of genotype gN4 with long-term sequelae (29) and genotype gB3 being found more often among congenitally CMV-infected than in postnatally infected children (40). In our study, no clear association between specific CMV gB and gH genotypes and severity of disease was observed, though the sample numbers were low.
Genotyping of CMV has been shown to enable the discrimination of reactivation of latent virus from reinfection with new CMV strains in transplant patients and has enabled the assessment of donor-to-recipient transmission patterns (16, 24). Data from Manuel et al. suggest that, in seropositive transplant recipients, approximately half of the infecting CMV strains originate from the organ donor and the other half are reactivated endogenous strains (24). Though CMV is more frequently transmitted to the fetus in preconceptionally seronegative women, recent calculations have demonstrated that the majority of congenitally CMV-infected children in the United States are born from seroimmune women (37). This major role of recurrent maternal infections emphasizes the convenience of a sensitive and rapid CMV genotyping assay, suitable for usage on DBS, in order to compare potentially mixed genotypes present in maternal blood with CMV strains in the newborn. The rapid and sensitive genotyping tool described in this study may support a better definition of mother-to-fetus transmission patterns and may lead to enhanced insight into transmission risk and outcome of congenital CMV infections. The implications of this increased insight into transmission risks for preventive and therapeutic strategies, including CMV vaccine research, may be significant.
ACKNOWLEDGMENT
We thank Eric Claas (Department of Medical Microbiology, LUMC, Leiden, the Netherlands) for critically reading the manuscript.
Footnotes
Published ahead of print 23 November 2011
REFERENCES
- 1. Aquino VH, Figueiredo LT. 2000. High prevalence of renal transplant recipients infected with more than one cytomegalovirus glycoprotein B genotype. J. Med. Virol. 61:138–142 [PubMed] [Google Scholar]
- 2. Arav-Boger R, et al. 2002. Polymorphisms of the cytomegalovirus (CMV)-encoded tumor necrosis factor-alpha and beta-chemokine receptors in congenital CMV disease. J. Infect. Dis. 186:1057–1064 [DOI] [PubMed] [Google Scholar]
- 3. Atkinson C, et al. 2009. Use of stored dried blood spots for retrospective diagnosis of congenital CMV. J. Med. Virol. 81:1394–1398 [DOI] [PubMed] [Google Scholar]
- 4. Barbi M, et al. 2001. CMV gB genotypes and outcome of vertical transmission: study on dried blood spots of congenitally infected babies. J. Clin. Virol. 21:75–79 [DOI] [PubMed] [Google Scholar]
- 5. Barbi M, Binda S, Primache V, Luraschi C, Corbetta C. 1996. Diagnosis of congenital cytomegalovirus infection by detection of viral DNA in dried blood spots. Clin. Diagn. Virol. 6:27–32 [DOI] [PubMed] [Google Scholar]
- 6. Boppana SB, et al. 2010. Dried blood spot real-time polymerase chain reaction assays to screen newborns for congenital cytomegalovirus infection. JAMA 303:1375–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choi KY, et al. 2009. Detection of cytomegalovirus DNA in dried blood spots of Minnesota infants who do not pass newborn hearing screening. Pediatr. Infect. Dis. J. 28:1095–1098 [DOI] [PubMed] [Google Scholar]
- 8. Coaquette A, et al. 2004. Mixed cytomegalovirus glycoprotein B genotypes in immunocompromised patients. Clin. Infect. Dis. 39:155–161 [DOI] [PubMed] [Google Scholar]
- 9. De Vries JJC, Claas ECJ, Kroes ACM, Vossen ACTM. 2009. Evaluation of DNA extraction methods for dried blood spots in the diagnosis of congenital cytomegalovirus infection. J. Clin. Virol. 46(Suppl. 4):S37–S42 [DOI] [PubMed] [Google Scholar]
- 10. De Vries JJC, Vossen ACTM, Kroes ACM. 2010. Screening newborns for congenital cytomegalovirus infection. JAMA 304:407. [DOI] [PubMed] [Google Scholar]
- 11. de Vries JJ, et al. 2011. Congenital cytomegalovirus (CMV) infection in the Netherlands: birth prevalence and risk factors. J. Med. Virol. 83:1777–1782 [DOI] [PubMed] [Google Scholar]
- 12. Fan J, et al. 2009. Monitoring of human cytomegalovirus glycoprotein B genotypes using real-time quantitative PCR in immunocompromised Chinese patients. J. Virol. Methods 160:74–77 [DOI] [PubMed] [Google Scholar]
- 13. Gandhi MK, Khanna R. 2004. Human cytomegalovirus: clinical aspects, immune regulation, and emerging treatments. Lancet Infect. Dis. 4:725–738 [DOI] [PubMed] [Google Scholar]
- 14. Gorzer I, Guelly C, Trajanoski S, Puchhammer-Stockl E. 2010. Deep sequencing reveals highly complex dynamics of human cytomegalovirus genotypes in transplant patients over time. J. Virol. 84:7195–7203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gorzer I, et al. 2008. Virus load dynamics of individual CMV-genotypes in lung transplant recipients with mixed-genotype infections. J. Med. Virol. 80:1405–1414 [DOI] [PubMed] [Google Scholar]
- 16. Gorzer I, Kerschner H, Redlberger-Fritz M, Puchhammer-Stockl E. 2010. Human cytomegalovirus (HCMV) genotype populations in immunocompetent individuals during primary HCMV infection. J. Clin. Virol. 48:100–103 [DOI] [PubMed] [Google Scholar]
- 17. Halwachs-Baumann G, et al. 2002. Human cytomegalovirus load in various body fluids of congenitally infected newborns. J. Clin. Virol. 25(Suppl. 3):S81–S87 [DOI] [PubMed] [Google Scholar]
- 18. Humar A, Kumar D, Gilbert C, Boivin G. 2003. Cytomegalovirus (CMV) glycoprotein B genotypes and response to antiviral therapy, in solid-organ-transplant recipients with CMV disease. J. Infect. Dis. 188:581–584 [DOI] [PubMed] [Google Scholar]
- 19. Jin H, Wang X, Li S. 2007. Human cytomegalovirus glycoprotein B genotype correlates with different symptoms of infected infants. Intervirology 50:219–223 [DOI] [PubMed] [Google Scholar]
- 20. Kalpoe JS, et al. 2004. Validation of clinical application of cytomegalovirus plasma DNA load measurement and definition of treatment criteria by analysis of correlation to antigen detection. J. Clin. Microbiol. 42:1498–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kenneson A, Cannon MJ. 2007. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev. Med. Virol. 17:253–276 [DOI] [PubMed] [Google Scholar]
- 22. Korver AM, et al. 2009. DECIBEL study: congenital cytomegalovirus infection in young children with permanent bilateral hearing impairment in the Netherlands. J. Clin. Virol. 46(Suppl. 4):S27–S31 [DOI] [PubMed] [Google Scholar]
- 23. Lukacsi A, et al. 2001. Human cytomegalovirus gB genotype 1 is dominant in congenital infections in South Hungary. J. Med. Virol. 65:537–542 [PubMed] [Google Scholar]
- 24. Manuel O, et al. 2009. An assessment of donor-to-recipient transmission patterns of human cytomegalovirus by analysis of viral genomic variants. J. Infect. Dis. 199:1621–1628 [DOI] [PubMed] [Google Scholar]
- 25. Novak Z, et al. 2008. Cytomegalovirus strain diversity in seropositive women. J. Clin. Microbiol. 46:882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pang X, Humar A, Preiksaitis JK. 2008. Concurrent genotyping and quantitation of cytomegalovirus gB genotypes in solid-organ-transplant recipients by use of a real-time PCR assay. J. Clin. Microbiol. 46:4004–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pignatelli S, Dal MP, Landini MP. 2001. gpUL73 (gN) genomic variants of human cytomegalovirus isolates are clustered into four distinct genotypes. J. Gen. Virol. 82:2777–2784 [DOI] [PubMed] [Google Scholar]
- 28. Pignatelli S, et al. 2003. Human cytomegalovirus glycoprotein N (gpUL73-gN) genomic variants: identification of a novel subgroup, geographical distribution and evidence of positive selective pressure. J. Gen. Virol. 84:647–655 [DOI] [PubMed] [Google Scholar]
- 29. Pignatelli S, et al. 2010. Cytomegalovirus gN genotypes distribution among congenitally infected newborns and their relationship with symptoms at birth and sequelae. Clin. Infect. Dis. 51:33–41 [DOI] [PubMed] [Google Scholar]
- 30. Puchhammer-Stockl E, et al. 2006. Emergence of multiple cytomegalovirus strains in blood and lung of lung transplant recipients. Transplantation 81:187–194 [DOI] [PubMed] [Google Scholar]
- 31. Renzette N, Bhattacharjee B, Jensen JD, Gibson L, Kowalik TF. 2011. Extensive genome-wide variability of human cytomegalovirus in congenitally infected infants. PLoS Pathog. 7:e1001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosen HR, Corless CL, Rabkin J, Chou S. 1998. Association of cytomegalovirus genotype with graft rejection after liver transplantation. Transplantation 66:1627–1631 [DOI] [PubMed] [Google Scholar]
- 33. Shen Z, Shang SQ, Zou CC, Zheng JY, Yu ZS. 2010. The detection and clinical features of human cytomegalovirus infection in infants. Fetal Pediatr. Pathol. 29:393–400 [DOI] [PubMed] [Google Scholar]
- 34. Shepp DH, et al. 1996. Cytomegalovirus glycoprotein B groups associated with retinitis in AIDS. J. Infect. Dis. 174:184–187 [DOI] [PubMed] [Google Scholar]
- 35. Soetens O, et al. 2008. Evaluation of different cytomegalovirus (CMV) DNA PCR protocols for analysis of dried blood spots from consecutive cases of neonates with congenital CMV infections. J. Clin. Microbiol. 46:943–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vauloup-Fellous C, et al. 2007. Evaluation of cytomegalovirus (CMV) DNA quantification in dried blood spots: retrospective study of CMV congenital infection. J. Clin. Microbiol. 45:3804–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang C, Zhang X, Bialek S, Cannon MJ. 2011. Attribution of congenital cytomegalovirus infection to primary versus non-primary maternal infection. Clin. Infect. Dis. 52:e11–e13 [DOI] [PubMed] [Google Scholar]
- 38. Yamamoto AY, et al. 2007. Human cytomegalovirus glycoprotein B genotypes in Brazilian mothers and their congenitally infected infants. J. Med. Virol. 79:1164–1168 [DOI] [PubMed] [Google Scholar]
- 39. Yamamoto AY, Mussi-Pinhata MM, Pinto PC, Figueiredo LT, Jorge SM. 2001. Usefulness of blood and urine samples collected on filter paper in detecting cytomegalovirus by the polymerase chain reaction technique. J. Virol. Methods 97:159–164 [DOI] [PubMed] [Google Scholar]
- 40. Yan H, et al. 2008. Genetic variations in the gB, UL144 and UL149 genes of human cytomegalovirus strains collected from congenitally and postnatally infected Japanese children. Arch. Virol. 153:667–674 [DOI] [PubMed] [Google Scholar]
- 41. Yu ZS, Zou CC, Zheng JY, Zhao ZY. 2006. Cytomegalovirus gB genotype and clinical features in Chinese infants with congenital infections. Intervirology 49:281–285 [DOI] [PubMed] [Google Scholar]