Abstract
Occidiofungin is a cyclic glyco-lipopeptide produced by Burkholderia contaminans. MICs against Candida species were between 0.5 and 2.0 μg/ml. Occidiofungin retains its in vitro potency in the presence of 5% and 50% human serum with a minimal lethal concentration (MLC) of 2 and 4 μg/ml, respectively. Time-kill and postantifungal effect (PAFE) experiments of occidiofungin against Candida albicans were performed. The results demonstrate that occidiofungin is fungicidal. Occidiofungin was also found to be a very stable molecule. It is resistant to extreme temperatures and pH and maintains its activity following exposure to gastric proteases.
INTRODUCTION
Novel antifungals are needed because of the importance of fungal infections in organ transplant patients and the limitations of currently available antifungal agents regarding their spectra of activity and toxicities. Furthermore, there is a demand for new antifungals, given the prevalence of azole-resistant fungal pathogens (1, 8). There are four major therapeutic groups of antifungal agents: polyenes, azoles, allylamines, and echinocandins. The first three groups primarily target ergosterol production or bind to ergosterol, disrupting the fungal membrane (10, 18, 24). Ergosterol, much like cholesterol found in mammalian cells, is important for maintaining proper cell permeability and fluidity. The echinocandins, the fourth group, are synthetically modified lipopeptides that originate from the natural compounds produced by fungi (15, 16). The antifungal activity of echinocandins is attributed to selective inhibition of 1,3-β-glucan synthesis by their function as noncompetitive inhibitors of 1,3-β-glucan synthase (5, 9, 14, 17, 27). There is a need to identify unique antifungals for possible development of new therapeutics.
Occidiofungin is an antifungal peptide produced by the bacterium Burkholderia contaminans MS14. This bacterial strain was isolated from soil that suppressed brown patch disease of lawn grass. A substantial amount of work has been done to characterize the genetic locus and regulatory elements of the antifungal compound (11–13). Structural characterization determined that occidiofungin is a cyclic glyco-lipopeptide (11, 25). Four structural variants of the antifungal peptide, named occidiofungin A to D, have been identified. They have masses of 1,200.39 Da, 1216.41 Da, 1234.17 Da, and 1250.41 Da, which correspond to the addition of oxygen and/or chlorine to the first compound. The target and the mechanism of action of occidiofungin are still unknown. The antifungal has been shown to inhibit a wide array of fungi (25), such as Alternaria alternata, Aspergillus fumigatus, Aspergillus niger, Fusarium oxysporum, Geotrichum candidum, Macrophomina phaseolina, Microsporum gypseum, Penicillium sp., Rhizoctonia solani MSCOT-1, and Trichophyton mentagrophytes. In addition, two Pythium species were sensitive at nanomolar concentrations: Pythium spinosum and Pythium ultimum. Toxicological evaluation of occidiofungin has been performed (W. Tan, et al., submitted for publication). B6C3F1 mice were given a single dose of occidiofungin up to 20 mg/kg of body weight or a daily dose for 5 days at 2 mg/kg of body weight. Key effects were a reduction in body and organ weights. Histological examination of treated mice did not exhibit any signs of organ-specific toxicity. In this study, we tested the sensitivity of Candida species to occidiofungin with and without the presence of human serum. We also performed simultaneous time-kill curves and postantifungal effect (PAFE) experiments on Candida albicans (ATCC 66027). Lastly, we tested occidiofungin's chemical stability against extreme pH and temperature, as well as its stability against gastric proteases.
MATERIALS AND METHODS
In vitro susceptibility testing.
Occidiofungin was purified as previously described (12). Microdilution broth susceptibility testing was performed in duplicate according to the CLSI M27-A3 method in RPMI 1640 growth medium(buffered to a pH of 7.0 with morpholinepropanesulfonic acid [MOPS]). Stock solutions (100×) of occidiofungin were prepared in dimethyl sulfoxide (DMSO). MIC endpoints for occidiofungin were determined by visual inspection and were based on the wells that had no visible growth (an optically clear well) after 24 h of incubation. Susceptibility testing was done in duplicates. DMSO containing no antifungal agent was used as a negative control.
Serum MICs.
Susceptibility testing, as described above, was performed in duplicate according to the CLSI M27-A3 method in RPMI medium or YPD (yeast extract-peptone-dextrose) growth medium in the presence of 5% and 50% (vol/vol) human serum (Sigma-Aldrich, St. Louis, MO) (29). Given the turbidity of 50% serum, visual MIC determination was not possible. Therefore, 50 μl of cell suspension in each well of the microtiter well was plated to determine the minimum lethal concentration (MLC). MLC is defined in terms of a well that has no viable cells (colonies) in the 50 μl spread on a fresh agar plate. This is in accordance with the fungicidal activity being defined as a 99.9% reduction in the number of CFU compared to the starting inoculum, which is approximately 1,000 CFU per ml (19).
Time-kill experiments and PAFE.
Time-kill and PAFE experiments provided important information about the activity of occidiofungin. These studies determined whether occidiofungin is fungicidal or fungistatic and its rate of activity. Fungicidal activity is defined as a 3-log reduction in cell count (20). The PAFE experiments determined the relative effect occidiofungin has on cells exposed for a short time period. This provides information about the affinity occidiofungin has to the cellular target and whether the antifungal activity can be washed away following exposure. Time-kill assays are essentially a measurement of CFU count following the addition of the antifungal to the yeast. Time-kill and PAFE experiments were done according to a method reported by Clancy et al. (2). Candida albicans (ATCC 66027) colonies on 24-h-old YPD plates were suspended in 9 ml of sterile water. The density was adjusted to a 0.5 McFarland standard (2, 6, 7, 19, 20) and was diluted 10-fold with RPMI 1640 medium to a final volume of 10 ml containing a final concentration of 16, 4, 2, 1, and 0 μg/ml of occidiofungin. The cultures were incubated at 35°C with agitation. For the time-kill experiment, 100-μl samples were drawn, serially diluted, and plated on YPD medium for colony counts. PAFE experiments were performed in a similar manner, except that following a 1-h exposure to the antifungal compound, the cells were pelleted (1,400 × g for 10 min), washed three times and resuspended with 10 ml of prewarmed (35°C) RPMI 1640 medium, and then returned to 35°C. Similarly, 100-μl samples at times 0, 2, 4, 8, 12, and 24 h were drawn and plated for counting CFU. Each experiment was completed at least twice for each sample.
Temperature stability.
A temperature stability assay was performed following a method similar to that reported by Wu et al. (30), except that the procedure was optimized for determining the MIC using the CLSI M27-A3 method. Occidiofungin was suspended in 0.5 ml of DMSO at a concentration of 400 μg/ml. Subsequently, 60 μl (24 μg) was added to 540 μl of 0.1 M sodium phosphate buffer (pH 7.0) in a 1.8-ml Eppendorf tube. Each tube was then placed in a water bath at 50, 60, 70, and 100°C for 30 min. Then, 200 μl from each tube was added to the first well of each row of a 48-well microtiter plate and serially diluted 2-fold in 100 μl of RPMI medium. Lastly, C. albicans (ATCC 66027) suspension was prepared using the CLSI protocol described above, and 900 μl was added to each well. A negative control was made by combining 1 ml of DMSO with 9 ml of 0.1 M sodium phosphate buffer and subsequently treated in the same manner as the occidiofungin samples. The microtiter plate was placed in the incubator for 24 h at 35°C. The MIC was determined as described above.
pH stability.
A pH stability assay was performed following a method similar to that reported by Wu et al. (30), except that the procedure was optimized for determining the MIC using the CLSI M27-A3 method. Occidiofungin (160 μg) was dried in eight separate 1.8-ml Eppendorf tubes ranging from pH 2 to pH 9. The dried samples were resuspended in 1 ml of RPMI 1640 medium. Using stock solutions of 6 M HCl and 6 M NaOH, the pHs of the samples were adjusted accordingly, and then samples were left at room temperature for 2 h. After a 2-h exposure, the pH was readjusted to pH 7.0. The 160 μg/ml solution was subsequently diluted 2-fold, resulting in concentrations of 160, 80, 40, 20, 10, 5, and 2.5 μg/ml. Using a 48-well microtiter plate, 100 μl of antifungal solution from each of these tubes was added to its corresponding well. A suspension of C. albicans (ATCC 66027) following the CLSI methodology described above was prepared, and 900 μl of the suspension was added to each well containing the antifungal solution. MICs were determined as described above.
Protease stability.
A protease stability assay was performed following a method similar to that reported by Wu et al. (30), except that the procedure was optimized for determining the MIC using the CLSI M27-A3 method. Occidiofungin's stability was tested against the digestive proteases trypsin, chymotrypsin, and pepsin. For the trypsin assay, a 2× stock solution of sodium phosphate buffer (0.134 M, pH 7.6) and a 10× trypsin stock solution (5.19 mg of trypsin in 1 ml of a 1 mM HCl solution) were used. For the chymotrypsin assay, a 2× stock solution of Tris buffer containing calcium chloride (200 mM Tris-HCl and 20 mM calcium chloride, pH 8.0) and a 10× chymotrypsin stock solution (5.6 mg of chymotrypsin in 1 ml of 1 mM HCl–2 mM calcium chloride) were used. For the pepsin assay, a 2× stock solution of 20 mM HCl and a 10× pepsin stock solution (0.8 mg of pepsin in 1 ml of sterile deionized water) were used. The reaction mixtures having a final volume of 250 μl were prepared and consisted of 25 μl of 10× protease stock solution, 125 μl of 2× stock solution, 97.5 μl of sterile water, and 2.5 μl of DMSO. Then, 100 μl of the reaction solution was added to two 1.8-ml Eppendorf tubes containing 8 μg of occidiofungin and incubated at 35°C for 30 min. A suspension of C. albicans (ATCC 66027) following the CLSI methodology described above was prepared, and 900 μl of the suspension was added to each tube containing the antifungal solution and incubated for 4 h at 35°C. The number of CFU was counted after 4 h of incubation by serial dilution and plating. The plates were incubated at 35°C for 24 h, and CFU counts were determined. The assays were repeated for 60- and 120-min exposures to the gastric proteases. Controls consisted of 10× stock solutions containing no protease, no occidiofungin, and protease inhibitors for trypsin (phenylmethylsulfonyl fluoride [PMSF]), chymotrypsin, and pepsin (0.5 μM E-64 [Sigma], 0.1 μM leupeptin, 0.7 μM pepstatin A, 0.6 μM bestatin, 37.5 μM [4-(2-aminoethyl)benzenesulfonyl fluoride] AEBSF, 1 mM PMSF, and 3 U of aprotinin [Trasylol]). Each experiment was completed in duplicate.
Protease activity of trypsin, chymotrypsin, and pepsin was confirmed using bovine serum albumin (BSA) as the substrate in place of occidiofungin in the reaction mixtures described above at a final concentration of 4.4 μg/μl. After 0, 30, 60, and 120 min at 35°C, 20-μl aliquots were removed, and the reaction was stopped by addition of 80 μl of SDS loading buffer. A volume equivalent to 4 μg of BSA was electrophoresed on a 12% SDS-PAGE gel, and proteins were visualized by Coomassie blue staining.
RESULTS
Susceptibility testing.
The occidiofungin MIC for C. albicans, C. glabrata, C. tropicalis, and C. parapsilosis was determined to be within a range of 2.0 to 0.5 μg/ml (Table 1). A caspofungin- and fluconazole-resistant strain was tested. The MIC for the caspofungin-resistant strain, C. albicans 11034, was 2 μg/ml, while the MIC for the fluconazole-resistant strain, C. albicans 2677, was 1.0 μg/ml. The most direct approach to determine the effect of protein binding on the antifungal activity of occidiofungin is to test the compound in the presence of serum. This provides information as to the amount of unbound antifungal compound that is present in blood. This information is also important for future formulations for efficacy studies in an animal model. Serum MICs were determined using 5% and 50% (vol/vol) human serum in YPD medium (Table 2) because C. albicans (ATCC 66027) and C. glabrata (ATCC 66032) would not grow in the presence of 50% serum in RPMI 1640 medium. There was no difference in the MIC values following the standard CLSI protocol against these two strains using YPD medium and RPMI 1640 medium. Given the turbidity of 50% serum, 50 μl of cell suspension from each well of the microtiter plate was plated to determine the minimum lethal concentration (MLC). Activity of occidiofungin was moderately inhibited by 5% and 50% human serum. The MIC at 5% human serum was 1 μg/ml, while the MLC was determined to be 2 μg/ml for C. albicans (ATCC 66027) and C. glabrata (ATCC 66032). The MLC at 50% human serum was 4 μg/ml. Presumably, the 2- to 4-fold reduction in activity in the presence of serum is attributed to protein binding. A moderate decrease in activity in the presence of serum is not uncommon for lipopeptides (3, 29).
Table 1.
Occidiofungin MICs
| Isolate | Occidiofungin MIC (μg/ml) |
|---|---|
| C. albicans TE | 0.5 |
| C. albicans LL | 0.5 |
| C. albicans 66027 | 1.0 |
| C. albicans 24067 | 1.0 |
| C. albicans 11034a | 2.0 |
| C. albicans 2677b | 1.0 |
| C. glabrata 66032 | 0.5 |
| C. glabrata 200989 | 0.5 |
| C. glabrata 2001 | 1.0 |
| C. tropicalis 66029 | 1.0 |
| C. tropicalis 13803 | 1.0 |
| C. parapsilosis 90018 | 1.0 |
| C. parapsilosis 34136 | 2.0 |
| C. parapsilosis 90875 | 2.0 |
Caspofungin-resistant strain (MIC > 8 μg/ml).
Fluconazole-resistant strain (MIC > 64 μg/ml).
Table 2.
Occidiofungin serum MICs and MLCsa
| Isolate | % Human serum | MIC (μg/ml) | MLC (μg/ml) |
|---|---|---|---|
| C. albicans 66027 | 5 | 1.0 | 2.0 |
| C. albicans 66027 | 50 | 4.0 | |
| C. glabrata 66032 | 5 | 1.0 | 2.0 |
| C. glabrata 66032 | 50 | 4.0 |
Only the MLC is reported for 50% serum because of its turbidity.
Time-kill and PAFE experiments.
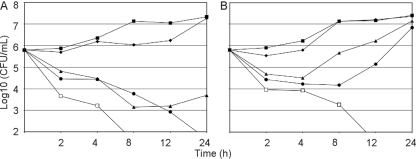
The occidiofungin MICs for the time-kill experiments were 2 μg/ml at 24 h (Fig. 1A). The higher MIC is most probably attributed to the 2- to 3-log increase in cell density compared to the cell density in the CLSI protocol. Time-kill experiments revealed that occidiofungin is fungicidal (20). There was a log decrease in cell density after a 2-h exposure with 1× MIC and 2× MIC and a 2-log decrease in cell density following a 2-h exposure to occidiofungin with 8× MIC. There was greater than a 3-log reduction by 8 h and 24 h with 8× MIC and 2× MIC, respectively. A 2-log decrease in cell density persisted for 24 h with 1× MIC dose. Interestingly, there appeared to be only a moderate effect on the rate of growth at 0.5× MIC in these experiments but no reduction from the starting inoculum. This suggests that a threshold concentration is required for fungicidal activity and that the fungal target needs to be saturated for effect. The rapid drop in cell density after a 2-h exposure suggests that the target for occidiofungin is critical for survival, while the slow gradual decrease in cell density after a 2-h exposure may be attributed to either the chance that these cells have not been saturated with occidiofungin or possibly that they were in a different cellular growth phase that is less susceptible to occidiofungin's effects.
Fig 1.
Kill curves for time-kill (A) and postantifungal effect (B) experiments on Candida albicans ATCC 66027. Symbols are as follows: □, 8× MIC (16 μg/ml); ●, 2× MIC (4 μg/ml); ▴, 1× MIC (2 μg/ml); ♦, 0.5× MIC (1 μg/ml); and ■, control (0 μg/ml).
In the PAFE experiments, the fungicidal activity of occidiofungin at 2× MIC was washed away (Fig. 1B). However, the effect of exposure was still visible 3 h and 7 h after occidiofungin was removed at 1× MIC and 2× MIC, respectively. Moreover, 8× MIC resulted in a greater than 3-log reduction in the cell count at 12 h. The fungicidal effect of a 1-h exposure with 8× MIC was permanent and was delayed only by 4 h compared to the time-kill experiments. Presumably, occidiofungin has a strong interaction with the fungal cells, given that three washes were not enough to eliminate its fungicidal activity at 8× MIC. Another possibility is that occidiofungin may have an intracellular target. Irreversible cellular damage following a 1-h exposure is not probable, given that viable cell counts were present 7 h after exposure in the PAFE experiments.
Chemical stability experiments.
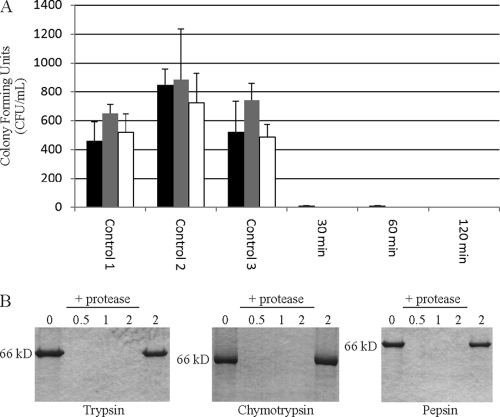
Occidiofungin was exposed to 50, 60, 70, and 100°C for 30 min. In this temperature stability experiment, the MICs of occidiofungin against C. albicans 66027 were 1 μg/ml for all temperatures. This suggests that occidiofungin is stable following exposure to extreme temperatures. Occidiofungin was incubated at pH 2, 3, 4, 5, 6, 7, 8, and 9 for 2 h. No loss in antifungal activity against C. albicans 66027 was observed in this study. The MICs for all pH samples were 1 μg/ml. Lastly, occidiofungin's stability against the gastric proteases trypsin, chymotrypsin, and pepsin was evaluated (Fig. 2). Occidiofungin's ability to kill following exposure to each of these proteases for 30, 60, and 120 min was measured. The potency of occidiofungin did not change following exposure to these proteases under the optimal conditions for their proteolytic activity. Presumably, the cyclic nature of occidiofungin provides protection against proteolytic cleavage.
Fig 2.
(A) Bioactivity assay assessing proteolytic stability of occidiofungin. Activity is reflective of the CFU count/ml. Occidiofungin (8 μg/ml) was exposed to trypsin (black), chymotrypsin (gray), and pepsin (white) for 30, 60, and 120 min. Control 1, control 2, and control 3 are the reaction buffer with protease, the reaction buffer alone, and the reaction buffer with protease and protease inhibitors, respectively. (B) Confirmation of protease activity using BSA as a target substrate. BSA was exposed to protease for 30, 60, and 120 min, followed by separation through a 12% SDS-PAGE gel. The first and last lanes are controls and contain BSA in reaction buffer without added protease for 0 and 120 min, respectively.
DISCUSSION
In vitro potency of occidiofungin against Candida species is in the submicromolar range and is not drastically inhibited by the presence of serum. The reduction in activity in the presence of serum is similar to the reduction observed for anidulafungin (29). In addition, occidiofungin is fungicidal against C. albicans. The initial 1- to 2-log drop in cell density following exposure to occidiofungin is different from the results for caspofungin in the time-kill and PAFE experiments (2). The differences in activities in the time-kill and PAFE experiments suggest that occidiofungin has a separate target from the lipopeptide caspofungin and, presumably, from other echinocandins. These experiments support the need for efficacy studies aimed at understanding occidiofungin's ability to treat systemic Candida infections.
Azoles are the only oral bioavailable drug for the treatment of fungal infections. Lipophilic drugs generally have good colonic absorption, but the oral bioavailability of caspofungin and anidulafungin is <10% (4, 22, 26). Low oral bioavailability of caspofungin is attributed to drug permeability limitations to absorption (22). However, in the same study caspofungin was reported to have some instability at an acidic pH. A detailed analysis of the chemical stability of echinocandins, caspofungin, anidulafungin, or micafungin is not available. Occidiofungin is stable under extreme temperature and pH. Furthermore, the compound is not inactivated by the gastric proteases trypsin, chymotrypsin, and pepsin. Given occidiofungin's chemical stability, studies aimed at understanding occidiofungin's oral bioavailability are warranted.
Occidiofungin is a structurally unique antifungal peptide sharing homology to cepacidine A (21, 23) and the recently discovered burkholdines (28). Occidiofungin is the first antifungal from this group to demonstrate its physical stability and in vitro activity against a human fungal pathogen. These data suggest that further studies on occidiofungin and presumably other antifungals within this small group of antifungals are necessary. These compounds may provide a new line of treatment for life-threatening fungal infections.
ACKNOWLEDGMENTS
We are grateful to Thomas D. Edlind, Drexel University College of Medicine, for providing many of the Candida isolates used in the study. We also thank Mahmoud Ghannoum, University Hospitals Case Medical Center, for the fluconazole- and caspofungin-resistant isolates tested in this study.
This work was supported in part by P20RR016476 (National Center for Research Resources) through the Mississippi INBRE at The University of Southern Mississippi to D.G.
Footnotes
Published ahead of print 21 November 2011
REFERENCES
- 1. Charlier C, et al. 2006. Fluconazole for the management of invasive candidiasis: where do we stand after 15 years? J. Antimicrob. Chemother. 57:384–410 [DOI] [PubMed] [Google Scholar]
- 2. Clancy CJ, Huang H, Cheng S, Derendorf H, Nguyen MH. 2006. Characterizing the effects of caspofungin on Candida albicans, Candida parapsilosis, and Candida glabrata isolates by simultaneous time-kill and postantifungal-effect experiments. Antimicrob. Agents Chemother. 50:2569–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cota J, et al. 2006. In vitro pharmacodynamics of anidulafungin and caspofungin against Candida glabrata isolates, including strains with decreased caspofungin susceptibility. Antimicrob. Agents Chemother. 50:3926–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Denning DW. 2003. Echinocandin antifungal drugs. Lancet 362:1142–1151 [DOI] [PubMed] [Google Scholar]
- 5. Douglas CM, et al. 1994. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-d-glucan synthase. Proc. Natl. Acad. Sci. U. S. A. 91:12907–12911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ernst EJ, Klepser ME, Ernst ME, Messer SA, Pfaller MA. 1999. In vitro pharmacodynamic properties of MK-0991 determined by time-kill methods. Diagn. Microbiol. Infect. Dis. 33:75–80 [DOI] [PubMed] [Google Scholar]
- 7. Ernst EJ, Klepser ME, Pfaller MA. 2000. Postantifungal effects of echinocandin, azole, and polyene antifungal agents against Candida albicans and Cryptococcus neoformans. Antimicrob. Agents Chemother. 44:1108–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Espinel-Ingroff A. 2008. Mechanisms of resistance to antifungal agents: yeasts and filamentous fungi. Rev. Iberoam. Micol. 25:101–106 [DOI] [PubMed] [Google Scholar]
- 9. Garcia-Effron G, Lee S, Park S, Cleary JD, Perlin DS. 2009. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-beta-d-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob. Agents Chemother. 53:3690–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghannoum MG, Rice LB. 1999. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 12:501–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gu G, Smith L, Liu A, Lu SE. 2011. Genetic and biochemical map for the biosynthesis of occidiofungin, an antifungal produced by Burkholderia contaminans strain MS14. Appl. Environ. Microbiol. 77:6189–6198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gu G, Smith L, Wang N, Wang H, Lu SE. 2009. Biosynthesis of an antifungal oligopeptide in Burkholderia contaminans strain MS14. Biochem. Biophys. Res. Commun. 380:328–332 [DOI] [PubMed] [Google Scholar]
- 13. Gu G, Wang N, Chaney N, Smith L, Lu SE. 2009. AmbR1 is a key transcriptional regulator for production of antifungal activity of Burkholderia contaminans strain MS14. FEMS Microbiol. Lett. 297:54–60 [DOI] [PubMed] [Google Scholar]
- 14. Ha Y-S, Covert SF, Momany M. 2006. FsFKS1, the 1,3-beta-glucan synthase from the caspofungin-resistant fungus Fusarium solani. Eukaryot. Cell 5:1036–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hashimoto S. 2009. Micafungin: a sulfated echinocandin. J. Antibiot. 62:27–35 [DOI] [PubMed] [Google Scholar]
- 16. Ikeda F, et al. 2007. Role of micafungin in the antifungal armamentarium. Curr. Med. Chem. 14:1263–1275 [DOI] [PubMed] [Google Scholar]
- 17. Kanasaki R, et al. 2006. FR220897 and FR220899, novel antifungal lipopeptides from Coleophoma empetri no. 14573. J. Antibiot. 59:149–157 [DOI] [PubMed] [Google Scholar]
- 18. Kavanagh K. 2007. New insights in medical mycology. Springer, New York, NY [Google Scholar]
- 19. Klepser ME, Ernst EJ, Lewis RE, Ernst ME, Pfaller MA. 1998. Influence of test conditions on antifungal time-kill curve results: proposal for standardized methods. Antimicrob. Agents Chemother. 42:1207–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klepser ME, et al. 2001. Multi-center evaluation of antifungal time-kill methods. J. Infect. Dis. Pharmacother. 5:29–41 [Google Scholar]
- 21. Lee CH, et al. 1994. Cepacidine A, a novel antifungal antibiotic produced by Pseudomonas cepacia. I. Taxonomy, production, isolation and biological activity. J. Antibiot. (Tokyo) 47:1402–1405 [DOI] [PubMed] [Google Scholar]
- 22. Li C, et al. 2001. Regional-dependent intestinal absorption and meal composition effects on systemic availability of LY303366, a lipopeptide antifungal agent, in dogs. J. Pharm. Sci. 90:47–57 [DOI] [PubMed] [Google Scholar]
- 23. Lim Y, et al. 1994. Cepacidine A, a novel antifungal antibiotic produced by Pseudomonas cepacia II. Physico-chemical properties and structure elucidation. J. Antibiot. 47:1406–1416 [DOI] [PubMed] [Google Scholar]
- 24. Lorian V. 2005. Antibiotics in laboratory medicine. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 25. Lu SE, et al. 2009. Occidiofungin, a unique antifungal glycopeptide produced by a strain of Burkholderia contaminans. Biochemistry 48:8312–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Raasch RH. 2004. Anidulafungin: review of a new echinocandin antifungal agent. Expert Rev. Anti Infect. Ther. 2:499–508 [DOI] [PubMed] [Google Scholar]
- 27. Radding JA, Heidler SA, Turner WW. 1998. Photoaffinity analog of the semisynthetic echinocandin LY303366: identification of echinocandin targets in Candida albicans. Antimicrob. Agents Chemother. 42:1187–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tawfik KA, et al. 2010. Burkholdines 1097 and 1229, potent antifungal peptides from Burkholderia ambifaria 2.2N. Org. Lett. 12:664–666 [DOI] [PubMed] [Google Scholar]
- 29. Wiederhold NP, et al. 2007. In vivo efficacy of anidulafungin and caspofungin against Candida glabrata and association with in vitro potency in the presence of sera. Antimicrob. Agents Chemother. 51:1616–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu XC, et al. 2010. Isolation and partial characterization of antibiotics produced by Paenibacillus elgii B69. FEMS Microbiol. Lett. 310:32–38 [DOI] [PubMed] [Google Scholar]