Abstract
Molecular investigations performed following the emergence of sulfadoxine-pyrimethamine (SP) resistance in Plasmodium falciparum have allowed the identification of the dihydrofolate reductase (DHFR) enzyme as the target of pyrimethamine. Although clinical cases of Plasmodium malariae are not usually treated with antifolate therapy, incorrect diagnosis and the high frequency of undetected mixed infections has probably exposed non-P. falciparum parasites to antifolate therapy in many areas. In this context, we aimed to assess the worldwide genetic diversity of the P. malariae dhfr gene in 123 samples collected in Africa and Asia, areas with different histories of SP use. Among the 10 polymorphic sites found, we have observed 7 new mutations (K55E, S58R, S59A, F168S, N194S, D207G, and T221A), which led us to describe 6 new DHFR proteins. All isolates from African countries were classified as wild type, while new mutations and haplotypes were recognized as exclusive to Madagascar (except for the double mutations at nucleotides 341 and 342 [S114N] found in one Cambodian isolate). Among these nonsynonymous mutations, two were likely related to pyrimethamine resistance: S58R (corresponding to C59R in P. falciparum and S58R in Plasmodium vivax; observed in one Malagasy sample) and S114N (corresponding to S108N in P. falciparum and S117N in P. vivax; observed in three Cambodian samples).
INTRODUCTION
Currently, in many parts of the globe, the emergence and spread of malaria parasites resistant to various antimalarial drugs recommended by the international organizations remain major factors threatening control efforts (41). Among the five Plasmodium species that affect humans (40), resistant Plasmodium falciparum parasites were first selected by chloroquine (CQ), a highly effective, fast-acting, and inexpensive 4-aminoquinoline widely used for several decades, in Southeast Asia or South America (29, 44) in the 1960s. CQ-resistant parasites of the other Plasmodium species emerged much later: in 1989 for Plasmodium vivax (Papua New Guinea) (32) and in 2002 for Plasmodium malariae (Indonesia) (26). Following the introduction of the sulfadoxine-pyrimethamine combination (SP) to replace CQ as the first-line treatment for uncomplicated P. falciparum malaria, the same scenario was observed. SP-resistant P. falciparum parasites were first detected in the 1980s (9, 19). Molecular investigations, based on laboratory and field isolates, demonstrated later that the resistance of P. falciparum to pyrimethamine was mediated by specific point mutations in the dihydrofolate reductase gene (dhfr) (30, 31). Currently, it is assumed that pyrimethamine resistance is conferred by the stepwise selection of a series of nonsynonymous point mutations (codons 50, 51, 59, and 164) from the S108N single mutant allele (24). Parasites with the triple mutant allele (N51I C59R S108N) have markedly reduced in vitro susceptibility to pyrimethamine, and the presence of this allele in a P. falciparum-infected patient increases the risk of SP therapeutic failure (15). The additional mutation I164L confers on the quadruple mutant a high level of resistance to pyrimethamine (22) and abrogates the clinical efficacy of SP, as observed in Southeast Asia, South America (31), and Africa (25). Although P. vivax and P. malariae infections are not usually treated directly with SP, the high frequency of mixed infections, such as P. falciparum-P. vivax or P. falciparum-P. malariae infections, that are not detected by microscopy examination (27) has inevitably exposed a large number of non-P. falciparum parasites to antifolate therapy in many areas.
Comparison of the dihydrofolate reductase (DHFR) enzymes in P. falciparum and P. vivax showed that the DHFR domains were 77.3% identical and the active-site regions were strongly conserved (23). Moreover, sequencing of the dhfr gene in numerous P. vivax isolates collected in areas where SP was used to treat P. falciparum and alignment of these alleles with P. falciparum dhfr alleles have demonstrated that mutations in codons 57, 58, 61, 117, and 173 were involved in resistance to pyrimethamine and corresponded to codons 51, 59, 108, and 164 in P. falciparum (for a review, see reference 17). Heterologous expression studies later confirmed the role of these mutations in the resistance of P. vivax to pyrimethamine (17).
More recently, Tanomsing et al. conducted a study that aimed to determine whether SP pressure had selected pyrimethamine-resistant P. malariae parasites (37). By cloning, sequencing, and alignment of 35 Plasmodium malariae dihydrofolate reductase (Pmdhfr) sequences with the Plasmodium brasilianum dihydrofolate reductase (Pbdhfr) sequence, they observed several nonsynonymous mutations in five different codons (H22Q, K48E, N50K, S114N/G, and I170M). Three of these (codons 50, 114, and 170) corresponded to analogous positions known to be associated with pyrimethamine resistance in P. falciparum (residues 51, 108, and 164) and P. vivax (residues 117 and 173) (37). After this finding, Choowongkomon et al. demonstrated by computational analysis of interactions between DHFR inhibitors and the modeled structure of malaria parasite DHFR enzymes that certain residues in P. malariae DHFR (PmDHFR) were associated with significant reductions in binding energy for pyrimethamine (I13, L45, N53, S117, and I170) (8). However, the data acquired on PmDHFR polymorphism remain patchy; they are based on a few isolates, collected essentially in Southeast Asian countries (30/35 isolates), where SP has not been used for several decades. In this context, this study presents additional comprehensive data on the genetic diversity of PmDHFR by sequencing a large set of isolates (n = 123) collected in areas with different histories of SP use (mainland Africa, Madagascar, and Cambodia).
MATERIALS AND METHODS
Sample collection.
P. malariae samples were collected in Cambodia, Madagascar, and other countries in Africa. Isolates from Cambodia (venipuncture blood and dried blood spots) were obtained from symptomatic or asymptomatic individuals at different sites in 2001, 2004, and 2007, following microscopy examination. Samples from Madagascar were collected, as part of the surveillance of antimalarial drug resistance, between 2006 and 2008 from symptomatic patients before treatment in eight different health centers, located in areas exhibiting the four epidemiological patterns of malaria transmission (2). Other samples, obtained in 2001 to 2009 from malaria-infected travelers of African origin returning to France from various African countries (Comoros, Côte d'Ivoire, Cameroon, Mali, Gabon, and Togo), were provided by the National Reference Center for Malaria (NRCM), Paris, France.
DNA extraction, PCR amplification, and sequencing.
Parasite DNA was extracted directly from blood samples and dried blood spots by using a DNA blood kit (Qiagen, Germany) or the InstaGene matrix (Bio-Rad, Marnes la Coquette, France) according to the manufacturer's instructions. The genomic DNA samples were stored at −20°C until use. Parasite species were confirmed by nested PCR (35) (Cambodian samples) or by real-time PCR (10) (Malagasy samples and samples from the NRCM).
Nested-PCR approaches were used to increase the sensitivity of amplification. First rounds of PCR were performed in 25 μl (final volume) of a reaction buffer containing 2.5 μl DNA, 0.25 μM each primer (PmDHFR_PCR_F [5′-ATTCGACATATATGCCATCTG-3′] and PmDHFR_PCR_R [5′-CCTTCTTGCGTTTACTGAAA-3′]), 250 μM each deoxynucleoside triphosphate (dNTP) (Solis BioDyne), 2.5 mM MgCl2, and 1.25 U Taq polymerase (FirePol DNA polymerase I; Solis BioDyne). Amplifications were performed under the following conditions: 94°C for 5 min, followed by 40 cycles of 94°C for 30 s, 52°C for 90 s, and 72°C for 120 s. DNA synthesis was achieved by final extension at 72°C for 10 min (770 bp). Nested PCRs were carried out in 55 μl (final volume) of a reaction buffer containing 2 μl of PCR products, 0.4 μM each primer (PmDHFR_Nest_F [5′-GCTTGCTGTAAAGTGCCAAA-3′] and PmDHFR_Nest_R [5′-TTACAAAGTCTAACGACGTGCAG-3′]), 250 μM each dNTP, 2.5 mM MgCl2, and 2.5 U Taq polymerase (FirePol DNA polymerase I; Solis BioDyne). Amplifications were performed under the following conditions: 94°C for 5 min; 35 cycles of 94°C for 30 s, 57°C for 90 s, and 72°C for 90 s; and a final extension at 72°C for 10 min (681 bp).
PCR products were purified, using a polyacrylamide gel (Bio-Gel P-100; Bio-Rad, Marnes-la-Coquette, France), by 96-well plate filtration (Millipore, St. Quentin en Yvelines, France). Sequencing reactions were performed using the ABI Prism BigDye Terminator cycle sequencing ready reaction kit and were run on a 3730xl genetic analyzer (Applied Biosystems, Courtaboeuf, France). Electrophoregrams were visualized and analyzed with CEQ 2000 Genetic Analysis System software (Beckman Coulter). Amino acid sequences were compared with the wild-type sequence (GenBank accession no. AY846633 for Pmdhfr) using the BioEdit Sequence Alignment Editor (16). The presence of single nucleotide polymorphisms (SNP) was confirmed by reads through both forward and reverse strands. The Pmdhfr gene polymorphisms identified among our isolates were compared to dhfr gene sequences of P. malariae available in GenBank.
Statistical analysis.
Raw data were input into Microsoft Excel 2007 software and were checked and analyzed using MedCalc software, version 9.1.0.1 (MedCalc Software, Mariakerke, Belgium). Continuous variables were compared by using the independent-sample t test. Sequence polymorphism analysis, including the calculation of haplotype diversity (h) and nucleotide diversity (π), was performed using DnaSP software, version 5.10.01 (33). Departures from selective neutrality were assessed by the ratio of nonsynonymous to synonymous substitutions (dN/dS ratio) in the P. malariae dhfr gene. Significance (probability of rejecting the null hypothesis of strict neutrality [dN = dS]) was determined by using the two-tailed Z-test (bootstrap method with 10,000 replicates using the Nei-Gojobori method) as implemented in MEGA, version 5.0.5 (36). In all tests performed, P values of <0.05 were considered statistically significant at the 5% level.
Ethical approval.
Ethical clearance for the samples used in this study was obtained from the Ethics Committee of the Cambodian Ministry of Health and the Ethics Committee of the Ministry of Health of Madagascar (007/SANPF/2007; registration number ISRCTN36517335). Informed written consent was provided by all patients or their parents/guardians before inclusion in the study.
Nucleotide sequence accession numbers.
The sequences for the new haplotypes of the Pmdhfr gene identified here have been deposited in GenBank under accession numbers JN038065 to JN038071.
RESULTS
P. malariae dihydrofolate reductase polymorphisms.
A 633-bp fragment (spanning the region from codon 15 to codon 225 in the Pmdhfr gene) was sequenced from 123 clinical P. malariae isolates: 26 from Cambodia (including 5 P. falciparum-P. malariae and 3 P. vivax-P. malariae mixed infections), 53 from Madagascar (including 13 mixed infections with P. falciparum and P. malariae and 1 with P. vivax and P. malariae), and 44 from various African countries (19 from Côte d'Ivoire, 17 from Cameroon, 4 from Comoros, 2 from Mali, 1 from Gabon, and 1 from Togo) (including 9 P. falciparum-P. malariae mixed infections).
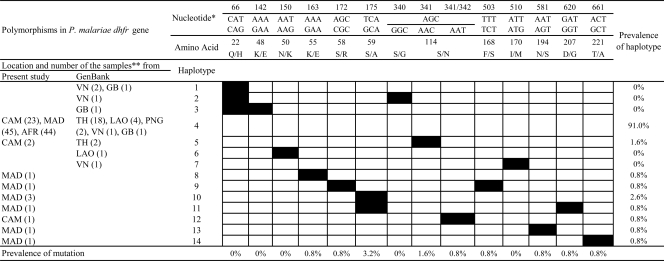
A total of 623 monomorphic and 10 polymorphic sites were found. Polymorphic sites included those with 2 synonymous substitutions (V60V [GTA to GTG]; S114N [AGC to AAT] [a nonsynonymous substitution was also found at this site]) and 8 nonsynonymous substitutions. The 2 silent mutations were observed only once (in one Cambodian isolate and one Malagasy sample). Of the 8 sites with nonsynonymous substitutions, all were dimorphic: K55E, S58R, S59A, S114N, F168S, N194S, D207G, and T221A. Most of the nonsynonymous substitutions (6/8 [75%]) were nonconservative, with changes of the physicochemical family of the amino acid. The substitutions observed at codons 55, 58, 168, 194, 207, and 221 were found once (0.81%), while those affecting codon 114 were observed in three Cambodian isolates (2.4%) and those affecting codon 59 in four Malagasy samples (3.2%). The polymorphisms in the Plasmodium malariae dhfr gene (including those previously described by Tanomsing et al. [37]) are detailed in Fig. 1.
Fig 1.
dhfr polymorphisms identified in 123 Plasmodium malariae isolates from Cambodia (CAM), Madagascar (MAD), and various African countries (AFR) (including Comoros, Côte d'Ivoire, Guinea, Cameroon, Mali, Gabon, and Togo) or described previously by Tanomsing et al. (37) and available in GenBank. *, nucleotide and amino acid numbers are given according to the wild-type sequence for Pmdhfr (GenBank accession no. AY846633); **, sample locations (with the numbers of isolates given in parentheses) are abbreviated as follows: VN, Vietnam; GB, Guinea-Bissau; TH, Thailand; LAO, Lao People's Democratic Republic; PNG, Papua New Guinea.
By following the numbering of haplotypes used by Tanomsing et al. (37), the polymorphisms observed in our samples were arranged into nine different haplotypes (H4, H5, and H8 to H14) and eight Plasmodium malariae DHFR proteins (H4, H5/H12, H8 to H11, H13, and H14) (Fig. 1). Haplotype H4 was the most prevalent (112/123 [91.0%]) and was considered the wild-type sequence (see Discussion). The other eight haplotypes were classified as either single mutant (H10 [S59A], H5 and H12 [S114N], H14 [T221A], H13 [N194S], and H8 [K55S]) or double mutant (H11 [S59A D207S] and H9 [S58R F168S]) types. By comparison of the polymorphisms identified in our sample population (n = 123) to sequences available in GenBank (n = 35), seven new mutations (K55E, S58R, S59A, F168S, N194S, D207G, and T221A) and seven new haplotypes (H8 to H13), leading to six new DHFR proteins, were observed (Fig. 1).
All new mutations and haplotypes observed in our sample set were recognized as exclusive to Madagascar (Fig. 1), except for the double mutations at nucleotides 341 and 342 (S114N) found in one Cambodian isolate. This isolate, classified as a new haplotype (H12), leads to a DHFR protein described previously in Thailand (H5). Only three isolates from Cambodia were classified as mutant type (S114N), while all isolates from African countries were classified as wild type.
The molecular diversity indices and neutrality tests for each P. malariae population are given in Table 1. The overall nucleotide diversity and the mean haplotype diversity in our sample population were 0.0004 and 0.171, respectively. Haplotype and nucleotide diversities were significantly higher in the Malagasy than in the Cambodian P. malariae population (P, 0.005 and 0.001, respectively) and significantly higher among Cambodian P. malariae sequences than among African P. malariae sequences (P, <0.0001). Analyses of the dN/dS ratio showed a negative ratio for Cambodian samples (−0.94), indicative of purifying selection, while Malagasy samples had a ratio greater than 1 (1.02), suggesting positive selection. For African samples, in which no synonymous mutations were detected, the dN/dS ratio was zero. However, in all three populations, the probability of rejecting the null hypothesis of strict neutrality (dN = dS) was nonsignificant.
Table 1.
dhfr polymorphisms identified in 123 Plasmodium malariae isolates from Cambodia, Madagascar, and various African countries
| Characteristic | Value for: |
|||
|---|---|---|---|---|
| Cambodia | Madagascar | African countries | All sites | |
| Sample size | 26 | 53 | 44 | 123 |
| No. of nucleotide sites observed | 633 | 633 | 633 | 633 |
| Monomorphic | 631 | 625 | 633 | 623 |
| Polymorphic | 2 | 8 | 0 | 10 |
| No. of changes | ||||
| Synonymous | 1 | 1 | 0 | 2 |
| Nonsynonymous | 1 | 7 | 0 | 8 |
| No. of haplotypes | 3 | 7 | 1 | 9 |
| Haplotype diversity (h) (SD) | 0.218 (0.103) | 0.279 (0.08) | 0 (0) | 0.171 (0.046) |
| Nucleotide diversity (π) (SD) | 0.00048 (0.00024) | 0.00067 (0.00024) | 0 (0) | 0.0004 (0.00013) |
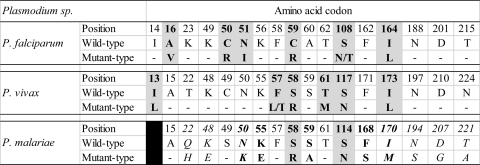
The correspondences between polymorphisms in the P. malariae dhfr gene and equivalent residues already known to be related to pyrimethamine resistance in the P. falciparum and P. vivax dhfr genes are presented in Fig. 2. Among the eight mutations described in the P. malariae dhfr gene, two were likely related to pyrimethamine resistance: S58R (H9; corresponding to C59R in P. falciparum and S58R in P. vivax; observed in one Malagasy sample) and S114N (H5 and H12; corresponding to S108N in P. falciparum and S117N in P. vivax; observed in three Cambodian samples). Three other mutations close to SNP involved in pyrimethamine resistance were also found: K55E (H8; observed in one Malagasy sample), S59A (H10 and H11; observed in four Malagasy samples), and F168S (H9; in association with S58R, observed in one Malagasy sample).
Fig 2.
Correspondence between polymorphisms in the P. malariae dhfr gene and equivalent residues known to be related to pyrimethamine resistance in the P. falciparum and P. vivax dhfr genes. For P. falciparum and P. vivax, codons resulting from nonsynonymous mutations related to pyrimethamine resistance are indicated by boldface and shading. For P. malariae, codons likely related to pyrimethamine resistance are indicated by boldface and shading, while those close to SNP involved in P. falciparum or P. vivax pyrimethamine resistance are shown in boldface only. P. malariae codons in italics correspond to mutations described previously by Tanomsing et al. (37).
DISCUSSION
Although the distribution of P. malariae infection is reported as patchy, it has been observed in all major regions of the world where malaria is endemic (18). More common in sub-Saharan Africa and the southwest Pacific (3, 5, 6, 12, 38), P. malariae is also detected in Asia (7, 11, 13, 14, 21), the Middle East (1), and South and Central America (34, 39). Responsible for quartan malaria, P. malariae is usually considered a benign pathogen compared to human sympatric P. falciparum and P. vivax. Severe clinical symptoms are rarely associated with the acute infection, except in cases of chronic nephropathy. To date, data from drug susceptibility studies on this species are scare, while treatments for P. falciparum and P. vivax malaria are often used in a context of mixed infections with P. malariae. In vitro assays of P. malariae are not currently available, and therapeutic efficacy studies are very difficult to conduct. Molecular investigations focused on putative molecular markers remain the only means of assessing the impact of antimalarial drugs on this species, especially by extrapolating to what we have already observed for P. falciparum or P. vivax parasites.
In this study, we have analyzed a total of 123 sequences from clinical P. malariae isolates from Cambodia, Madagascar, and various African countries. Among these, 10 polymorphic sites, including 2 sites with synonymous substitutions and 8 with nonsynonymous substitutions (codons 55, 58, 59, 114, 168, 194, 207, and 221), were found. One of the most interesting results was that glutamine (Q) was the most prevalent amino acid at position 22. This amino acid was previously considered mutant type (H22Q) by Tanomsing et al., who had used the P. brasilianum dhfr sequence as the wild-type reference sequence, despite the fact that more than 85% (30/35) of their sequences had a glutamine at this position (37). Their main rationale for using P. brasilianum as the wild-type sequence was that given the widespread use of SP and the apparent ease in which P. malariae acquires resistance to pyrimethamine (42, 43), they suspected that all the sequences obtained from P. malariae isolates could already carry mutations, unlike the laboratory-maintained P. brasilianum strain. However, several indications seem to show otherwise: (i) the high prevalence of the haplotype carrying a glutamine at position 22, (ii) the very high susceptibility of this haplotype to pyrimethamine by use of the classical in vitro assay (50% inhibitory concentration [IC50], <7 ng/ml) (37), (iii) the lack of equivalent residues in Pfdhfr and Pvdhfr previously associated with pyrimethamine resistance, (iv) the absence of a significant reduction in the energy of binding between pyrimethamine and the modeled PmDHFR enzyme observed in computational analysis (8), and (v) the difference in amino acid composition at codon 206 between sequences of Plasmodium brasilianum (valine) and Plasmodium malariae (isoleucine). All these indications lead us to suggest that the Pmdhfr wild-type sequence carries a glutamine at residue 22.
Compared to previous data, seven new mutations (at positions 55, 58, 59, 168, 194, 207, and 221) were observed exclusively in Malagasy samples, while the African isolates presented no mutations and the Cambodian samples presented only one mutation (S114N), already found in Thai samples. In total, nine haplotypes were found among our samples: two were shared with the sequences of Tanomsing et al. (37) (the wild-type haplotype and the S114N haplotype shared by Cambodian and Thai P. malariae sequences), and seven (exclusively from Malagasy samples) have never been described previously. However, the prevalences of these new haplotypes were very low (from 0.80% to 3.25%) compared to that of the wild type (91.05%). Among the new haplotypes, two were classified as double mutant type (S59A D207S and S58R F168S), while the others were classified as single mutant type (S59A, S114N, T221A, N194S, and K55S). Of the four mutations (leading to changes in residues 50, 58, 114, and 170) previously found to be associated with pyrimethamine resistance in P. falciparum and P. vivax (Fig. 2), only two were observed in our samples: S58R in one Malagasy sample (0.80%) and S114N in three Cambodian isolates (2.40%). We did not find mutations at position 50 (observed in one Lao People's Democratic Republic sample) or 170 (observed in one Vietnamese sample) in our set of samples. Recent data from in silico experiments on binding between modeled Pmdhfr mutant sequences and antifolates revealed that certain residues (I13, L45, D53, S117, and I170) appear to play important roles in binding with pyrimethamine (8). Among the mutations (N50K, S58R, S114N, I170M) found in field isolates from their previous work (37) that correlate with those seen in PfDHFR and PvDHFR, the authors showed that only the Pmdhfr I170M mutant type was associated with a significant reduction in docking energy for pyrimethamine, suggesting differences between species in terms of the ability of the DHFR enzyme to tolerate mutations. Three other mutations close to SNP involved in pyrimethamine resistance (K55E, S59A, and F168S) were also found in Malagasy samples. These mutations were not directly studied by Choowongkomon et al. (8) in their experiments, but they could have indirect implications for ligand binding by DHFR enzymes. For example, we cannot exclude the possibility that the S59A mutation was selected by antifolate drug pressure, since it is closely related to the S58R mutation, or that the mutation observed at codon F168S, which is closely related to I170M, would be an important target for the selection pressure of pyrimethamine resistance against PmDHFR. Further studies involving heterologous expression of the PmDHFR mutants found in our study would be useful in addressing this issue.
Comparing P. malariae dhfr sequences in Cambodian, Malagasy, and African populations, we observed greater haplotype and nucleotide diversity among the Malagasy P. malariae population. These data could be explained partly by the fact that the prevalence of P. malariae infections is about 10-fold higher in Madagascar than in Cambodia (4, 20). In both populations, wild-type P. malariae isolates were largely predominant (around 85%). In the Cambodian population, only one mutant type haplotype (114N) that correlates with P. falciparum or P. vivax mutant type haplotypes was observed, suggesting that this allele was selected when SP was used as an antimalarial in the 1980s (19). In Madagascar, P. malariae DHFR mutant type alleles are more polymorphic, a pattern likely related to the pyrimethamine pressure exerted with the increased use of SP since the implementation of intermittent preventive treatment (IPT) for pregnant women (28).
In conclusion, polymorphism in the Plasmodium malariae dhfr gene appears to be low compared to that in its sympatric Plasmodium species. However, according to our data from Cambodian and Malagasy isolates, local epidemiology and SP drug pressure history influence the selection of DHFR mutations, as is the case for P. falciparum and P. vivax. Additional studies comparing dhfr sequences in Plasmodium species from the same population or from patients with mixed infections would be worthwhile.
ACKNOWLEDGMENTS
We thank the patients and health care workers involved in the studies performed in Cambodia, Madagascar, and France. We are especially grateful to Sandra Incardona and Nicolas Steenkeste (Cambodia), Céline Barnadas, and Valérie Andriantsoanirina (Madagascar).
This study was supported by grants from Natixis and the Genomics Platform, Pasteur Génopôle, Pasteur Institute, France. Sample collection was funded in Cambodia by the European Community (ResMalChip project), the French Foreign Ministry (FSP-RAI project), and the Global Fund project, round 6 (grant CAM-607-G10M-CNM3), and in Madagascar by the Global Fund project, round 3 (grant MDG-304-G05-M). Financial support for the Master MIVA training of N. Khim (Biodiversity and Interactions of Microbial and Parasitic Organisms, Sciences for the Environment, University of Montpellier 2, Montpellier, France) was provided by the Pasteur Institute, Paris, France (Bourse d'Étude, Programme Calmette), and the Pasteur Institute in Cambodia. D. Ménard was supported by the French Ministry of Foreign Affairs during this work.
Footnotes
Published ahead of print 28 November 2011
REFERENCES
- 1. Al-Maktari MT, et al. 2003. Malaria status in Al-Hodeidah Governorate, Yemen: malariometric parasitic survey and chloroquine resistance P. falciparum local strain. J. Egypt Soc. Parasitol. 33:361–372 [PubMed] [Google Scholar]
- 2. Andriantsoanirina V, et al. 2009. Plasmodium falciparum drug resistance in Madagascar: facing the spread of unusual pfdhfr and pfmdr-1 haplotypes and the decrease of dihydroartemisinin susceptibility. Antimicrob. Agents Chemother. 53:4588–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anthony RL, et al. 1992. Heightened transmission of stable malaria in an isolated population in the highlands of Irian Jaya, Indonesia. Am. J. Trop. Med. Hyg. 47:346–356 [DOI] [PubMed] [Google Scholar]
- 4. Barnadas C, et al. 2007. Prevalence and chloroquine sensitivity of Plasmodium malariae in Madagascar. Am. J. Trop. Med. Hyg. 77:1039–1042 [PubMed] [Google Scholar]
- 5. Bonnet S, et al. 2002. Level and dynamics of malaria transmission and morbidity in an equatorial area of South Cameroon. Trop. Med. Int. Health 7:249–256 [DOI] [PubMed] [Google Scholar]
- 6. Browne EN, et al. 2000. Malariometric update for the rainforest and savanna of Ashanti region, Ghana. Ann. Trop. Med. Parasitol. 94:15–22 [PubMed] [Google Scholar]
- 7. Cabrera BD, Arambulo PV., III 1977. Malaria in the Republic of the Philippines. A review. Acta Trop. 34:265–279 [PubMed] [Google Scholar]
- 8. Choowongkomon K, et al. 2010. Computational analysis of binding between malarial dihydrofolate reductases and anti-folates. Malar. J. 9:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Darlow B, Vrbova H, Stace J, Heywood P, Aalpers M. 1980. Fansidar-resistant falciparum malaria in Papua New Guinea. Lancet ii:1243. [DOI] [PubMed] [Google Scholar]
- 10. de Monbrison F, Angei C, Staal A, Kaiser K, Picot S. 2003. Simultaneous identification of the four human Plasmodium species and quantification of Plasmodium DNA load in human blood by real-time polymerase chain reaction. Trans. R. Soc. Trop. Med. Hyg. 97:387–390 [DOI] [PubMed] [Google Scholar]
- 11. Dhangadamajhi G, Kar SK, Ranjit MR. 2009. High prevalence and gender bias in distribution of Plasmodium malariae infection in central east-coast India. Trop. Biomed. 26:326–333 [PubMed] [Google Scholar]
- 12. Genton B, et al. 1995. The epidemiology of malaria in the Wosera area, East Sepik Province, Papua New Guinea, in preparation for vaccine trials. I. Malariometric indices and immunity. Ann. Trop. Med. Parasitol. 89:359–376 [DOI] [PubMed] [Google Scholar]
- 13. Ghosh SK, Yadav RS. 1995. Naturally acquired concomitant infections of Bancroftian filariasis and human plasmodia in Orissa. Indian J. Malariol. 32:32–36 [PubMed] [Google Scholar]
- 14. Gordon DM, et al. 1991. Significance of circumsporozoite-specific antibody in the natural transmission of Plasmodium falciparum, Plasmodium vivax, and Plasmodium malariae in an aboriginal (Orang Asli) population of central peninsula Malaysia. Am. J. Trop. Med. Hyg. 45:49–56 [DOI] [PubMed] [Google Scholar]
- 15. Gregson A, Plowe CV. 2005. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol. Rev. 57:117–145 [DOI] [PubMed] [Google Scholar]
- 16. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 17. Hawkins VN, Joshi H, Rungsihirunrat K, Na-Bangchang K, Sibley CH. 2007. Antifolates can have a role in the treatment of Plasmodium vivax. Trends Parasitol. 23:213–222 [DOI] [PubMed] [Google Scholar]
- 18. Haworth J. 1988. The global distribution of malaria and the present control effort, p 1379–1420 In Wernsdorfer WH, McGregor I. (ed), Malaria: principles and practice of malariology. Churchill Livingstone, Edinburgh, United Kingdom [Google Scholar]
- 19. Hurwitz ES, Johnson D, Campbell CC. 1981. Resistance of Plasmodium falciparum malaria to sulfadoxine-pyrimethamine (‘Fansidar') in a refugee camp in Thailand. Lancet i:1068–1070 [DOI] [PubMed] [Google Scholar]
- 20. Incardona S, et al. 2007. Large-scale malaria survey in Cambodia: novel insights on species distribution and risk factors. Malar. J. 6:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kawamoto F, Liu Q, Ferreira MU, Tantular IS. 1999. How prevalent are Plasmodium ovale and P. malariae in East Asia? Parasitol. Today 15:422–426 [DOI] [PubMed] [Google Scholar]
- 22. Kiara SM, et al. 2009. In vitro activity of antifolate and polymorphism in dihydrofolate reductase of Plasmodium falciparum isolates from the Kenyan coast: emergence of parasites with Ile-164-Leu mutation. Antimicrob. Agents Chemother. 53:3793–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kongsaeree P, et al. 2005. Crystal structure of dihydrofolate reductase from Plasmodium vivax: pyrimethamine displacement linked with mutation-induced resistance. Proc. Natl. Acad. Sci. U. S. A. 102:13046–13051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lozovsky ER, et al. 2009. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc. Natl. Acad. Sci. U. S. A. 106:12025–12030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lynch C, et al. 2008. Emergence of a dhfr mutation conferring high-level drug resistance in Plasmodium falciparum populations from southwest Uganda. J. Infect. Dis. 197:1598–1604 [DOI] [PubMed] [Google Scholar]
- 26. Maguire JD, et al. 2002. Chloroquine-resistant Plasmodium malariae in south Sumatra, Indonesia. Lancet 360:58–60 [DOI] [PubMed] [Google Scholar]
- 27. Mayxay M, Pukrittayakamee S, Newton PN, White NJ. 2004. Mixed-species malaria infections in humans. Trends Parasitol. 20:233–240 [DOI] [PubMed] [Google Scholar]
- 28. Ménard D, et al. 2008. Assessment of the efficacy of antimalarial drugs recommended by the National Malaria Control Programme in Madagascar: up-dated baseline data from randomized and multi-site clinical trials. Malar. J. 7:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moore DV, Lanier JE. 1961. Observations on two Plasmodium falciparum infections with an abnormal response to chloroquine. Am. J. Trop. Med. Hyg. 10:5–9 [DOI] [PubMed] [Google Scholar]
- 30. Peterson DS, Milhous WK, Wellems TE. 1990. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc. Natl. Acad. Sci. U. S. A. 87:3018–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Plowe CV, Kublin JG, Doumbo OK. 1998. P. falciparum dihydrofolate reductase and dihydropteroate synthase mutations: epidemiology and role in clinical resistance to antifolates. Drug Resist. Updat. 1:389–396 [DOI] [PubMed] [Google Scholar]
- 32. Rieckmann KH, Davis DR, Hutton DC. 1989. Plasmodium vivax resistance to chloroquine? Lancet ii:1183–1184 [DOI] [PubMed] [Google Scholar]
- 33. Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497 [DOI] [PubMed] [Google Scholar]
- 34. Scopel KK, Fontes CJ, Nunes AC, Horta MF, Braga EM. 2004. High prevalence of Plasmodium malariae infections in a Brazilian Amazon endemic area (Apiacas-Mato Grosso State) as detected by polymerase chain reaction. Acta Trop. 90:61–64 [DOI] [PubMed] [Google Scholar]
- 35. Steenkeste N, et al. 2009. Towards high-throughput molecular detection of Plasmodium: new approaches and molecular markers. Malar. J. 8:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tanomsing N, et al. 2007. Genetic analysis of the dihydrofolate reductase-thymidylate synthase gene from geographically diverse isolates of Plasmodium malariae. Antimicrob. Agents Chemother. 51:3523–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Trape JF, et al. 1994. The Dielmo project: a longitudinal study of natural malaria infection and the mechanisms of protective immunity in a community living in a holoendemic area of Senegal. Am. J. Trop. Med. Hyg. 51:123–137 [DOI] [PubMed] [Google Scholar]
- 39. Warren M, Collins WE, Jeffery GM, Skinner JC. 1975. The seroepidemiology of malaria in Middle America. II. Studies on the Pacific coast of Costa Rica. Am. J. Trop. Med. Hyg. 24:749–754 [DOI] [PubMed] [Google Scholar]
- 40. White NJ. 2008. Plasmodium knowlesi: the fifth human malaria parasite. Clin. Infect. Dis. 46:172–173 [DOI] [PubMed] [Google Scholar]
- 41. World Health Organization 2010. Global report on antimalarial efficacy and drug resistance: 2000–2010. World Health Organization, Geneva, Switzerland [Google Scholar]
- 42. Young MD. 1957. Resistance of Plasmodium malariae to pyrimethamine (daraprim). Am. J. Trop. Med. Hyg. 6:621–624 [DOI] [PubMed] [Google Scholar]
- 43. Young MD. 1957. The response of Plasmodium malariae infections to pyrimethamine (daraprim). Am. J. Trop. Med. Hyg. 6:223–224 [DOI] [PubMed] [Google Scholar]
- 44. Young MD, Contacos PG, Stitcher JE, Millar JW. 1963. Drug resistance in Plasmodium falciparum from Thailand. Am. J. Trop. Med. Hyg. 12:305–314 [DOI] [PubMed] [Google Scholar]