Abstract
We conducted population-based surveillance for pneumococcal bacteremia within a 5-county region surrounding Philadelphia from October 2001 through September 2008, the period following introduction of the seven-valent pneumococcal conjugate vaccine. Erythromycin resistance increased from 14.7% in 2001-2002 to 20.3% in 2007-2008, while the resistance rate to penicillin (MIC, ≥2 μg/ml) decreased from 7.2% to 4.2% during the same period. The most predominant serotypes associated with erythromycin resistance in 2007-2008 included 19A (29.7%), 15A (29.2%), 6C (10.1%), 3 (5.6%), and 6A (4.5%). The molecular mechanisms for the increasing erythromycin resistance were mainly due to the growing presence of mef(A) negative erm(B)+ and mef(A)+ erm(B)+ genotypes, which increased from 20.0% to 46.1% and from 1.8% to 19.1%, respectively, from 2001-2002 to 2007-2008. However, mef(A)-mediated erythromycin resistance decreased from 72.7% in 2001-2002 to 34.8% in 2007-2008. Serotypes related to the erm(B) gene were 15A (45.6%), 19A (20.9%), 3 (10.1%), and 6B (6.3%); serotypes related to the mef(A) gene were 6A (18.6%), 19A (15.0%), 6C (9.3%), and 14(8.4%); serotypes associated with the presence of both erm(B) and mef(A) were 19A (81.5%), 15A (7.7%), and 19F (6.2%). Pulsed-field gel electrophoresis analysis demonstrated that erythromycin-resistant isolates within the 19A serotype were genetically diverse and related to several circulating international clones. In contrast, erythromycin-resistant isolates within the 15A serotype consisted of clonally identical or closely related isolates.
INTRODUCTION
Streptococcus pneumoniae is a major pathogen that causes pneumonia, bacteremia, and meningitis in humans (9, 17). The surface capsular polysaccharide is one of the most important virulence factors and is the basis for all licensed pneumococcal vaccine strategies (21), with more than 90 immunologically distinct serotypes. S. pneumoniae is notable for its ability to switch serotypes and acquire antimicrobial drug resistance genes, reflecting an ability to incorporate foreign DNA (13). Antimicrobial resistance, which is often multidrug resistance, among clinical isolates of S. pneumoniae is common, limiting options for effective antimicrobial therapy.
In particular, macrolide resistance among Streptococcus pneumoniae isolates has risen in recent years worldwide (8, 10, 11, 14). Erythromycin resistance is mainly due to the presence of mef(A) and erm(B) genes (4, 25). Mef A is an efflux pump that removes most intracellular macrolides, resulting in low- to intermediate-level macrolide resistance, termed the M phenotype. Erythromycin resistance may also be mediated by the presence of an erythromycin-ribosomal methylase, which is encoded by the erm(B) gene. erm(B)-encoded methylation of adenine at position 2059 in the 23S rRNA blocks the binding of macrolides (e.g., erythromycin), lincosamides (e.g., clindamycin), and streptogramin B (e.g., dalfopristin) and results in high-level resistance to these antibiotics (MLSB phenotype), with high erythromycin MICs (≥256 μg/ml). In rare cases, macrolide resistance may also be caused by mutations in 23S rRNA (A2059G) or ribosomal proteins L4 and L22 (6, 25). The erm(B) gene is the most prevalent genotype globally, accounting for the majority of clinical isolates in Europe (5). In the United States, mef(A) is a common genotype (66% in 2001 to 2004 to 54% in 2005-2006) (11), and the presence of the erm(B) genotype has remained relatively low. However, recent reports have noted an increase in the percentage of isolates carrying both erm(B) and mef(A) (12, 15).
The introduction of the seven-valent pneumococcal conjugate vaccine (PCV-7) in 2000 had a profound impact on the seroepidemiology of pneumococcal disease, with significant declines observed in pediatric and adult disease due to vaccine serotypes (16, 26). Initial reports indicated that the decline in vaccine serotypes was also associated with a decline in the frequency of drug resistance, due to the fact that the serotypes targeted by PCV-7 were among the more common drug-resistant types in the prevaccine era (16, 26). However, the emergence of nonvaccine serotypes in recent years has been driven, in part, by antimicrobial drug selection pressures (12, 15) and is changing the epidemiology of pneumococcal drug resistance.
We investigated the prevalence and molecular epidemiology of macrolide resistance among invasive pneumococcal isolates in the post-PCV-7 era, with the aim of understanding the antimicrobial susceptibility profile, serotype distributions, and the genetic relatedness among macrolide-resistant isolates. We were especially interested in examining the prevalence of non-PCV-7 serotypes among erythromycin-resistant isolates, including those that would and would not be covered by the newly introduced 13-valent pneumococcal conjugate vaccine (PCV-13).
MATERIALS AND METHODS
Population-based surveillance for invasive pneumococcal disease.
Data were collected as part of a population-based surveillance for bacteremic pneumococcal disease within the 5-county region surrounding Philadelphia (Bucks, Chester, Delaware, Montgomery, and Philadelphia Counties). Adult (age, ≥18 years) population surveillance was initiated in October 2001 as part of a larger study of risk factors for community-acquired bacteremic pneumococcal disease. The surveillance network currently encompasses 48 of the 49 acute care hospitals that serve the 3.7 million residents of the five counties. The one nonparticipating hospital is a small hospital that is closed to external studies and accounted for <2% of all cases in the region.
Subjects were identified through the microbiology laboratories at all hospitals. Hospital personnel were contacted by study personnel on a regular basis throughout the surveillance period in order to ensure complete capture of new cases. We confirmed the total number of eligible cases through contact with laboratory directors and review of their log books on an annual basis, as well as comparison with data from the City of Philadelphia Health Department, which mandates reporting of cases of pneumococcal bacteremia for sites within the city (19, 20).
Eligible patients were identified based on the parent study and included hospitalized adults residing in the five-county region with at least one set of blood cultures positive for S. pneumoniae drawn within 48 h of hospitalization and no prior hospitalization within 10 days of the episode of pneumococcal bacteremia, in order to exclude hospital-acquired infections.
Bacterial isolates and identification.
All pneumococcal isolates were transported to the central laboratory at the Hospital of the University of Pennsylvania. Bacteria were cultured on blood agar medium in a 5% CO2 atmosphere at a temperature of 35°C and identified using conventional methods, including bile solubility testing.
Antibiotic susceptibility testing.
All the isolates were examined for antibiotic susceptibility against a panel of drugs, including oxacillin (for penicillin), erythromycin, tetracycline, clindamycin, penicillin, and trimethoprim-sulfamethoxazole (cotrimoxazole) by the disk-diffusion method (BD Diagnostics) and verified for resistant strains using the Etest (AB Biodisk, Solna, Sweden) for penicillin and erythromycin resistance. Testing for inducible erm(B) was conducted using apposed clindamycin and erythromycin disks placed ∼10 mm apart. Interpretations of disk diffusion and Etest MICs were in accord with the Clinical and Laboratory Standards Institute (CLSI) 2007 criteria (3), with the exception of Etest erythromycin MICs, which were interpreted per AB Biodisk guidelines (1). An isolate with an erythromycin MIC of ≥2 μg/ml was considered resistant based on a 2-fold-higher MIC of erythromycin with the Etest and growth in 5 to 10% CO2 versus results with ambient air agar dilution susceptibility testing. A penicillin MIC of ≥2 μg/ml was considered resistant based on the 2007 CLSI criteria and not the more recent 2010 criteria (MIC, ≥8 μg/ml). We used the earlier MIC breakpoint for penicillin in order to better compare results with those from previous studies during the same surveillance period. Erythromycin-nonsusceptible isolates were identified as the M phenotype if they were susceptible to clindamycin and the MLSB phenotype if they were nonsusceptible to clindamycin.
Detection of erythromycin resistance genes.
All erythromycin-resistant isolates were analyzed for the presence of the macrolide resistance genes erm(B) and mef(A) by using a SYBR green-based real-time PCR method. DNA was prepared by boiling bacterial cultures. The primers used to screen for the presence of erm(B) and mef(A) are available on request from the authors. PCR conditions followed the protocol described by J. Sutcliffe et al. (24). The PCR products for positive mef(A) and erm(B) isolates were confirmed by DNA sequencing for the first 10 positive isolates from each gene.
Identification of the 23S rRNA A2059G point mutation.
All macrolide-resistant isolates were screened for the presence of the 23S rRNA A2059G point mutation by using a TaqMan single-nucleotide polymorphism (SNP) genotyping assay. There are four copies of 23S rRNA in S. pneumoniae, and the level of resistance to macrolides depends on the copy number of the mutated gene. The probes and primers for detecting the 23S rRNA A2059G mutation were designed by Applied Biosystems using the TaqMan SNP genotyping assay. The assay reagent consisted of a 40× mix of unlabeled PCR primers (23SRNA_F, GACTCGGTGAAATTTTAGTATCTGTGAAGA; 23SRNA_R, TCAATATCAAACTGCAGTAAAGCTCCAT) and TaqMan MGB probes (labeled with the fluorochrome dyes 6-carboxyfluorescein [FAM] and VIC; FAM-AGGACGGAGAGACC and VIC-AGGACGGAAAGACC). The TaqMan real-time PCR was performed according to the method provided by the manufacturer. To evaluate the accuracy of the assay, we selected 20 clinical macrolide-resistant isolates and 2 additional strains with a known 23S rRNA A2059G mutation (provided generously by A. Tait-Kanradt). The accuracy and specificity of the TaqMan SNP method were 100% identical to those for DNA sequencing. The primers for amplifying each of the four alleles of the 23S rRNA are available upon request from the authors. All PCR-positive isolates for the A2059 mutation were then confirmed by sequencing.
Serotyping.
All pneumococcal isolates were serotyped by the Quellung reaction using antisera from the Staten Serum Institut (SSI; Copenhagen, Denmark) (2, 23).
PFGE.
Chromosomal DNA for pulsed-field gel electrophoresis (PFGE) was prepared as described by M. C. McEllistrem et al. (18) and was then digested with the restriction enzyme SmaI. The DNA fragments were resolved in a CHEF-Mapper apparatus (Bio-Rad) at 6.0 V/cm for 20 h with pulse times of 1 to 30 s, followed by another 6 h with pulse times of 5 to 9 s. We completed PFGE analysis on erythromycin-resistant isolates collected from 2005 to 2008 because we received funding for this component during the second half of the surveillance period. Seven international clones identified in the Pneumococcal Molecular Epidemiology Network were purchased from American Type Culture Collection and used for comparison.
Statistical analyses.
We calculated descriptive statistics for all cases, using means and medians as appropriate and geometric means for susceptibility results. We compared the frequency of macrolide resistance phenotypes and genotypes by using chi-square test statistics. We analyzed linear trends in proportions over time using the Mantel-Haenszel chi-square test for trend. We calculated population rates of disease for individual serotypes by using adult population estimates from the U.S. Census population intercensal estimates for 2001 to 2007. These county-level population estimates are based on the 2000 decennial Census, with annual population adjustments based on sampling and boundary adjustments (http://www.census.gov/popest/archives/2000s/vintage_2008/). Since our analyses focused on 1 October through 30 September analysis periods, we used the estimated population denominator of the year at the start of the observation period for each annual incidence rate calculation. We analyzed linear trends in the incidence of infection due to each genotype by using linear regression. The PFGE profiles were analyzed with the Fingerprinting II Informatix software (Bio-Rad).
RESULTS
Trends in antimicrobial resistance among invasive pneumococcal isolates.
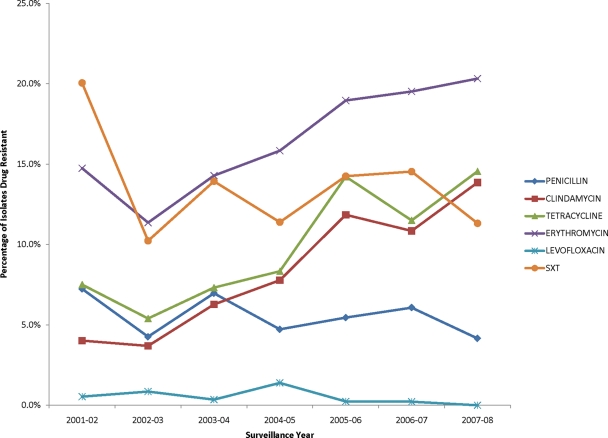
A total of 2,688 clinical isolates of S. pneumoniae were collected from October 2001 to September 2008 from blood cultures from adult patients admitted to any of the 46 hospitals within the surveillance region. The number of isolates per year was as follows: 373 (2001-2002), 352 (2002-2003), 287 (2003-2004), 360 (2004-2005), 422 (2005-2006), 461 (2006-2007), and 433 (2007-2008). Overall, the percentage of isolates resistant to each antimicrobial drug was as follows: penicillin, 5.5% (n = 148), erythromycin, 17.2% (n = 462), clindamycin, 8.7% (n = 234), tetracycline, 10.2% (n = 274), cotrimoxazole, 13.7% (n = 368), and levofloxacin, 0.5% (n = 13). Figure 1 displays the trends in antimicrobial resistance patterns over the study period for each of the antimicrobial drugs. An increasing trend in antimicrobial resistance was observed from the 2001-2002 season to the 2007-2008 season for erythromycin (14.7% to 20.3%; P = 0.0004), clindamycin (4.0% to 13.9%; P < 0.0001), and tetracycline (7.5% to 14.5%; P < 0.0001). In contrast, resistance to penicillin decreased over the years (7.2% to 4.2%; P = 0.23), and resistance to cotrimoxazole also decreased (20.1% to 11.3%; P = 0.06). Applying the newer 2010 CLSI criteria for penicillin susceptibility, the percentage of resistant isolates decreased from 3.7% to 2.8% (P = 0.44).
Fig 1.
Antimicrobial resistance profiles of bacteremic S. pneumoniae isolates by year from 2001 to 2008. The percentages of clinical isolates in each year with resistance to each of the six antimicrobial drugs are plotted. Each year includes the period from October through September, to encompass a single respiratory season.
Mechanisms of resistance among macrolide-resistant isolates.
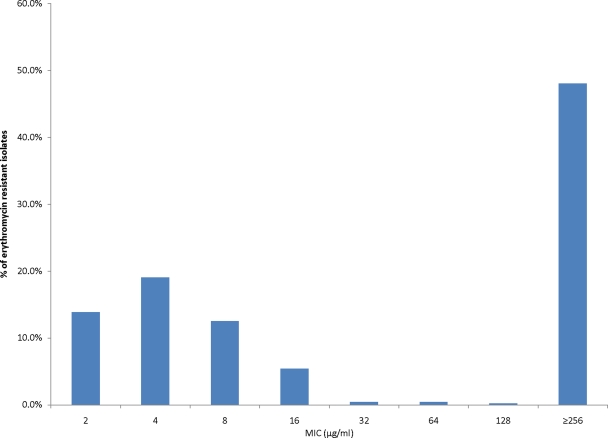
Among 462 erythromycin-resistant isolates, the penicillin, tetracycline, and cotrimoxazole resistance rates were 26.0%, 53.7%, and 49.6%, respectively. Approximately half (n = 226) of the erythromycin-resistant isolates displayed the MLSB phenotype (i.e., clindamycin resistant), and of these, 98.2% (n = 222) had high levels of erythromycin resistance (MIC, ≥256 μg/ml) (Fig. 2).
Fig 2.
Distribution of erythromycin MICs among S. pneumoniae isolates collected from October 2001 to September 2009. Among a total of 2,688 isolates, 2,226 isolates were susceptible (MIC, ≤1 μg/ml). Among these, three isolates with MICs of 1 μg/ml were classified as susceptible according to the 2007 CLSI guideline. A total of 462 isolates were resistant (MIC, ≥2 μg/ml), and the MIC distribution is plotted.
The genotype distribution of macrolide-resistant isolates was as follows: mef(A)+ erm(B) negative, 48.9%; mef(A) negative erm(B)+, 34.2%; mef(A)+ erm(B)+, 14.1%. Among those isolates with the mef(A)+ erm(B)-negative genotype, 98.7% displayed the M phenotype. In contrast, among the isolates with the mef(A)-negative erm(B)+ genotype, 97.5% displayed the MLSB phenotype. No isolates with an inducible erm(B) phenotype were detected. In addition, a higher proportion of the mef(A)+ erm(B)+ isolates demonstrated resistance to penicillin (76.9%) than did the mef(A)-negative erm(B)+ isolates (9.5% penicillin resistant) or the mef(A)+ erm(B)-negative isolates (23.5% penicillin resistant).
In terms of secular trends, erythromycin resistance mediated by mef(A) was the most common genotype (72.7%) in 2001-2002, and this percentage fell to 34.8% in 2007-2008 (Table 1). In contrast, erythromycin-resistant isolates expressing the erm(B) gene alone increased from 20.0% to 46.1%, and mef(A)+ erm(B)+ isolates increased significantly from 1.8% to 19.1% over the same time period. From a population perspective, the annual rate of infection due to isolates with the mef(A)+ erm(B)+genotype increased from 0.03 cases per 100,000 adults to 0.6 cases per 100,000 adults over the study period (P = 0.003 for trend). The rate of infection due to isolates with the mef(A)-negative erm(B)+ genotype increased from 0.4 to 1.4 cases per 100,000 adults (P = 0.002 for trend), and the rate of infection due to isolates with the mef(A)+ erm(B)-negative genotype declined from 1.4 cases to 1.0 cases per 100,00 adults (P = 0.97 for trend).
Table 1.
Genotype distributions among erythromycin-resistant S. pneumoniae isolates collected from 2001 to 2008
| Genotypea | % of isolates with indicated genotype during period |
P valueb | ||||||
|---|---|---|---|---|---|---|---|---|
| 2001-2002 (n = 55) | 2002-2003 (n = 41) | 2003-2004 (n = 42) | 2004-2005 (n = 57) | 2005-2006 (n = 84) | 2006-2007 (n = 93) | 2007-2008 (n = 89) | ||
| mef(A)+erm(B) negative | 72.7 | 70.7 | 52.4 | 50.9 | 40.5 | 44.1 | 34.8 | <0.0001 |
| mef(A) negative erm(B)+ | 20.0 | 26.8 | 26.2 | 36.8 | 40.5 | 31.2 | 46.1 | 0.01 |
| erm(B)+mef(A)+ | 1.8 | 0.0 | 9.5 | 10.5 | 17.9 | 23.7 | 19.1 | <0.0001 |
| 23S rRNA (A2059G) | 3.6 | 2.4 | 7.1 | 0.0 | 1.2 | 1.1 | 1.1 | 0.17 |
Isolates with an unknown genotype constituted 1.8%, 0%, 4.8%, 1.8%, 0%, 1.1%, and 0% in each year, respectively.
Determined using the Mantel-Haenszel chi-square test for trend.
There were a total of nine strains (1.8%) with the 23S rRNA point mutation. Two mef(A)+ erm(B)-negative strains were detected with the A2059G mutation in all four copies of the 23S rRNA. The other seven isolates with the A2059G mutation were mef(A) negative erm(B) negative. Among these, five had the A2059 mutation in all four gene copies and two had the mutation in two copies. Strains with the mutation in four copies of the 23S rRNA displayed an MLSB phenotype and high-level resistance to erythromycin (MIC, ≥256 μg/ml). Two isolates, each with two copies of the A2059G mutation, exhibited the M phenotype and low-level erythromycin resistance. In addition, there were five erythromycin-resistant isolates harboring no detectable erm(B) or mef(A) genes or A2059G mutations. Further DNA sequence analyses of the 23S rRNA and ribosomal protein L4 and L22 for these five isolates did not identify any mutations in these genes.
Serotype patterns among erythromycin-resistant isolates.
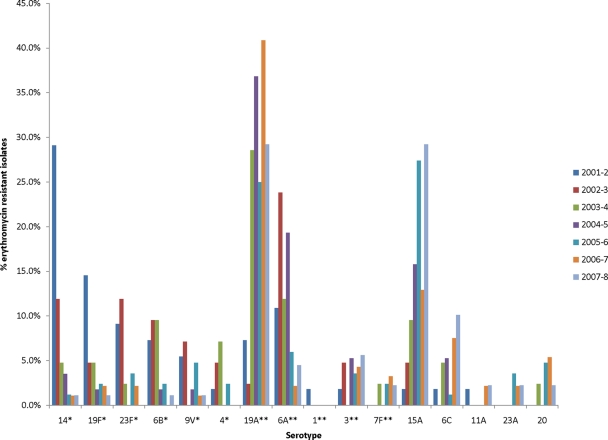
Of the 462 erythromycin-resistant isolates, 448 isolates were typeable and comprised 30 different serotypes. The overall serotype distribution among these erythromycin-resistant isolates was as follows: serotype 19A (27.5%), 15A (17.2%), 6A (9.2%), and 14 (6.3%) (Table 2). PCV-7 serotypes 14, 19F, 6B, and 23F were the most frequent serotypes associated with erythromycin resistance in 2001-2002. These serotypes decreased significantly over the study period (Fig. 3). By the end of the study period, the most common non-PCV-7 serotypes, 19A, 15A, 6C, 3, and 6A, accounted for 29.7%, 29.2%,10.1%, 5.6%, and 4.5% of the erythromycin-resistant isolates, respectively, compared with 7.3%, 1.8%,1.8%, 1.8%, and 10.9% in 2001-2002. Among all of the serotypes represented in Fig. 3, the proportion of isolates with macrolide resistance demonstrated a statistically significant increase over the surveillance period only for serotypes 9V (from 11.5% to 100%; P = 0.02) and 19A (from 13.8% to 27.4%; P = 0.02). The proportion of isolates with macrolide resistance within each of the remaining serotypes did not change significantly over the surveillance period. For example, 100% of 15A isolates were macrolide resistant at the start and end of the surveillance period, and 25% of 6C isolates were macrolide resistant at both time points.
Table 2.
Serotype distributions, overall and by genotype, among erythromycin-resistant isolates
| Serotype | % of isolates in genotype group with the indicated serotype |
|||
|---|---|---|---|---|
| All resistant isolates (n = 448) | mef(A)+erm(B) negative | mef(A) negative erm(B)+ | mef(A)+erm(B)+ | |
| 19Aa | 27.57 | 15.0 | 20.9 | 81.5 |
| 15A | 17.2 | 0.4 | 45.6 | 7.7 |
| 6Aa | 9.2 | 18.6 | 0.6 | 0.0 |
| 14 | 6.3 | 8.4 | 5.7 | 0.0 |
| 6C | 4.7 | 9.3 | 0.6 | 0.0 |
| 19Fb | 3.8 | 5.8 | 0.6 | 6.2 |
| 3a | 3.6 | 0.4 | 10.1 | 0.0 |
| 6Bb | 3.6 | 1.8 | 6.3 | 0.0 |
| 33F | 3.6 | 5.8 | 1.3 | 1.5 |
| 23Fb | 3.4 | 4.9 | 1.3 | 0.0 |
| 9Vb | 2.9 | 4.9 | 0.6 | 0.0 |
| 20 | 2.7 | 5.3 | 0.0 | 0.0 |
| 7Fa | 1.8 | 3.5 | 0.0 | 0.0 |
| 4b | 1.8 | 3.1 | 0.0 | 0.0 |
| 23A | 1.6 | 1.8 | 1.9 | 0.0 |
| 23B | 1.1 | 2.2 | 0.0 | 0.0 |
| 11A | 0.9 | 1.3 | 0.6 | 0.0 |
| 35B | 0.7 | 0.9 | 0.0 | 0.0 |
| Othersc | 3.5 | 6.6 | 6.9 | 3.1 |
Serotype that was included only in the PCV-13 vaccine.
Serotype that was included in the PCV-7 and PCV-13 vaccines.
Other serotypes identified included the following (number of isolates shown in parentheses): 1 (1), 7C (1), 9A (2), 9C (1), 10A (2), 12F (2), 15 (1), 15C (2), 15F (1), 22F (3), and 25A (1).
Fig 3.
Serotype distribution of erythromycin-resistant S. pneumoniae isolates from October 2001 to September 2008. There were a total of 29 serotypes distributed among the 462 erythromycin-resistant isolates. The percentage of resistant isolates in each year of each serotype is depicted. Serotypes marked with an asterisk are PCV-7 serotypes; serotypes marked with double asterisks are PCV-13 serotypes. The following isolates were not included in this figure: 33F (3.5%; n = 16), 23B (1.1%; n = 5), 35B (0.6%; n = 3), 22F (0.6%; n = 3), 15C (0.4%; n = 2), 12F (0.4%; n = 2), 10A (0.4%; n = 2), 8 (0.4%; n = 2), 9L (0.2%; n = 1), 7C (0.2%; n = 1), 25A (0.2%; n = 1), 15F (0.2%; n = 1), 15B (0.2%; n = 1).
The most common serotypes among mef(A)-negative erm(B)+ strains included serotypes 15A, 19A, 3, and 6B, which comprised 45.6%, 20.9%, 10.1%, and 6.3% of the mef(A)-negative erm(B)+ strains (Table 2). Serotype 19A was the most common serotype among the mef(A)+ erm(B)+ isolates (81.5%).
Genetic relatedness analysis by PFGE.
We performed PFGE analysis on the 293 erythromycin-resistant strains collected from 2005 to 2008. There were a total of 12 major clusters (Table 3, A to L). Serotype 19A was the largest group of the macrolide-resistant isolates, which was distributed into six major clusters: isolates carrying both the erm(B) and mef(A) genes were related to international clone Taiwan 19F; isolates harboring the mef(A) gene alone were related to international clones Tennessee-23F and England14-9; isolates expressing the erm(B) gene alone were related to international clones Spain 23F-1 and North Carolina 6A-23. Serotype15A was the second largest group among macrolide-resistant isolates, and it appeared highly clonal. The majority of serotype15A isolates displayed close relatedness to the North Carolina 6A-23 clone.
Table 3.
PFGE analysis of erythromycin-resistant S. pneumoniae isolates collected from 2005 to 2008c
| Cluster | Related clone(s) [genotype]a | Serotype | No. of strains | Susceptibility patternb | Genotype |
|---|---|---|---|---|---|
| A | NA | 6A | 10 | Clins Peni Tets Sxts | mef(A)+ erm(B) negative |
| B | NA | 33F | 8 | Clins Pens Tets Sxtr | mef(A)+ erm(B) negative |
| C | N. Carolina6A-23 [mef(A)+ erm(B) negative] | 15A | 60 | Clinr Peni Tetr Sxts | mef(A) negative erm(B)+ (55),mef(A)+ erm(B)+ (5) |
| 19A | 7 | Clinr Peni Tetr Sxts | mef(A) negative erm(B)+ | ||
| D | NA | 6C | 15 | Clins Peni Tets Sxtr | mef(A)+ erm(B) negative |
| 3 | 10 | Clinr Pens Tetr Sxts | mef(A) negative erm(B)+ | ||
| E | Tennessee23F-4 [mef(A)+ erm(B) negative] | 23B | 3 | Clins Penr Tets Sxts | mef(A)+erm(B) negative |
| 23A | 4 | Clins Pens Tets Sxts | mef(A)+ erm(B) negative | ||
| 19A | 15 | Clins Pens Tets Sxts | mef(A)+ erm(B) negative | ||
| F | England14-9 [mef(A)+ erm(B) negative], Spain 23F-1 [mef(A) negative erm(B) negative] | 14 | 3 | Clins Pens Tets Sxts | mef(A)+ erm(B) negative |
| 19A | 5 | Clins Peni Tets Sxtr | mef(A)+ erm(B) negative | ||
| 23F | 4 | Clins Pens Tetr Sxtr | mef(A)+ erm(B) negative | ||
| G | NA | 11A | 3 | Clins Pens Tets Sxts | mef(A)+ erm(B) negative |
| 15A | 2 | Clinr Pens Tetr Sxts | mef(A) negative erm(B)+ | ||
| 19A | 10 | Clinr Peni Tetr Sxtr | mef(A) negative erm(B)+ | ||
| H | NA | 20 | 11 | Clins Pens Tets Sxtr | mef(A)+ erm(B) negative |
| I | Spain 9V-3 [mef(A) negative erm(B) negative] | 9V | 6 | Clins Penr Tets Sxtr | mef(A)+ erm(B) negative |
| J | NA | 7F | 7 | Clins Pens Tets Sxts | mef(A)+ erm(B) negative |
| K | S. Africa 19A-13 [mef(A) negative erm(B)+] | 23A | 3 | Clinr Pens Tetr Sxts | mef(A) negative erm(B)+ |
| 19A | 2 | Clinr Peni Tetr Sxts | mef(A) negative erm(B)+ | ||
| L | Taiwan 19F-14 [mef(A)+ erm(B) negative] | 19A | 44 | Clinr Penr Tetr Sxts | mef(A)+ erm(B)+ |
| 19F | 4 | Clinr Penr Tetr Sxtr | mef(A)+ erm(B)+ |
Referenced from the Pneumococcal Molecular Epidemiology Network (http://www.sph.emory.edu/PMEN/). NA, not available.
Clin, clindamycin; Pen, penicillin; Tet, tetracycline; Sxt, trimethoprim-sulfamethoxazole.
Isolates (and counts) not listed in the above clusters: cluster A [23B (1), 23F (1), 19A (1), untypeable (1)]; cluster C [6C (1), 23F (1), 15C (2), 22F (1)]; cluster D [6A (1), 6B (1), 22F (1), 19A (1)]; cluster E [15A (1), 4 (2), 3 (1), 14 (1)]; cluster F [10A (1), 19F (1)]; cluster H [7C (1)]; cluster I [19A (1), 15F (1)]; cluster J [6B (1), 35B (2)]; cluster K [6B (1), 12F (1)].
DISCUSSION
Population-based surveillance for bacteremic pneumococcal disease among adults in the Philadelphia region between 2001 and 2008 demonstrated increasing antimicrobial resistance to erythromycin, tetracycline, and clindamycin, while resistance to penicillin declined over the same period. In particular, erythromycin resistance increased steadily from 14.7% in 2001-2002 to 20.3% in 2007-2008. Among all erythromycin-resistant isolates, the proportion of isolates from serotypes included in PCV-7 fell between 2001-2002 and 2007-2008, from 67.3% to 4.5%, and the proportion from serotypes included in PCV-13 fell from 89.1% to 46.1%. In 2001-2002, of the serotypes found in PCV-7, types 14, 19F, 23F, and 6B were the most prevalent erythromycin-resistant serotypes. In 2007-2008, types19A, 15A, and 6C emerged as the most common serotypes among erythromycin-resistant isolates. The increase in macrolide resistance over the surveillance period represented both an expansion of serotypes, with high macrolide resistance at the start of the period (e.g., 15A), and acquisition of macrolide resistance within specific serotypes (e.g., 19A), either through capsular switching or introduction of novel clones into the region.
We observed that the genotype distribution patterns of erythromycin resistance shifted during the study period. An efflux pump mediated by the mef(A) gene was the predominant mechanism responsible for macrolide resistance at the beginning of the study period. mef(A)-mediated macrolide resistance gradually dropped from its peak of 72.7% in 2001-2002, to 40.5% in 2004-2005, and to 34.8% in 2007-2008. In parallel, erm(B)-positive strains increased from 20.0% at the start of surveillance to 46.1%, becoming the most prevalent macrolide resistance genotype in 2007-2008. The most notable shift was the increase of mef(A)+ erm(B)+ isolates from 1.8% in 2001-2002 to 19.1% in 2007-2008. The higher prevalence of the erm(B) genotype was primarily related to the higher frequency of erm(B)+ serotype 15A, a serotype not covered by the recently introduced PCV-13 vaccine. Our data are consistent with those of PROTEKT US nationwide S. pneumoniae surveillance, which showed the prevalence of the mef(A)+ erm(B)+ genotype increased from 9.7% in 2000-2001 to 24.1% in 2005-2006 among clinical isolates and that erm(B)+ genotype prevalence increased slightly from 16.5% to 18.8%, while mef(A)+ genotype prevalence decreased from 65.7% to 53.8% over the same time period (12).
Serotype19A has been widely recognized as the key emerging serotype in the post-PCV-7 era, notably for its multidrug-resistant phenotype (7). Of note, 19A is a component of PCV-13, which was introduced in 2010, and so future surveillance will need to determine the impact on this serotype. Since serotypes 15A, 23A, and 6C are not included in the PCV-13 vaccine, a future increase in cases caused by these serotypes may be expected, particularly in response to ongoing drug selection for antimicrobial-resistant isolates. It is very likely that these serotypes could play important roles in the expansion of macrolide resistance in the post-PCV-13 era, especially type 15A. The majority (96.3%) of 15A isolates displayed high-level macrolide resistance [erm(B) encoded] and also displayed multidrug resistance. In addition, a small percentage of 15A isolates expressed both the erm(B) and mef(A) genes and were genetically related, as demonstrated by PFGE. This suggests that serotype 15A acquired erm(B) and mef(A) genes through a horizontal gene transfer event within serogroup 15 rather than capsular switching with other serogroup strains positive for the erm(B) and mef(A) genes.
We did not detect a high frequency of the 23S rRNA A2059G point mutation. Among all serotypes, 23F isolates displayed the highest frequency of the A2509G mutation (14% of 23F macrolide-resistant isolates). This could have resulted from clonal spread or could have arisen from independent mutation events. Joloba et al. demonstrated that serotype 23F cannot be naturally transformed in vitro under inducing conditions (13). This may explain the relatively high rate of the 23S rRNA A2059G mutation as a mechanism for resistance among serotype 23F isolates.
Our work builds on prior studies demonstrating an increase in macrolide resistance among pneumococcal isolates, adding additional data on the serotypes and molecular types responsible for this increase. Taken together, surveillance of invasive S. pneumoniae in the Philadelphia region highlights the upward trend of macrolide resistance, especially of high-level macrolide resistance mediated by erm(B) alone or both the erm(B) and mef(A) genes. This study has identified several emerging serotypes associated with macrolide resistance, specifically, serotypes 15A, 6C, and 23A, which are not covered by the PCV-13 vaccine. It has been speculated that serotype 6A in PCV-13 may provide some cross-protection against 6C (22). Future surveillance studies will help assess the degree of cross-protection observed in practice. Moreover, it is imperative to monitor whether the existing non-PCV-13 serotypes will spread further or whether additional replacement serotypes will emerge following the introduction of PCV-13.
ACKNOWLEDGMENTS
We acknowledge the valuable contributions of Linda Crossette for coordinating the activities of this study and the staff at the Clinical Microbiology Laboratory of the Hospital of the University of Pennsylvania for the microbiology testing.
This project was supported by grants R01-AI46645 (J.P.M.), K24-AI073957 (J.P.M.), and K24-AI080942 (E.L.) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
The funding agency had no role in the design and conduct of the study; collection, management, analysis and interpretation of the data; or preparation, review, or approval of the manuscript.
E. Lautenbach has received research funding from 3M, AstraZeneca, and Cubist. None of the other authors have any conflicts of interest or financial disclosures.
The Delaware Valley Case Control Network includes the following hospitals, listed with their respective physician coinvestigators and laboratory directors: Abington Memorial Hospital, Robert R. Dee and Herbert Auerbach; Albert Einstein Medical Center, Jerry Zuckerman and Nancy Young; Brandywine Hospital, John H. Bartels and Stephen B. Chasko; Bryn Mawr Hospital, Peter Spitzer; Chester County Hospital, John Roberts and Jim Heald; Chestnut Hill Hospital, Lawrence Livornese and Andrew So; Children's Hospital of Philadelphia, Susan E. Coffin and Karin McGowan; Crozer-Chester Medical Center, Springfield Hospital, and Taylor Hospital, William D. Ravreby and Harvey B. Spector; Delaware County Memorial Hospital, Jackeline Iaccovella and Lawrence M. Matthews; Doylestown Hospital, David Loughran and Robert Trotta; Elkins Park Hospital (closed during the course of the project), Donald Marcus and Xiaoli Chen; Episcopal Hospital (closed during the course of the project), Peter Axelrod and Allan Truant; Fox Chase Cancer Center, Peter Axelrod; Aria Health Bucks County, Frankford and Torresdale campuses, Donald Marcus and Peter Farano; Graduate Hospital (closed during the course of the project), Milchael Silverman and Fernando Garcia; Grand View Hospital, Abby Huang and Irwin Hollander; Hahnemann University, Mashiul Chowdhury and Christopher Emery; Holy Redeemer Hospital and Medical Center, Robert R. Dee and Pantaleon Fagel; Hospital of the University of Pennsylvania, Joshua P. Metlay and Paul Edelstein; Jeanes Hospital, Richard Tepper and Irma Palazzo; Jennersville Regional Medical Center, John H. Bartels and James Monihan; Lankenau Hospital, Lawrence Livornese, Olarae Giger, and Albert Keshgegian; Lansdale Hospital, Abby Huang; Lower Bucks Hospital, Donald Marcus and Tatiana Chernova; Mercy Community Hospital (closed during the course of the project), Mercy Fitzgerald Hospital, and Mercy Hospital of Philadelphia, William McNamee and Lorenzo Galindo; Mercy Suburban Hospital, Wayne Miller; Methodist Hospital, Robert Measley and Harvey Bellin; Montgomery Hospital, Hazel Bluestien and Paul H. Belser; Northeastern Hospital, Jerry Zuckerman; Paoli Memorial Hospital, David Trevino; Parkview Hospital (closed during the course of the project), Jerry Zuckerman; Pennsylvania Hospital, Michael Braffman, John Stern, and Julieta Barroeta; Phoenixville Hospital, Raymond Kovalski and Leonas Bekeris; Pottstown Memorial Medical Center, Raymond Kovalski and Dante DiMarzio; Presbyterian Medical Center, Vincent LoRe; Riddle Memorial Hospital, Marc Gilbert and Susan Yaron; Roxborough Memorial Hospital, Lawrence Livornese and Pradeep Bhagat; St. Agnes Medical Center (closed during the course of the project), Robert Measley and John McCormick; St. Christopher's Hospital for Children, Jane Gould; St. Joseph's Hospital, David Loughran and Alberto Millos; St. Luke's Quakertown Hospital, Abby Huang and David Anderson; St. Mary Medical Center, Donald Marcus and Zenon Gibas; Temple University Hospital, Peter Axelrod and Carmelita Flores; Thomas Jefferson University Hospital, Michael Baram and Stephen Peiper; Veterans Affairs Medical Center, Darren Linkin and Laura Chandler; Warminster Hospital (closed during the course of the project), David Loughran and Manjula Balasubramanian.
Footnotes
Published ahead of print 28 November 2011
REFERENCES
- 1. ABI Biodisk. 2004. Etest technical manual. ABI Biodisk, Solna, Sweden [Google Scholar]
- 2. Austrian R. 1976. The Quellung reaction, a neglected microbiologic technique. Mt. Sinai J. Med. 43:699–709 [PubMed] [Google Scholar]
- 3. Clinical and Laboratory Standards Institute 2007. Performance standards for antimicrobial susceptibility testing: 17th informational supplement. M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 4. Edelstein PH. 2004. Pneumococcal resistance to macrolides, lincosamides, ketolides, and streptogramin B agents: molecular mechanisms and resistance phenotypes. Clin. Infect. Dis. 38(Suppl. 4):S322–S327 [DOI] [PubMed] [Google Scholar]
- 5. Farrell DJ, Couturier C, Hryniewicz W. 2008. Distribution and antibacterial susceptibility of macrolide resistance genotypes in Streptococcus pneumoniae: PROTEKT year 5 (2003–2004). Int. J. Antimicrob. Agents 31:245–249 [DOI] [PubMed] [Google Scholar]
- 6. Farrell DJ, et al. 2003. Macrolide resistance by ribosomal mutation in clinical isolates of Streptococcus pneumoniae from the PROTEKT 1999–2000 study. Antimicrob. Agents Chemother. 47:1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Farrell DJ, Klugman KP, Pichichero M. 2007. Increased antimicrobial resistance among nonvaccine serotypes of Streptococcus pneumoniae in the pediatric population after the introduction of 7-valent pneumococcal vaccine in the United States. Pediatr. Infect. Dis. J. 26:123–128 [DOI] [PubMed] [Google Scholar]
- 8. Gay K, et al. 2000. The emergence of Streptococcus pneumoniae resistant to macrolide antimicrobial agents: a 6-year population-based assessment. J. Infect. Dis. 182:1417–1424 [DOI] [PubMed] [Google Scholar]
- 9. Harwell JI, Brown RB. 2000. The drug-resistant pneumococcus: clinical relevance, therapy, and prevention. Chest 117:530–541 [DOI] [PubMed] [Google Scholar]
- 10. Hyde TB, et al. 2001. Macrolide resistance among invasive Streptococcus pneumoniae isolates. JAMA 286:1857–1862 [DOI] [PubMed] [Google Scholar]
- 11. Jenkins SG, Brown SD, Farrell DJ. 2008. Trends in antibacterial resistance among Streptococcus pneumoniae isolated in the USA: update from PROTEKT US years 1–4. Ann. Clin. Microbiol. Antimicrob. 7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jenkins SG, Farrell DJ. 2009. Increase in pneumococcus macrolide resistance, United States. Emerg. Infect. Dis. 15:1260–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joloba ML, et al. 2010. Comparison of transformation frequencies among selected Streptococcus pneumoniae serotypes. Int. J. Antimicrob. Agents 36:124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jones RN, Jacobs MR, Sader HS. 2010. Evolving trends in Streptococcus pneumoniae resistance: implications for therapy of community-acquired bacterial pneumonia. Int. J. Antimicrob. Agents 36:197–204 [DOI] [PubMed] [Google Scholar]
- 15. Jones RN, Sader HS, Moet GJ, Farrell DJ. 2010. Declining antimicrobial susceptibility of Streptococcus pneumoniae in the United States: report from the SENTRY antimicrobial surveillance program (1998–2009). Diagn. Microbiol. Infect. Dis. 68:334–336 [DOI] [PubMed] [Google Scholar]
- 16. Lexau CA, et al. 2005. Changing epidemiology of invasive pneumococcal disease among older adults in the era of pediatric pneumococcal conjugate vaccine. JAMA 294:2043–2051 [DOI] [PubMed] [Google Scholar]
- 17. Mandell LA. 2004. Epidemiology and etiology of community-acquired pneumonia. Infect. Dis. Clin. North Am. 18:761–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McEllistrem MC, Stout JE, Harrison LH. 2000. Simplified protocol for pulsed-field gel electrophoresis analysis of Streptococcus pneumoniae. J. Clin. Microbiol. 38:351–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Metlay JP, et al. 2006. Macrolide resistance in adults with bacteremic pneumococcal pneumonia. Emerg. Infect. Dis. 12:1223–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Metlay JP, Lautenbach E, Li Y, Shults J, Edelstein PH. 2010. Exposure to children as a risk factor for bacteremic pneumococcal disease: changes in the post-conjugate vaccine era. Arch. Intern. Med. 170:725–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Obaro SK. 2002. The new pneumococcal vaccine. Clin. Microbiol. Infect. 8:623–633 [DOI] [PubMed] [Google Scholar]
- 22. Park IH, et al. 2008. Differential effects of pneumococcal vaccines against serotypes 6A and 6C. J. Infect. Dis. 198:1818–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sorensen UB. 1993. Typing of pneumococci by using 12 pooled antisera. J. Clin. Microbiol. 31:2097–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tait-Kamradt A, et al. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Whitney CG, et al. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348:1737–1746 [DOI] [PubMed] [Google Scholar]