Abstract
Pseudomonas aeruginosa possesses complex regulatory networks controlling virulence and survival under adverse conditions, including antibiotic pressure, which are interconnected and share common regulatory proteins. Here, we screen a panel of 13 mutants defective in intracellular proteases and demonstrate that, in addition to the known alterations in Lon and AsrA mutants, mutation of three protease-related proteins PfpI, ClpS, and ClpP differentially affected antibiotic resistance, swarming motility, and biofilm formation.
TEXT
Pseudomonas aeruginosa is a Gram-negative bacterium that can be found in a wide range of environments. In humans, it is an important opportunistic pathogen, being a leading cause of nosocomial infections in elderly, immunocompromised, and severely burned individuals, as well as a major contributor to morbidity and mortality in cystic fibrosis patients (9, 11). Infections with P. aeruginosa are especially difficult to treat due to this organism's high intrinsic antibiotic resistance (3). Additionally, this microbe can become less susceptible through the acquisition or mutation of resistance markers and through adaptation to subinhibitory antibiotic concentrations and environmental stimuli (3, 7). Good examples of environmental adaptations are social activities, like swarming motility and biofilms, which have been associated with increased resistance to antibiotics (11, 15, 19).
The success of this organism in occupying a variety of niches is related in part to its complex regulatory circuits that allow efficient adaptation to changes in environmental conditions. Indeed, almost 10% of all open reading frames (ORFs) in the P. aeruginosa genome encode regulatory proteins (23). Furthermore, recent evidence indicates that these networks are interconnected via common regulatory proteins, which coordinately control phenotypes related to both virulence and resistance to antimicrobials (3). One such protein is the ATP-dependent protease Lon, which participates in resistance to ciprofloxacin, as well as in swarming motility and biofilm formation (2, 17). Also, the AsrA protease from P. aeruginosa mediates the adaptive response to the aminoglycoside tobramycin by controlling heat shock responses (13). Bacterial intracellular proteases are known to play essential roles in orchestrating cellular activities via regulation of the levels of chaperones, labile regulators, and stress-related proteins, as well as by degrading misfolded proteins (10). For this reason, we set out to explore the participation of other intracellular proteases in virulence and antibiotic resistance in P. aeruginosa. Mutants from the P. aeruginosa PA14 transposon library carrying transposon insertions in genes encoding intracellular proteases (Table 1) were screened for altered phenotypes related to antibiotic resistance, motility (swarming, swimming, and twitching), and biofilm formation (16).
Table 1.
P. aeruginosa PA14 transposon mutants used in this study
| Mutant from PA14 librarya | PAO1 ortholog (gene name) | Product of gene |
|---|---|---|
| PAMr_nr_mas_12_1:C6 | PA0355 (pfpI) | Protease PfpI |
| PAMr_nr_mas_02_3:G5 | PA0372 | Probable zinc protease |
| PAMr_nr_mas_03_2:C2 | PA0459 | Probable ClpA/B protease ATP binding subunit |
| PAMr_nr_mas_14_4:E8 | PA0779 (asrA)b | ATP-dependent protease |
| PAMr_nr_mas_08_4:A5 | ||
| PAMr_nr_mas_12_4:E7 | PA1801 (clpP) | ATP-dependent Clp protease proteolytic subunit |
| PAMr_nr_mas_08_1:F11 | PA1802 (clpX) | ATP-dependent Clp protease ATP binding subunit |
| PAMr_nr_mas_04_1:A9 | PA1803 (lon)b | ATP-dependent Lon protease |
| PAMr_nr_mas_08_4:C11 | PA2620 (clpA) | ATP-dependent Clp protease ATP binding subunit |
| PAMr_nr_mas_06_2:F9 | PA2621 (clpS) | ATP-dependent Clp protease adaptor protein |
| PAMr_nr_mas_11_1:C10 | PA3326 | Probable Clp-family ATP-dependent protease |
| PAMr_nr_mas_11_1:G12 | PA3535 | Probable serine protease |
| PAMr_nr_mas_04_1:G10 | PA4576 | Probable ATP-dependent protease |
Antibiotic resistance of protease mutants.
As mentioned above, previous studies demonstrated the participation of the ATP-dependent proteases Lon and AsrA in the resistance to antibiotics of P. aeruginosa (2, 4, 13). Here, the involvement of other intracellular proteases in antibiotic resistance was determined. MICs to different antimicrobials were assessed using the broth microdilution method according to CLSI guidelines, with the exception that we used LB broth, as well as polypropylene 96-well plates and cation-adjusted Mueller-Hinton medium for polymyxin B (25). None of the mutants showed any differences in susceptibility to the aminoglycoside tobramycin or the peptide polymyxin B. The pfpI and clpP mutants, respectively, exhibited a 4-fold increase and a 2-fold decrease in resistance to the fluoroquinolone ciprofloxacin. This altered fluoroquinolone resistance phenotype could be complemented by introducing wild-type copies of the genes clpP and pfpI (Table 2). In the case of the clpS mutant, we observed 2- and 4-fold increases in resistance to piperacillin, a β-lactam, in strains PA14 and PAO1, respectively (Table 2). ClpS mutants also showed increased resistance to other tested β-lactams, specifically to aztreonam, ceftazidime, and imipenem (Table 2). To assess whether the alteration in β-lactam susceptibility in the clpS mutant was due to altered β-lactamase production, we carried out the nitrocefin assay as previously described (1) for samples grown in the presence or absence of subinhibitory (0.25× MIC) concentrations of piperacillin, ceftazidime, or imipenem. We observed no significant differences in rate of nitrocefin hydrolysis between the wild type and the clpS mutant with or without antibiotic induction of β-lactamase activity. Therefore, it seems unlikely that increased β-lactamase production is the explanation for the resistance phenotype of this mutant. With regard to ciprofloxacin resistance, Breidenstein et al. (4) previously observed changes in susceptibility in the clpP and pfpI mutants, as part of a study of the ciprofloxacin resistome of P. aeruginosa PA14. In the case of pfpI, since the mutation could be complemented, the previously described antimutator function reported by Rodriguez-Rojas and Blazquez (22) might not be responsible for resistance, and in that study no difference in antibiotic resistance to diverse antimicrobial agents, including ciprofloxacin, was observed. Overall, our results reinforce the notion that intracellular proteases participate in the regulation of resistance to antimicrobials. This was shown previously for the Lon and AsrA proteases, which participate in intrinsic resistance to ciprofloxacin and adaptive resistance to aminoglycosides, respectively (2, 13).
Table 2.
MICs of selected mutants to various antibiotics
| Strain or genotype | MIC (μg/ml) |
||||||
|---|---|---|---|---|---|---|---|
| Ciprofloxacin | Piperacillin | Tobramycin | Polymyxin B | Imipenem | Aztreonam | Ceftazidime | |
| PA14 (wild type) | 0.1 | 8 | 2 | 1 | 0.5 | 4 | 2 |
| pfpI | 0.4 | 8 | 2 | 1 | —a | — | — |
| pfpI+ | 0.05 | 8 | 2 | 1 | — | — | — |
| clpS | 0.1 | 16 | 2 | 1 | 1 | 16 | 16 |
| clpP | 0.05 | 8 | 2 | 1 | — | — | — |
| clpP+ | 0.1 | 8 | 2 | 1 | — | — | — |
| PA01 (wild type) | 0.1 | 4 | 2 | 1 | 1 | 4 | 2 |
| UW-clpS | 0.1 | 16 | 2 | 1 | 2 | 8 | 4 |
—, not determined.
Motility phenotypes of different P. aeruginosa protease mutants.
P. aeruginosa exhibits different types of motility depending on the composition and viscosity of its environment. The three most characterized to date are pilus-mediated twitching on solid surfaces, flagellum-mediated swimming in aqueous environments, and swarming on semisolid surfaces. Both flagella and pili, as well as the production of rhamnolipids, are required for swarming (14), which is a complex adaptation that is associated with increased antibiotic resistance and production of virulence factors (15, 19, 26). The transposon mutants described in Table 1 were analyzed for these three motility types in at least four independent experiments.
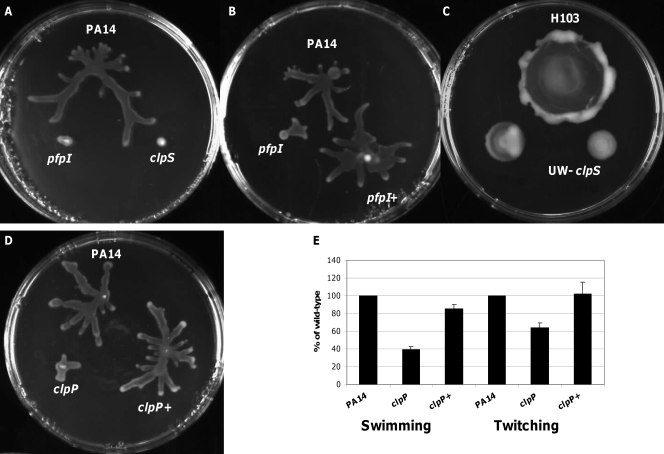
Swarming motility was examined on BM2-glucose plates containing 0.5% (wt/vol) agar and 0.1% or 0.5% (wt/vol) Casamino Acids instead of (NH4)2SO4 for PA14- or PA01-derived strains, respectively, as described previously (19). One-μl aliquots from mid-logarithmic-phase (optical density at 600 nm [OD600] of 0.5 to 0.6) cultures grown in liquid BM2-glucose medium were spotted onto the plates. After approximately 24 h of incubation at 37°C, swarming colonies were visually inspected for altered swarming phenotypes. Three PA14 transposon mutants, namely, pfpI (PA0355), clpS (PA2621), and clpP (PA1801) mutants, like the lon mutant (17), displayed a strongly impaired ability to swarm (Fig. 1A, B, and D). For complementation, the clpP gene, together with its own promoter, was amplified by PCR from wild-type P. aeruginosa PA14 and cloned into the broad-host-range high-copy vector pUCP19 (24). The resulting hybrid plasmid, pUCP19::clpP, was electroporated into the clpP mutant, leading to complete restoration of swarming motility (Fig. 1D). Complementation of the pfpI mutant was accomplished by introduction of the plasmid pBBR-pfpI-14 (22), carrying a wild-type copy of gene pfpI (Fig. 1B). We were unable to complement the swarming-deficient phenotype of the PA14 clpS mutant, perhaps due to overexpression lethality (data not shown). Therefore, we analyzed the swarming capabilities of independently isolated PAO1 mutant phoAwp08q1A11 from the University of Washington library, harboring a transposon insertion in clpS (UW-clpS) (12). This mutant also had a significant loss of swarming motility (Fig. 1C). pfpI or clpS mutants had not previously been related to motility defects in other microorganisms, whereas a Pseudomonas fluorescens clpP mutant was swarming deficient (5). However, unlike the results shown here, in P. fluorescens, swarming motility could not be fully complemented with the cloned gene, and it was suggested that this defect was due to a deficiency in production of the cyclic lipopeptide massetolide, which P. aeruginosa does not synthesize.
Fig 1.
Motility phenotypes of P. aeruginosa mutants in intracellular proteases. (A) Swarming of the PA14 wild type and pfpI and clpS mutants. (B) Swarming of the PA14 wild type, pfpI mutant, and complemented pfpI mutant (pfpI+). (C) Swarming of the PAO1 wild type and the UW-clpS mutant. The pictures shown are representative of at least four independent experiments with the same results. (D) Swarming of the PA14 wild type, clpP mutant, and complemented clpP mutant (clpP+). (E) Swimming and twitching motility analysis of the PA14 wild type, PA14 clpP transposon insertion mutant, and clpP complemented strain (clpP+). The results represent the average percentages of wild-type motility and standard deviations from four independent experiments.
Swimming motility was evaluated on BM2-glucose plates containing 0.25% (wt/vol) agar (19). The diameters of the swimming zones were measured after 24 h of incubation at 37°C, and the clpP mutant was the only mutant with a substantial 60% defect, which could be restored by complementation (Fig. 1E). The clpS mutant also showed a very slightly reduced ability to swim, with an approximately 25% decrease in swimming zone in strain PA14 but not in strain PAO1 (data not shown). Twitching motility, analyzed on LB plates containing 1.5% (wt/vol) agar, indicated that only the clpP mutant showed a modest 40% twitching defect, which was complemented by introduction of the wild-type gene (Fig. 1E).
To test the possibility that the motility defects were due to altered growth rates, the growth at 37°C under shaking conditions of all strains was monitored with a spectrophotometer by determining the absorbance at 600 nm every hour for 10 h and then at 24 h postinoculation. Of all the mutants tested, only the clpP mutant showed a growth defect, approximately a 2-h delay, that could be complemented in trans (data not shown). However, it seems unlikely that this would result in a complete abolishment of swarming motility. Nonetheless, it is possible that the moderate impairments in swimming and twitching were to some extent due to the clpP growth defect.
Biofilm assays.
Like swarming, biofilms represent a social activity of bacteria and are also known to participate in pathogenesis and antibiotic resistance. Abiotic solid-surface mature biofilm formation of the PA14 protease mutants was analyzed using 96-well polystyrene microtiter plates as described previously (8, 18). Of all the mutant strains tested, only the pfpI, clpS, and clpP mutants showed any difference in biofilm formation, demonstrating approximately 65%, 70%, and 35% less than the level for the wild type, respectively. Coincidentally, these were also the strains that displayed a swarming defect. The mutation of clpP had the most dramatic effect on the production of mature biofilms. Complementation of the clpP mutant was successful in restoring the wild-type phenotype (Fig. 2A). The ClpP protease has previously been related to biofilm formation in P. fluorescens (5, 18). In P. aeruginosa, ClpP is known to participate in the regulation of alginate production, but its role in biofilms had not been reported before (21). The clpS mutant formed slightly less biofilm than the wild type. In the case of the pfpI mutant, there was also a general reduction in biofilm formation, although this was not always the case as some of the biological repeats tested actually displayed increased biofilm-forming ability of up to 70%. Nevertheless, in all cases we observed a complementation of the altered phenotype in the mutant strain carrying plasmid pBBR-pfpI-14 (22).
Fig 2.
Biofilm formation and Congo red binding. (A) Analysis of mature biofilm formation of the PA14 wild type, PA14 clpP transposon insertion mutant, and clpP complemented strain (clpP+). The results represent the average percentages of wild-type ability and standard deviations from four independent experiments. (B) Results of the Congo red binding assay for the PA14 wild type, PA14 clpP transposon insertion mutant, and clpP complemented strain (clpP+). A more intense gray tone at the edge of the colony can be observed in the wild-type PA14 and the complemented strain, while in the clpP mutant an epigenetic phenomenon was evident, whereby dense staining was observed in discrete areas.
To determine whether the differences in formation of mature biofilms were due to a defect during the initial stages of biofilm formation, we evaluated the rapid attachment capabilities of the pfpI, clpS, and clpP mutants by assessing abiotic biofilm formation after 1 h as previously described (8, 18). The pfpI and clpP mutants had no changes in rapid attachment compared to that for the wild type, while the clpS mutant had a modest 20% reduction, which could partly explain its defect in mature biofilm formation.
Additionally, we performed Congo red binding assays with the PA14 pfpI, clpS, and clpP mutants (8). Congo red has been shown to bind to the exopolysaccharide synthesized by the products of the pel genes. This extracellular matrix is known to participate in biofilm and pellicle formation. All three mutants showed a lower Congo red binding ability than the wild type (data not shown), which was restored in the clpP complemented mutant (Fig. 2B). These results are in good agreement with the lower biofilm-forming capacity of the three mutants and indicate that, at least to some extent, this defect is due to a diminished production of exopolysaccharide. Intriguingly, the clpP mutant demonstrated an epigenetic phenomenon whereby discrete spots of the colony were very highly stained; this was also complementable.
Concluding remarks.
Previously, we had shown that the related Lon and AsrA proteases play major roles in regulating diverse phenotypes associated with virulence and/or antibiotic resistance in P. aeruginosa. Here, we demonstrate that strains carrying disruptions in the genes pfpI, clpS, and clpP, all of which encode proteins involved in intracellular protease complexes, had altered phenotypes in antibiotic susceptibility, swarming motility, and biofilm formation. Previous studies identified two other ATP-dependent proteases, Lon and AsrA (2, 13, 17), with distinct phenotypic properties. However, the patterns of changes varied when these 5 individual ATP-dependent proteases were mutated, suggesting that they were independently determined, likely through the processing of one or more regulatory factors by each protease. Overall, this reinforces the notion that virulence and antibiotic susceptibility in P. aeruginosa are regulated in a coordinated manner and that these 5 ATP-dependent proteases are involved in this coordination. Moreover, our results emphasize the importance of the regulatory function carried out by intracellular proteases in this pathogen, which goes beyond their involvement in the regulation of stress responses.
ACKNOWLEDGMENTS
The work described in this paper was funded by a grant from Cystic Fibrosis Canada (CFC). E.B.M.B. and D.S. were supported by a scholarship and a studentship from CFC, respectively. R.E.W.H. holds a Canada Research Chair.
We thank A. Rodriguez-Rojas and J. Blazquez for kindly providing plasmid pBBR-pfpI-14 for the complementation of the pfpI mutant.
Footnotes
Published ahead of print 28 November 2011
REFERENCES
- 1. Alvarez-Ortega C, Wiegand I, Olivares J, Hancock REW, Martínez JL. 2010. Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to beta-lactam antibiotics. Antimicrob. Agents Chemother. 54:4159–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brazas MD, Breidenstein EBM, Overhage J, Hancock REW. 2007. Role of Lon, an ATP-dependent protease homolog, in resistance of Pseudomonas aeruginosa to ciprofloxacin. Antimicrob. Agents Chemother. 51:4276–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Breidenstein EBM, de la Fuente-Núñez C, Hancock REW. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 19:419–426 [DOI] [PubMed] [Google Scholar]
- 4. Breidenstein EBM, Khaira BK, Wiegand I, Overhage J, Hancock REW. 2008. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob. Agents Chemother. 52:4486–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Bruijn I, Raaijmakers JM. 2009. Regulation of cyclic lipopeptide biosynthesis in Pseudomonas fluorescens by the ClpP protease. J. Bacteriol. 191:1910–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reference deleted.
- 7. Fernández L, Breidenstein EBM, Hancock REW. 2011. Creeping baselines and adaptive resistance to antibiotics. Drug Resist. Updat. 14:1–21 [DOI] [PubMed] [Google Scholar]
- 8. Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675–690 [DOI] [PubMed] [Google Scholar]
- 9. Gibson RL, Burns JL, Ramsey BW. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918–951 [DOI] [PubMed] [Google Scholar]
- 10. Gottesman S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465–506 [DOI] [PubMed] [Google Scholar]
- 11. Hutchison ML, Govan JR. 1999. Pathogenicity of microbes associated with cystic fibrosis. Microbes Infect. 1:1005–1014 [DOI] [PubMed] [Google Scholar]
- 12. Jacobs MA, et al. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kindrachuk KN, Fernández L, Bains M, Hancock REW. 2011. Involvement of an ATP-dependent protease, PA0779/AsrA, in inducing heat shock in response to tobramycin in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55:1874–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kohler T, Curty LK, Barja F, van Delden C, Pechere JC. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signalling and requires flagella and pili. J. Bacteriol. 182:5990–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lai S, Tremblay J, Déziel E. 2009. Swarming motility: a multicellular behaviour conferring antimicrobial resistance. Environ. Microbiol. 11:126–136 [DOI] [PubMed] [Google Scholar]
- 16. Liberati NT, et al. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marr AK, Overhage J, Bains M, Hancock REW. 2007. The Lon protease of Pseudomonas aeruginosa is induced by aminoglycosides and is involved in biofilm formation and motility. Microbiology 153:474–482 [DOI] [PubMed] [Google Scholar]
- 18. O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 19. Overhage J, Bains M, Brazas MD, Hancock REW. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 190:2671–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reference deleted.
- 21. Qiu D, Eisinger VM, Head NE, Pier GB, Yu HD. 2008. ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiology 154:2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rodriguez-Rojas A, Blazquez J. 2009. The Pseudomonas aeruginosa pfpI gene plays an antimutator role and provides general stress protection. J. Bacteriol. 191:844–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stover CK, et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 24. West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81–86 [DOI] [PubMed] [Google Scholar]
- 25. Wiegand I, Hilpert K, Hancock REW. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3:163–175 [DOI] [PubMed] [Google Scholar]
- 26. Yeung AT, et al. 2009. Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J. Bacteriol. 191:5592–5602 [DOI] [PMC free article] [PubMed] [Google Scholar]