Abstract
Two closely related Enterobacter aerogenes isolates presented a new identical aac(6′)-Ib-cr genetic environment, including IS26. One isolate showed lower MICs of ciprofloxacin, norfloxacin, tobramycin, and amikacin and decreased expression of aac(6′)-Ib-cr, which might be related to a 12-bp deletion causing a displacement of the −10 box upstream of the aac(6′)-Ib-cr gene.
TEXT
The aac(6′)-Ib-cr gene encodes an aminoglycoside 6′-acetyltransferase able to modify not only aminoglycosides but also quinolones with a piperazinyl substituent (such as ciprofloxacin and norfloxacin) by acetylation of the amino nitrogen on this chemical group (12). This gene has been detected in the variable regions of integrons and in several structures commonly associated with mobile genetic elements, especially the IS26 insertion sequence (11, 13).
The aim of this work was to analyze the expression of the aac(6′)-Ib-cr gene in two clonally related Enterobacter aerogenes clinical isolates (C2653 and C2657) obtained from two patients in different wards of the same hospital (both isolates were recovered in 2009) and to correlate this expression with fluoroquinolone and aminoglycoside susceptibilities. In addition, other antimicrobial resistance mechanisms and plasmid content were investigated.
Both isolates showed a closely related XbaI-digested pulsed-field gel electrophoresis (PFGE) pattern (data not shown). Susceptibility testing to 23 antimicrobial agents was carried out by disk diffusion and agar dilution methods or Etest (5), and 34 genes conferring resistance to quinolones, β-lactams, carbapenems, aminoglycosides, tetracycline, chloramphenicol, sulfonamides, and trimethoprim were tested by PCR with subsequent sequencing of all amplicons obtained.
The C2653 isolate showed higher MIC values of ciprofloxacin and norfloxacin (but not levofloxacin), aminoglycosides, and carbapenems than the C2657 isolate did (Table 1). Both isolates carried the blaCTX-M-15, blaTEM-1b, blaOXA-1, aph(3)-Ia, sul1, sul3, and dfrA12 genes. The presence of class 1 and class 2 integrons was determined, the characterization of their variable regions were analyzed (14), and a new gene cassette array, which was deposited in GenBank under accession number JF729199, was identified in both isolates (Table 1). The amino acid change D86Y was identified in the quinolone resistance-determining region (QRDR) of the GyrA protein, while the wild sequence was demonstrated in the ParC protein, as determined by PCR and sequencing of the corresponding genes in both isolates (3, 15).
Table 1.
Resistance phenotype and genotypes of E. aerogenes donor isolates, the E. coli recipient strain, and the transformant strain
| Straina | MIC (mg/liter)b |
Additional resistanceb,c | Resistance genesd | Amino acid change in GyrA QRDRe | ParC QRDR | Class 1 integron variable region | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAL | CIP | LEV | NOR | TOB | AMK | KAN | GEN | AMP | CTX | ERT | IMP | MER | DOR | ||||||
| C2657 | 1,024 | 1 | 2 | 8 | 1 | 2 | >256 | 0.5 | >256 | 128 | 0.5 | 1 | 0.5 | 0.5 | AMC, CAZ, FOX, AZT, STR (I), SUL, SXT, CHL, FFM (I) | aac(6′)-Ib-cr, blaCTX-M-15, blaTEM-1b,blaOXA-1, aph(3)-Ia, sul1, sul3, dfrA12 | D86Y | Wild | dfrA12-orf-ΔaadA2-IS6100f |
| C2653 | 512 | 8 | 2 | 64 | 64 | 32 | >256 | 2 | >256 | 256 | >32 | >32 | 2 | 4 | AMC, CAZ, FOX, AZT, IMP (I), ERT, STR (I), SUL (I), SXT (I), FFM (I) | aac(6′)-Ib-cr, blaCTX-M-15, blaTEM-1b,blaOXA-1, aph(3)-Ia, sul1, sul3, dfrA12 | D86Y | Wild | dfrA12-orf-ΔaadA2-IS6100 |
| Tf-C2653 | 1 | 0.004 | 0.004 | 0.03 | 16 | 8 | 256 | 0.5 | >256 | 128 | 0.06 | 0.5 | 0.125 | 0.1 | AMC, CAZ (I), STR (I) | aac(6′)-Ib-cr, blaCTX-M-15, blaTEM-1b,blaOXA-1, aph(3)-Ia, dfrA12 | dfrA12-orf-ΔaadA2-IS6100 | ||
| E. coli DH10B (recipient strain) | 0.25 | 0.002 | 0.004 | 0.007 | 0.125 | 0.03 | 1 | 0.125 | 4 | 0.03 | 0.03 | 0.25 | 0.03 | 0.05 | |||||
Strain Tf-C2653 is a transformant of isolate C2653.
NAL, nalidixic acid; CIP, ciprofloxacin; LEV, levofloxacin; NOR, norfloxacin; TOB, tobramycin; AMK, amikacin; KAN, kanamycin; GEN, gentamicin; AMP, ampicillin; CTX, cefotaxime; ERT, ertapenem; IMP, imipenem; MER, meropenem; DOR, doripenem; AMC, amoxicillin-clavulanic acid; CAZ, ceftazidime; FOX, cefoxitin; AZT, aztreonam; STR, streptomycin; SUL, sulfamethoxazole; SXT, trimethoprim-sulfamethoxazole; CHL, chloramphenicol; FFM, fosfomycin.
(I), intermediate resistance.
The resistance genes screened were qnrA, qnrB, qnrS, aac(6′)-Ib-cr, qepA, oqxAB, blaCTX-M, blaOXA, blaSHV, blaTEM, blaVIM, blaIMP, blaSPM, blaGIM, blaSIM, blaACC, blaCIT, blaDHA, blaEBC, blaFOX, blaMOX, aac(3)-I, aac(3)-II, aac(3)-III, aac(3)-IV, aph(3′)-Ia, aph(3′)-II, aadA1 or aadA2, aadA5, ant(2′), dfrA1 to dfrA17, dfrB1, dfrB2, and dfrB3.
QRDR, quinolone resistance-determining region.
The variable region of this integron is new and has been deposited in GenBank with accession number JF729199.
Mutations in omp35 and omp36 porin genes of the two E. aerogenes isolates were analyzed by PCR and sequencing (6) and outer membrane proteins (OMPs) were obtained and visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (9). The carbapenem-resistant isolate C2653 did not express the two major porins. A deletion of the nucleotides TT at positions 71 and 72 of the omp36 porin gene was detected, while omp35 gene presented the wild sequence. Since these characteristics were not identified in the carbapenem-susceptible C2657 isolate, porin alteration might be responsible for the high MIC of carbapenems in the C2653 isolate.
Plasmids from the two E. aerogenes isolates were extracted, and genetic transfer of the aac(6′)-Ib-cr gene was carried out by transformation into Escherichia coli DH10B. Conjugation and transformation assays were carried out using aminoglycosides, quinolones, and β-lactams for selection. A transformant of E. aerogenes isolate C2653 could be obtained (selected on Mueller-Hinton agar plates supplemented with tobramycin [6 μg/ml]), but conjugation and transformation assays from the C2657 isolate were unsuccessful. Resistance phenotypes and genotypes of the donor, transformant, and recipient isolates are shown in Table 1. The acquisition of the aac(6′)-Ib-cr gene from isolate C2653 by the transformant was associated with an increase in the MICs of quinolones (except levofloxacin) and aminoglycosides.
The plasmids of donor and transformant isolates were classified according to their incompatibility group using the PCR-based replicon typing method (7), and their number and size was determined by a PFGE assay with the total DNA digested by S1 nuclease (1). Eight plasmid addiction systems were studied by PCR as previously described (10). The plasmid locations of the aac(6′)-Ib-cr, blaCTX-M-15, and intI1 genes were analyzed by transferring S1 DNA digested PFGE gels onto nylon membranes by Southern blotting and hybridized with specific probes. Hybridization was performed by using the digoxigenin (DIG) high prime DNA labeling and detection starter kit I (Roche Applied Science, Barcelona, Spain). The chromosomal location of the aac(6′)-Ib-cr gene was also determined by Southern hybridization following genomic DNA digestion with I-CeuI nuclease as previously described (8). Only a plasmidic location of the aac(6′)-Ib-cr gene was observed. Results of plasmid characterization are shown in Table 2. Both E. aerogenes isolates carried a high-molecular-weight plasmid of approximately 200 kb that carried the aac(6′)-Ib-cr, intI1, and blaCTX-M-15 genes, and it was acquired by the Tf-C2653 isolate (Tf stands for transformant).
Table 2.
Plasmid characterization and plasmid location of aac(6′)-Ib-cr, blaCTX-M-15, and intI1 genes
| Straina | Detected plasmid |
No. of plasmids detected | Size (kb) of plasmid detected | S1-PFGE-hybridizationc |
|||
|---|---|---|---|---|---|---|---|
| Replicon typeb | Addiction systems | Plasmid size (kb) | Replicon typeb | Detected genes | |||
| C2653 | IncR, ColE | pemK/I | 3 | 9, 120, 200 | 200 | ND | aac(6′)-Ib-cr, intI1, blaCTX-M-15 |
| Tf-C2653 | ND | pemK/I | 1 | 200 | 200 | ND | aac(6′)-Ib-cr, intI1, blaCTX-M-15 |
| C2657 | IncR | pemK/I | 2 | 9, 200 | 200 | ND | aac(6′)-Ib-cr, intI1, blaCTX-M-15 |
Strain Tf-C2653 is a transformant of isolate C2653.
ND, could not be determined.
Total DNA was digested by S1 nuclease, and then PFGE and hybridization were performed.
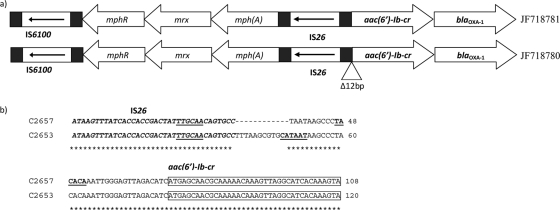
Determination of the genetic environment of the aac(6′)-Ib-cr gene was performed by cloning. The total DNA of E. aerogenes C2657 was extracted with QIAmp DNA minikit (Qiagen Science Inc., MD) and partially digested by Sau3AI restriction enzyme for 10 min at 37°C. The digestion products were ligated in the BamHI site of the pUC19 cloning vector, and they were introduced into E. coli DH10B by electroporation. The transformants harboring the recombinant plasmids were selected on Mueller-Hinton agar supplemented with ticarcillin (100 μg/ml) and tobramycin (6 μg/ml). Finally, inserts were analyzed by PCR and sequencing with M13 universal primers. PCR mapping and a primer walking sequencing method were used to elucidate a larger surrounding region of the aac(6′)-Ib-cr gene in the two E. aerogenes isolates based on the results obtained after cloning and the previously reported structures (16). The genetic environments of the aac(6′)-Ib-cr gene detected in the two isolates were the same except for a deletion of 12 bp between the IS26 insertion sequence and the aac(6′)-Ib-cr gene observed in E. aerogenes C2657. These surrounding regions of aac(6′)-Ib-cr gene in both isolates had not been previously reported and were deposited in GenBank with accession numbers JF718780 and JF718781 (Fig. 1). The level of expression of the aac(6′)-Ib-cr gene was determined by real-time PCR (RTi-PCR) in the two E. aerogenes isolates following a methodology described before (4). Primers used for RTi-PCR were aac6-RTi-F (F stands for forward) (5′-TGCATCACAACTGGGCAAAGGCT-3′) and aac6-RTi-R (R stands for reverse) (5′-ACACGGCTGGACCATATGGGGT-3′) for the aac(6′)-Ib-cr gene and rrsKp-F (5′-CAGGCGGTCTGTCAGTCGGAT-3′) and rrsKp-R (5′-CGCACCTGAGCGTCAGTCTTTG-3′) for the rrs gene. The amplicon sizes of the aac(6′)-Ib-cr and rrs genes were 191 and 184 bp, respectively. The rrs gene was used as an internal reference to normalize the relative amount of RNA. The expression of the aac(6′)-Ib-cr gene was 365 times higher in E. aerogenes C2653 than in E. aerogenes C2657, reflecting the almost complete absence of expression of the gene in the latter strain. The genetic environment of aac(6′)-Ib-cr was identical in both isolates, but in the case of isolate C2567, a deletion of 12 bp was identified between IS26 and the aac(6′)-Ib-cr gene (Fig. 1).
Fig 1.
Genetic environment of the aac(6′)-Ib-cr gene of E. aerogenes isolates. (a) Complete structure surrounding the aac(6′)-Ib-cr gene with their GenBank accession numbers. mphR-mrx-mph(A) is a macrolide inactivation gene cluster. (b) Alignment of the nucleotide sequence between the IS26 insertion sequence and the aac(6′)-Ib-cr gene of the two E. aerogenes isolates. The IS26 sequence is shown in italic type, the aac(6′)-Ib-cr gene is boxed, and the putative −10 and −35 boxes are underlined. Gaps introduced to maximize alignment are indicated by dashes.
These results totally agree with the fact that the level of resistance to ciprofloxacin, norfloxacin, tobramycin, and amikacin of E. aerogenes C2653 was clearly higher than that of isolate C2657. As expected, the level of resistance to levofloxacin, which does not have a piperazinyl nitrogen, was the same in both isolates, confirming that fluoroquinolones lacking that piperazinyl group are not affected by the AAC(6′)-Ib-cr enzymatic variant. It is important to point out that some authors had suggested that there was probably a −10 box, which could be involved in the expression of the aac(6′)-Ib-cr gene located downstream of IS26 (2, 11). Assuming that there is a −10 box, the −35 box would be located 17 bp upstream (TTGCAA), which was detected in both isolates. In that case, the −10 box of strain C2657 could be displaced 12 bp (TACACA) and is probably involved in the lack of expression of aac(6′)-Ib-cr gene.
Nucleotide sequence accession numbers.
Nucleotide sequence data reported in this paper are available in the GenBank nucleotide database under accession numbers JF729199, JF718780 (E. aerogenes C2657), and JF718781 (E. aerogenes C2653).
ACKNOWLEDGMENTS
This work was supported by the Gobierno de la Rioja of Spain that awarded Elena Ruiz a predoctoral fellowship and the Project SAF2009-08570 from the Ministry of Education and Science of Spain and FEDER. A.A.O.-S. and L.M.-M. are supported by the Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III - FEDER, Spanish Network for the Research in Infectious Diseases (REIPI RD06/0008).
Footnotes
Published ahead of print 21 November 2011
REFERENCES
- 1. Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235–240 [DOI] [PubMed] [Google Scholar]
- 2. Boyd DA, et al. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 48:3758–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brisse S, Verhoef J. 2001. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int. J. Syst. Evol. Microbiol. 51:915–924 [DOI] [PubMed] [Google Scholar]
- 4. Cabot G, et al. 2011. Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa from blood stream infections: prevalence and linkage to resistance in a Spanish multicenter study. Antimicrob. Agents Chemother. 55:1906–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement. M100-S21 CLSI, Wayne, PA [Google Scholar]
- 6. Doumith M, Ellington MJ, Livermore DM, Woodford N. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J. Antimicrob. Chemother. 63:659–667 [DOI] [PubMed] [Google Scholar]
- 7. GarcíA-Fernández A, Fortini D, Veldman K, Mevius D, Carattoli A. 2009. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J. Antimicrob. Chemother. 63:274–281 [DOI] [PubMed] [Google Scholar]
- 8. Liu SL, Hessel A, Sanderson KE. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:6874–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Martínez-Martínez L, et al. 2000. Activities of imipenem and cephalosporins against clonally related isolates of Escherichia coli hyperproducing chromosomal beta-lactamase and showing altered porin profiles. Antimicrob. Agents Chemother. 44:2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mnif B, et al. 2010. Molecular characterization of addiction systems of plasmids encoding extended-spectrum beta-lactamases in Escherichia coli. J. Antimicrob. Chemother. 65:1599–1603 [DOI] [PubMed] [Google Scholar]
- 11. Partridge SR, Tsafnat G, Coiera E, Iredell JR. 2009. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33:757–784 [DOI] [PubMed] [Google Scholar]
- 12. Robicsek A, et al. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83–88 [DOI] [PubMed] [Google Scholar]
- 13. Ruiz E, et al. 2011. New genetic environments of aac(6′)-Ib-cr gene in a multiresistant Klebsiella oxytoca strain causing an outbreak in a pediatric intensive care unit. Diagn. Microbiol. Infect. Dis. 69:236–238 [DOI] [PubMed] [Google Scholar]
- 14. Sáenz Y, et al. 2004. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob. Agents Chemother. 48:3996–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weigel LM, Steward CD, Tenover FC. 1998. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob. Agents Chemother. 42:2661–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Woodford N, et al. 2009. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 53:4472–44782 [DOI] [PMC free article] [PubMed] [Google Scholar]