Abstract
The current treatment of chronic hepatitis C is based on pegylated alpha interferon (PEG-IFN-α) and ribavirin. The aim of this study was to identify biological and clinical variables related to IFN therapy that could predict patient outcome. The study enrolled 47 patients treated with PEG-IFN and ribavirin combined therapy. The interferon concentration was measured in serum by a bioassay. The expression of 93 interferon-regulated genes in peripheral blood mononuclear cells was quantified by real-time quantitative reverse transcription-PCR (RT-PCR) before and after 1 month of treatment. The interferon concentration in the serum was significantly lower in nonresponders than in sustained virological responders. Moreover, a significant correlation was identified between interferon concentration and interferon exposition as well as body weight. The analysis of interferon-inducible genes in peripheral blood mononuclear cells among the genes tested did not permit the prediction of treatment outcome. In conclusion, the better option seems to be to treat patients with weight-adjusted PEG-IFN doses, particularly for patients with high weight who are treated with PEG-IFN-α2a. Although the peripheral blood mononuclear cell samples are the easiest to obtain, the measurement of interferon-inducible genes seems not be the best strategy to predict treatment outcome.
INTRODUCTION
Hepatitis C virus (HCV) infection is a major health problem worldwide, affecting more than 170 million people (29). HCV infection is a common cause of chronic liver disease, which may progress to hepatocellular carcinoma, and it is the most common indication of liver transplantation (28). Current treatment is based on the association between pegylated interferon (PEG-IFN) and ribavirin (RBV). This treatment is effective in about 55% of patients (15, 23).
Treatment outcome has been shown to be influenced by viral factors such as the HCV RNA baseline or HCV genotype (35), as well as by host factors such as obesity, cirrhosis, ethnic background, or fibrosis (17). Recently, a genetic polymorphism near the interleukin-28B gene encoding IFN-λ3 has been associated with the response to treatment (26, 33).
The early identification of patients who do not respond to PEG-IFN and RBV is a real challenge given the morbid side effects and cost efficacy of the treatment. It has been demonstrated that a rapid virological response (RVR; defined as the achievement of an undetectable HCV RNA level after 4 weeks of treatment) can accurately predict the sustained virologic response (SVR) (24). A short duration of treatment has been proposed for these patients (12, 38). In addition, the lack of early virological response (EVR; defined as a 2-log reduction in HCV RNA after 12 weeks of treatment) is predictive of a nonresponse (NR) with 97 to 98% accuracy. For these patients, a prolonged treatment of up to 72 weeks has been proposed (4).
Other parameters derived from the treatment can influence the response, such as RBV doses or plasma concentrations. Indeed, it is now firmly accepted that the body weight adjustment of RBV doses increases the EVR and RVR rates (3). Hence, the study of the pharmacokinetic parameters of RBV (such as RBV exposition or RBV concentration in serum) suggests that they can predict the treatment outcome (22, 25). For PEG-IFN, two molecules are currently available, PEG-IFN-α2a, which has a large branched PEG moiety and is administered at a fixed dose of 180 μg/week, and PEG-IFN-α2b, which has a small linear PEG structure and is administered at a dose of 1.5 μg/kg of body weight/week. Unlike the case for RBV, the importance of the IFN dose and/or concentration in the treatment response has not been deeply investigated (6, 7, 13).
In this study, we have focused on identifying IFN-related factors that could influence treatment outcome. We have analyzed the IFN concentrations in serum 1 month after the initiation of treatment and the expression of IFN-inducible genes in peripheral blood mononuclear cells (PBMCs) before and during treatment. We showed that the IFN concentration in the serum could influence treatment outcome and is dependent on the IFN exposition, particularly for high-weight patients. The expression of interferon-related genes in the PBMC among our set of genes could not predict the treatment outcome.
MATERIALS AND METHODS
Clinical protocol and patients.
The study enrolled 56 patients who were eligible for therapy, and they were recruited between September 2005 and August 2007. These patients have established diagnoses of chronic hepatitis C virus with detectable HCV antibodies and detectable HCV RNA in serum (COBAS TaqMan HCV test; Roche Diagnostics, Meylan, France). Exclusion criteria were the presence of other hepatitis viruses, other hepatic diseases, HIV coinfection, and other medical complications. All participants gave their informed consent, and the study was validated by the local ethics committee (Comité de Protection de la Personne Nord-Ouest II; number 04H21). Only 47 patients have been included in the present study; among patients who were excluded, 6 stopped the treatment prematurely without performing the follow-up, 1 was spontaneously cured of virus, and PBMCs were not collected for 2 patients.
HCV treatment was based on a combination of PEG-IFN and RBV. The doses were 180 μg/week for PEG-IFN-α2a (n = 32) and 1.5 μg/kg of body weight/week for PEG-IFN-α2b (n = 15). For patients infected with HCV genotype 1 or 4, RBV doses were adjusted to the body weight (doses ranged from 800 to 1,200 mg) during 48 weeks. For HCV patients infected with HCV genotype 2 or 3, a fixed dose (800 mg) of RBV was given during 24 weeks.
Sample collection, HCV RNA quantitation, and serum IFN concentration.
PBMCs were collected with PAXgene RNA tubes (BD Diagnostics, Le Pont de Claix, France), which contain an RNA stabilization reagent, and were frozen at −80°C until use. They were collected before the initiation of treatment (d0; the time of pretreatment consultation) and 1 month after the first injection (M1). To avoid variations in IFN concentrations due to the time of the injection, PBMCs were collected 3 days after the last injection at M1. HCV RNA was quantified from serum before the initiation of treatment (d0) and at M1, 3 months after the first injection (M3), and 6 months after the end of the treatment as described above. Interferon concentrations were determined from serum at M1 as previously described (14).
RNA isolation and real-time reverse transcription-PCR (RT-PCR).
RNA were extracted using the PAXgene blood RNA kit (Qiagen, Courtaboeuf, France) as recommended by the manufacturer. RNA concentration and integrity were determined with a Nanodrop (Thermoscientific, Illkirch, France) and an Agilent Bioanalyzer (Agilent Technologies, Massy, France). Reverse transcription was performed using random hexamers and the high-capacity cDNA kit (Applied Biosystems, Courtaboeuf, France) according to the manufacturer's instructions. Real-time PCR experiments were performed using an ABI Prism 7900 sequence detection system (Applied Biosystems) and Microfluidic card technology (Applied Biosystems). In the present study, the cards were configured into two 96-gene sets, which enabled the analysis of gene expression in two different conditions (see Table S1 in the supplemental material). Gene expression values were normalized according to the level of the β-actin gene, which was determined as the best endogenous control by the software GeNorm (34). Differential gene expression was determined using the ΔΔCT (cyclic threshold) method.
Statistical analysis.
A correlation analysis between IFN concentration, body weight, and IFN exposition was performed using the Mann-Whitney, Kruskal-Wallis, or Pearson statistical test. A multivariate logistic regression model was used to explore the independent factors that could be used to predict a virological response. Statistical analysis was performed using GraphPad Prism5 and R software. Results were considered significant when P < 0.05.
IFN-regulated gene expression in PBMCs was analyzed using R-based BRB-ArrayTools software, version 3.5.0, developed by Richard Simon and the BRB-ArrayTools development team (32). Differentially expressed genes were identified by a univariate two-sample t test with a random variance model (8). Permutation P values for significant genes (P < 0.05) were computed based on 10,000 random permutations with a false discovery rate of <1% and with 95% confidence. Class prediction analysis was based on the compound covariate predictor, diagonal linear discriminant analysis, nearest neighbor classification, and support vector machines with the linear kernel. The prediction models incorporated genes that were differentially expressed among genes at the 0.01 significance level as assessed by a random-variance t test. The leave-one-out cross-validation method was used to compute the misclassification rate. Binary tree prediction was based on the compound covariate predictor algorithm by incorporating genes that were differentially expressed among classes at the 0.05 significance level as assessed by the random-variance t test. For all statistical analyses, PCR duplicates were used separately to increase the relevance of the results; only genes with a similar regulation of the two probes were considered significant.
RESULTS
Baseline characteristics of study groups.
For each patient included in the study, HCV RNA was measured before the initiation of the treatment (d0), after 1 (M1) and 3 (M3) months, at the end of the treatment (EOT), and 6 months after EOT (designated follow-up). Fifteen patients (32%) exhibited a sustained virological response (SVR), 15 patients (32%) did not respond to treatment (nonresponder [NR]), and 17 patients (36%) had a relapse (responder relapser [RR]). Patients who did not have a 2 log decrease of HCV RNA levels at M3 (EVR−) all were nonresponders. Among patients with undetectable HCV RNA at M1 (RVR+), 75% exhibited an SVR. Several parameters influencing the response have been studied. As shown in Table 1, the degrees of fibrosis or IFN exposition (defined as the dose of IFN per kg of body weight) were significantly different compared to the complete response. On the contrary, body weight, serum alanine aminotransferase (ALT) level, and RBV dose were not significantly different between the types of responses.
Table 1.
Characteristics of the cohortd
| Parameter | Result by response type |
P value | ||
|---|---|---|---|---|
| NR | RR | SVR | ||
| No. of patients | 15 | 17 | 15 | |
| Body wt, kg (means [SD]) | 82.73 (15.34) | 83.53 (14.05) | 76.56 (11.88) | 0.3789 |
| HCV RNA baseline, log10 IU/ml (means [SD]) | 6.39 (0.42) | 6.43 (0.49) | 5.59 (1.01) | 0.0763 |
| ALT, xN (means [SD])e | 1.85 (1.18) | 2.14 (1.67) | 1.45 (0.80) | 0.3777 |
| Fibrosis score (means [SD]) | 2.73 (1.12) | 2.88 (1.06) | 1.53 (1.25) | 0.0130a |
| IFN exposition, μg/kg/wk (means [SD]) | 1.70 (0.36) | 2.05 (0.48) | 2.34 (0.44) | 0.0027b |
| Ribavirin dose, mg/kg/day (means [SD]) | 12.54 (2.10) | 12.34 (1.38) | 12.88 (1.69) | 0.4055 |
| Age, yr (means [SD]) | 48 (9.71) | 52 (12.34) | 43 (7.65) | 0.1245 |
| Sex (male/female) | 10/5 | 13/4 | 12/3 | 0.6811 |
| Genotype 1-4/other genotype | 15/0 | 12/5 | 10/5 | 0.0488c |
For NR versus RR, P = 0.7626; for RR versus SVR, P = 0.0073; for NR versus SVR, P = 0.0265.
For NR versus RR, P = 0.0149; for RR versus SVR, P = 0.0858; for NR versus SVR, P = 0.0026.
By chi-squared test.
NR, nonresponder patients; RR, responder relapser; SVR, sustained virological responder. Several parameters have been studied according to the treatment response. P values were the results of Kruskal-Wallis tests. When this P value was significant, a nonparametric statistical test was performed between each condition.
N, normal.
Treatment response depends on serum IFN concentration.
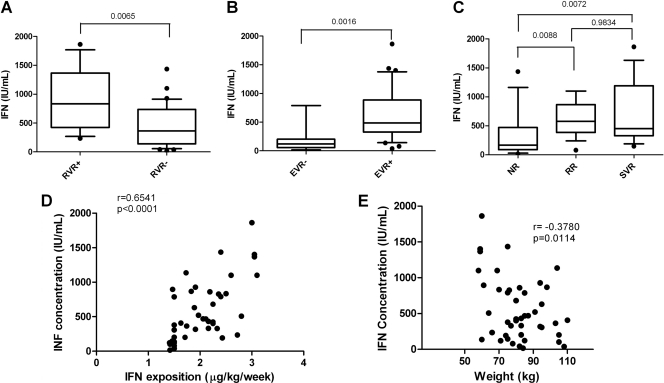
For each patient, the IFN concentration was measured in the serum at M1. Given the great variability in IFN concentration between patients, a statistical analysis was performed to determine if the IFN concentration could be correlated with the treatment response. As shown in Fig. 1A, IFN concentration at month 1 was significantly higher for patients having an RVR (median RVR−, 364 IU/ml; RVR+, 834 IU/ml). Moreover, patients without EVR (Fig. 1B) or that were NR (Fig. 1C) had a lower serum IFN concentration (median EVR+, 488 IU/ml; EVR−, 121 IU/ml; NR, 164 IU/ml; RR, 576 IU/ml; SVR, 451 IU/ml).
Fig 1.
IFN concentration and response to treatment. For each patient, the IFN concentration was measured at M1. Box plots represent the IFN concentration according to the antiviral response at M1 (RVR+, n = 11; RVR−; n = 33) (A), at M3 (EVR−, n = 8; EVR+, n = 36) (B), and 6 months after the end of the treatment (NR, n = 14; RR, n = 17; SVR, n = 14) (C). P values were the results of Mann-Whitney tests. The correlations between the IFN concentration and IFN exposition (D) and body weight (E) are represented on the graphs. r and P values were the results of Pearson correlation tests.
To evaluate if IFN concentration could be related to IFN exposition, a correlation analysis between these two parameters was performed. As shown in Fig. 1D, these two parameters were positively and significantly correlated (r = 0.65; P < 10−5), suggesting that body weight influenced the interferon concentration in serum. This effect was more pronounced with IFN-α2a, which was administered independently of body weight (data not shown). Moreover, we found that body weight was negatively correlated with interferon concentration (Fig. 1E) (r = −0.37; P = 0.01).
Multivariate analysis was performed on treatment outcomes using the two most significant variables (i.e., fibrosis and IFN exposition). Treatment outcome was characterized as response (SVR patients) or nonresponse (RR and NR patients). Statistical analysis showed that fibrosis and IFN exposition were two independent factors (P = 0.0143 and 0.0289, respectively). The cutoff was 2.06 μg/kg/week. The odds ratio for IFN exposition was 5.862 (1.200 to 28.636) with a sensitivity of 87% and a specificity of 69%.
IFN-regulated gene signature during the first month of treatment.
Since differences in IFN concentration were observed during treatment, we analyzed the expression of a set of IFN-induced genes in PBMCs. Ninety-three genes were selected according to the bibliographic survey and were further analyzed by RT-quantitative PCR (9). Gene expression ratios between d0 and M1 were analyzed to identify specific gene expression patterns related to clinically relevant groups. We first studied the complete response to treatment. Class comparison analysis identified only one probe (STAT2) that differs significantly between the SVR and NR patients. One probe (STAT5A) was significantly altered between RR and SVR patients, and 7 probes (4 genes) were significantly altered between NR and RR groups (Table 2). However, the expression of these genes was unable to predict the treatment response with great confidence, as revealed by class prediction analysis.
Table 2.
Gene induction according to response
| Gene and comparison | Induction (fold change) |
P value |
|
|---|---|---|---|
| Parametric | Permutation | ||
| NR versus SVR | |||
| STAT2 | 0.77 | 0.0442 | 0.0483 |
| SVR versus RR | |||
| STAT5A | 1.17 | 0.0469 | 0.0471 |
| NR versus RR | |||
| IRF2a | 0.74 | 0.019 | 0.0174 |
| GZMAa | 0.64 | 0.0301 | 0.0305 |
| IRF8 | 0.78 | 0.0381 | 0.0368 |
| ISGF3G | 1.24 | 0.0419 | 0.0435 |
For these genes the two probes were significantly regulated, and the P value is indicated for the most significant probe.
Prediction of treatment outcome.
The early prediction of treatment outcome during therapy is of both clinical and economical interest. Thus, we performed a class comparison and prediction analysis on the gene expression data generated at d0 or at M1.
The first analysis concerned the prediction of treatment response before the initiation of the treatment by analyzing the mRNA levels at d0. Class comparison analysis identified two genes (ISGF3G and IFNAR2) that differ significantly between the SVR and NR patients, four genes between SVR and RR patients (APOL1, CD4, BAK1, and NMI), and six genes between NR and RR patients (ISGF3G, FAS, PSME1, IRF8, BAK1, and IFNAR2) (Table 3). The induction of IRF8, IFNAR2, and ISGF3G and the repression of BAK1 were more pronounced for RR patients, suggesting that there were more differences in gene expression between RR and NR or SVR patients than between SVR and NR groups. This observation was confirmed by a binary tree classification in which NR patients clustered near the SVR group. RR patients were identified in another cluster. As previously observed, class prediction analysis using gene expression in PBMCs did not allow a strongly reliable prediction of treatment response (SVR from NR patients, 53% correct classification by the compound covariate predictor algorithm [P = 0.41] using one probe; NR from RR patients, 65% correct classification by 1-nearest neighbor algorithm [P = 0.14] using six probes).
Table 3.
Gene expression before treatment according to responsea
| Gene and comparison | Expression (fold change) |
P value |
|
|---|---|---|---|
| Parametric | Permutation | ||
| NR versus SVR | |||
| ISGF3G | 1.3 | 0.004 | 0.0051 |
| IFNAR2 | 0.81 | 0.018 | 0.0181 |
| SVR versus RR | |||
| APOL1 | 1.28 | 0.007 | 0.0081 |
| CD4 | 0.78 | 0.017 | 0.0162 |
| BAK1 | 3.72 | 0.019 | 0.0219 |
| NMI | 0.78 | 0.03 | 0.03 |
| NR versus RR | |||
| ISGF3G | 1.18 | 0.0031 | 0.0037 |
| FAS | 0.68 | 0.0034 | 0.0034 |
| PSME1 | 0.75 | 0.05 | 0.0049 |
| IRF8 | 0.62 | 0.008 | 0.006 |
| BAK1 | 3.82 | 0.02 | 0.03 |
| IFNAR2 | 0.81 | 0.029 | 0.03 |
For all genes the two probes were significantly regulated, and the P value is indicated for the most significant probe.
A similar analysis was performed from RNA abundances determined after 1 month of treatment. Class comparison analysis identified two genes that differ significantly between NR and SVR patients (FAS and IFNAR2), five genes that differ significantly between SVR and RR patients (MX1, STAT1, TRIM21, BAK1, and SP110), and eight genes that differ significantly between NR and SVR patients (PSME1, IFNAR2, STAT1, FAS, RELB, BAK1, TRIM21, and IRF8) (Table 4). As previously observed, BAK1 was repressed for RR patients, and FAS, IFNAR2, and TRIM21 were repressed in NR patients. Recapitulating our previous observation, binary tree prediction could not distinguish NR patients from SVR patients. Accordingly, no prediction of treatment outcome was possible with great confidence (data not shown). The genotype (1, 4, and 5 versus 2 and 3) did not influence gene expression (data not shown).
Table 4.
Gene expression at M1 according to responsea
| Gene and comparison | Expression (fold change) |
P value |
|
|---|---|---|---|
| Parametric | Permutation | ||
| NR versus SVR | |||
| FAS | 0.78 | 0.007 | 0.0089 |
| IFNAR2 | 0.87 | 0.035 | 0.0351 |
| SVR versus RR | |||
| MX1 | 0.71 | 0.001 | 0.0017 |
| STAT1 | 0.78 | 0.013 | 0.0137 |
| TRIM21 | 0.81 | 0.014 | 0.0137 |
| SP110 | 0.73 | 0.03 | 0.0128 |
| BAK1 | 2.9 | 0.03 | 0.03 |
| NR versus RR | |||
| PSME1 | 0.74 | 0.006 | 0.0054 |
| IFNAR2 | 0.81 | 0.006 | 0.0048 |
| STAT1 | 0.79 | 0.007 | 0.0051 |
| FAS | 0.76 | 0.012 | 0.0117 |
| RELB | 1.5 | 0.014 | 0.015 |
| BAK1 | 3.4 | 0.017 | 0.02 |
| TRIM21 | 0.8 | 0.025 | 0.024 |
| IRF8 | 0.77 | 0.039 | 0.038 |
For all genes the two probes were significantly regulated, and the P value is indicated for the most significant probe.
DISCUSSION
We showed that IFN concentrations in sera during treatment were higher in patients presenting an SVR, an RVR, or an EVR than in nonresponders or patients with neither RVR or EVR. Moreover, IFN exposition, calculated as the dose of interferon per kg of weight, and the weight also are correlated with the IFN concentration. This observation suggests that the weight of the patients influences the pharmacokinetics of PEG-IFN, and that high-weight patients had a lower IFN concentration in serum. This observation is particularly obvious for patients treated with PEG-IFN-α2a, for which dose is not weight adjusted, because the volume of distribution was more restricted with PEG-IFN-α2a (18), which increases its availability in serum and target organs. However, our results suggest that for overweight patients, this explanation is not sufficient, and that the administration of higher doses of IFN could increase the SVR rate. For PEG-IFN-α2b, for which the dose is weight adjusted, the weight had less influence on IFN concentration (data not shown). These results confirmed a previous study that compared the pharmacokinetic parameters of the two PEG-IFNs. A decrease in drug exposure was observed with an increase in body weight for PEG-IFN-α2a only (31).
The small number of patients who presented a lack of RVR or EVR in our study can influence the statistical results, but for patients with a lack of EVR, all patients but one had an IFN concentration lower than 200 IU/ml. For these patients, the measure of the IFN concentration could be useful for adapting the doses of IFN and modifying the outcome.
This hypothesis was supported by a clinical study that compared responses following PEG-IFN-α2a treatments of 180 to 270 μg/week in obese patients (5). In that study, a dose of 270 μg/week increased the IFN concentration in serum, and the SVR rate was 14%. Few studies suggested that IFN concentration was important for the response independently of weight. This was demonstrated with true nonresponder patients infected by HCV genotype 1 treated with higher doses of PEG-IFN-α2a (11). The area under the serum concentration-time curve of PEG-IFN-α2a increased in a dose-dependent manner and was associated with an SVR. Moreover, PEG-IFN-α2a concentration was associated with the SVR in HCV/HIV coinfected patients (21). Nevertheless, this result was not confirmed by Asahina et al. in a cohort of HCV-infected patients (1). The study was realized with weight-adjusted PEG-IFN-α2b. This measure of interferon activity gives additional information on treatment efficacy and perhaps is more exact than an immunological assay.
Singular results were obtained by studying the expression of IFN-regulated genes in PBMCs at M1. As the response was correlated with IFN concentration, it was legitimate to assume that the serum IFN concentration could influence the IFN-regulated gene expression. In fact, the gene expression in PBMCs was not directly correlated with the interferon concentration (data not shown) and was not easily available to predict the response with great confidence.
Other groups described that gene expression in PBMCs was predictive of treatment outcome (10, 36). One group demonstrated that the expression of STAT6 and SOCS1 was predictive of the SVR in pretreatment samples (36). These genes were not included in our assay. A second group identified four genes (TNFAIP6, MT2A, IFIT2, and CCRL2) regulated early during the treatment (12 h after the initiation of therapy) that could be predictive of the response (10). These results were not confirmed by another group (27), and we did not confirm these results for IFIT2. The great variability in the results could be explained in part by differences in the timing of the analysis, i.e., before or during treatment, after 1 month, or earlier.
Indeed, IFN gene expression in the liver was more informative than that in PBMCs (2, 27, 30), suggesting that the treatment of chronic HCV infection has a strong local hepatic effect on the IFN system. This could be because the IFN concentration (always >100 UI/ml in serum) was sufficient to induce the interferon-stimulated genes, and the difference of induction between 100 and 1,000 IU/ml did not change the induction in PBMCs. However, the concentration in serum could influence the concentration in the liver and then the induction of ISG in hepatocytes.
In conclusion, it would be interesting to treat patients with weight-adjusted doses of PEG-IFN, particularly for overweight patients, to increase the IFN concentration in the serum. This concentration could influence antiviral efficacy in the liver. Gene induction in PBMCs did not reflect the antiviral efficacy in the liver. A confirmation of this study with a greater number of patients is necessary, but these results are already significant.
Two direct-acting antiviral agents (DAAs) are available. Boceprevir and telaprevir are two inhibitors of the NS3 protease and are administered in combination with PEG-IFN and RBV, increasing the response rate. Many other DAAs (anti-NS3, anti-NS5B, and anti-NS5A) are in trial alone or in combination with IFN and RBV. Several trials report that a combination of DAAs could efficiently decrease HCV replication without IFN and RBV (16, 19, 37). Nevertheless, the long-term efficacy seems to be related to the presence of PEG-IFN and RBV, at least to avoid resistance to DAA (20).
Supplementary Material
ACKNOWLEDGMENTS
We thank Pauline-Eva Pecquet for her technical assistance and Momar Diouf for statistical analyses of IFN concentrations.
This work was supported by the Programme Hospitalier de Recherche Clinique de Picardie (PHRC, 2004). C.C. is a recipient of a fellowship from Association pour la Recherche sur le Cancer, France.
We declare that we do not have anything to disclose regarding funding from industries or conflicts of interest with respect to the manuscript.
Footnotes
Published ahead of print 28 November 2011
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Asahina Y, et al. 2007. Pharmacokinetics and enhanced PKR response in patients with chronic hepatitis C treated with pegylated interferon alpha-2b and ribavirin. J. Viral Hepat. 14:396–403 [DOI] [PubMed] [Google Scholar]
- 2. Asselah T, et al. 2008. Liver gene expression signature to predict response to pegylated interferon plus ribavirin combination therapy in patients with chronic hepatitis C. Gut 57:516–524 [DOI] [PubMed] [Google Scholar]
- 3. Bain VG, et al. 2008. Clinical trial: exposure to ribavirin predicts EVR and SVR in patients with HCV genotype 1 infection treated with peginterferon alpha-2a plus ribavirin. Aliment. Pharmacol. Ther. 28:43–50 [DOI] [PubMed] [Google Scholar]
- 4. Berg T, et al. 2006. Extended treatment duration for hepatitis C virus type 1: comparing 48 versus 72 weeks of peginterferon-alfa-2a plus ribavirin. Gastroenterology 130:1086–1097 [DOI] [PubMed] [Google Scholar]
- 5. Bressler B, Wang K, Grippo JF, Heathcote EJ. 2009. Pharmacokinetics and response of obese patients with chronic hepatitis C treated with different doses of PEG-IFN alpha-2a (40KD) (PEGASYS). Br. J. Clin. Pharmacol. 67:280–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bruno R, et al. 2004. Viral dynamics and pharmacokinetics of peginterferon alpha-2a and peginterferon alpha-2b in naive patients with chronic hepatitis c: a randomized, controlled study. Antivir. Ther. 9:491–497 [PubMed] [Google Scholar]
- 7. Bruno R, et al. 2007. Pharmacodynamics of peginterferon alpha-2a and peginterferon alpha-2b in interferon-naive patients with chronic hepatitis C: a randomized, controlled study. Aliment. Pharmacol. Ther. 26:369–376 [DOI] [PubMed] [Google Scholar]
- 8. Coulouarn C, Factor VM, Thorgeirsson SS. 2008. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology 47:2059–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Der SD, Zhou A, Williams BR, Silverman RH. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95:15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Devitt E, Lawless MW, Sadlier DABJ, Walsh C, Crowe J. 2010. Early viral and peripheral blood mononuclear cell responses to pegylated interferon and ribavirin treatment: the first 24 h. Eur. J. Gastroenterol. Hepatol. 22:1211–1220 [DOI] [PubMed] [Google Scholar]
- 11. Diago M, et al. 2007. Clinical trial: pharmacodynamics and pharmacokinetics of re-treatment with fixed-dose induction of peginterferon alpha-2a in hepatitis C virus genotype 1 true non-responder patients. Aliment. Pharmacol. Ther. 26:1131–1138 [DOI] [PubMed] [Google Scholar]
- 12. Ferenci P, et al. 2008. Peginterferon alfa-2a and ribavirin for 24 weeks in hepatitis C type 1 and 4 patients with rapid virological response. Gastroenterology 135:451–458 [DOI] [PubMed] [Google Scholar]
- 13. Formann E, Jessner W, Bennett L, Ferenci P. 2003. Twice-weekly administration of peginterferon-alpha-2b improves viral kinetics in patients with chronic hepatitis C genotype 1. J. Viral Hepat. 10:271–276 [DOI] [PubMed] [Google Scholar]
- 14. François C, et al. 2005. Quantification of different human alpha interferon subtypes and pegylated interferon activities by measuring MxA promoter activation. Antimicrob. Agents Chemother. 49:3770–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fried MW, et al. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975–982 [DOI] [PubMed] [Google Scholar]
- 16. Gane EJ, et al. 2010. Oral combination therapy with a nucleoside polymerase inhibitor (RG7128) and danoprevir for chronic hepatitis C genotype 1 infection (INFORM-1): a randomised, double-blind, placebo-controlled, dose-escalation trial. Lancet 376:1467–1475 [DOI] [PubMed] [Google Scholar]
- 17. Ghany MG, Strader DB, Thomas DL, Seeff LB. 2009. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology 49:1335–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Glue P, et al. 2000. Pegylated interferon-alpha2b: pharmacokinetics, pharmacodynamics, safety, and preliminary efficacy data. Hepatitis C Intervention Therapy Group. Clin. Pharmacol. Ther. 68:556–567 [DOI] [PubMed] [Google Scholar]
- 19. Lawitz E, et al. 2011. Once daily dual-nucleotide combination of PSI-938 and PSI-7977 provides 94% HCV RNA < load at day 14: first purine/pyrimidine clinical combination data (the NUCLEAR study). J. Hepatol. 54(Suppl):S543 [Google Scholar]
- 20. Lok A, et al. 2011. Quadruple therapy with BMS-790052, BMS-650032 and PEG-IFN/RBV for 24 weeks results in 100% SVR12 in HCV genotype 1 null responders. J. Hepatol. 54(Suppl):S536. [Google Scholar]
- 21. Lopez-Cortes LF, et al. 2008. Role of pegylated interferon-alpha-2a and ribavirin concentrations in sustained viral response in HCV/HIV-coinfected patients. Clin. Pharmacol. Ther. 84:573–580 [DOI] [PubMed] [Google Scholar]
- 22. Loustaud-Ratti V, et al. 2008. Ribavirin exposure after the first dose is predictive of sustained virological response in chronic hepatitis C. Hepatology 47:1453–1461 [DOI] [PubMed] [Google Scholar]
- 23. Manns MP, et al. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958–965 [DOI] [PubMed] [Google Scholar]
- 24. Martinot-Peignoux M, et al. 2009. Virological response at 4 weeks to predict outcome of hepatitis C treatment with pegylated interferon and ribavirin. Antivir. Ther. 14:501–511 [PubMed] [Google Scholar]
- 25. Maynard M, Pradat P, Gagnieu MC, Souvignet C, Trepo C. 2008. Prediction of sustained virological response by ribavirin plasma concentration at week 4 of therapy in hepatitis C virus genotype 1 patients. Antivir. Ther. 13:607–611 [PubMed] [Google Scholar]
- 26. McCarthy JJ, et al. 2010. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology 138:2307–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sarasin-Filipowicz M, et al. 2008. Interferon signaling and treatment outcome in chronic hepatitis C. Proc. Natl. Acad. Sci. U. S. A. 105:7034–7039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seeff LB. 2002. Natural history of chronic hepatitis C. Hepatology 36:S35–S46 [DOI] [PubMed] [Google Scholar]
- 29. Seeff LB, Hoofnagle JH. 2003. Appendix: the National Institutes of Health Consensus Development Conference Management of Hepatitis C 2002. Clin. Liver Dis. 7:261–287 [DOI] [PubMed] [Google Scholar]
- 30. Selzner N, et al. 2008. Hepatic gene expression and prediction of therapy response in chronic hepatitis C patients. J. Hepatol. 48:708–713 [DOI] [PubMed] [Google Scholar]
- 31. Silva M, et al. 2006. A randomised trial to compare the pharmacokinetic, pharmacodynamic, and antiviral effects of peginterferon alfa-2b and peginterferon alfa-2a in patients with chronic hepatitis C (COMPARE). J. Hepatol. 45:204–213 [DOI] [PubMed] [Google Scholar]
- 32. Simon R, et al. 2007. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 3:11–17 [PMC free article] [PubMed] [Google Scholar]
- 33. Tanaka Y, et al. 2009. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat. Genet. 41:1105–1109 [DOI] [PubMed] [Google Scholar]
- 34. Vandesompele J, et al. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wohnsland A, Hofmann WP, Sarrazin C. 2007. Viral determinants of resistance to treatment in patients with hepatitis C. Clin. Microbiol. Rev. 20:23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Younossi ZM, et al. 2009. Early gene expression profiles of patients with chronic hepatitis C treated with pegylated interferon-alfa and ribavirin. Hepatology 49:763–774 [DOI] [PubMed] [Google Scholar]
- 37. Zeuzem S, et al. 2010. Strong antiviral activity and safety of IFN-sparing treatment with the protease inhibitor BI 201335, the HCV polymerase inhibitor BI207127 and ribavirin in patients with chronic HCV. Hepatology 52(Suppl.):876a [Google Scholar]
- 38. Zeuzem S, et al. 2006. Efficacy of 24 weeks treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C infected with genotype 1 and low pretreatment viremia. J. Hepatol. 44:97–103 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.