Abstract
Human cathelicidin LL-37 is a critical cationic antimicrobial peptide for host defense against infection, immune modulation, and wound healing. This article elucidates the functional roles of the cationic side chains of the major antimicrobial region of LL-37, corresponding to residues 17 to 32 (designated GF-17). Antimicrobial assays, killing kinetics studies, and vesicle leakage experiments all indicate that a conversion of lysines to arginines affected the ability of the peptide to kill the Gram-positive Staphylococcus aureus strain USA300. Alanine scanning experiments show that S. aureus is less sensitive than Escherichia coli to a single cationic residue mutation of GF-17. Among the five cationic residues, R23 appears to be somewhat important in killing S. aureus. However, R23 and K25 of GF-17 are of prime importance in killing the Gram-negative organism E. coli. In particular, R23 is essential for (i) rapid recognition, (ii) permeation of the E. coli outer membrane, (iii) clustering of anionic lipids in a membrane system mimicking the E. coli inner membrane, and (iv) membrane disruption. Bacterial aggregation (i.e., rapid recognition via charge neutralization) is the first step of the peptide action. Structurally, R23 is located in the interface (i.e., the first action layer), a situation ideal for the interactions listed above. In contrast, residues K18, R19, and R29 are on the hydrophilic surface of the amphipathic helix and play only a secondary role. Mapping of the functional spectrum of cationic residues of GF-17 provides a solid basis for engineering bacterium-specific antimicrobials using this highly potent template.
INTRODUCTION
Natural antimicrobial peptides (AMPs) are universal host defense molecules that have remained potent through millions of years of evolution (4, 18, 46, 64). More than 1,770 AMPs from bacteria, fungi, plants, and animals were registered in the Antimicrobial Peptide Database (APD) (http://aps.unmc.edu/AP/main.html) (58, 60) as of August 2011. Many of these peptides can rapidly eliminate invading bacteria, frequently by disrupting membranes, making it difficult for bacteria to develop resistance. Therefore, natural AMPs are promising candidates for developing a new generation of antimicrobials to meet the challenge of antibiotic resistance in our era.
AMPs can adopt a variety of 3-dimensional (3D) structures (11), which have recently been classified into four major families: α, β, αβ, and non-αβ (57). The α family consists of AMPs that form helical structures. In contrast, the β family is characterized by the formation of a β-sheet. While the αβ family contains both α-helical and β-sheet structures, the non-αβ family has neither α-helices nor β-sheet structures. AMPs from the helical family have been widely utilized as model systems for the study of structure-activity relationships (9, 49, 53, 59). These studies led to the recognition of both cationic and hydrophobic residues as important elements for antimicrobial activity. Indeed, certain synthetic peptides consisting merely of hydrophobic leucines and cationic lysines are active against bacteria (3, 23, 41). Database analysis revealed that leucines and lysines are most abundant (>10%) in 274 helical peptides annotated in the APD (6), supporting the choice of lysines and leucines in the studies mentioned above. Likewise, synthetic acyl-lysyl polymers are antimicrobial (47). Depending on the peptide sequence and the membrane type, these peptides may disrupt bacterial membranes either by generating a pore (27, 34, 36) or via the carpet mechanism (49). More recently, anionic lipid clustering in the inner membranes has also been proposed as a mechanism against Gram-negative bacteria (14).
LL-37, a host defense peptide encoded by the only human cathelicidin gene, is a representative member of the helical family. The biological and medicinal significance of human LL-37 can be traced back more than 100 years. Interestingly, in 1903, Niels Finsen received his Nobel Prize for using light therapy to treat tuberculosis; however, it was not until the 21st century that the mechanism of this light therapy was elucidated. Light triggers the synthesis of vitamin D, which induces the expression and release of human LL-37, which kills the bacterium (32, 61). Furthermore, the protective role of LL-37 against bacterial infection has been demonstrated in clinical and animal models (28, 39, 42).
A consensus view on the mechanism of action of LL-37 is that it eliminates bacteria by targeting membranes (21, 40). This cationic peptide preferentially binds to vesicles containing phosphatidylglycerols (PGs) (38, 65), suggesting the importance of electrostatic interactions. To elucidate the structural basis of its activity, LL-37 has been investigated in different biological states. In water at an acidic pH, LL-37 is disordered, and the charged residues confer solubility on the peptide; however, at physiological pH, LL-37 forms oligomers (21). Our nuclear magnetic resonance (NMR) studies indicate that residues 2 to 36 of the peptide are involved in this oligomerization. In this case, cationic side chains participate in salt bridges that lock the peptide into a tetramer (57). Upon association with anionic membranes, LL-37 is in the monomeric state (40), allowing the peptide to associate with either the outer or the inner membrane of Gram-negative bacteria. Our structural determination reveals that the long helical domain covering residues 2 to 31 of LL-37 is responsible for binding to lipopolysaccharides (LPS) or PGs (56), which are major anionic components of the outer and inner membranes of Gram-negative bacteria, respectively (14). In addition, NMR studies detected direct interactions between aromatic side chains and PGs as well as between arginines and PGs, underscoring the importance of both hydrophobic and electrostatic interactions for targeting bacterial membranes (55, 56).
As a complementary approach to understanding the structure-activity relationship of LL-37, several active fragments were identified using synthetic peptide libraries, in silico design, or protease digestion (5, 31, 37, 50, 51). Among these fragments, GF-17, corresponding to residues 17 to 32 of LL-37 (Fig. 1), was discovered on the basis of NMR structural studies (31). We found that GF-17, in contrast to intact LL-37, was highly potent against methicillin-resistant Staphylococcus aureus (MRSA) in vitro. However, the molecular basis for the antimicrobial potency of GF-17 has not yet been elucidated. The current study defines the functional roles of the cationic side chains of GF-17 by utilizing an array of biochemical and biophysical approaches. The use of this potent peptide region removes the potential complications due to the N-terminal chemotaxis signal of LL-37, as well as its self-oligomerization at pH ∼7, contributed by both terminal regions (21, 40, 57). Our study reveals the importance of select basic residues of AMPs in killing bacteria. In the case of S. aureus, arginines appear to be more important than lysines, while R23 and K25 of GF-17 are critical for activity against Escherichia coli. We discuss these results in the context of the 3D structure of GF-17, and we propose that rapid bacterial capture via electrostatic attractions (as revealed by bacterial aggregation) is the first step of cationic AMP attack and that multiple actions are involved in the killing of bacteria by the peptide.
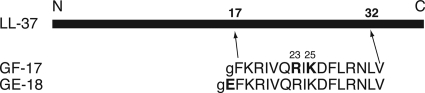
Fig 1.
GF-17 and GE-18 correspond to residues 17 to 32 and 16 to 32 of human LL-37, respectively. To better mimic natural antimicrobial peptides in the database (60), a glycine was added to the N terminus of each of these peptides and an amide group (NH2) was appended at the C terminus. Critical residues identified in this study are shown in boldface.
MATERIALS AND METHODS
Peptide design.
Two groups of peptides (GF-17 and GE-18) were designed (Fig. 1). In the GF-17 group, the residues of the cationic side chains of the peptide were mutated to alanine, one at a time. GE-18 was obtained by inserting one additional glutamate after the N-terminal glycine of GF-17. Two more peptides were generated for the GE-18 group by changing either all lysines of the peptide to arginines (GE-18R) or all arginines to lysines (GE-18K). All of the peptides (for sequences, see Table 1) were chemically synthesized and purified to homogeneity (>95% pure) by reverse-phase high-pressure liquid chromatography (HPLC) (Genemed Synthesis, Inc., TX).
Table 1.
Antimicrobial activity (MIC in μM) of GF-17, GE-18 and their variants
| Peptide | Sequencea | MIC (μM) for: |
|
|---|---|---|---|
| E. coli K-12 | S. aureus USA300 | ||
| GF-17 | GFKRIVQRIKDFLRNLV-NH2 | 7.5 | 7.5 |
| K18A | GFARIVQRIKDFLRNLV-NH2 | 15 | 7.5 |
| R19A | GFKAIVQRIKDFLRNLV-NH2 | 15 | 7.5 |
| R23A | GFKRIVQAIKDFLRNLV-NH2 | 60 | 15 |
| K25A | GFKRIVQRIADFLRNLV-NH2 | 60 | 7.5 |
| R29A | GFKRIVQRIKDFLANLV-NH2 | 15 | 7.5 |
| GE-18 | GEFKRIVQRIKDFLRNLV-NH2 | 60 | 60 |
| GE-18K | GEFKKIVQKIKDFLKNLV-NH2 | 60 | 120 |
| GE-18R | GEFRRIVQRIRDFLRNLV-NH2 | 60 | 30 |
Altered residues are underlined and in boldface.
Bacterial strains.
The strains of bacteria chosen for the assays were the Gram-negative organism E. coli K-12 and the Gram-positive organism S. aureus USA300, a community-onset MRSA isolate.
Antibacterial activity assays.
The antibacterial activities of the peptides were determined using the standard broth microdilution method (59) with modifications. In brief, a 2-ml culture was grown overnight. A fresh culture (2 ml) was inoculated with a small aliquot of the overnight culture and was incubated at 37°C (240 rpm) until the bacterial growth reached the logarithmic stage (i.e., optical density at 600 nm [OD600], ≈0.5). The culture was then diluted to an OD600 of 0.001, and 90-μl aliquots were placed in a 96-well polystyrene microplate with ∼105 CFU per well. A series of peptide solutions (10 μl), each at a 2-fold dilution, was added. The assays were performed in duplicate or triplicate for each peptide and were also repeated on different dates. The microplate was incubated at 37°C and was read on a ChroMate 4300 microplate reader at 630 nm (GMI, Ramsey, MN) after overnight incubation. The MIC was defined as the lowest peptide concentration that fully inhibited bacterial growth. In addition, microplates were also read at 0, 2, and 4 h after antimicrobial agent application. These early readings help to identify nonnormal optical readings due to bacterial aggregation, allowing more-accurate determination of MICs for each peptide.
Killing kinetics.
Killing kinetics experiments were set up in the same manner as the antibacterial assays described above with the following additions. Aliquots of cultures (1 × 105 CFU) were taken at 15, 30, 50, 90, and 120 min, diluted 100-fold, and plated, and colonies were counted after overnight incubation at 37°C.
Flow cytometry.
Membrane permeation was measured by using a BacLight bacterial membrane potential kit (Molecular Probes, Invitrogen) and TO-PRO3 according to the manufacturer's guidelines on a FACSAria flow cytometer (BD Biosciences, San Jose, CA). In brief, exponential-phase cultures of S. aureus USA300 (OD600, 0.6 to 1.0) were washed twice and were resuspended to the original volume in filter-sterilized (pore size, 0.2 μm) phosphate-buffered saline (PBS). The bacteria (1 × 108 cells per ml) were then treated with appropriate concentrations of AMPs and were incubated at 37°C with shaking at 250 rpm. Bacteria were then stained for 10 min with a 3,3′-diethyloxacarbocyanine iodide [DiOC 2(3)] (30 μM)–TO-PRO-3 (100 nM) dye mixture, and flow cytometry measurements were carried out at a flow rate of approximately 1,000 cells per s. Bacteria were distinguished from the background by using a combination of forward-scatter and side-scatter parameters. In total, 10,000 events were collected for each sample, and fluorescent signals were measured using logarithmic amplifications. DiOC2(3) was excited at 488 nm, and fluorescent emissions in the green and red spectra were detected using 530- and 610-nm band-pass filters, respectively, each with a 20-nm band pass. The DNA-binding dye TO-PRO-3 was excited at 633 nm, and emissions were detected by a 660- to 20-nm band-pass filter. Ratiometric data were generated by normalizing red fluorescence signals acquired at 610 nm (membrane potential) and 695 nm (membrane permeation) to their corresponding green signals at 530 nm (bacterial size). Data were analyzed with FlowJo software (Tree Star, Inc., Ashland, OR).
Membrane permeation.
Membrane permeation experiments were performed using a mutant E. coli strain developed by Lehrer et al. (30) and kindly provided to us by R. I. Lehrer. E. coli strain ML-35p was engineered specifically to monitor the permeation of both the inner and the outer membrane (OM) of the same bacterial strain (29). E. coli ML35p is constitutive for cytoplasmic β-galactosidase, lacks Lac permease, and expresses a plasmid-encoded periplasmic β-lactamase. Two chromogenic reporter molecules are used to monitor the permeabilization of outer and inner membranes in a single assay (15, 29, 30). Nitrocefin, a chromogenic cephalosporin, cannot cross the OM and is excluded from the periplasmic space. However, permeabilization of the OM allows nitrocefin to enter the periplasm, where it is cleaved by a β-lactamase and produces a color change that can be monitored spectrophotometrically at 486 nm. Since there is no lactose permease in this strain of E. coli, o-nitrophenylgalactose (ONPG) cannot traverse its inner membrane to be cleaved by cytoplasmic β-galactosidase to o-nitrophenol unless permeabilization of the inner membrane occurs. ONPG cleavage produces a color change that can be measured spectrophotometrically at 420 nm.
Bacteria were grown from a single colony, overnight, in tryptic soy broth (TSB) at 37°C. After 3 washes in phosphate buffer (pH 7.4) (10 mM phosphate buffer, 100 mM NaCl), the culture was diluted to ∼106 CFU/ml in incubation buffer (phosphate buffer [pH 7.4] containing 0.3 mg/ml TSB) and was added to all wells in a non-culture-treated polystyrene microplate, together with increasing concentrations of peptide variants or melittin solutions (0 to 50 μg/ml), in phosphate buffer. Each well also contained 30 μM nitrocefin or 2.5 mM ONPG in phosphate buffer. Absorbance was followed simultaneously at 490 nm and at 450 nm (these were closest to the maximum value that the fixed-wavelength microplate allowed us to use) for 36 min, by taking readings every 2 min at 37°C, with a few seconds of shaking prior to each reading. OM permeabilization was followed for only 20 min in order to avoid artifacts due to spontaneous hydrolysis after longer times. A SpectraMax Pro microplate reader equipped with MaxSoft Plus software (Molecular Devices, Sunnyvale, CA) was used.
Preparation of MLVs.
Lipids were dissolved in chloroform-methanol (2:1, vol/vol). The solvent was then evaporated with nitrogen to deposit a film on the wall of a glass test tube. Final traces of solvent were removed for 2 to 3 h in an evacuated chamber. The films were suspended in the indicated buffer by vortexing at room temperature to form multilamellar liposome vesicles (MLVs). Peptides were mixed with the lipid by dissolving them in the same buffer that was used to hydrate the lipid films.
Differential scanning calorimetry (DSC).
Measurements were made in a Nano II differential scanning calorimeter (Calorimetry Sciences Corporation, Lindon, UT). Lipid films, consisting of 1-palmitoyl-2-oleoylphosphatidylethanolamine (POPE)-tetraoleoyl cardiolipin (TOCL) (75:25), were hydrated at room temperature with peptide solutions in piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) buffer (20 mM PIPES, 140 mM NaCl, 1 mM EDTA [pH 7.4]) or with PIPES buffer alone, vortexed vigorously to make MLVs, and then degassed before being loaded into the sample cell of the calorimeter. Controls using peptide solutions in the sample cell in the absence of lipid showed no transition. Degassed PIPES buffer was placed in the reference cell. The concentration of phospholipids in the samples was maintained at 2.5 mg/ml and the molar ratio of lipid to peptide at 10. The cell volume was 0.34 ml. Samples were equilibrated in the calorimeter at 0°C, and successive heating and cooling scans were carried out between 0 and 35°C at a scan rate of 1.0°C/min, with a delay of 5 min between sequential scans in a series to allow for thermal equilibration. The resulting curves were analyzed by using the fitting program provided by Microcal Inc. (Northampton, MA) and were plotted with Origin, version 7.0.
Lipid-vesicle lysis.
Lipid films were hydrated with a solution containing 12.5 mM 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS), 45 mM P-xylene-bis-pyridinium bromide (DPX), and 10 mM HEPES adjusted to pH 7.4 (25). A solution of NaCl was added to adjust the final osmolarity to 300 mOsm using a freezing point osmometer (model 3300; Advanced Instruments, Inc., Norwood, MA). The lipid suspensions were then freeze-thawed five times and were extruded 10 times through two stacked polycarbonate filters (pore diameter, 100 nm) to make visually transparent solutions of large unilamellar vesicles (LUV) with diameters of 100 nm. The LUV were put through a 20- by 1.5-cm Sephadex G-75 column (Pharmacia, Uppsala, Sweden) preequilibrated with a buffer containing 10 mM HEPES, 1 mM EDTA, and NaCl adjusted to 300 mOsm and pH 7.4 (HEPES buffer). LUV were collected in the void volume, and the concentration of lipid was determined by phosphate analysis (1).
An aliquot of LUV was placed in a quartz cuvette containing HEPES buffer at 37°C to give a final lipid concentration of 50 μM, and fluorescence was recorded for a few seconds. Then an aliquot of a peptide solution was added, and the increase in fluorescence was followed for 3 to 4 min. To determine 100% release of the entrapped fluorophore, 20 μl of 20% Lubrol XT was added at the end of the leakage measurement. Excitation was set at 360 nm and emission at 530 nm, with a 500-nm cutoff filter in the emission monochromator and excitation slits set at 8 nm and emission slits set at 4 nm. Measurements were performed at 37°C in an Aminco Bowman SLM-II spectrofluorimeter.
NMR spectroscopy.
The NMR sample (0.6 ml) contained 2 mM GF-17 or GE-18, 80 mM deuterated sodium dodecyl sulfate (SDS) (Cambridge Isotope Laboratories, Andover, MA), and 10% D2O as the field-locking signal. The pH of the NMR sample was adjusted to 5.4. NMR data were recorded on a 600-MHz Varian Inova NMR spectrometer equipped with a triple-resonance cryogenic probe with a z axis gradient capability. A set of 2D NMR spectra, including nuclear Overhauser effect spectroscopy (NOESY), total correlation spectroscopy (TOCSY), and double-quantum-filtered correlation spectroscopy (DQF-COSY) (2, 20, 43) spectra, was recorded for signal assignments. In 2D 1H NMR spectra, the spectral width in both dimensions was 8,510.6 Hz. In addition, gradient-enhanced heteronuclear single-quantum correlation (HSQC) spectra (22), between 1H and 15N or between 1H and 13C, were recorded at natural abundance. The 1H, 15N, and 13C carriers were set at 4.77, 118.27, and 36.37 ppm, respectively. Typically, 30 increments (128 scans) and 80 increments (256 scans) were collected for the 15N (spectral width, 2,200 Hz) and aliphatic 13C (spectral width, 12,000 Hz) dimensions, respectively. Chemical shifts were referenced as described previously (35). Data were processed on a Silicon Graphics Octane workstation using NMRPipe (10) and were analyzed by PIPP (17).
To measure the temperature coefficients of backbone amide protons, TOCSY spectra were recorded at various temperatures ranging from 10 to 25°C at every 5°C. Each spectrum was subsequently peak-picked to extract chemical shifts. A linear fit between the amide chemical shifts and the temperature led to temperature coefficients. Amide protons with temperature coefficients of >−4.5 × 10−3 ppm/°C were predicted to be hydrogen bonded (6).
Structure calculations.
For structural calculations, major NMR restraints were derived from 2D NOESY spectra (62). In brief, the cross-peak intensities of the NOESY spectrum at a mixing time of 100 ms were detected by PIPP (17) and were converted to distance restraints of 1.8 to 2.8, 1.8 to 3.8, 1.8 to 5.0, and 1.8 to 6.0 Å, corresponding to strong, medium, weak, and very weak types of NOE peaks, respectively (7). Based on 1Hα, 15N, 13Cα, and 13Cβ chemical shifts, backbone angles were predicted by using an updated version of the TALOS program (8). Hydrogen bond restraints were introduced based on the backbone amide temperature coefficients and the NOE-derived structures. An extended covalent structure was used as starting coordinates. In total, 100 structures were calculated by using the simulated annealing protocol in the Xplor-NIH program (48). Twenty structures were accepted based on the following criteria: no NOE-derived distance violations greater than 0.30 Å, backbone dihedral angle violations of <5o, root mean square deviation (RMSD) for bond deviations from ideality of <0.01 Å, and RMSD for dihedral angle deviations from ideality of <5o. The structures were viewed and analyzed using MOLMOL (24) and PROCHECK (26).
Protein structure accession number.
The coordinates for the structure of GF-17 in complex with SDS micelles were deposited in the RCSB Protein Data Bank (PDB entry 2L5M).
RESULTS
Concentration-dependent antimicrobial activity.
To evaluate the functional roles of the cationic residues in GF-17, variants of this peptide in each of which one of the cationic amino acids was mutated to an alanine were synthesized (Table 1) and tested. Antimicrobial activity assays revealed that alanine substitutions had a more dramatic impact on the activity of GF-17 against E. coli than against S. aureus strains (Table 1). In particular, the R23A and K25A mutations increased the MIC of this peptide against E. coli 8-fold. In contrast, these substitutions had only minimal effects on S. aureus killing. To further evaluate the effect of charged residues, we also extended the peptide sequence to obtain GE-18 (Fig. 1) by including the acidic residue E16 of LL-37. The inclusion of E16 was detrimental, since GE-18 exhibited lower activity against E. coli and S. aureus than GF-17. However, GE-18 became slightly more active against S. aureus USA300 when the two lysines were changed to arginines (GE-18R) but less active against both strains when the three arginines were replaced with lysines (GE-18K) (Table 1). These results indicate that the type of cationic residue (K versus R) in GE-18 influences its antibacterial activity against S. aureus.
Concentration-dependent bacterial aggregation.
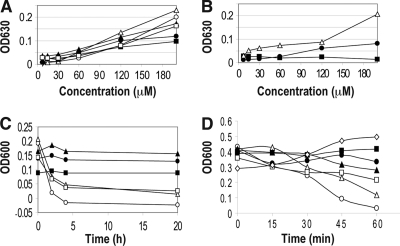
Notably, during antibacterial activity assays, the addition of a peptide appeared to cause rapid bacterial aggregation, especially at elevated peptide concentrations (>30 μM) (Fig. 2A and B). The amount of visible aggregates in the wells was proportional to optical readings of microplates immediately after peptide treatment. For both E. coli (Fig. 2A) and S. aureus (see Fig. S1A in the supplemental material), we ranked the peptides in descending order of aggregation ability, as follows: the K18A peptide and GF-17, the R19A peptide, the R29A and R23A peptides, and the K25A peptide. In the GE-18 series, GE-18R caused more bacterial aggregation than GE-18 (Fig. 2B). However, there were no visible aggregates with GE-18K throughout the concentration range (7.5 to 200 μM) for either bacterial species (Fig. 2; see also Fig. S1 in the supplemental material). Thus, the arginines of GE-18 were also more effective than lysines in aggregating bacteria. Interestingly, cell aggregates of S. aureus USA300 disappeared in 4 h with GF-17, the K18A peptide, and the R29A peptide, but not with the R19A, R23A, or K25A peptide (Fig. 2C). A similar phenomenon was observed with E. coli K-12 (data not shown). Note that peptide-induced bacterial aggregation was also observed when E. coli K12 was grown in a minimal growth medium.
Fig 2.
GF-17 and its variants can aggregate and lyse bacteria. (A) Concentration-dependent bacterial aggregation was observed due to treatment of E. coli K-12 with GF-17 or its variants. Open circles, GF-17; open triangles, K18A peptide; filled triangles, R19A peptide; filled circles, R23A peptide; filled squares, K25A peptide; open squares, R29A peptide. (B) Concentration-dependent bacterial aggregation was observed due to treatment of E. coli K-12 with GE-18 and its variants. Filled circles, GE-18; filled squares, GE-18K; open triangles, GE-18R. (C) Kinetics of visible bacterial (S. aureus USA300) aggregates in a microplate with 200 μM peptide without shaking. (D) Time-dependent changes in the OD600 of E. coli K-12 incubated with shaking after treatment with 100 μM peptide. In panels C and D, symbols are as explained for panel A.
Time-dependent killing.
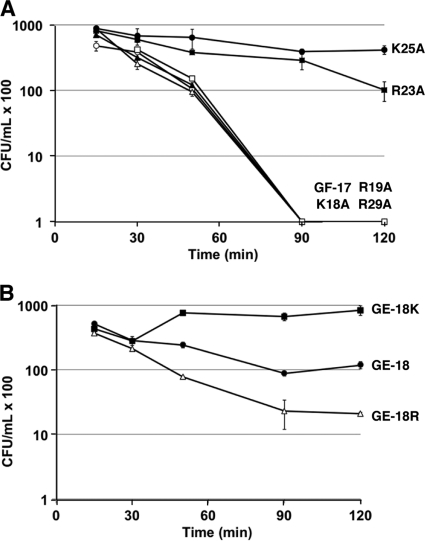
To further substantiate the bactericidal properties of these peptides, we also followed their killing kinetics at a peptide concentration of 30 μM or less. This concentration was chosen based on the results presented in Fig. 2A and B, which showed concentration-dependent bacterial aggregation as described above. We reasoned that the use of lower concentrations would avoid a potential complication due to bacterial aggregation: reduction of bacterial counts on petri dishes. For E. coli K-12, most of the GF-17 peptides displayed rapid killing within 90 min, except for the R23A and K25A mutants, which still caused a reduction in cell viability but at a lower rate (Fig. 3A). These results indicated that the positively charged residues, R23 and K25 of GF-17, were critical for bactericidal activity against E. coli. For S. aureus USA300, only the R23A mutant showed slightly slower killing at 30 μM than the other variants (see Fig. S2 in the supplemental material). The GE-18 peptide series was ranked in descending order of ability to kill S. aureus, as follows: GE-18R, GE-18, GE-18K (Fig. 3B). Overall, these results agree nicely with the MICs of these peptides (Table 1).
Fig 3.
Time-dependent bacterial killing. (A) Killing kinetics of GF-17 (15 μM) and its variants (30 μM) against E. coli K-12. Symbols are as explained for Fig. 2A. (B) Killing kinetics of GE-18 (filled circles), GE-18K (filled squares), and GE-18R (open triangles) at 30 μM against S. aureus USA300. The error bars represent the standard errors of duplicated experiments.
To further resolve the relationship between aggregation and killing, we also conducted bacterial killing experiments at an elevated peptide concentration. When the E. coli K-12 cultures (OD600, ≈0.3) were treated with GF-17 or its variants at 100 μM, we immediately observed an increase in the OD600 at 0 min over that of the untreated culture (Fig. 2D). Note that this increase in the OD600 at 0 min also occurred at a lower peptide concentration (50 μM), and subsequent killing of GF-17 was observed as well. However, the bacteria were more sensitive to killing at 100 μM because of high density (3 × 108 CFU/ml). Based on the results presented in Fig. 2A, we could attribute the increase in the OD600 at 0 min to peptide-induced bacterial aggregation. Subsequently, the OD600 of the culture treated with GF-17 decreased rapidly in 60 min, leading to clearing of the culture (Fig. 2D), indicative of bacterial lysis. The GF-17 peptide series was ranked in descending order of ability to lyse bacteria, as follows: GF-17, K18A peptide, R29A peptide, R19A peptide, R23A peptide, K25A peptide. We also compared the effects of GF-17 and the R23A peptide on optical density; again, the R23A peptide exhibited lower lytic activity than GF-17 (see Fig. S3 in the supplemental material). It appeared that R23 and K25 of GF-17 were more important than other cationic residues in both aggregating (Fig. 2A) and lysing (Fig. 2D) E. coli. For S. aureus USA300, only R23 of GF-17 appeared to be more important than other cationic residues in killing (see Fig. S3 in the supplemental material).
Membrane targeting.
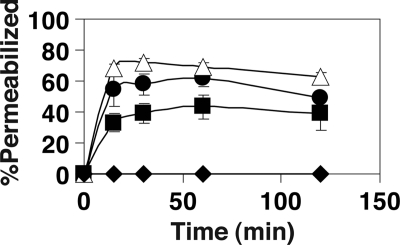
To further understand the difference between arginines and lysines in killing S. aureus (Table 1), we measured the effects of the GE-18 peptide group on membrane permeation by using flow cytometry. When cells were permeabilized and killed, the nonpermeant DNA-binding probe TO-PRO-3 could enter bacteria to associate with DNA and fluoresce. We found that GE-18R was most potent at permeabilizing bacterial membranes. GE-18 killed S. aureus USA300 more rapidly than GE-18K (Fig. 4), fully consistent with the MIC data in Table 1 as well as the killing kinetics in Fig. 3B.
Fig 4.
Populations of permeabilized S. aureus USA300 after treatment with 15 μM GE-18 (solid circles), GE-18K (solid squares), GE-18R (open triangles), or phosphate buffer (solid diamonds) as followed by flow cytometry. The populations of the cells in each quadrant were normalized and plotted. The error bars represent the standard errors from three experiments. A similar killing trend was observed when the bacterium was treated with 30 μM peptide.
Membrane penetration abilities of the GF-17 variants.
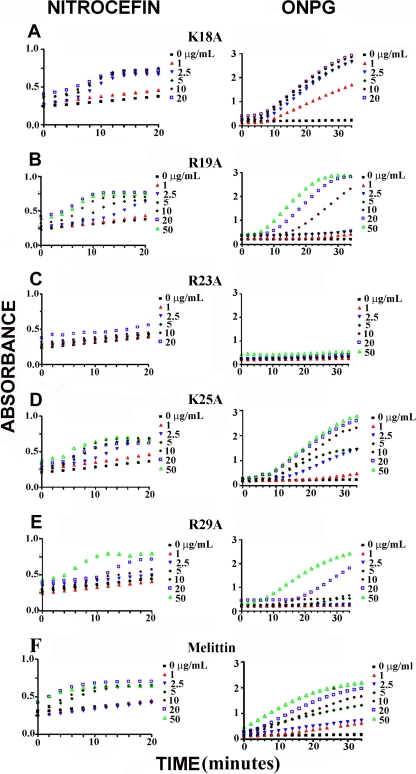
Unlike S. aureus, E. coli possesses an OM that may influence the ability of a membrane-targeting AMP to reach the inner cytoplasmic membrane. To test the possible effect of the OM, E. coli strain ML35p was used. This strain is constitutive for cytoplasmic β-galactosidase, lacks lactose permease, and expresses a plasmid-encoded periplasmic β-lactamase. Thus, the release of these two enzymes, as measured by cleavage of the two chromogenic reporter molecules ONPG and nitrocefin, respectively, was used to monitor the permeabilization of the bacterial outer and inner membranes in a single assay (30). The time-dependent response curves of the GF-17 variants are provided in Fig. 5. As a positive control, we used melittin, which has the same MIC against E. coli as GF-17 (63). All of the GF-17 variants caused membrane permeability increases except for the R23A mutant. Because the R23A peptide did not permeate the outer and inner membranes of this E. coli strain, R23 of GF-17 was considered a critical residue for membrane permeation. Further investigation of the response curves (Fig. 5) revealed that these peptide variants achieved similar levels of membrane permeation at different concentrations (see Table S1 in the supplemental material). In particular, higher concentrations of peptide variants were needed when one of the three arginines of GF-17 was mutated, indicating an important role for arginines in membrane penetration. The higher efficiency of arginines than of lysines at membrane penetration might be due to the bifurcated chemical structure of arginine side chains, which enabled the formation of multiple hydrogen bonds or salt bridges with anionic membrane components (44, 45). We propose that the membrane permeation ability of R23 of GF-17 was so strong that it might have masked the mutational effect of K25 on membrane permeation.
Fig 5.
Outer and inner membrane permeation assays with E. coli strain ML35p in the presence of the K18A (A), R19A (B), R23A (C), K25A (D), and R29A (E) GF-17 variants. Melittin (F) was used as a positive control. Nitrocefin absorbance was measured at 490 nm, and ONPG absorbance was measured at 450 nm. Peptides were added at increasing concentrations, starting from zero up to a final concentration of 50 μg/ml, in the assay.
Lipid clustering by GF-17, GE-18, and their variants.
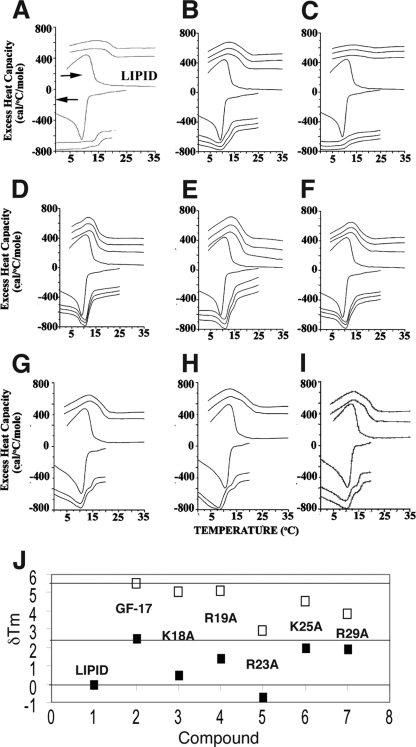
More than a dozen AMPs or their mimics have been shown to be able to cluster anionic lipids (14). Lipid clustering by KR-12, the smallest antibacterial peptide derived from LL-37 (56), was supported by solid-state NMR, DSC, and freeze fracture electron microscopy (12, 13). GF-17 itself (Fig. 6A) was previously found to induce lipid clustering (12). It was not clear whether the effect was due to all the cationic side chains or to a select set of them. To address this issue, we also conducted DSC experiments on these GF-17 variants, using the membrane model system described in Materials and Methods and used previously (14). When anionic TOCL was clustered in the membranes of TOCL-POPE, the observed phase transition of the lipid mixture shifted to a temperature closer to that of the pure zwitterionic POPE component. Thus, the variants of GF-17 caused shifts in the melting temperatures (Tm) of lipids to varying degrees, indicative of the formation of a new localized domain enriched in POPE, due to the removal of TOCL from the lipid mixture by clustering. In contrast, no shifts in Tm were observed in the presence of GE-18 or its analogs (Fig. 6G to I), consistent with their poor activity against E. coli K-12.
Fig 6.
DSC analysis (A to I) and melting temperature changes (J) as indications of the lipid-clustering abilities of GF-17, GE-18, and their variants. (A) GF-17; (B) K18A peptide; (C) R19A peptide; (D) R23A peptide; (E) K25A peptide; (F) R29A peptide; (G) GE-18; (H) GE-18K; (I) GE-18R. Data for GF-17 were taken from reference 19. Heating and cooling scans of the lipid mixture are shown at the center of each panel. Other scans are for samples containing the peptide at a lipid-to-peptide molar ratio of 10. Two successive cycles of heating (positive values) and cooling (negative values) are presented for each peptide from 0 to 35°C at a scan rate of 1°/min. The lipid mixture is POPE-TOCL (75:25), and peptide was added at a lipid/peptide ratio of 10. (J) Graphic view of the deviations in the melting temperatures (δTm) of the model membrane system as a result of peptide treatment. The major and minor components from the deconvoluted DSC curves of GF-17 and its variants (A to F) are represented by filled and open squares, respectively (see the text).
To better compare the lipid-clustering abilities of these peptides, the DSC curves were deconvoluted, and the deviations in the Tm (δTm) of lipids with and without these peptides are plotted in Fig. 6J. In each case, two δTm values, corresponding to major and minor components, are shown. Compared to GF-17, the variants showed slightly reduced lipid-clustering ability (i.e., smaller δTm relative to the Tm of the lipids alone). The major components of the K18A and R19A variants moved closer to zero, whereas it was the minor components of the K25A and R29A variants that clearly moved down relative to that of GF-17. In the case of R23A, the δTm values of both components shifted substantially. Therefore, the GF-17 variants were ranked in descending order of lipid-clustering abilities as follows: the K25A and R29A peptides, the R19A and K18A peptides, and the R23A peptide. In other words, R23, K18, and R19 played a more important role in clustering anionic lipids than K25 or R29.
Vesicle lysis.
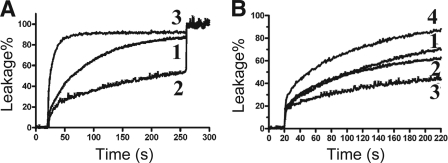
To further substantiate the membrane targeting of the GE-18 peptide series, we also investigated their effects on vesicles (POPG-TOCL, 60:40) that mimic Gram-positive bacteria (12). The peptides were ranked in descending order of ability to induce liposome leakage at 10 μM, as follows: GE-18R, GE-18, GE-18K (Fig. 7A). Consistent with strong activity against S. aureus (Table 1), GF-17 and its variants all showed strong vesicle lysis (see Fig. S4 in the supplemental material). Interestingly, as with the bacteria, we also found that GE-18 and GE-18R could aggregate anionic vesicles, leading to a drop in fluorescence (see Fig. S5 in the supplemental material), whereas GE-18K could not. To verify that the aggregation resulted from charge neutralization (16), we also conducted the same experiments using zwitterionic vesicles. In this case, no aggregation formed, despite the observation that these peptides still lysed the vesicles (Fig. 7B). These peptides were ranked in descending order of ability to lyse the zwitterionic vesicles, as follows: GE-18R, GE-18 and GF-17, GE-18K. It appeared that the all-arginine analog of GE-18 was more effective at vesicle lysis or aggregation of anionic vesicles than its all-lysine analog GE-18K.
Fig 7.
(A) Anionic vesicle lysis by GE-18 (curve 1), GE-18K (curve 2), and GE-18R (curve 3). The concentrations of the peptide and lipid vesicles (DOPG-TOCL, 60:40) are 10 and 50 μM, respectively. (B) Zwitterionic vesicle lysis by GF-17 (curve 1), GE-18 (curve 2), GE-18K (curve 3), and GE-18R (curve 4). The zwitterionic 1-palmitoyl-2-oleoyl phosphatidylcholine (POPC) vesicle system was utilized with 50 μM lipid and 40 μM peptide.
Three-dimensional structures.
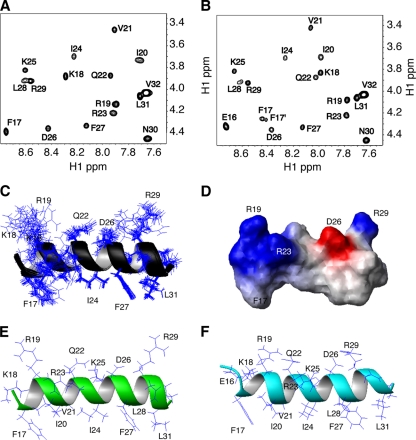
To provide structural insight into peptide-membrane interactions, we determined the 3D structure of membrane-bound GF-17 by solution NMR spectroscopy. Structural studies of membrane proteins are normally conducted in membrane-mimetic environments, such as micelles (19, 54, 57). Previously, we found identical structures for a bacterial membrane anchor, an analog of amphibian aurein 1.2 and human LL-37 in anionic SDS and dioctanoyl PG (D8PG) micelles (56, 59). Therefore, we chose deuterated SDS micelles in this study. At a peptide/SDS molar ratio of 1:40, the peptide was predominantly in the bound state. The amide proton signals of GF-17 in SDS micelles were all resolved in the 2D TOCSY spectrum (Fig. 8A). The proton signals of the peptide were assigned by using the Wüthrich method (62). In brief, the spin systems for the amino acid residues of the peptide were identified in the TOCSY spectrum. These spin systems were then linked using cross peaks in the NOESY spectra. Signal assignments of long side chains were substantiated using DQF-COSY or HSQC spectra at natural abundance (59).
Fig 8.
NMR spectra and solution structures of GF-17 and GE-18. (A and B) Fingerprint regions of the TOCSY spectra (mixing time, 75 ms) of GF-17 (A) and GE-18 (B) in complex with deuterated micelles (peptide/SDS molar ratio, 1:40) at 25°C and pH 5.4. Cross peaks for each residue of both peptides are labeled. Note the double sets of peaks for F17 of GE-18 (B). (C) Side chain view of the final 20 accepted structures of G-17, with the backbone atoms N, Cα, and C O of residues 17 to 31 superimposed and represented by ribbons. (D) Potential surface view of GF-17 with the basic (blue) and acidic (red) residues labeled. (E) A representative structure of GF-17 illustrates its amphipathic nature: hydrophilic side chains above the ribbon, hydrophobic side chains underneath the ribbon, and interfacial side chains in front of (e.g., R23) or behind (e.g., K25) the ribbon. (F) A representative structure of GE-18 illustrates the amphipathic nature in the same manner as that for GF-17 (E).
O of residues 17 to 31 superimposed and represented by ribbons. (D) Potential surface view of GF-17 with the basic (blue) and acidic (red) residues labeled. (E) A representative structure of GF-17 illustrates its amphipathic nature: hydrophilic side chains above the ribbon, hydrophobic side chains underneath the ribbon, and interfacial side chains in front of (e.g., R23) or behind (e.g., K25) the ribbon. (F) A representative structure of GE-18 illustrates the amphipathic nature in the same manner as that for GF-17 (E).
For determination of the 3D structure of GF-17, the NOESY spectra were fully analyzed, and 216 distance restraints were obtained as described in Materials and Methods. To refine the distance-derived structure, we also obtained 13C and 15N chemical shifts for this unlabeled peptide by recording 2D HSQC spectra at natural abundance (59). Based on 1Hα, 15N, 13Cα, and 13Cβ chemical shifts of the micelle-bound GF-17, we obtained 30 peptide backbone dihedral angles (ϕ and ψ) by using the updated TALOS program (8). In addition, we identified seven hydrogen bonds based on the temperature coefficients of the backbone amide protons. A summary of the NMR restraints is given in Table 2, and an ensemble of 20 structures of GF-17 is presented in Fig. 8C. The structures were accepted based on the criteria defined in Materials and Methods. When the backbone atoms of residues 17 to 31 of GF-17 were superimposed, the RMSD was 0.24 Å. According to the PROCHECK analysis (26), 96.7% of the residues were located in the most favored region of the Ramachandran plot and 3.3% were located in the additionally allowed region. Detailed structural statistics are given in Table 2. Taken together, these criteria indicate that the 3D structure of micelle-bound GF-17 was of high quality.
Table 2.
Structural statistics of GF-17 or GE-18 bound to deuterated sodium dodecyl sulfate micellesa at pH 5.4 and 25°C
| Statistic | Value for: |
|
|---|---|---|
| GF-17 | GE-18 | |
| Structural restraints | ||
| NOE restraints (total) | 216 | 228 |
| Intraresidue | 73 | 84 |
| Sequential | 63 | 72 |
| Short range | 80 | 72 |
| Backbone angles (ϕ/ψ)b | 30 | 30 |
| Hydrogen bond restraintsc | 14 | 14 |
| Total | 260 | 272 |
| Structural quality | ||
| Backbone RMSD (Å)d | 0.24 | 0.36 |
| NOE-derived distance violations (Å) | <0.3 | <0.3 |
| TALOS-derived angle violations (°) | <5 | <5 |
| Ramachandran plote | ||
| % of residues in most favored region | 96.7 | 96.1 |
| % of residues in additional allowed region | 3.3 | 3.9 |
Peptide/SDS ratio, 1:40.
Predicted by using the updated version of the TALOS program (8).
Two distance restraints are applied for each hydrogen bond.
Calculated by using the MOLMOL program (24) when the backbone atoms for residues 17 to 31 of 20 structures are superimposed.
Average values were calculated by using the Procheck software for the ensemble of 20 structures (26).
The precision of side chain locations in the structure determined can be viewed in Fig. 8C. While the hydrophobic side chains of F17, I24, F27, and L31 (bottom) were well defined, the long hydrophilic side chains of K18, R19, and R29 (top) were frayed, implying flexibility. The potential surface of the peptide is displayed in Fig. 8D. The first half of the peptide was highly basic (blue), and the second half (C terminus) contained a pair of basic (R29) and acidic (D26) residues. To better delineate the locations of different side chains of micelle-bound GF-17, only one structure is shown in Fig. 8E. It is evident that the hydrophobic surface is composed of residues F17, I20, V21, I24, F27, L28, and L31, whereas residues K18, R19, Q22, D26, and R29 are located on the hydrophilic surface. Note that the side chains of R23 and K25 are located in the interfacial regions, ideal for interactions with bacterial membranes. Indeed, alterations of either of these two residues to an alanine substantially reduced the activity of GF-17 (Table 1). This effect was due primarily to the absence of a particular cationic side chain of GF-17, because the same helical conformation was retained in these peptide variants as that indicated by the typical circular dichroism spectra in membrane-mimetic micelles (see Fig. S6 in the supplemental material).
To provide insight into the substantial activity difference between GF-17 and GE-18 (Table 1), we also determined the 3D structure of GE-18 in the same membrane-mimetic model under identical conditions. As can be seen from the fingerprint region of the TOCSY spectrum of GE-18, the cross peaks were well resolved (Fig. 8B). In addition, most of the peaks of GE-18 appeared at spectral regions similar to those for GF-17, and only the amide protons of residues F17 and K18 were significantly shifted (>0.2 ppm), indicating a local perturbation. This was substantiated by NOESY spectra, where nearly identical cross peaks existed. Thus, GE-18 had a structure similar to that of GF-17. In total, 228 NOE cross peaks were picked and integrated for GE-18. The structural statistics of GE-18 are provided in Table 2. Because more than 96% of the residues were located in the most favored region of the Ramachandran plot, the structure of GE-18 was also of high quality. As shown in Fig. 8F, GE-18 indeed adopted a similar amphipathic helical structure, with the same side chains occupying almost the same locations as those observed in the 3D structure of GF-17 (Fig. 8E). The addition of E16 did not change the helical structure of GE-18 but did alter its interaction with the membrane-mimetic micelle. In particular, F17 obtained a second conformational state in both the TOCSY (Fig. 8B) and NOESY (see Fig. S7 in the supplemental material) spectra, presumably due to electrostatic repulsions of E16 with the negatively charged micelle surface. This could have reduced the antibacterial activity of GE-18 against E. coli (Table 1) substantially, as well as its lipid-clustering ability (Fig. 6G).
DISCUSSION
Using a systematic approach, this study investigated the functional roles of cationic side chains, taking the major antimicrobial region of LL-37 as a model. In the case of S. aureus USA300, we utilized GE-18, a helical AMP, as a peptide model (Fig. 8F) to evaluate the effect of lysines versus arginines on antimicrobial activity. GE-18R, the arginine variant, was found to be more effective at killing than GE-18K, the lysine variant. Remarkably, GE-18R was more effective at membrane permeation, vesicle lysis, and aggregation than GE-18K (Fig. 2, 4, and 7). Zou et al. (66) found that the change from lysines to arginines in defensins, a β-sheet AMP, also enhanced antimicrobial activity and killing efficiency. It remains to be established whether this Lys-to-Arg mutational effect is universal.
In the case of E. coli, we focused on GF-17, because its single basic residue variants displayed more-variable activity against this bacterium (Table 1). In membrane-mimetic micelles, GF-17 adopted a classic amphipathic structure, where R23 and K25 are located in the hydrophobic and hydrophilic interface (Fig. 8C to E). These two basic residues are essential for the antimicrobial activity of GF-17 against E. coli K-12 (Table 1). They might also play an important role in rapidly capturing negatively charged bacteria, leading to bacterial aggregation above 30 μM (Fig. 2A; see also Fig. S1 in the supplemental material). Interestingly, a recent study revealed that LL-37 at 8 μM could quickly saturate the outer membrane of E. coli K-12 and reached the inner membrane, halting bacterial growth in 5 to 25 min (52). Our killing experiments, performed at an elevated concentration (Fig. 2D), revealed a similar mechanism of action. In other words, instant bacterial aggregation due to charge neutralization could be regarded as the first step of peptide action (i.e., rapid recognition), rather than another mechanism for bacterial killing as proposed elsewhere (33). The GF-17 peptide subsequently lysed bacterial membranes in less than 120 min (Fig. 2 to 4). Our current study identified R23 as a residue critical for the ability of GF-17 to attach rapidly to bacteria, to penetrate the outer membrane (Fig. 5), and to lyse cell membranes. In addition, interfacial R23 of GF-17 was also critical for clustering anionic lipids (Fig. 6J). Using a similar peptide, we previously observed, by solution NMR spectroscopy, direct arginine-PG lipid contacts for R23, but not for R19 or R29 (55). All of these observations firmly established the critical role of R23 of GF-17 in recognizing, penetrating, and disrupting bacterial membranes.
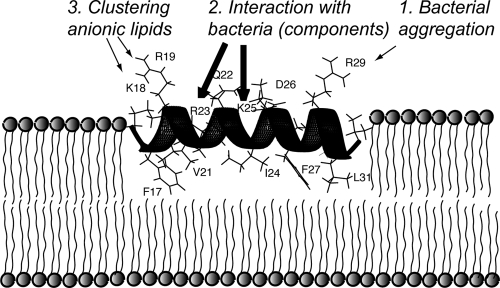
Figure 9 summarizes the functional spectrum of cationic side chains of GF-17 against E. coli. Considering the significance of interfacial cationic side chains R23 and K25, we define the hydrophobic surface flanked by these two critical cationic side chains as the core antimicrobial structure of GF-17, since they are in direct contact with bacterial membranes, thereby constituting the first action layer of this cationic AMP. Depending on a particular physical process, this core structure could recruit additional cationic side chains (K18, R19, or R29 in the second action layer) to optimize the interactions. For bacterial aggregation, the importance of R29 was only secondary to that of the core cationic side chains R23 and K25 (Fig. 2A; see also Fig. S1 in the supplemental material). For bacterial lysis, the importance of R19 was secondary to that of the core residues (Fig. 2D). However, K18 and R19 became secondary only to R23 during anionic lipid clustering (Fig. 6J).
Fig 9.
Cartoon view of the GF-17-membrane interaction. Thick arrows indicate the cationic side chains that play the major roles in bacterial recognition, aggregation, membrane penetration, and lipid clustering, while thin arrows indicate those of secondary importance. See the text for additional details. This plot suggests that each cationic side chain of this peptide could contribute to peptide-bacterium interactions, depending on the particular mechanism along the pathway of microbial killing.
The core structure provided a basis for the design of new AMPs with selective activity. First, mutations of the core cationic side chains of GF-17 led to the R23A and K25A variants, which were poor at killing E. coli but remained active against S. aureus (Table 1). Second, the hydrophobic surface of the core structure could also be modified by either sequence truncation or partial incorporation of d-amino acids, leading to peptides that are inhibitory only to E. coli, not to S. aureus USA300 (31, 56). Thus, the wide-spectrum GF-17, the major antimicrobial peptide of human LL-37, could be tailored into different AMPs that kill primarily either Gram-negative or Gram-positive pathogenic bacteria. Such peptides with selective killing effects are vital for the prevention of the simultaneous elimination of probiotic bacteria during treatment of a particular type of bacterial infection.
In summary, human LL-37 is a multifunctional host defense peptide. The concentration of the peptide, although difficult to measure under physiological conditions, may be an important factor in determining the function of LL-37. The physiological concentration of LL-37 was reported as in the range of 0.4 to 1 μM in the airways and 1 to 3 μM in pulmonary infections, and it could reach 300 μM in the skin lesions of psoriasis patients (reviewed in reference 39). It is reasonable to speculate that a low level of LL-37 is expressed primarily as a signaling molecule and that a high level (e.g., above the MIC) of the peptide is produced mainly for pathogen elimination and clearance (for the structural basis of the LL-37 action, see the introduction). Although intact LL-37 was also observed to aggregate bacteria, it is not clear whether peptide-induced bacterial aggregation observed in vitro plays a role in bacterial clearance in vivo as well. Our observation that intact LL-37 is inactive against community-associated S. aureus USA300 in vitro is worrisome, since it would imply that vitamin D therapy might not be effective for this particular bacterial strain. As a consequence, our current characterization of the structure and activity of GF-17, the major antimicrobial region of LL-37 identified through NMR studies (31), is of prime importance for the development of a surrogate for LL-37 to boost human innate immunity. Indeed, GF-17 was capable of eliminating both Gram-positive and Gram-negative bacteria, such as S. aureus USA300 and E. coli K-12, in vitro. Interestingly, we showed that S. aureus was not sensitive to an alanine scan of cationic side chains of GF-17. In contrast, E. coli was susceptible. Among the five cationic lysines and arginines, R23 and K25 of GF-17 were essential for antibacterial activity against E. coli. It seems that the mechanism of action of GF-17 against E. coli is more complex than once thought. Through our study, it is now clear that there were multiple steps in the pathway to killing by the peptide, starting with rapid recognition of bacteria via electrostatic interactions, which caused visible bacterial aggregation with an increase in peptide concentration. Our observation was consistent with a recent fluorescence study that revealed saturation of the bacterial surface in seconds (52). GF-17 could then penetrate the outer membrane of E. coli by interacting with negatively charged components, such as LPS (56). This peptide could also interact with anionic PGs in the inner membrane. Such interactions could induce the clustering of anionic lipids in the bacterial membranes. GF-17 was so potent that it subsequently caused damage to bacterial membranes, leading to cell lysis. In these processes, R23 was found to play a critical role from initial recognition to the final membrane lysis. The high potency of arginines, higher than that of lysines, in these cases is related to the bifurcated side chain of arginines (44, 45). Remarkably, the significance of R23 is fully consistent with the 3-dimensional structure of GF-17 determined in membrane-mimetic micelles. R23 is located in the interface of the amphipathic helix (Fig. 8D). Such a position is ideal for performing the actions mentioned above, ranging from membrane binding to lysis. Indeed, we previously observed direct interactions between the R23 side chain (but not R19 or R29) and PGs, the first such example provided by solution NMR spectroscopy (55). This NMR observation validated our new structure-derived concept of action layers in AMPs. In conclusion, GF-17 could target different molecules of E. coli, and in these interactions, R23 in the first action layer was indispensable. The same arginine residue appeared also to play an important role in recognizing S. aureus, followed by membrane permeation and lysis. Moreover, the all-arginine variant was more effective than the all-lysine analog in aggregating S. aureus bacteria, penetrating the membrane, and killing. This study enriches our understanding of the mechanisms of action of natural AMPs against different bacteria and guides the design of novel AMPs with desired properties.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Megan Michalak and Victoria Smith of the UNMC Cell Analysis Facility for technical assistance with flow cytometry, to Edward Ezell of the UNMC NMR Core Facility for maintaining the spectrometer, and to Kari L. Nelson for editing the final version of the manuscript.
This work is supported by NIH funding to G.W. (R56AI081975) and K.W.B. (P01AI83211 and R01AI038901) and by the Canadian Institutes of Health Research (grant MOP 86608, to R.M.E.).
Footnotes
Published ahead of print 14 November 2011
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Ames BN. 1966. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8:115–118 [Google Scholar]
- 2. Bax A, Davis DG. 1985. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 65:355–360 [Google Scholar]
- 3. Blondelle SE, Houghten RA. 1992. Design of model amphipathic peptides having potent antimicrobial activities. Biochemistry (N.Y.) 31:12688–12694 [DOI] [PubMed] [Google Scholar]
- 4. Boman HG. 1995. Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 13:61–92 [DOI] [PubMed] [Google Scholar]
- 5. Braff MH, et al. 2005. Structure-function relationships among human cathelicidin peptides: dissociation of antimicrobial properties from host immunostimulatory activities. J. Immunol. 174:4271–4278 [DOI] [PubMed] [Google Scholar]
- 6. Cierpicki T, Otlewski J. 2001. Amide proton temperature coefficients as hydrogen bond indicators in proteins. J. Biomol. NMR 21:249–261 [DOI] [PubMed] [Google Scholar]
- 7. Clore GM, Gronenborn AM. 1998. Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol. 16:22–34 [DOI] [PubMed] [Google Scholar]
- 8. Cornilescu G, Delaglio F, Bax A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13:289–302 [DOI] [PubMed] [Google Scholar]
- 9. Dathe M, Wieprecht T. 1999. Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells. Biochim. Biophys. Acta 1462:71–87 [DOI] [PubMed] [Google Scholar]
- 10. Delaglio F, et al. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277–293 [DOI] [PubMed] [Google Scholar]
- 11. Epand RM, Vogel HJ. 1999. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta 1462:11–28 [DOI] [PubMed] [Google Scholar]
- 12. Epand RF, Wang G, Berno B, Epand RM. 2009. Lipid segregation explains selective toxicity of a series of fragments derived from the human cathelicidin LL-37. Antimicrob. Agents Chemother. 53:3705–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Epand RM, et al. 2010. Lipid clustering by three homologous arginine-rich antimicrobial peptides is insensitive to amino acid arrangement and induced secondary structure. Biochim. Biophys. Acta 1798:1272–1280 [DOI] [PubMed] [Google Scholar]
- 14. Epand RM, Epand RF. 2010. Biophysical analysis of membrane targeting antimicrobial peptides: membrane properties and design of peptides specifically targeting Gram-negative bacteria, p 116–127 In Wang G. (ed), Antimicrobial peptides: discovery, design and novel therapeutic strategies. CABI, Wallingford, Oxfordshire, England [Google Scholar]
- 15. Ericksen B, Wu Z, Lu W, Lehrer RI. 2005. Antibacterial activity and specificity of the six human α-defensins. Antimicrob. Agents Chemother. 49:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferre R, et al. 2009. Synergistic effects of the membrane actions of cecropin-melittin antimicrobial hybrid peptide BP100. Biophys. J. 96:1815–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garrett DS, Powers R, Gronenborn AM, Clore GM. 1991. A common sense approach to peak picking in two-, three-, and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson. 95:214–220 [DOI] [PubMed] [Google Scholar]
- 18. Hancock RE, Sahl HG. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24:1551–1557 [DOI] [PubMed] [Google Scholar]
- 19. Haney EF, Hunter HN, Matsuzaki K, Vogel HJ. 2009. Solution NMR studies of amphibian antimicrobial peptides: linking structure to function? Biochim. Biophys. Acta 1788:1639–1655 [DOI] [PubMed] [Google Scholar]
- 20. Jeener J, Meier BH, Bachmann P, Ernst RR. 1979. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 71:4546–4553 [Google Scholar]
- 21. Johansson J, Gudmundsson GH, Rottenberg ME, Berndt KD, Agerberth B. 1998. Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J. Biol. Chem. 273:3718–3724 [DOI] [PubMed] [Google Scholar]
- 22. Kay LE, Keifer P, Saarinen T. 1992. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114:10663–10665 [Google Scholar]
- 23. Kim YS, Cha HJ. 2010. Disperse distribution of cationic amino acids on hydrophilic surface of helical wheel enhances antimicrobial peptide activity. Biotechnol. Bioeng. 107:216–223 [DOI] [PubMed] [Google Scholar]
- 24. Koradi R, Billeter M, Wüthrich K. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51–55 [DOI] [PubMed] [Google Scholar]
- 25. Ladokhin AS, Wimley WC, White SH. 1995. Leakage of membrane vesicle contents: determination of mechanism using fluorescence requenching. Biophys. J. 69:1964–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477–486 [DOI] [PubMed] [Google Scholar]
- 27. Lee M, Hung W, Chen F, Huang HW. 2008. Mechanism and kinetics of pore formation in membranes by water-soluble amphipathic peptides. Proc. Natl. Acad. Sci. U. S. A. 105:5087–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee PHA, et al. 2005. Expression of an additional cathelicidin antimicrobial peptide protects against bacterial skin infection. Proc. Natl. Acad. Sci. U. S. A. 102:3750–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lehrer RI, Barton A, Ganz T. 1988. Concurrent assessment of inner and outer membrane permeabilization and bacteriolysis activity in E. coli by multiple-wavelength spectroscopy. J. Immunol. Methods 108:153–158 [DOI] [PubMed] [Google Scholar]
- 30. Lehrer RI, et al. 1989. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Invest. 84:553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li X, Li Y, Han H, Miller DW, Wang G. 2006. Solution structures of human LL-37 fragments and NMR-based identification of a minimal membrane-targeting antimicrobial and anticancer region. J. Am. Chem. Soc. 128:5776–5785 [DOI] [PubMed] [Google Scholar]
- 32. Liu PT, et al. 2006. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science 311:1770–1773 [DOI] [PubMed] [Google Scholar]
- 33. Loit E, Hincke MT, Altosaar I. 2010. Synthetic antimicrobial peptide L8 (MHLHKTSRVTLYLL) has membrane permeabilisation and bacterial aggregation activity. Int. J. Antimicrob. Agents 35:410–416 [DOI] [PubMed] [Google Scholar]
- 34. Ludtke SJ, et al. 1996. Membrane pores induced by magainin. Biochemistry 35:13723–13728 [DOI] [PubMed] [Google Scholar]
- 35. Markley JL, et al. 1998. Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the Standardization of Data Bases of Protein and Nucleic Acid Structures Determined by NMR Spectroscopy. J. Biomol. NMR 12:1–23 [DOI] [PubMed] [Google Scholar]
- 36. Matsuzaki K. 1999. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta 1462:1–10 [DOI] [PubMed] [Google Scholar]
- 37. Nell MJ, et al. 2006. Development of novel LL-37 derived antimicrobial peptides with LPS and LTA neutralizing and antimicrobial activities for therapeutic application. Peptides 27:649–660 [DOI] [PubMed] [Google Scholar]
- 38. Neville F, et al. 2006. Lipid headgroup discrimination by antimicrobial peptide LL-37: insight into mechanism of action. Biophys. J. 90:1275–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nijnik A, Hancock REW. 2009. The roles of cathelicidin LL-37 in immune defences and novel clinical applications. Curr. Opin. Hematol. 16:41–47 [DOI] [PubMed] [Google Scholar]
- 40. Oren Z, Lerman JC, Gudmundsson GH, Agerberth B, Shai Y. 1999. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity. Biochem. J. 341:501–513 [PMC free article] [PubMed] [Google Scholar]
- 41. Papo N, Shai Y. 2003. Can we predict biological activity of antimicrobial peptides from their interactions with model phospholipid membranes? Peptides 24:1693–1703 [DOI] [PubMed] [Google Scholar]
- 42. Putsep K, Carlsson G, Boman HG, Andersson M. 2002. Deficiency of antibacterial peptides in patients with morbus Kostmann: an observation study. Lancet 360:1144–1149 [DOI] [PubMed] [Google Scholar]
- 43. Rance M, et al. 1983. Improved spectral resolution in COSY 1H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun. 117:479–485 [DOI] [PubMed] [Google Scholar]
- 44. Rothbard JB, Jessop TC, Lewis RS, Murray BA, Wender PA. 2004. Role of membrane potential and hydrogen bonding in the mechanism of translocation of guanidinium-rich peptides into cells. J. Am. Chem. Soc. 126:9506–9507 [DOI] [PubMed] [Google Scholar]
- 45. Sakai N, Matile S. 2003. Anion-mediated transfer of polyarginine across liquid and bilayer membranes. J. Am. Chem. Soc. 125:14348–14356 [DOI] [PubMed] [Google Scholar]
- 46. Sambhara S, Lehrer RI. 2007. The innate immune system: a repository for future drugs? Expert Rev. Anti Infect. Ther. 5:1–5 [DOI] [PubMed] [Google Scholar]
- 47. Sarig H, et al. 2010. A miniature mimic of host defense peptides with systemic antibacterial efficacy. FASEB J. 24:1904–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. 2003. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160:65–73 [DOI] [PubMed] [Google Scholar]
- 49. Shai Y. 1999. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1462:55–70 [DOI] [PubMed] [Google Scholar]
- 50. Sieprawska-Lupa M, et al. 2004. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob. Agents Chemother. 48:4673–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sigurdardottir T, et al. 2006. In silico identification and biological evaluation of antimicrobial peptides based on human cathelicidin LL-37. Antimicrob. Agents Chemother. 50:2983–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sochacki KA, Barns KJ, Bucki R, Weisshaar JC. 2011. Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proc. Natl. Acad. Sci. U. S. A. 108:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tossi A, Sandri L, Giangaspero A. 2000. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 55:4–30 [DOI] [PubMed] [Google Scholar]
- 54. Wang G. 2006. Structural biology of antimicrobial peptides by NMR spectroscopy. Curr. Org. Chem. 10:569–581 [Google Scholar]
- 55. Wang G. 2007. Determination of solution structure and lipid micelle location of an engineered membrane peptide by using one NMR experiment and one sample. Biochim. Biophys. Acta 1768:3271–3281 [DOI] [PubMed] [Google Scholar]
- 56. Wang G. 2008. Structures of human host defense cathelicidin LL-37 and its smallest antimicrobial peptide KR-12 in lipid micelles. J. Biol. Chem. 283:32637–32643 [DOI] [PubMed] [Google Scholar]
- 57. Wang G. 2010. Structural studies of antimicrobial peptides provide insight into their mechanisms of action, p 141–168 In Wang G. (ed), Antimicrobial peptides: discovery, design and novel therapeutic strategies. CABI, Wallingford, Oxfordshire, England [Google Scholar]
- 58. Wang G, Li X, Wang Z. 2009. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 37:D933–D937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang G, Li Y, Li X. 2005. Correlation of three-dimensional structures with the antibacterial activity of a group of peptides designed based on a nontoxic bacterial membrane anchor. J. Biol. Chem. 280:5803–5811 [DOI] [PubMed] [Google Scholar]
- 60. Wang Z, Wang G. 2004. APD: the Antimicrobial Peptide Database. Nucleic Acids Res. 32:D590–D592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. White JH, Bitton AJ. 2010. Role of vitamin D in the enhancement of antimicrobial peptide gene expression, p 181–194 In Wang G. (ed), Antimicrobial peptides: discovery, design and novel therapeutic strategies. CABI, Wallingford, Oxfordshire, England [Google Scholar]
- 62. Wüthrich K. 1986. NMR of proteins and nucleic acids. Wiley, New York, NY [Google Scholar]
- 63. Yan H, Li S, Sun X, Mi H, He B. 2003. Individual substitution analogs of Mel(12–26), melittin's C-terminal 15-residue peptide: their antimicrobial and hemolytic actions. FEBS Lett. 554:100–104 [DOI] [PubMed] [Google Scholar]
- 64. Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395 [DOI] [PubMed] [Google Scholar]
- 65. Zhang X, et al. 2010. Dual functions of the human antimicrobial peptide LL-37-target membrane perturbation and host cell cargo delivery. Biochim. Biophys. Acta 1798:2201–2208 [DOI] [PubMed] [Google Scholar]
- 66. Zou G, et al. 2007. Toward understanding the cationicity of defensins. Arg and Lys versus their noncoded analogs. J. Biol. Chem. 282:19653–19665 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.