Abstract
Most research on HIV transmission and microbicides focuses on the inhibition of cell-free virus (CFV) present in genital secretions. However, an effective microbicide should also block the transmission of cell-associated virus (CAV) originating from seminal T cells and macrophages. Because inhibition of CAV remains controversial, especially for viral entry inhibitors, we developed a novel in vitro assay to evaluate the activities of different classes of candidate microbicides against cell-free HIV and HIV-infected leukocytes (i.e., resting peripheral blood mononuclear cells [PBMC], activated PBMC, and monocyte-derived macrophages). The assay is based on two CD4+ CXCR4+ T-cell lines (R5MaRBLE and X4MaRBLE) that both contain a firefly luciferase reporter gene but differ in the expression of the CCR5 coreceptor. Consequently, the quantification of the luciferase activities and the Gag p24 concentrations in cocultures of R5-tropic HIV-infected leukocytes with each cell line separately allowed us to discriminate between the infection of the cell lines (i.e., target cells), the ongoing infection in the HIV-infected leukocytes (i.e., effector cells), and the total infection of the coculture (i.e., effector plus target cells). All 14 antiretrovirals tested were able to block target cell infection by all three sources of CAV, although a small decrease in activity (2- to 18-fold) was observed for all entry inhibitors. On the other hand, the production of Gag p24 by the infected effector cells could be blocked only by protease inhibitors. Overall, these results show that entry and protease inhibitors are eligible drug classes for inclusion in future combination microbicides.
INTRODUCTION
Most of the 3 million new HIV infections each year occur in women who often have no control over condom use by their sexual partners (30). Vaginal microbicides could empower women to protect themselves from sexual transmission and are thus urgently needed (45). To be effective, these candidate microbicides should prevent infection of vaginal target cells by HIV in human semen. Although the nature of these first target cells remains elusive, CD4+ T cells are currently considered prime suspects, among other cell types, such as macrophages, dendritic cells (DCs), and Langerhans cells (25). The seminal source of HIV, on the other hand, remains unknown, as human semen contains not only cell-free virus (CFV), but also virus associated with HIV-infected leukocytes, such as T lymphocytes and macrophages (i.e., cell-associated virus [CAV]). Over the past 28 years, evidence has accumulated that seminal leukocytes can cross the vaginal barrier and reach uninfected permissive target cells within the epithelium and/or submucosal tissue (16, 18, 30, 42). As HIV can be transferred very efficiently from cell to cell in vitro (47), it is plausible that the cell-associated virus in human semen represents a major source of HIV transmission in vivo (4).
During cell-to-cell spread, a tight adhesive junction, termed the virological synapse (VS), is formed in which Env (on the infected effector cell) and CD4 and coreceptors (on the uninfected target cell) are recruited to the site of contact (32). As a result, virion release is concentrated and polarized toward the susceptible target cell (27). The actual transfer of virus at the VS occurs through different mechanisms, such as the conventional budding of virions from the effector cell, followed by CD4 and coreceptor binding and subsequent virion fusion with the target cell. However, other mechanisms of viral transfer at the VS have also been reported, including formation of nanotubes (48) or filopodia (44), fusion of cells into syncytia, and endocytosis of budding virions (32). Although the dominant mechanism of cell-to-cell spread remains to be determined, this mode of viral dissemination has been suggested as an immune evasion mechanism offering protection from viral entry inhibitors and neutralizing antibodies (15, 43). However, most research on microbicides has been focusing on the inhibition of cell-free HIV; thus, it is uncertain whether the activity of candidate microbicides against CFV observed in vitro also implies activity against CAV (4).
A nonpolarized coculture of HIV-infected effector cells with uninfected permissive target cells would allow the study of HIV cell-to-cell spread from different cell sources, such as infected T cells or macrophages. However, in contrast to CFV stocks, HIV-infected cells cannot be separated from the assays' target cells, thus complicating viral titration, which is required when comparing different viral stocks. Readout of extracellular Gag p24 to assess productive infection of the target cells will be blurred by the presence of p24-producing effector cells. Moreover, the presence of target cells can influence the infection of the effector cells, and vice versa. Consequently, although primary cells would be the most relevant in vitro target cells, they are not suited to the rapid screening of multiple microbicides against CAV. One study by Buffa et al. (10) circumvented these problems by using the expression of firefly luciferase (FL) to assess target cell infection. To this end, HIV-infected PM-1 T cells were cocultured with TZM-bl reporter cells containing a firefly luciferase reporter gene. However, it is questionable whether the cervical-carcinoma-derived TZM-bl cells are a suitable model for the in vivo target cells in the vaginal mucosa. Furthermore, as it is a single-cycle assay, it does not support the evaluation of antiretrovirals that inhibit late stages of the viral life cycle, such as protease inhibitors (PIs). Finally, because the TZM-bl model uses the polycation DEAE-dextran to enhance CFV infection, a reliable comparison between the inhibition of CFV and CAV might be troublesome.
Therefore, in this study, we developed an improved in vitro coculture assay based on this FL approach to evaluate the efficacy of candidate microbicides against cell-to-cell transmission of HIV compared to CFV transmission. The model represents a “worst-case scenario,” where the HIV-infected seminal cells can reach uninfected permissive cells within the epithelium and submucosal tissue. The use of two T-cell lines (R5MaRBLE [R5M] and X4MaRBLE [X4M]) (14) and the quantification of both luciferase activity and the Gag p24 concentration allowed us not only to measure the infection of the target cells, but also to discriminate between the ongoing infection in the effector cells and the total infection of the coculture (effector plus target cells). In this model, the reverse transcriptase inhibitors dapivirine (TMC120), UC781, and tenofovir (TFV); the protease inhibitors lopinavir (LPV) and saquinavir (SQV); the specific entry inhibitors enfuvirtide (T-20), M48-U1, CD4-immunoglobulin G2 (CD4-IgG2), b12, BMS-806, C34-cholesterol (C34-chol), griffithsin (GRFT), and maraviroc (MVC); and the integrase inhibitor raltegravir (RAL) were evaluated for their activities against different R5-tropic HIV-infected leukocytes derived from primary cells of a healthy donor. Our results indicate that all antiretrovirals were able to block CAV, although all entry inhibitors showed a small decrease in activity compared to CFV transmission.
MATERIALS AND METHODS
Antiretroviral compounds.
The gp41-mediated fusion inhibitor T-20 (20), the CCR5 antagonist MVC (20), the protease inhibitor LPV (20), and the integrase inhibitor RAL (20) were obtained through the NIH AIDS Research and Reference Reagent Program, Germantown, PA. The nucleotide reverse transcriptase inhibitor (NtRTI) TFV (20) and the nonnucleoside reverse transcriptase inhibitor (NNRTI) UC781 (6) were kindly provided by Jan Balzarini (Rega Institute, Leuven, Belgium), while the NNRTI dapivirine (TMC120) (6) and the small-molecule CD4-binding site inhibitor BMS-806 (6) were donated by Tibotec BVBA, Beerse, Belgium. The protease inhibitor SQV (20) was obtained through the National Institute for Biological Standards and Control (NIBSC) Centralized Facility for AIDS Reagents. The fusion inhibitor C34-chol (26), the glycan binding lectin GRFT (35), and the CD4-binding site inhibitor M48-U1 (51), respectively, were kindly provided by Michael Miller (Merck Research Laboratories, West Point, PA), Kenneth Palmer (University of Louisville, Louisville, KY), and Loïc Martin (CEA, Paris, France). The monoclonal antibody b12 (11) and CD4-IgG2 (3), respectively, were obtained from Polymun Scientific GmbH (Austria) and Progenics Pharmaceuticals (New York, NY) (Table 1). Compounds were dissolved in dimethyl sulfoxide (DMSO) or water and used at noncytotoxic concentrations as determined by the WST-1-tetrazolium cytotoxicity assay (Roche Diagnostic, Mannheim, Germany) as described by Gali et al. (21).
Table 1.
Overview of antiretroviral compounds
| Compound | Abbreviation | Drug class | Target | Molecular wt | Chemical type |
|---|---|---|---|---|---|
| BMS-806 | BMS-806 | Entry inhibitor | CD4-binding site on gp120 | 406 | Small molecule |
| M48-U1 | M48-U1 | Entry inhibitor | CD4-binding site on gp120 | 3,048 | Miniprotein |
| CD4-immunoglobulin | CD4-IgG2 | Entry inhibitor | CD4-binding site on gp120 | 200,000 | Protein |
| Neutralizing antibody b12 | b12 | Entry inhibitor | CD4-binding site on gp120 | 150,000 | Protein |
| Griffithsin | GRFT | Entry inhibitor | Glycans on gp120 | 12,700 | Lectin |
| Enfuvirtide | T-20 | Entry inhibitor | gp41 | 4,492 | Peptide |
| C34-cholesterol | C34-chol | Entry inhibitor | gp41 | 5,021 | Peptide plus cholesterol |
| Maraviroc | MVC | Entry inhibitor | CCR5 coreceptor | 514 | Small molecule |
| Saquinavir | SQV | Protease inhibitor | Protease | 767 | Small molecule |
| Lopinavir | LPV | Protease inhibitor | Protease | 629 | Small molecule |
| Dapivirine | TMC120 | Nonnucleoside reverse transcriptase inhibitor | Reverse transcriptase | 329 | Small molecule |
| UC781 | UC781 | Nonnucleoside reverse transcriptase inhibitor | Reverse transcriptase | 336 | Small molecule |
| Tenofovir | TFV | Nucleotide reverse transcriptase inhibitor | Reverse transcriptase | 287 | Small molecule |
| Raltegravir | RAL | Integrase strand transfer inhibitor | Integrase | 444 | Small molecule |
Cell lines used as non-HIV-infected target cells.
The T-cell lines X4M and R5M (14) naturally express CXCR4 and CD4 but were engineered to express the FL reporter protein under HIV-1 long terminal repeat (LTR) promoter regulation. In addition, the R5M cell line was genetically modified to express the CCR5 receptor, allowing R5-tropic HIV infection (14). Both X4M and R5M cells were maintained at 0.1 × 106 to 2 × 106/ml in RPMI 1640 containing 10% heat-inactivated fetal calf serum (FCS), 100 U/ml penicillin, 100 μg/ml streptomycin (Lonza, Verviers, Belgium), 250 μg/ml Geneticin (Invitrogen, Merelbeke, Belgium), and 0.1 μg/ml puromycin (Sigma-Aldrich, Bornem, Belgium) with (R5M) or without (X4M) 150 μg/ml hygromycin B (Roche Diagnostics, Mannheim, Germany). Saturated cultures were split 1:3 or 1:4 every 3 to 4 days.
In vitro generation of HIV-infected effector cells.
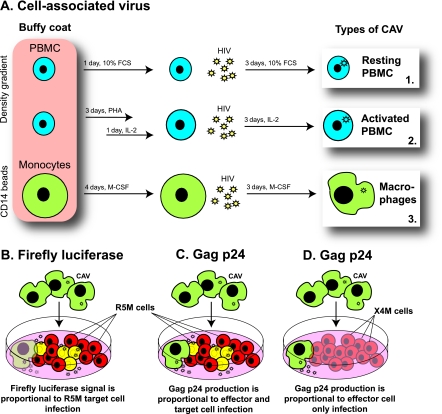
Lymphoprep density gradient centrifugation (Lucron, Sint Martens-Latem, Belgium) was used to separate human peripheral blood mononuclear cells (PBMC) from buffy coats of healthy donors provided by the Antwerp, Belgium, Red Cross Transfusion Center. These PBMC were aliquoted and cryopreserved in FCS with 10% DMSO. Monocytes (MO) and CD4+ T cells were isolated from the remaining PBMC by magnetic isolation using CD14 and CD4 microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions and also aliquoted and cryopreserved in FCS with 10% DMSO. At the start of each experiment, two PBMC aliquots and one MO aliquot were thawed at 37°C and seeded at 1 × 106 to 2 × 106/ml in a 6-well plate (Becton Dickinson Biosciences, Franklin Lake, NJ). The PBMC aliquots were either incubated for 5 days in 10% FCS medium (RPMI 1640 containing 10% FCS and 50 μg/ml gentamicin) or stimulated for 48 h in phytohemagglutinin (PHA) medium (RPMI 1640 containing 10% FCS and 50 μg/ml gentamicin [Lonza, Verviers, Belgium] supplemented with 2 μg/ml Polybrene [Sigma-Aldrich, Bornem, Belgium] and 2 μg/ml PHA [Remel, Kent, United Kingdom]) and subsequently activated for 24 h in interleukin-2 (IL-2) medium (RPMI 1640 medium with 15% FCS and 50 μg/ml gentamicin supplemented with 1 ng/ml IL-2 [Gentaur, Brussels, Belgium], 2 μg/ml Polybrene, and 5 μg/ml hydrocortisone [Calbiochem, Leuven, Belgium]) (Fig. 1A). Similarly, the aliquots with CD4+ T cells were either activated in PHA and IL-2 medium or left untreated in 10% FCS medium to be used as a control. The MO aliquot was incubated for 7 days in 10% FCS medium containing 50 ng/ml human macrophage colony-stimulating factor (M-CSF) (PeproTech, London, United Kingdom) in order to induce differentiation into macrophages. During the last 3 days of incubation, the MO and PBMC cultures were infected with the R5 subtype B strain BaL at a multiplicity of infection (MOI) of 10−3, resulting in three sources of cell-associated virus (i.e., HIV-infected effector cells), which we refer to here as resting PBMC, activated PBMC, and macrophages (Fig. 1A). At the end of the incubation period, the infected cells were gently scraped (Greiner Bio-One) from the 6-well plate and extensively washed to remove the cell-free virus in the supernatant.
Fig 1.
Experimental setup. (A) In vitro generation of HIV-infected effector cells. Human PBMC were either activated with PHA and IL-2 or left untreated, while monocytes from the same donor were differentiated to macrophages. During the last 3 days of culture, all three cell types were infected with the R5-tropic subtype B strain BaL, resulting in three sources of HIV-infected effector cells: resting PBMC, activated PBMC, and macrophages. (B) Measuring infection of target cells. In a coculture of BaL-infected effector cells (green cells) with a firefly-luciferase-containing target T-cell line, R5M (red cells), luminescence originates only from infected R5M target cells (yellow cells). (C) Measuring infection of the coculture. In a coculture of BaL-infected effector cells with the HIV-permissive T-cell line R5M, the Gag p24 concentration in the supernatant quantifies the infection of both effector and target cells. (D) Measuring infection of effector cells. In a coculture of BaL-infected effector cells with the X4M T-cell line (blue cells), which lacks the coreceptor CCR5, the Gag p24 concentration in the supernatant quantifies the infection of the effector cells only, as the X4M cells do not become productively infected.
Phenotyping of HIV-infected effector cells.
HIV-infected effector cells were stained for 15 min at 4°C with the following antibodies: anti-CD4 (phycoerythrin [PE]), anti-CD3 (fluorescein isothiocyanate [FITC] or PE), anti-CD13 (PE), and anti-CD14 (FITC) (all antibodies were purchased from BD Biosciences), subsequently washed in phosphate-buffered saline (PBS) with 1% bovine serum albumin, and finally fixed in PBS with 1% paraformaldehyde. The samples were analyzed using a FACScan (BD Biosciences), and data analysis was performed with FlowJo version 8.8.4 (Tree Star Inc., San Carlos, CA).
TCID50 determination of cell-free HIV and HIV-infected effector cells.
The 50% tissue culture infective dose (TCID50) was determined for cell-free BaL, as well as for BaL-infected effector cells (i.e., resting PBMC, activated PBMC, and macrophages) using the R5M cell line as HIV target cells. The FL reporter enzyme is transcribed and expressed under the influence of Tat-long terminal repeat (LTR) interaction and is therefore restricted to infected R5M cells. Light emission after substrate addition, therefore, allows the specific quantification of de novo infection of these target cells and does not measure virus production by the HIV-infected effector cells. In practice, six replicates of 5 × 104 R5M cells in 10% FCS culture medium were incubated in a 96-well plate with serially diluted cell-associated or cell-free virus for 7 days at 37°C, 5% CO2. Subsequently, 75 μl cell suspension was added to 75 μl Steadylite HTS (Perkin Elmer, Life Sciences, Zaventem, Belgium) to quantify luciferase activity from lysed R5M cells using a TriStar LB941 luminometer (Berthold Technologies GmbH and Co. KG, Bad Wildbad, Germany). Viral titers were calculated using the Reed and Muench method and expressed as infectious units per milliliter (for cell-free HIV) or as the number of cells needed for 50% infection (for HIV-infected effector cells) (41).
Integrated proviral DNA quantification.
DNA was extracted from cells using the QiaAmp DNA minikit (Qiagen) according to the manufacturer's instructions. Subsequently, a two-step PCR amplification was performed according to a protocol adapted from O'Doherty et al. (37). An initial preamplification was done using the following primers: genomic Alu forward (5′-GCC TCC CAA AGT GCT GGG ATT ACA-3′) and HIV-1 Gag reverse (5′-GCT CTC GCA CCC ATC TCT CTC C-3′). The reactions were done in a 50-μl volume: 10 mM Tris-HCl, 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphates (dNTPs), 100 nM Alu forward primer, 600 nM Gag reverse primer, and 2.6 U of enzyme (Expand High Fidelity PCR system; Roche Diagnostics). The thermal cycling was done using an S1000 Thermal Cycler (Bio-Rad) and consisted of a 2-min initial denaturation step at 94°C, followed by 20 cycles of denaturation at 93°C for 30 s, annealing at 50°C for 1 min, and extension at 70°C for 1 min 40 s. The second-round real-time protocol was done using 5 μl of the preamplification material. The sequences of the primers and the probe were as follows: LTR forward, 5′-GCC TCA ATA AAG CTT GCC TTG A-3′; LTR reverse, 5′-TCC ACA CTG ACT AAA AGG GTC TGA-3′; and LTR beacon probe, 5′- 6-carboxyfluorescein (FAM)-GCG AGT GCC CGT CTG TTG TGT GAC TCT GGT AAC TAG CTC GC-4-(4′-dimethylaminophenylazo) benzoic acid (DABCYL)-3′. Reactions were performed in a volume of 50 μl using the TaqMan PCR master mix (Applied Biosciences) with 250 nM LTR forward primer, 250 nM LTR reverse primer, and 200 nM LTR probe. The thermal cycling was as follows: an initial denaturation step at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 15 s and extension at 60°C for 1 min (ABI 7500; Applied Biosystems). Each sample was tested in duplicate. Viral loads were calculated using an external curve for HIV proviral DNA prepared by serially diluting 8E5 human T-lymphocytes, each cell containing one copy of integrated HIV DNA. The threshold of detection was 100 copies/106 cells.
Abilities of candidate microbicides to inhibit cell-free HIV versus cell-associated HIV.
Fourteen antiretrovirals of different classes (Table 1) were evaluated for their activities against the cell-free BaL virus and BaL-infected effector cells (i.e., resting PBMC, activated PBMC, and macrophages). The firefly luciferase activity and p24 concentration were measured as endpoints for antiviral activity on two different non-HIV-infected target cell lines, namely, R5M and X4M. Both cell lines contain a firefly luciferase reporter gene and differ only in the expression of the CCR5 receptor, which allows R5-tropic HIV infection. Consequently, in a coculture of HIV-infected effector cells with the R5M target cells, de novo infection of the target cells can be quantified by measuring firefly luciferase activity (Fig. 1B), whereas the total infection of the coculture (i.e., effector plus target cells) can be quantified as the p24 concentration in the supernatant (Fig. 1C). Furthermore, since X4M cells are not permissive for the R5-tropic BaL virus, the ongoing infection in the effector cells can be quantified by measuring p24 concentrations in the supernatant of a coculture of X4M target cells with the HIV-infected effector cells (Fig. 1D). To this end, either 5 × 104 R5M or 5 × 104 X4M cells were preincubated in triplicate wells of a 96-well plate for 30 min at 37°C, 5% CO2 in the presence or absence of serial dilutions of the respective compound. Subsequently, 100 TCID50 of either cell-free BaL virus, BaL-infected resting PBMC, activated PBMC, or macrophages was added to each well up to a total volume of 200 μl in 10% FCS medium. The cultures were incubated for 7 days, and subsequently, the cumulative p24 production of the effector plus R5M or effector plus X4M coculture was quantified using an in-house p24 antigen capture enzyme-linked immunosorbent assay (ELISA) (8). Briefly, 50 μl culture supernatant was pooled from the triplicate wells for each compound dilution and serially diluted in NP-40 to bypass the limited range of the in-house ELISA. Absolute p24 concentrations were then calculated and plotted against the compound concentrations. Finally, the luciferase activity of the effector plus R5M cocultures was quantified with a TriStar LB941 luminometer after the addition of 75 μl Steadylite HTS to 75 μl cell suspension. Percentages of viral inhibition were calculated compared to untreated control wells and plotted against the compound concentrations. Nonlinear regression analysis was used to calculate the 50% effective concentration (EC50) based on at least three independent measurements and using GraphPad Prism version 5.03 for Windows (GraphPad Software, San Diego, CA). Significant differences were calculated using a Z test with Bonferroni correction.
Relative importance of cell-to-cell spread in a coculture of R5M target cells with HIV-infected effector cells.
Briefly, 1 × 105 and 3 × 104 HIV-infected effector cells were physically separated from 2.4 × 105 R5M target cells by a virion-permeable membrane (3-μm pore size) in a total volume of 1 ml using a 24-well Transwell system (Corning, NY) with the effector cells placed in the apical compartment and the target cells in the basal compartment. Similarly, cocultures of effector and target cells were set up in the same system by adding the effector cells directly to the R5M target cells in the basal compartment while adding only 10% FCS medium to the apical compartment. After 5 days of incubation, 75 μl of cell suspension from the basal compartment was used to determine target cell infection by measuring firefly luciferase activity. Cell-to-cell spread of HIV through the virion-permeable membrane was subsequently calculated as the target cell infection in the separated cultures relative to the target cell infection in the corresponding cocultures and was compared to cell-free HIV spread in the same system. Additionally, the TCID50 of the infected effector cells was determined as described above, but this time using a 96-well Transwell system with the effector cells placed in the apical compartment and the target cells in the basal compartment. Viral titers were calculated using the Reed and Muench method and expressed as a percentage relative to the titer obtained in cocultures of effector and target cells.
RESULTS
In vitro generation of HIV-infected effector cells.
As the infectious potential of real seminal leukocytes from HIV+ patients is too low for anti-HIV activity screening in vitro, infected T cells and macrophages were generated from PBMC as a model for infected seminal leukocytes. To this end, PBMC purified from healthy donors were either activated with PHA/IL-2 or left untreated in medium without activation stimulus. Monocyte-derived macrophages were generated using M-CSF. Because R5-tropic viruses are predominantly transmitted in vivo (31), all three cell types were infected with the R5 subtype B strain BaL, resulting in three sources of cell-associated virus (i.e., HIV-infected effector cells), which are referred to here as resting PBMC, activated PBMC, and macrophages. As a control for the titration experiments, CD4+ T cells were also obtained from the same donor and treated similarly to the activated and resting PBMC. Fluorescence-activated cell sorting (FACS) analysis confirmed that the resting PBMC population comprised a mixture of mainly T cells and monocytes, whereas the activated PBMC population primarily contained CD4+ T cells (60%) and, to a lesser extent, CD4− T cells (30%). The infected macrophages were confirmed to be a CD13+/CD14+ homogeneous population, which was weakly CD4+ (data not shown).
Infectious potential of cell-free HIV and HIV-infected effector cells.
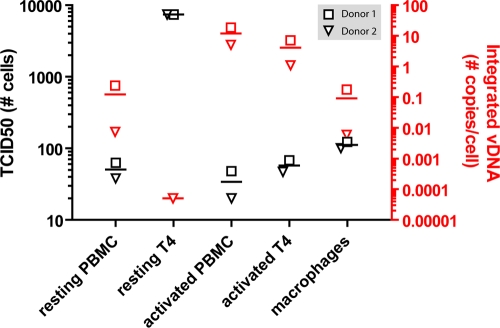
A dilution series of virus (either cell-free HIV or HIV-infected effector cells) was added to R5M target cells and incubated for 7 days. De novo infection of the target cells was subsequently quantified using the firefly luciferase reporter enzyme. During their in vitro generation, the various effector cell types had been exposed to the same concentration of infectious BaL virions. However, activated and resting PBMC infected with these virions showed a higher infectious potential (TCID50s of 34 and 50 cells, respectively) on R5M cells than on macrophages (TCID50, 123 cells) (Fig. 2, black symbols). Furthermore, the activated PBMC showed titers similar to those of activated CD4+ T cells purified from the same buffy coat, suggesting that CD4+ T cells within the activated PBMC population account for the observed titers. On the other hand, resting CD4+ T cells showed a severe decrease in infectious potential (TCID50, 7,400 cells) compared to resting PBMC (Fig. 2, black symbols).
Fig 2.
Infectious titer and proviral DNA load of HIV-infected effector cells. The TCID50 of BaL-infected effector cells (i.e., resting PBMC/CD4+ T cells, activated PBMC/CD4+ T cells, and macrophages) was determined in a 7-day coculture with R5M target cells. Firefly luciferase was used to assess target cell infection, and the TCID50 was calculated using the method of Reed and Muench, expressed as the number of cells needed for 50% infection and plotted on the left y axis (black symbols). The integrated proviral DNA load was determined using a two-step Alu-Gag PCR. Each sample was tested in duplicate. Viral loads were calculated using an external curve for HIV proviral DNA by serially diluting 8E5 T cells and were plotted on the right y axis (red symbols). The threshold of detection was 100 copies/106 cells. The triangles and squares represent two PBMC donors from which the effector cells were generated. Horizontal lines represent the means of the depicted points.
All three types of HIV-infected effector cells produced p24 protein over time, which suggests that these cells are productively infected. However, the observed p24 increase could also be caused by the passive release of virions from intracellular compartments. We therefore performed Alu-Gag PCR on the effector cells to measure HIV integration. Overall, the infectious potentials of the cell populations mirror the proviral DNA load, with activated PBMC having the highest (10 copies/cell) and macrophages having the lowest (0.1 copies/cell) copy numbers of integrated viral DNA. Again, activated CD4+ T cells had copy numbers similar to those of activated PBMC, whereas resting CD4+ T cells contained an amount of integrated HIV DNA several orders of magnitude smaller than that of resting PBMC (Fig. 2, red symbols).
Abilities of candidate microbicides to prevent the infection of target cells.
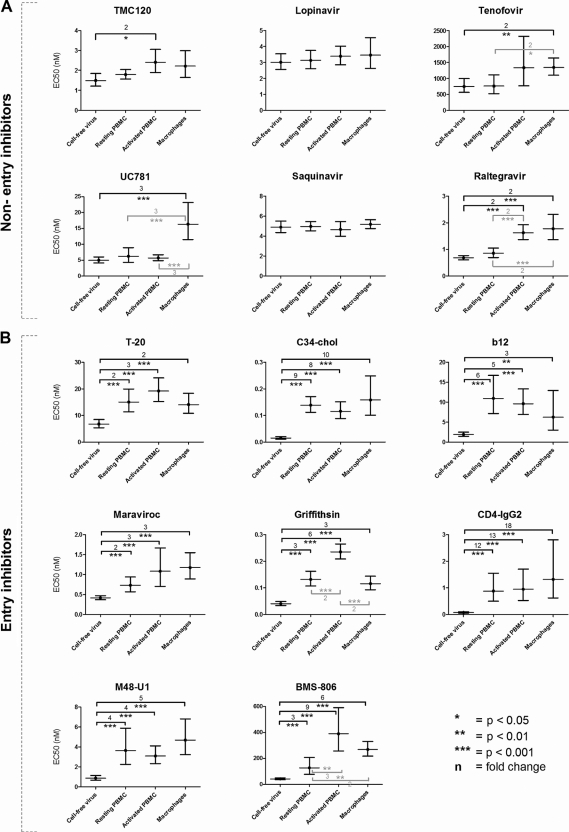
Fourteen antiretrovirals of different classes (the entry inhibitors MVC, T-20, C34-chol, BMS-806, CD4-IgG2, b12, M48-U1, and GRFT; the NNRTIs TMC120 and UC781; the NtRTI TFV; the integrase inhibitor RAL; and the protease inhibitors LPV and SQV) (Table 1) were evaluated for the ability to prevent infection of target cells by cell-free BaL and the BaL-infected effector cells. The selected entry inhibitors differed in their modes of action (binding to CCR5, gp41, the CD4 binding site, or glycans on gp120), chemical classes (neutralizing antibodies, small-molecule inhibitors, or peptides), and molecular masses (0.4 to 200 kDa). Briefly, 100 TCID50 of either cell-free virus, resting PBMC, activated PBMC or macrophages was added to R5M target cells and incubated for 7 days in the presence of serial dilutions of the respective compound, and firefly luciferase was used to assess target cell infection. For the antiretrovirals tested, the EC50 values obtained in R5M cells against CFV were similar to those generally obtained in PBMC (data not shown). Moreover, all these compounds were able to block infection of the R5M target cells by all three sources of cell-associated BaL (Fig. 3). In general, compared to the inhibition of CFV, most non-entry inhibitors retained their activity against CAV. However, a small decrease in activity against macrophage-associated HIV was observed for the NNRTI UC781 (3-fold), the NtRTI tenofovir (2-fold), and the integrase inhibitor RAL (2-fold). Against activated PBMC, activity was reduced for the NNRTI TMC120 and the integrase inhibitor RAL (2-fold) (Fig. 3A). On the other hand, all entry inhibitors showed more clear-cut losses in activity against all sources of cell-associated virus ranging from 2- to 18-fold. The largest differences were observed for the CD4-binding site inhibitor CD4-IgG2 and the fusion inhibitor C34-chol and the smallest for the CCR5 antagonist MVC and the fusion inhibitor T-20 (Fig. 3B). Interestingly, the entry inhibitors C34-chol and GRFT were the most potent of all tested compounds, showing EC50 values in the pM range regardless of the viral source (Fig. 3).
Fig 3.
EC50 values of 14 antiretrovirals for cell-free HIV and HIV-infected effector cells using firefly luciferase measurement on R5M cells. One hundred TCID50 of either cell-free virus, resting PBMC, activated PBMC, or macrophages was added to R5M target cells and incubated for 7 days in the presence of a serial dilution of the respective compound. Firefly luciferase was used to assess target cell infection, and the EC50 was calculated using nonlinear regression analysis on data from at least three to six independent experiments (GraphPad Prism). Nonentry inhibitors (A) and entry inhibitors (B) are shown (Table 1). The error bars depict the 95% confidence intervals, while the numbers and brackets represent the fold changes between cell-free virus and cell-associated virus (black) or between two types of cell-associated virus (gray). Significant differences were calculated using a Z test with Bonferroni correction; ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001.
Abilities of candidate microbicides to inhibit ongoing productive infection in the effector cell populations.
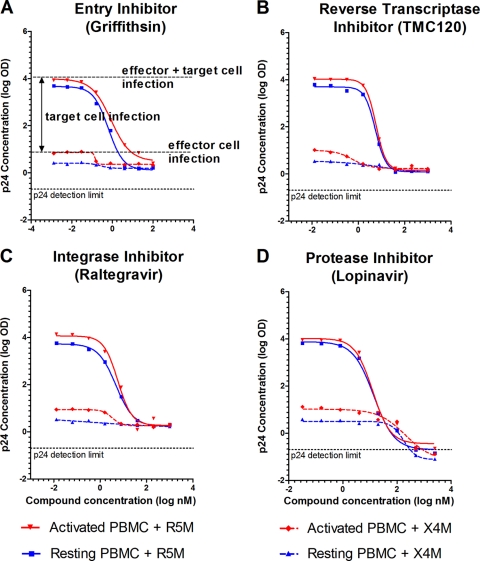
Whereas firefly luciferase is used to assess target cell infection, the Gag p24 concentration can be used to discriminate between the total productive infection of the coculture (i.e., effector plus target cells) and the ongoing infection in the effector cells. As R5M cells are permissive for R5-tropic HIV infection, the cumulative p24 output in cocultures of R5M plus effector cells represents the infection of both effector and target cells (Fig. 1C). However, the infection of solely the effector cells can also be quantified by measuring the p24 concentration in X4M plus effector cell cocultures. The X4M cells will not be infected by R5-tropic BaL and control for cell-cell interactions that might influence virus production by the effector cells (Fig. 1D). Consistent with the firefly luciferase data, all tested antiretrovirals inhibited total p24 production of the R5M plus effector cell cocultures (Fig. 4). Interestingly, the p24 production of the X4M plus effector cell cocultures was also inhibited by all the antiretrovirals, including the entry inhibitors (Fig. 4A). As expected, no firefly luciferase activity was measured in the CCR5-negative X4M reporter cell line. Clearly, all antiretrovirals not only block viral spread to the target cells, but also inhibit viral spread within the effector cell population. Of all compounds tested, only the protease inhibitors SQV and LPV were able to fully inhibit (i.e., below the detection limit) p24 production by the effector cells (Fig. 4D).
Fig 4.
Dose-response curves of four antiretrovirals for cell-free HIV and HIV-infected effector cells using Gag p24 measurement on R5M and X4M cells. One hundred TCID50 of resting PBMC (blue) or activated PBMC (red) was added to either R5M or X4M target cells and incubated for 7 days in the presence of a serial dilution of the respective compound. Cumulative Gag p24 was subsequently measured in the supernatant of R5M cocultures to assess effector plus target cell infection (full regression lines) or in the supernatants of X4M cocultures to assess ongoing infection in the effector cells (dashed regression lines) and plotted against the compound concentrations. Nonlinear regression was performed on data from one representative experiment (GraphPad Prism). As similar results were obtained for the other eight compounds, only one representative inhibitor of each antiretroviral class is depicted. OD, optical density.
Relative importance of cell-to-cell spread in a coculture of R5M target cells with HIV-infected effector cells.
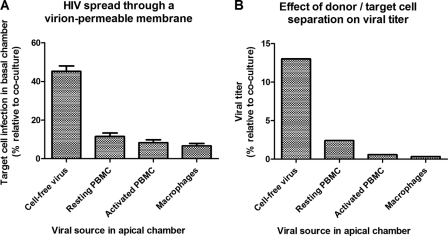
As the observed differences between inhibition of CFV and CAV were not spectacular (1.8- to 18-fold), it could be argued that cell-to-cell spread of HIV is not the main route of viral spread in our coculture model and that instead mainly cell-free virus is transmitted between effector and R5M cells. Therefore, we assessed the relative importance of cell-to-cell versus cell-free spread in our system by physically separating effector and target cells with a virion-permeable membrane. The R5M target cell infection was then assessed using the firefly luciferase readout and compared to the target cell infection obtained when target and effector cells are cocultured in a single compartment (Fig. 5A). To prevent cell migration over the virus-permeable membrane, we first used membranes with a pore size of 0.4 μm. These membranes appeared inadequate, as they reduced the CFV infectivity by almost 90% (data not shown), suggesting that most virions were retained in the apical compartment. Therefore, experiments were conducted using a more permeable membrane (3-μm pore size), which reduced the CFV target cell infection by only 50%. However, when the HIV-infected resting PBMC, activated PBMC, and macrophages were separated from the R5M target cells, target cell infection was reduced by 89, 92, and 94%, respectively (Fig. 5A). Because we used an R5-tropic HIV strain, no syncytia were microscopically observed to appear in the cocultures. Hence, these figures suggest that only 6 to 11% of the infection in coculture is due to CFV, although this might be an overestimation, as small numbers of effector cells migrated through the virion-permeable membrane. Corresponding to the relative target cell infection, higher numbers of resting PBMC than of activated PBMC and macrophages could be observed crossing the membrane barrier (data not shown), which might be explained by their cell size (resting PBMC < activated PBMC < macrophages). Similar results were obtained when the viral titer was assessed instead of target cell infection. Although the infectious potential of CFV was reduced to 12.5% when using the virion-permeable membrane, resting PBMC, activated PBMC, and macrophages retained only 2.4, 0.6, and 0.3% of their infectious potential in coculture (Fig. 5B).
Fig 5.
Relative importance of cell-to-cell spread in a coculture of R5M target cells with HIV-infected effector cells. (A) Cell-free HIV or HIV-infected effector cells (x axis) were physically separated from the R5M target cells using a virion-permeable membrane with 3-μm pores in a 24-well Transwell system with the cell-free virus or effector cells placed in the apical compartment and the target cells in the basal compartment. In parallel, cocultures of cell-free virus or effector cells with R5M target cells were set up in the basal compartment. After 5 days of incubation, R5M infection was assessed using firefly luciferase. Values for target cell infection by cell-free HIV or HIV-infected effector cells when separated from the target cells are expressed as percentages of those obtained in the respective cocultures (y axis). Mean percentages ± standard errors of the mean (SEM) of three independent experiments are depicted. (B) Additionally, the TCID50 of the infected effector cells was determined as described in the legend to Fig. 2 using a 96-well Transwell system with the effector cells placed in the apical compartment and the target cells in the basal compartment. Viral titers were calculated using the method of Reed and Muench and expressed as percentages of the titer obtained in cocultures of effector and target cells. One experiment is represented.
DISCUSSION
In this study, we describe a new in vitro assay to evaluate the efficacies of candidate microbicides against cell-to-cell and cell-free HIV spread and subsequently apply this method to assess the antiviral efficacies of a series of inhibitors with different mechanisms of action. We show that all tested antiretrovirals, including the entry inhibitors, are able to block target cell infection by all three sources of CAV. This opposes the hypothesis that virological synapse-mediated viral spread is an immune or entry inhibitor evasion mechanism (43). Some studies on DC-T- and T-T-cell synapses indeed suggested that cell-to-cell spread is resistant to neutralizing monoclonal antibodies, such as 2F5 (13, 22), and other specific entry inhibitors, such as the fusion inhibitor T-20 (7, 13, 22, 50) and the CCR5 inhibitor TAK779 (15, 50). However, the same compounds were shown to block cell-to-cell spread in other studies (28, 33, 34, 40). A possible explanation for these discrepant results has been provided by Puigdomenech et al. (40) by distinguishing “HIV transfer” from “HIV transmission.” The authors suggest that primary cells are highly sensitive to the physical passage of virions across synapses in a fusion- and coreceptor-independent manner called “HIV transfer.” Once the virions are transferred, they can infect the target cells through a fusion- and coreceptor-dependent mechanism termed “HIV transmission.” Thus, whereas “HIV transfer” comprises just the physical transfer of virions between cells, “HIV transmission” also includes the productive infection following the initial transfer. Although HIV transmission is a more relevant endpoint, earlier studies on T-cell-to-T-cell spread measured HIV transfer rather than HIV transmission by determining intracellular Gag p24 concentrations (13, 15). Using a different approach based on real-time PCR, quantifying HIV integration as a measure of HIV transmission, antibodies and other entry inhibitors, such as TAK779 and T-20, were shown to interfere efficiently with cell-to-cell spread (33, 34). Electron tomography also revealed that virological synapses are relatively permeable structures that can be accessed by entry inhibitors (33). The observations in our assay, which uses a firefly luciferase approach to measure HIV transmission, are consistent with these recent studies. However, other groups quantified Gag p24 in the culture supernatant, yet another measure of HIV transmission, and observed a severe loss of T-20 activity (7, 50). In these studies, the compounds were removed after a certain period, which could explain the conflicting results. Terrazas et al. (50) used freshly isolated resting PBMC, which were incubated overnight with HIV as a model for CAV. As these cells do not contain integrated viral DNA at the start of the experiment, while the average HIV-1 generation time is estimated to be 2.6 days (39), T-20 will not be able to prevent target cell infection when it is removed before its target site in gp41 becomes available. Recent experiments in our laboratory confirmed this hypothesis, as they showed that T-20 regains its activity when used continuously during culture. Similarly, Balzarini et al. (7) removed T-20 after loading dendritic cells with HIV. As HIV uptake by DCs occurs through a fusion- and coreceptor-independent mechanism (15), T-20 will not be able to prevent the transfer of HIV particles to an intracellular compartment within these DCs. When the cells are subsequently washed and added to the target cells, the virions will be released from the DCs, but T-20 will no longer be present to inhibit the gp41-mediated fusion with the target cells. Altogether, these studies underline the importance of assay parameters and endpoints when interpreting results.
Although we show that cell-to-cell spread is not inherently resistant to entry inhibition, a decrease in activity against CAV could be observed for all entry inhibitors. However, the gp120 CD4-binding site inhibitors CD4-IgG2, b12, BMS-806, and M48-U1 showed larger reductions in efficacy (3- to 18-fold) than the CCR5 antagonist MVC and the fusion inhibitor T-20 (2- to 3-fold). This is consistent with other studies that showed a loss of efficacy against CAV for inhibitors that interfere with CD4-gp120 binding (such as b12, Q4120, and CD4-IgG2), whereas CCR5 inhibitors (such as TAK779) and fusion inhibitors (such as T-20 and the monoclonal antibodies 2F5 and 4E10) largely remained unaffected by the mode of virus transmission (1, 33). The activity against cell-to-cell spread does not appear to be influenced by the molecular size and chemical type of the tested entry inhibitors, given the fact that BMS-806 is a small-molecule inhibitor, b12 a neutralizing antibody, and T-20 a peptide. Overall, the observed decrease in efficacy against cell-to-cell transmission is rather small, and the EC50s remain well below the toxic concentration for all entry inhibitors. The fusion inhibitor C34-chol and the glycan-binding lectin GRFT remain active even in the pM range. Hence, it is unlikely that these small reductions in efficacy will have an effect in vivo, when much higher concentrations will be applied. Because GRFT can be manufactured very cheaply in tobacco leaves (38) and because C34-chol has hydrophobic properties due to the cholesterol molecule attached to C34 (26), these observations add to the already increasing interest in listing both compounds as candidate vaginal microbicides.
Finally, using the X4M cell line, we showed that all the compounds were also able to prevent viral spread from infected to uninfected effector cells. However, only the protease inhibitors completely inhibited Gag p24 production originating from the infected effector cells. This observation can be explained by the fact that PIs prevent the maturation of virions by inhibiting the cleavage of p55 and p160 into p17 and p24. Furthermore, using the R5M cell line, the PIs were the only compounds that could inhibit all types of effector cells at equal concentrations. As they act on the late stages of the viral life cycle and cannot prevent cell-free HIV from entering target cells, PIs have only recently been considered possible microbicides (9, 19, 49). In fact, vaginal HIV transmission is currently assumed to occur through the formation of a small founder population of infected target cells that subsequently expands locally using the influx of new target cells recruited through outside-in signaling (25). Hence, although PIs cannot prevent the initial infection of target cells, they could use this window of opportunity to prevent further amplification of the initial infection (5). Moreover, within the context of cell-associated virus, PIs could have an advantage over other inhibitors by disabling virions budding from infected seminal cells. As other authors have shown that PIs can also block cell-to-cell endocytosis in dendritic cells (36), we believe that the addition of PIs to future combination microbicides could be favorable.
The in vitro assay that we describe in this study has several advantages over other in vitro models (29, 30) that were recently used to measure HIV transmission in cocultures of HIV-infected effector cells with uninfected target cells. The use of a firefly-luciferase-expressing target cell line allows the rapid screening of multiple microbicides in microtiter plates. Furthermore, the quantification of Gag p24 output enables discrimination between the productive infection of target cells, effector cells, or both. In contrast to the TZM-bl system, multiple viral life cycles are permitted, which allows the evaluation of protease inhibitors acting on the late stages of the viral life cycle and eliminates the need for DEAE-dextran to enhance CFV infection. Moreover, our assay models the in vivo situation more realistically, since it is based on a coculture of primary effector cells with a target T-cell line. However, as with all models, there are limitations relating to both target and effector cells. The R5M target cells in our assay are genetically engineered to express high levels of the CCR5 coreceptor. This might promote cell-free viral spread over cell-to-cell spread. Moreover, the formation of virological synapses might be altered due to differences in surface receptors between CD4+ T-cell lines and primary CD4+ T cells. Thus, although cell-to-cell spread has been shown to be the dominant mode of virus dissemination in primary cell cultures (47), it remains uncertain whether the same holds true for cocultures of primary cells with the R5M cell line. However, using virion-permeable membranes, we showed that cell-to-cell spread does occur in our system and is responsible for the majority (>80%) of new infections. The effector cells, on the other hand, were derived from human PBMC and subsequently infected with cell-free HIV because of the low numbers and limited infectious potential of real seminal leukocytes. As cell-to-cell spread is reported to be influenced by the type of effector cell, we generated three distinct effector cell populations to cover the most likely leukocyte populations in human semen (4). Monocyte-derived macrophages represent seminal macrophages, while activated PBMC and resting PBMC represent, respectively, seminal T cells or a mixture of seminal leukocytes. Surprisingly, the resting PBMC population became productively infected, although resting CD4+ T cells are known to restrict infection at or before reverse transcription (29). Indeed, our data confirm that CD4+ T cells purified from the same buffy coat are not easily infected (TCID50, 7,400 cells), although some integration does occur (1 copy per 20,000 cells). This is in line with the work of Agosto et al. and Dai et al. (2, 17), who showed that HIV-1 integration into naïve, resting CD4+ T cells does occur, even at low inocula, albeit with slower kinetics. However, removal of the CD4+ T cells from the infected resting PBMC population confirmed that both CD4+ T cells and the remaining cells within this population are highly infected with HIV and thus are both responsible for the high infectious potential (data not shown). Most likely, differentiating monocytes/macrophages within the resting PBMC population are infected during culture (46) while providing the necessary activation stimulus to the resting CD4+ T cells to become highly permissive for HIV infection (12, 24). Interestingly, although the different effector cell populations were infected with equal amounts of cell-free virus per cell, the resting PBMC and infected macrophages contained a 100-times-smaller amount of integrated viral DNA than the activated PBMC while showing only small differences in infectious titers (2.5- to 3.5-fold). This discrepancy might be explained by the fact that macrophages (present in both cell populations) are able to internalize large quantities of virus into an internal compartment without the macrophages themselves becoming productively infected. Upon synapse formation, these virions are redistributed to the site of contact and subsequently transferred to the target cells (12, 23), which might result in a high infectious potential but low viral copy number.
In conclusion, we developed an in vitro coculture assay to evaluate the efficacy of candidate microbicides against cell-to-cell and cell-free HIV spread using primary cells as effectors and two T-cell lines as targets. The assay allows the rapid screening of different drug classes targeting different stages of the viral life cycle. Moreover, through measurement of the Gag p24 concentration and/or firefly luciferase activity, it can discriminate between the infection of target cells, effector cells, or both. Using this assay, we showed that all tested antiretrovirals are able to block cell-to-cell spread, although small decreases in efficacy were observed for all entry inhibitors. Of all the compounds tested, only the protease inhibitors were able to fully inhibit the p24 production originating from the effector cells. Overall, these results show that entry inhibitors and protease inhibitors are eligible drug classes for inclusion in future combination microbicides.
ACKNOWLEDGMENTS
We are grateful to Wataru Sugiura, who made this work possible by providing us the R5M and X4M cell lines. We also thank J. Balzarini, K. Palmer, M. Miller, and L. Martin for their generous gifts of reagents and the Antwerp Red Cross Blood Transfusion Center for supplying the buffy coats.
P.S. is a predoctoral fellow of the Research Foundation-Flanders (FWO), Belgium, and the research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 242135 (CHAARM) and from the FWO (G.0125.06). We thank the EUROPRISE Network of Excellence for their support.
Footnotes
Published ahead of print 14 November 2011
REFERENCES
- 1. Abela IA. 2011. The role of neutralizing antibodies in inhibition of cell-to-cell transmission, abstr 101. Abstr. Keystone Symp. HIV Evol., Genomics Pathog., Whistler, Canada [Google Scholar]
- 2. Agosto LM, et al. 2007. HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration. Virology 368: 60–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allaway GP, et al. 1995. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res. Hum. Retrovir. 11: 533–539 [DOI] [PubMed] [Google Scholar]
- 4. Anderson DJ, et al. 2010. Targeting Trojan Horse leukocytes for HIV prevention. AIDS 24: 163–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ariën KK, Jespers V, Vanham G. 2011. HIV sexual transmission and microbicides. Rev. Med. Virol. 21: 110–133 [DOI] [PubMed] [Google Scholar]
- 6. Balzarini J, Van Damme L. 2007. Microbicide drug candidates to prevent HIV infection. Lancet 369: 787–797 [DOI] [PubMed] [Google Scholar]
- 7. Balzarini J, Van Herrewege Y, Vermeire K, Vanham G, Schols D. 2007. Carbohydrate-binding agents efficiently prevent dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN)-directed HIV-1 transmission to T lymphocytes. Mol. Pharmacol. 71: 3–11 [DOI] [PubMed] [Google Scholar]
- 8. Beirnaert E, et al. 1998. Design and evaluation of an in-house HIV-1 (group M and O), SIVmnd and SIVcpz antigen capture assay. J. Virol. Methods 73: 65–70 [DOI] [PubMed] [Google Scholar]
- 9. Brouwers J, Vermeire K, Grammen C, Schols D, Augustijns P. 2011. Early identification of availability issues for poorly water-soluble microbicide candidates in biorelevant media: a case study with saquinavir. Antiviral Res. 91: 217–223 [DOI] [PubMed] [Google Scholar]
- 10. Buffa V, et al. 2009. Cyanovirin-N potently inhibits human immunodeficiency virus type 1 infection in cellular and cervical explant models. J. Gen. Virol. 90: 234–243 [DOI] [PubMed] [Google Scholar]
- 11. Burton DR, Barbas CF., III 1994. Human antibodies from combinatorial libraries. Adv. Immunol. 57: 191–280 [DOI] [PubMed] [Google Scholar]
- 12. Carr JM, Hocking H, Li P, Burrell CJ. 1999. Rapid and efficient cell-to-cell transmission of human immunodeficiency virus infection from monocyte-derived macrophages to peripheral blood lymphocytes. Virology 265: 319–329 [DOI] [PubMed] [Google Scholar]
- 13. Chen P, Hubner W, Spinelli MA, Chen BK. 2007. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J. Virol. 81: 12582–12595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chiba-Mizutani T, et al. 2007. Use of new T-cell-based cell lines expressing two luciferase reporters for accurately evaluating susceptibility to anti-human immunodeficiency virus type 1 drugs. J. Clin. Microbiol. 45: 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clotet-Codina I, et al. 2009. HIV endocytosis after dendritic cell to T cell viral transfer leads to productive virus infection. Antiviral Res. 83: 94–98 [DOI] [PubMed] [Google Scholar]
- 16. Coombs RW, Reichelderfer PS, Landay AL. 2003. Recent observations on HIV type-1 infection in the genital tract of men and women. AIDS 17: 455–480 [DOI] [PubMed] [Google Scholar]
- 17. Dai J, et al. 2009. Human immunodeficiency virus integrates directly into naive resting CD4+ T cells but enters naive cells less efficiently than memory cells. J. Virol. 83: 4528–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. D'Cruz OJ, Uckun FM. 2006. Dawn of non-nucleoside inhibitor-based anti-HIV microbicides. J. Antimicrob. Chemother. 57: 411–423 [DOI] [PubMed] [Google Scholar]
- 19. Evans A. 2010. Protease inhibitors darunavir, lopinavir and ritonavir as potential microbicides, abstr 24. Abstr. Int. Microbicides Conf. [Google Scholar]
- 20. Field HJ, De Clercq E. 2008. Antiviral Chemistry and Chemotherapy's current antiviral agents FactFile: DNA viruses. Antivir. Chem. Chemother. 19: 51. [DOI] [PubMed] [Google Scholar]
- 21. Gali Y, et al. 2010. Development of an in vitro dual-chamber model of the female genital tract as a screening tool for epithelial toxicity. J. Virol. Methods 165: 186–197 [DOI] [PubMed] [Google Scholar]
- 22. Ganesh L, et al. 2004. Infection of specific dendritic cells by CCR5-tropic human immunodeficiency virus type 1 promotes cell-mediated transmission of virus resistant to broadly neutralizing antibodies. J. Virol. 78: 11980–11987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gousset K, et al. 2008. Real-time visualization of HIV-1 GAG trafficking in infected macrophages. PLoS Pathog. 4: e1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Groot F, Welsch S, Sattentau QJ. 2008. Efficient HIV-1 transmission from macrophages to T cells across transient virological synapses. Blood 111: 4660–4663 [DOI] [PubMed] [Google Scholar]
- 25. Haase AT. 2010. Targeting early infection to prevent HIV-1 mucosal transmission. Nature 464: 217–223 [DOI] [PubMed] [Google Scholar]
- 26. Ingallinella P, et al. 2009. Addition of a cholesterol group to an HIV-1 peptide fusion inhibitor dramatically increases its antiviral potency. Proc. Natil. Acad. Sci. U. S. A. 106: 5801–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jolly C. 2010. T cell polarization at the virological synapse. Viruses 2: 1261–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ketas TJ, et al. 2003. Human immunodeficiency virus type 1 attachment, coreceptor, and fusion inhibitors are active against both direct and trans infection of primary cells. J. Virol. 77: 2762–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kreisberg JF, Yonemoto W, Greene WC. 2006. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J. Exp. Med. 203: 865–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lederman MM, Offord RE, Hartley O. 2006. Microbicides and other topical strategies to prevent vaginal transmission of HIV. Nat. Rev. Immunol. 6: 371–382 [DOI] [PubMed] [Google Scholar]
- 31. Margolis L, Shattock R. 2006. Selective transmission of CCR5-utilizing HIV-1: the ‘gatekeeper’ problem resolved? Nat. Rev. Microbiol. 4: 312–317 [DOI] [PubMed] [Google Scholar]
- 32. Martin N, Sattentau Q. 2009. Cell-to-cell HIV-1 spread and its implications for immune evasion. Curr. Opin. HIV AIDS 4: 143–149 [DOI] [PubMed] [Google Scholar]
- 33. Martin N, et al. 2010. Virological synapse-mediated spread of human immunodeficiency virus type 1 between T cells is sensitive to entry inhibition. J. Virol. 84: 3516–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Massanella M, et al. 2009. Antigp41 antibodies fail to block early events of virological synapses but inhibit HIV spread between T cells. AIDS 23: 183–188 [DOI] [PubMed] [Google Scholar]
- 35. Mori T, et al. 2005. Isolation and characterization of Griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J. Biol. Chem. 280: 9345–9353 [DOI] [PubMed] [Google Scholar]
- 36. Muratori C, Ruggiero E, Sistigu A, Bona R, Federico M. 2009. Human immunodeficiency virus type 1 (HIV-1) protease inhibitors block cell-to-cell HIV-1 endocytosis in dendritic cells. J. Gen. Virol. 90: 2777–2787 [DOI] [PubMed] [Google Scholar]
- 37. O'Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. 2002. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J. Virol. 76: 10942–10950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Keefe BR, et al. 2009. Scaleable manufacture of HIV-1 entry inhibitor griffithsin and validation of its safety and efficacy as a topical microbicide component. Proc. Natl. Acad. Sci. U. S. A. 106: 6099–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271: 1582–1586 [DOI] [PubMed] [Google Scholar]
- 40. Puigdomenech I, Massanella M, Cabrera C, Clotet B, Blanco J. 2009. On the steps of cell-to-cell HIV transmission between CD4 T cells. Retrovirology 6: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27: 493–497 [Google Scholar]
- 42. Sallé B, et al. 2010. Infection of macaques after vaginal exposure to cell-associated simian immunodeficiency virus. J. Infect. Dis. 202: 337–344 [DOI] [PubMed] [Google Scholar]
- 43. Sattentau Q. 2008. Avoiding the void: cell-to-cell spread of human viruses. Nat. Rev. Microbiol. 6: 815–826 [DOI] [PubMed] [Google Scholar]
- 44. Sherer NM, et al. 2007. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat. Cell Biol. 9: 310–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith RJ, Bodine EN, Wilson DP, Blower SM. 2005. Evaluating the potential impact of vaginal microbicides to reduce the risk of acquiring HIV in female sex workers. AIDS 19: 413–421 [DOI] [PubMed] [Google Scholar]
- 46. Sonza S, et al. 1996. Human immunodeficiency virus type 1 replication is blocked prior to reverse transcription and integration in freshly isolated peripheral blood monocytes. J. Virol. 70: 3863–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sourisseau M, Sol-Foulon N, Porrot F, Blanchet F, Schwartz O. 2007. Inefficient human immunodeficiency virus replication in mobile lymphocytes. J. Virol. 81: 1000–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sowinski S, et al. 2008. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat. Cell Biol. 10: 211–219 [DOI] [PubMed] [Google Scholar]
- 49. Stefanidou M. 2010. Evaluation of saquinavir as a candidate microbicide compound in cellular and mucosal tissue models of HIV-1 infection, abstr 115. Abstr. Int. Microbicides Conf [Google Scholar]
- 50. Terrazas-Aranda K, Van Herrewege Y, Lewi PJ, Van Roey J, Vanham G. 2007. In vitro pre- and post-exposure prophylaxis using HIV inhibitors as microbicides against cell-free or cell-associated HIV-1 infection. Antivir. Chem. Chemother. 18: 141–151 [DOI] [PubMed] [Google Scholar]
- 51. Van Herrewege Y, et al. 2008. CD4 mimetic miniproteins: potent anti-HIV compounds with promising activity as microbicides. J. Antimicrob. Chemother. 61: 818–826 [DOI] [PubMed] [Google Scholar]