Abstract
Pseudomonas putida KT2440 is a chloramphenicol-resistant bacterium that is able to grow in the presence of this antibiotic at a concentration of up to 25 μg/ml. Transcriptomic analyses revealed that the expression profile of 102 genes changed in response to this concentration of chloramphenicol in the culture medium. The genes that showed altered expression include those involved in general metabolism, cellular stress response, gene regulation, efflux pump transporters, and protein biosynthesis. Analysis of a genome-wide collection of mutants showed that survival of a knockout mutant in the TtgABC resistance-nodulation-division (RND) efflux pump and mutants in the biosynthesis of pyrroloquinoline (PQQ) were compromised in the presence of chloramphenicol. The analysis also revealed that an ABC extrusion system (PP2669/PP2668/PP2667) and the AgmR regulator (PP2665) were needed for full resistance toward chloramphenicol. Transcriptional arrays revealed that AgmR controls the expression of the pqq genes and the operon encoding the ABC extrusion pump from the promoter upstream of open reading frame (ORF) PP2669.
INTRODUCTION
Chloramphenicol is a broad-spectrum antibiotic naturally produced by Streptomyces that, after decades of limited use, has been the focus of renewed interest due to the lack of new antibiotic agents and the appearance of resistance caused by the indiscriminate use of current antibiotics (31, 37, 56). In fact, at present a number of multiresistant clinical isolates from pathogenic bacteria are still sensitive to chloramphenicol, a fact that could be attributed to the limited use of this antibiotic in developed countries. Thus, chloramphenicol is being reconsidered as an option for treatment of certain infections in critically ill patients (20, 37, 43).
Environmental microorganisms represent the most relevant reservoir of resistance to antibiotics and other drugs. Antibiotic resistance genes are crucial in niche colonization since microbes need to combat antimicrobial compounds produced by other microbes and higher organisms in the environment. Horizontal gene transfer mechanisms and natural selection of resistant clones by antimicrobial pressure are relevant mechanisms for spreading antibiotic resistance traits (9, 11).
The most common mechanism of resistance to chloramphenicol in bacteria is its enzymatic inactivation by acetylation mainly via acetyltransferases or, in some cases, by chloramphenicol phosphotransferases (1, 56). Resistance to chloramphenicol may also be due to target site mutation/modification (39), decreased outer membrane permeability (10), and the presence of efflux pumps that often act as multidrug extrusion transporters, thereby reducing the effective intracellular drug concentration (15, 53).
Pseudomonas putida is an environmental bacterium that can survive in hospital settings because of its resistance to multiple antibiotics and can occasionally cause nosocomial infections in newborn, ill, or immunocompromised patients (32, 60). Genome annotation of several natural chloramphenicol-resistant P. putida strains failed to show any specific chloramphenicol-modifying enzymes (36, 41, 45, 67); however, Godoy and colleagues (23) showed that P. putida mutants in the multidrug TtgABC pump exhibited compromised growth in the presence of chloramphenicol (21). Further studies in P. putida DOT-T1E confirmed the role of the TtgABC efflux pump in tolerance to chloramphenicol and other toxic compounds while unveiling the molecular mechanisms involved in the regulation of the expression of this pump in this strain (14, 18, 58). However, no other insights on the overall mechanisms of chloramphenicol resistance and response to chloramphenicol in this species have been obtained.
To determine the molecular basis for chloramphenicol resistance in P. putida KT2440, we have established the transcriptional profile of this strain growing in the presence of the antibiotic and screened a genome-wide collection of mutants for their response and growth abilities in culture medium containing chloramphenicol. Our study confirms the absence of chloramphenicol-modifying enzymes in KT2440; however, it showed that chloramphenicol tolerance in this strain is the result of an orchestrated combination of microbial defenses that include transcriptional regulators, which allow the induction of multidrug extrusion pumps, ribosomal protein overproduction, and the activation of a stress-response program.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Pseudomonas putida KT2440R and its isogenic mutant derivatives (see Table S1 in the supplemental material) were grown in LB medium at 30°C or in modified M9 minimal medium with glucose (0.5% [wt/vol]) or citrate (16 mM) as a carbon source (2, 17). When required, antibiotics were added to reach the following final concentrations: chloramphenicol (Cm), 25 μg/ml (except when other concentrations are indicated); kanamycin (Km), 50 μg/ml; and rifampin (Rif), 10 μg/ml.
MIC.
Assays were performed in liquid medium (M9 with glucose) with serial 2-fold dilutions of chloramphenicol according to the guidelines of the Clinical and Laboratory Standards Institute (40). At least three independent experiments were carried out for each determination, and each experiment was run in triplicate. The MIC was determined as the lowest concentration of chloramphenicol that completely inhibited the growth of the strain as evaluated by lack of turbidity after incubation for 24 h under optimal growth conditions.
Screening of a P. putida KT2440 mini-Tn5 mutant collection for chloramphenicol-sensitive clones.
A collection of 8,064 independent transconjugants (17) was screened for chloramphenicol sensitivity using a Pick Up robot QPix2 (Genetix, Hampshire, United Kingdom). Clones were spotted on rectangular plates with LB without addition (control) or supplemented with 25 μg/ml of chloramphenicol. Chloramphenicol-sensitive clones were kept for further assays, including determination of the exact insertion point of the mini-Tn5 in each mutant, which was determined by DNA sequencing as previously described (17).
Chemical shock assays in liquid culture medium.
Cells were grown in M9 medium until the cultures reached the early-exponential growth phase (turbidity between 0.3 and 0.4 at 660 nm). Subsequently, the cultures were divided into two halves; one was kept as a control, while a known concentration of chloramphenicol was added to the other to reach the desired final concentration. The number of culturable cells was determined as CFU counts/ml before chloramphenicol was added and 15, 30 and 45 min later. Experiments were run in duplicate and repeated at least three times.
DNA techniques.
Chromosomal DNA was isolated using the Wizard genomic DNA purification kit (Promega, USA). PCR amplification and nucleic acid electrophoresis were carried out according to standard procedures (6). PCR products were purified from agarose gels by QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) and sequenced using the Sanger method by Secugen S.L. (Madrid, Spain). The oligonucleotides used in this study are listed in Table S2 in the supplemental material.
Antibiotic susceptibility testing.
The Kirby-Bauer disk diffusion technique was used as described previously (8). Luria-Bertani agar plates and Luria-Bertani agar plates supplemented with chloramphenicol (10 μg/ml) were overlaid with a suspension of P. putida strain KT2440R. Thereafter, antibiotic disks of ofloxacin (5 μg), pefloxacin (5 μg), ciprofloxacin (5 μg), norfloxacin (10 μg), nalidixic acid (30 μg), amoxicillin (25 μg), piperacillin (100 μg), ticarcillin (75 μg), ampicillin (10 μg), carbenicillin (100 μg), cefotaxime (30 μg), tetracycline (30 μg), imipenem (10 μg), gentamicin (10 μg), streptomycin (10 μg), neomycin (30 μg), kanamycin (30 μg), rifampin (30 μg), polymyxin B (300 μg), and colistin (50 μg) (bioMérieux, Spain) were placed on the plates. After 18 to 20 h at 30°C, the inhibition zone (in millimeters) was measured around each disk.
DNA microarrays.
The Pseudomonas putida array (Progenika, Spain) contains 5,539 gene-specific oligonucleotides (50-mer) spotted in duplicate onto γ-amino silane-treated microscope slides (25 by 75 mm) and bound to the slide with UV light and heat (69). Pseudomonas putida KT2440R cells were grown overnight on M9 minimal medium with glucose as a carbon source and were used to inoculate fresh medium with or without chloramphenicol at a concentration of 25 μg/ml, and cultures were incubated at 30°C until a turbidity of 0.4 to 0.5 at 660 nm was reached. Cells were then harvested and immediately subjected to RNA extraction.
Standard protocols were used for RNA isolation and the subsequent preparation of fluorescently labeled cDNA (19, 54). Hybridization conditions and data collection were carried out as previously described (19, 54, 59). Data were normalized by applying the LOWESS intensity-dependent normalization method (66) and statistically analyzed with Almazen System software (Alma Bioinformatics S.L, Spain). P values were calculated with Student's t test. An open reading frame (ORF) was considered differentially expressed when the fold change was at least 2 and the P value was ≤0.05. Each experiment was run in duplicate and repeated three times; therefore the results reported for each condition are the result of six arrays.
Microarray data accession number.
The microarray data were deposited in the Array Express Archive database www.ebi.ac.uk/arrayexpress/ (44), under accession numbers E-MEXP-2831, E-MEXP-3206, and E-MEXP-3203.
RESULTS
Pseudomonas putida KT2440R tolerance to chloramphenicol.
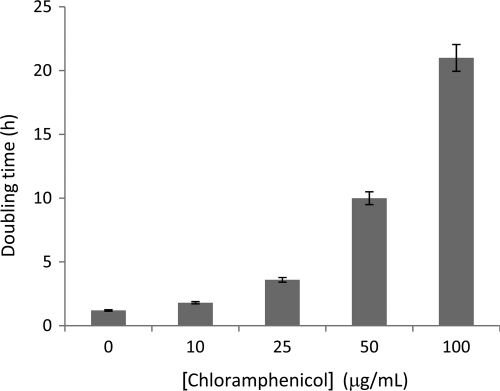
Growth of P. putida KT2440R was tested in liquid medium (M9 with glucose as a carbon source) with increasing concentrations of chloramphenicol. To this end the culture medium was inoculated with about 105 CFU/ml and growth monitored over time during the exponential phase. We found a correlation between an increase in antibiotic concentration and an increase in bacterial doubling time (Fig. 1): the effect of chloramphenicol on growth rate was noticeable at 10 μg/ml, and at a concentration of 25 μg/ml, the doubling time was 3 times longer than in chloramphenicol-free medium. At the highest tested concentration (100 μg/ml), the growth rate was almost 20 times slower than in the absence of the antibiotic. MIC was reached at 200 μg/ml (Table 1). Chloramphenicol treatment had no bactericidal effect when subinhibitory concentrations of chloramphenicol were applied (data not shown).
Fig 1.
Ability of P. putida KT2440R to grow in the presence of chloramphenicol. Doubling times (h) in M9 minimal medium with glucose as a C source and with increasing concentrations of chloramphenicol (μg/ml) were determined in triplicate, and a minimum of three independent experiments were carried out. Error bars show standard errors.
Table 1.
Minimal inhibitory concentration (MIC) of chloramphenicol for P. putida KT2440R and several isogenic mutants
| P. putida strain | MIC (μg/ml) |
|---|---|
| KT2440R | 200 |
| mut::ttgB | 50 |
| mut::pqqC | 50 |
| mut::pqqB | 50 |
| mut::PP2663 | 100 |
| mut::agmR | 100 |
| mut::2669 | 100 |
Gene expression changes in P. putida KT2440R growing in the presence of chloramphenicol.
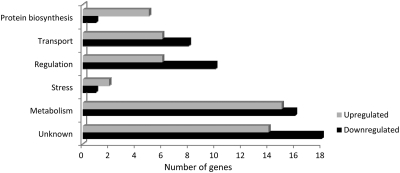
To study the cellular strategies used by KT2440R to grow, albeit at a slower pace, in the presence of chloramphenicol, we decided to analyze transcriptional changes in KT2440R cells growing in the presence of 25 μg/ml of this antibiotic. We found that 102 genes exhibited a significantly altered expression profile (fold change ≥2 or ≤−2; P value < 0.05) when cells were grown in the presence of chloramphenicol, representing a change in around 1.8% of the total number of genes. Of the 102 genes, 48 were upregulated (Table 2) and 54 were downregulated (Table 3). Functional annotation of these genes revealed that they fall within six main categories (Fig. 2): (i) metabolism and energy, (ii) response to cellular stress, (iii) transcriptional regulators, (iv) transporters and efflux pumps, (v) ribosomal proteins and related determinants, and (vi) proteins of unknown function or hypothetical proteins.
Table 2.
Upregulated genes in P. putida KT2440R grown in chloramphenicol-containing medium (25 μg/ml)a
| TIGR identifierb | Gene/description | Fold change | Functional categoryc |
|---|---|---|---|
| PP0378 (M) | pqqC (coenzyme PQQ synthesis protein C) | 2.8 | a |
| PP0379 (M) | pqqB (coenzyme PQQ synthesis protein B) | 2.2 | a |
| PP0380 | pqqA (coenzyme PQQ synthesis protein A) | 4.5 | a |
| PP0389 | rpsU (ribosomal protein S21) | 2.1 | e |
| PP0491 | Formate dehydrogenase cytochrome b556 subunit | 2 | a |
| PP0560 | aroQ-1-3-dehydroquinate dehydratase type II | 2.8 | a |
| PP0600 | rpsT (ribosomal protein S20) | 2.4 | e |
| PP0601 | Membrane protein MviN family | 2.7 | f |
| PP0626 | ndh-NADH dehydrogenase | 2.4 | a |
| PP1868 (M) | ATP-dependent RNA helicase DEAD box family | 3.4 | e |
| PP1911 | rpmF (ribosomal protein L32) | 2.2 | e |
| PP1935 | Transcriptional regulator Cro/CI family | 2.1 | c |
| PP2320 | Conserved hypothetical protein | 2.2 | f |
| PP2644 | Hypothetical protein | 2.3 | f |
| PP2645 | mgtB (magnesium-translocating P-type ATPase) | 2.4 | d |
| PP2663 (M) | FIST N sensory domain-containing protein | 10.3 | c |
| PP2664 | Sensory box histidine kinase/response regulator | 5 | c |
| PP2665 (M) | agmR (DNA-binding response regulator AgmR) | 1.5 | c |
| PP2666 | Hypothetical protein | 1.6 | f |
| PP2667 | ABC efflux transporter permease protein | 3.3 | d |
| PP2668 | ABC efflux transporter ATP-binding protein | 1.2 | d |
| PP2669 (M) | Outer membrane protein, putative | 3.4 | f |
| PP2670 | Hypothetical protein | 3 | f |
| PP2671 (M) | Sensor histidine kinase | 2.8 | c |
| PP2672 (M) | DNA-binding response regulator LuxR family | 2.1 | c |
| PP2673 | Pentapeptide repeat family protein | 1.3 | f |
| PP2674 (M) | qedH (quinoprotein ethanol dehydrogenase) | 2 | a |
| PP2675 (M) | Cytochromec-type protein | 5.3 | a |
| PP2676 | Periplasmic binding protein, putative | 3.2 | f |
| PP2677 (M) | Hypothetical protein | 5.1 | f |
| PP2678 | Hydrolase, putative | 1.4 | f |
| PP2679 | Quinoprotein ethanol dehydrogenase putative | 2.8 | a |
| PP2680 | Aldehyde dehydrogenase family protein | 5.2 | a |
| PP2681 | Coenzyme PQQ synthesis protein D, putative | 3.5 | a |
| PP2682 | Alcohol dehydrogenase iron-containing | 2.8 | a |
| PP2683 | Sensory box histidine kinase/response regulator | 1.2 | c |
| PP2688 | Conserved hypothetical protein | 2.7 | f |
| PP2694 (M) | Aldehyde dehydrogenase family protein | 2 | a |
| PP2695 (M) | Transcriptional regulator LysR family | 2.8 | c |
| PP2723 (M) | Oxidoreductase short-chain dehydrogenase/reductase | 3 | a |
| PP2821 | Conserved hypothetical protein | 2 | f |
| PP2936 | ABC transporter ATP-binding protein | 2 | d |
| PP2943 | Cytochrome c551 peroxidase, putative | 2.1 | b |
| PP3138 (M) | VirK domain protein | 2 | f |
| PP3155 (M) | Outer membrane ferric siderophore receptor, putative | 2 | d |
| PP3214 | Conserved hypothetical protein | 2 | f |
| PP3297 | Hypothetical protein | 2.1 | f |
| PP3316 | Chaperone-associated ATPase, putative | 2 | b |
| PP3455 (M) | Multidrug efflux RND membrane fusion protein | 2.2 | d |
| PP3797 (M) | Conserved hypothetical protein | 3.4 | f |
| PP4671 | Conserved hypothetical protein | 2.6 | f |
| PP4810 | nadD (nicotinic acid mononucleotide adenylyltransferase) | 2.2 | a |
| PP5066 (M) | Sodium/hydrogen exchanger family protein | 4 | d |
| PP5087 | rpmE (ribosomal protein L31) | 2 | e |
The largest upregulated region is shown in bold type. Genes from this region but not differentially expressed have also been included.
(M), mutant availability.
Functional category corresponds with the probable role of the gene in the cell: a, metabolism and energy; b, cellular stress; c, genetic regulation; d, transport and efflux; e, protein biosynthesis; and f, unknown.
Table 3.
Downregulated genes in P. putida KT2440R grown in chloramphenicol-containing medium (25 μg/ml)
| TIGR identifier | Gene/description | Fold change | Functional categorya |
|---|---|---|---|
| PP0030 | Sensor histidine kinase | −2 | c |
| PP0076 | Glycine betaine-binding protein, putative | −2 | d |
| PP0140 | Conserved hypothetical protein | −2.2 | f |
| PP0181 | Conserved hypothetical protein | −2.9 | f |
| PP0185 | pPrA-LytTR family two-component transcriptional regulator | −2 | c |
| PP0205 | Oxidoreductase, putative | −3.1 | a |
| PP0323 | soxB (sarcosine oxidase beta subunit) | −2.3 | a |
| PP0339 | aceE (pyruvate dehydrogenase E1 component) | −2 | a |
| PP0368 | Acyl-CoA dehydrogenase, putative | −2 | a |
| PP0504 | oprG (outer membrane protein OprG) | −2.5 | d |
| PP0529 | xseB (exodeoxyribonuclease VII small subunit) | −2.4 | b |
| PP0586 | Heavy metal translocating P-type ATPase | −2 | d |
| PP0588 | Copper-binding protein, putative | −2.2 | d |
| PP0613 | Amidase family protein | −2.4 | a |
| PP0620 | Transcriptional regulator GntR family | −2.1 | c |
| PP0741 | Conserved hypothetical protein | −3.2 | f |
| PP0742 | Conserved hypothetical protein | −2.2 | f |
| PP0765 | Conserved hypothetical protein | −2.4 | f |
| PP0766 | Conserved hypothetical protein | −2.6 | f |
| PP0951 | rpoX (sigma54 modulation protein) | −2.7 | e |
| PP1018 | Sugar ABC transporter ATP-binding subunit | −2.3 | d |
| PP1514 | Conserved domain protein | −2.1 | f |
| PP1516 | RND membrane fusion protein | −2.1 | f |
| PP2284 | Tail tubular protein B | −2.5 | f |
| PP2310 | Methyl-accepting chemotaxis transducer | −2 | c |
| PP2512 | folE (2-GTP cyclohydrolase I) | −2.4 | a |
| PP2736 | Conserved hypothetical protein | −2.2 | f |
| PP2738 | Transcriptional regulator, putative | −2 | c |
| PP3352 | Arylsulfatase, putative | −2.3 | a |
| PP3440 | Hypothetical protein | −2 | f |
| PP3656 | Aromatic compound-specific porin, putative | −2.5 | d |
| PP3657 | Nitrobenzoate reductase, putative | −4.4 | a |
| PP3761 | Sensor histidine kinase/response regulator | −2.1 | c |
| PP3781 | Oxygen-independent coproporphyrinogen III oxidase family | −2 | a |
| PP3782 | Hypothetical protein | −2.2 | f |
| PP3783 | Conserved hypothetical protein | −3.2 | f |
| PP3809 | Hypothetical protein | −2 | f |
| PP4051 | Malto-oligosyltrehalose trehalohydrolase | −2.6 | a |
| PP4250 | ccoN-1 (cytochrome c oxidase cbb3-type subunit I) | −3.5 | a |
| PP4252 | ccoQ-1 (cytochrome c oxidase cbb3-type CcoQ subunit) | −2 | a |
| PP4253 | ccoP-1 (cytochrome c oxidase cbb3-type subunit III) | −2 | a |
| PP4264 | hemN (oxygen-independent coproporphyrinogen III oxidase) | −2.4 | a |
| PP4265 | anr (transcriptional regulator Anr) | −2 | c |
| PP4623 | Hypothetical protein | −2 | f |
| PP4624 | Hydrolase alpha/beta fold family | −2.7 | a |
| PP4647 | Transcriptional regulator LuxR family | −2.2 | c |
| PP4863 | Branched-chain amino acid ABC transporter ATP-binding protein | −2.7 | d |
| PP5207 | ABC transporter ATP-binding protein/permease protein | −2.4 | d |
| PP5338 | aspA (aspartate ammonia-lyase) | −2.4 | a |
| PP5365 | Cyclopropane-fatty-acyl-phospholipid synthase, putative | −3.3 | a |
| PP5382 | Hypothetical protein | −4.8 | f |
| PP5383 | copR (transcriptional activator CopR) | −4.5 | c |
| PP5384 | copS (sensor protein CopS) | −2.9 | c |
| PP5390 | Hypothetical protein | −5.6 | f |
| PP5391 | Hypothetical protein | −4.7 | f |
Functional category corresponds with the probable role of the gene in the cell: a, metabolism and energy; b, cellular stress; c, genetic regulation; d, transport and efflux; e, protein biosynthesis; and f, unknown.
Fig 2.
Functional distribution of genes differentially expressed in P. putida KT2440R grown in chloramphenicol-containing medium. Shown are genes induced and downregulated in response to chloramphenicol. Genes were considered differentially expressed when their transcription level was changed 2-fold or more and had a P value of ≤0.05.
In silico analysis of the largest upregulated region.
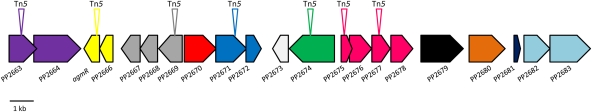
Approximately 70% of the differentially expressed genes were distributed throughout the genome of KT2440, but 30% of genes were concentrated in a 24-kb chromosomal region (Fig. 3) that comprised 21 genes (from PP2663 through to PP2683) of which 15 were upregulated. No elements associated with DNA islands were found in the analysis of adjacent DNA sequences. In addition, this 24-kb region is highly conserved in related P. putida strains (65), with 99% and 97% identity in the corresponding aligned sequences of P. putida F1 and GB-1 strains, respectively, and 98% identity with the corresponding region of the recently sequenced P. putida BIRD1 (36) (data not shown).
Fig 3.
Largest upregulated region scheme. The 24-kb DNA segment included 21 genes, 15 of which were upregulated in response to chloramphenicol (Table 1). Operons are indicated with genes in the same color. Triangles above an ORF indicate the knockout points. The complete genome sequence of P. putida KT2440 in the NCBI database was the source for the gene numbers and organization.
Operon prediction of genes in this region was first carried out according to the methodology of Price and colleagues (48) and confirmed by reverse transcription-PCR (RT-PCR) with primers based on the 3′ terminal end of genes and the 5′ terminal end of downstream adjacent genes. In Fig. 3 the genes of each of the identified operons are indicated with the same color. ORF PP2665 encodes the AgmR regulator, and it forms an operon with the gene that precedes it, ORF PP2666, which encodes a hypothetical protein of unknown function. It is worth noting that two ORFs which also form an operon, PP2663 and PP2664, encode potential sensor proteins, but the response regulator(s) that may form part of the corresponding two-component regulatory system is unknown. In this 24-kb region ORFs 2671 and the adjacent 2672 also form an operon where PP2671 is a histidine kinase that may work in conjunction with the LuxR regulator PP2672. The 24-kb region also contains a putative ABC efflux operon (PP2669/PP2668/PP2667), where the ORF PP2669 gene product is the 329-residue-long outer membrane protein of the YVTN family beta-propeller repeat-containing proteins of unknown function. The PP2668 gene product is a putative 246-residue ATP-binding protein belonging to an ABC efflux system that shares conserved domains with proteins related to resistance to daunorubicin (DrrA) (53% identity and 68% similarity; E value, 4e−71) and bacitracin (BcrA) (31% identity and 55% similarity; E value, 2e−36) resistance proteins (30, 47). The last gene, ORF PP2667, encodes a putative 263-residue-long ABC transporter membrane protein with domains that are also present in the daunorubicin resistance protein DrrB (30).
Several genes were predicted to encode quinoprotein ethanol dehydrogenases (PP2674, PP2675, and PP2679) and a gene (pqqD, ORF PP2681) encodes one of the coenzyme PQQ synthesis proteins. It should be noted that a distally located set of pqqABC genes (PP0378, PP0379, and PP0380) exist, which are also involved in coenzyme PQQ biosynthesis, and are also upregulated in response to chloramphenicol. Using further bioinformatic analysis, no obvious role could be attributed to the remaining genes clustered in this chromosomal region.
Phenotypic analysis of knockout mutants of upregulated genes in response to chloramphenicol.
A library of mini-Tn5 mutants of P. putida KT2440 is available at the Pseudomonas Reference Culture Collection (PRCC). The collection consists of more than 8,064 independent mutants (17). For further analysis we used all the available mutants in the collection that held insertions in the genes shown to be upregulated by chloramphenicol (Table 2).
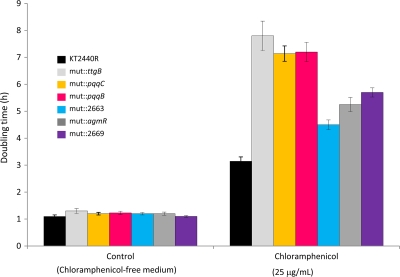
To rule out any effect of the ethanol used to dissolve chloramphenicol on the growth rate, the same amount of ethanol was added to control cultures. All but five mutant strains showed growth rates and yields similar to those of the controls (data not shown) in the presence of chloramphenicol. The mutants that exhibited compromised growth rates corresponded to the following genes: pqqC (PP0378), pqqB (PP0379) (Fig. 4), which are both involved in PQQ biosynthesis; PP2669, which encodes the inner-membrane element of the ABC efflux transporter; and PP2663 and the PP2665 regulator. These mutants had doubling times between 1.4- and 2.4-fold higher than the parental strain in the presence of chloramphenicol (Fig. 4). These five mutants also exhibited lower MICs (Table 1).
Fig 4.
Chloramphenicol tolerance of P. putida KT2440R and several isogenic knockout mutants. Doubling time in control M9 minimal medium or in the same medium with 25 μg/ml chloramphenicol.
All of the mutants which exhibited slower growth in the presence of chloramphenicol corresponded to those that bear insertions in genes in the 24-kb cluster or genes outside this cluster but related to the biosynthesis of PQQ, and one of them corresponds to an insertion in ORF PP2665 that encodes the AgmR regulator. In order to test if this regulator is directly involved in the control of the set of genes in the 24-kb cluster, we carried out comparative transcriptomic analysis in which we compared the expression patterns of all genes in the mutant versus the wild-type strain growing in the absence and in the presence of chloramphenicol. The results obtained are shown in Table 4. We found a set of 17 genes that were expressed at a lower level in the AgmR mutant strain with respect to the wild-type strain when the cells grew in the presence of chloramphenicol. As expected, all of these genes were upregulated in the parental strain when cells grew in the presence of chloramphenicol. This group of genes represents a set whose expression appears to be directly regulated by AgmR. This group includes the pqqA gene, the ABC efflux pump PP2667/2669 and the sodium exchanger PP5066, and some of the sensor proteins mentioned above. When the gene expression pattern of the AgmR mutant strain growing in the absence of chloramphenicol was compared with that of the parental strain, we found only genes from an operon (ORF 3781 through 3785) whose expression was lower than in the wild-type strain. This suggests that this operon is induced by AgmR regardless of chloramphenicol. No function has been assigned to the genes in this operon.
Table 4.
Microarray data comparison
| TIGR identifier | Gene/description | Fold change |
||
|---|---|---|---|---|
| Mutant vs wild typea | Expression differenceb | CHL comparisonc | ||
| PP0154 | Acetyl-coA hydrolase/transferase family | −2 | 1.7 | |
| PP0379 | pqqB (coenzyme PQQ synthesis protein B) | −3 | 2.2 | |
| PP0380 | pqqA (coenzyme PQQ synthesis protein A) | −2.6 | 4.5 | |
| PP2663 | FIST N sensory domain containing protein | −13.7 | 10.3 | |
| PP2664 | Sensory box histidine kinase/response regulator | −2.2 | 5 | |
| PP2666 | Hypothetical protein | −2.9 | 1.6 | |
| PP2667 | ABC efflux transporter permease protein | −2.2 | 3.3 | |
| PP2669 | Outer membrane protein, putative | −3 | 3.4 | |
| PP2674 | qedH (quinoprotein ethanol dehydrogenase) | −2 | 2 | |
| PP2675 | Cytochrome c-type protein | −3.7 | 5.3 | |
| PP2676 | Periplasmic binding protein, putative | −8.5 | 3.2 | |
| PP2677 | Hypothetical protein | −4.1 | 5.1 | |
| PP2679 | Quinoprotein ethanol dehydrogenase, putative | −2 | 2.8 | |
| PP2680 | Aldehyde dehydrogenase family protein | −2.6 | 5.2 | |
| PP2681 | Coenzyme PQQ synthesis protein D, putative | −5 | 3.5 | |
| PP2682 | Alcohol dehydrogenase iron-containing protein | −2.5 | 2.8 | |
| PP5066 | Sodium/hydrogen exchanger family protein | −2.2 | 3.8 | |
| PP3781 | Oxygen-independent coproporphyrinogen III oxidase | −2.3 | −2 | |
| PP3782 | Hypothetical protein | −2.2 | −2.3 | |
| PP3783 | Conserved hypothetical protein | −2.2 | −3.3 | |
| PP3784 | Conserved hypothetical protein | −2 | −1.8 | |
| PP3785 | Hypothetical protein | −2 | −2.2 | |
Fold change of genes differentially expressed in mutant versus wild-type agmR growing in the presence of 25 μg/ml chloramphenicol (CHL).
Fold change of genes differentially expressed in mutant agmR versus parental strain growing in chloramphenicol-free medium.
Fold change found in KT2440R growing with chloramphenicol (25 μg/ml) versus the same strain in chloramphenicol-free medium.
Effect of chloramphenicol exposure on KT2440 sensitivity to other antibiotics.
The above findings suggest chloramphenicol-mediated activation of unspecific resistance mechanisms in KT2440. Consequently, we decided to test the effect of chloramphenicol preexposure on bacterial sensitivity to other antibiotics. The disk diffusion test (see Materials and Methods) was carried out in LB agar plates both without chloramphenicol and with 10 μg/ml chloramphenicol. The results showed a significant reduction of at least 20% on the inhibition halo diameter in chloramphenicol-containing plates for 5 of the 17 antibiotics tested (Table 5); these include ofloxacin, ticarcillin, carbenicillin, tetracycline, and gentamicin.
Table 5.
Effect of chloramphenicol preexposure on the sensitivity of P. putida KT2440 to other antibioticsa
| Antibiotic | Chloramphenicol-free medium, inhibition halo diam (mm) (mean vs SD) | Result in medium with chloramphenicol |
|
|---|---|---|---|
| Total inhibition halo decrease (mm) (mean vs SD) | Inhibition halo decrease (%) | ||
| Ofloxacin | 21.7 ± 1.5 | 4.5 ± 0.20 | 20% |
| Ticarcillin | 13.5 ± 1.2 | 3.0 ± 0.01 | 22% |
| Carbenicillin | 10.0 ± 0.0 | 10.0 ± 0.0 | 100% |
| Tetracycline | 21.5 ± 1.0 | 5.2 ± 0.20 | 23.8% |
| Gentamicin | 17.7 ± 0.9 | 4.0 ± 0.15 | 22.2% |
Antibiotics whose inhibition halos were reduced by more than 20% when chloramphenicol was present in the culture medium. Analyses were carried out by a disk diffusion method performed both on LB agar plates without chloramphenicol and on LB agar plates containing 10 μg/ml chloramphenicol. Chemicals with inhibition halos not affected by preexposure to chloramphenicol were nalidixic acid, pefloxacin, ciprofloxacin, norfloxacin, amoxicillin, piperacillin, cefotaxime, imipenem, neomycin, kanamycin, polymyxin B, and colistin.
Identification of an RND efflux pump involved in chloramphenicol resistance whose expression does not change significantly in the presence of the antibiotic.
The existence of genes whose products are important for a particular process but which do not exhibit changes in transcriptional levels is not revealed by DNA microarray assays (21, 54). To overcome this limitation, the whole mutant library was screened on plates containing 25 μg/ml chloramphenicol and clones with impaired growth in the presence of the antibiotic were isolated. The mini-Tn5 insertion point was verified by sequencing of the adjacent DNA. The results allowed us to identify the ttgB (PP1385) gene, a component of the TtgABC multidrug resistance-nodulation-division (RND) transporter. The ttgB mutant exhibited a doubling time of almost 8 h in the presence of chloramphenicol (Fig. 4) versus around 3 h for the wild-type strain, and its MICs were reduced compared to that of the parental strain (Fig. 4; Table 1).
DISCUSSION
Chloramphenicol is known as a general inhibitor of translation in bacteria (55, 56). It is a bacteriostatic agent, occasionally also with bactericidal activity, that binds to the 50S ribosomal subunit and blocks the elongation of peptides during the biosynthesis of proteins (39, 56). The response to treatments with chloramphenicol in sensitive clinical strains such as Streptococcus pneumoniae (42), Bacillus subtilis (31), Yersinia pestis (51), and Enterococcus faecalis (1) has been analyzed, and it was shown that in addition to protein biosynthesis inhibition, chloramphenicol provokes oxidative stress in sensitive bacteria (1, 3).
Despite lacking cat genes or other genes encoding chloramphenicol-modifying enzymes, Pseudomonas putida KT2440 is able to grow in high concentrations of this antibiotic. We found that at a sublethal chloramphenicol concentration (25 μg/ml) an effect on growth rate is noticeable, and that the strain responds by altering the expression of 102 genes—48 of which are upregulated and 54 downregulated.
Our results show that exposure of P. putida to chloramphenicol mediates a complex response that combines different cellular defense strategies, including (i) antibiotic extrusion to reduce the intracellular concentration of the compound by efflux pumps, (ii) upregulation of relevant genes related to the biosynthesis of proteins, and (iii) the expression of oxidative stress-related genes, such as a peroxidase (encoded by PP2943), a chaperone (PP3316), and the coenzyme PQQ-encoding genes. The general character of the activated tolerance mechanisms agrees with our findings for the antibiotic susceptibility test of chloramphenicol-preexposed KT2440 cells, where this bacterium exhibited an increased resistance to different antimicrobials when cells were in chloramphenicol-containing medium. A more detailed examination of each of these functions is presented below.
Extrusion mechanisms. (i) ABC efflux system.
Among the upregulated genes was an ABC efflux transporter encoded by PP2669/PP2668/PP2667, where PP2668 encodes the ABC-binding protein and PP2667 the corresponding permease protein. A mutant at ORF PP2669 showed delayed growth in chloramphenicol-containing medium and was sensitive to an acute exposure to high concentrations of this antibiotic. Matilla and colleagues (35) found that expression of PP2669 was strongly induced when KT2440 was present in the rhizosphere of plants, an environment where oxidative stress induced by antimicrobial secondary metabolites is relevant (7, 64). PP2668 and PP2667 exhibited conserved sequences with respect to the DrrAB efflux system of Streptomyces that confers self-protection to producer strains against daunorubicin and doxorubicin, two anthracycline-type aromatic compounds (27, 30, 34). Thus, in KT2440, the efflux system consisting of PP2669/PP2668 and PP2667 may be involved in the direct efflux of chloramphenicol.
(ii) Multidrug extrusion pump.
In this study, we have isolated a mutant in the ttgB gene that exhibited increased sensitivity to chloramphenicol, confirming Godoy and colleagues' observations (23). The TtgABC efflux pump is a multidrug efflux system that expels chloramphenicol from the cell, as it is known to do for tetracycline, ethidium bromide, ampicillin, and other antibiotics (14, 18, 23, 58).
Differential transcription of genes involved in protein synthesis.
Since chloramphenicol acts directly at the ribosome by inhibiting peptide chain elongation, an increase in the ribosome biosynthesis machinery would be an expected survival mechanism to decrease the ratio between drug and target ribosomal particles. We found that KT2240R responds to chloramphenicol by increasing the level of transcription of certain ribosome-associated proteins (encoded by rpsU, rpsT, rpmF, and rpmE) and the expression of RNA helicase (PP1868), whereas rpoX (PP0951), encoding a ribosome-associated modulation/inhibition factor, was downregulated. Similar responses have been reported before in bacteria treated with inhibitors of protein biosynthesis (1, 31, 42, 51).
Oxidative stress-related mechanisms. Putative role of PQQ coenzyme.
Most of the genes involved in pyrroloquinoline quinone coenzyme (PQQ) biosynthesis (50) are clustered in the KT2440 chromosome at the pqqFABCDE (PP0381-PP0376) operon. An additional copy of the pqqD gene (PP2681) is unlinked (61). Many of these genes were upregulated when cells were grown in the presence of chloramphenicol.
Pyrroloquinoline quinone (PQQ) is an o-quinone that serves as a cofactor for a number of periplasmic as well as cytosolic prokaryotic dehydrogenases, known as quinoproteins or quinoenzymes (25, 33), which are involved in a variety of bacterial processes, such as mineral phosphate solubilization via the production of 2-ketogluconic acid from glucose (24), alkaloid lupanine degradation in P. putida DH2001 (29), ethanol oxidation in P. aeruginosa (22, 26), 2-phenylethylamine and 2-phenylethanol metabolism in P. putida U (5), and induction of systemic resistance by Enterobacter intermedium 60-2G (28). PQQ is also involved in cellular protection against oxidative stress, irradiation, and DNA-damaging agents in the extremophile Deinococcus radiodurans (38, 52). In Bradyrhizobium japonicum, desiccation stress induced several putative pyrroloquinoline quinone-containing alcohol dehydrogenases in addition to genes involved in PQQ biosynthesis (13). The authors of the study suggested that this set of genes was directly involved in the B. japonicum desiccation response by reducing oxidative stress in cells. Phenotypic analysis of knockout mutants in pqqC and pqqB revealed their increased sensitivity to chloramphenicol, which suggests that PQQ plays a role in the tolerance of KT2440 toward this antibiotic. Our results suggest that PQQ is involved in stress endurance in P. putida, which could be related to the associated oxidative stress response.
Transcriptional regulator AgmR.
We found that an agmR mutant exhibited impeded growth in chloramphenicol-containing medium. AgmR was found to be upregulated in tetracycline-exposed KT2440 cells (68), and it has been described as a regulator of the quinoprotein ethanol oxidation system in P. putida ATCC 11172 (63), P. putida HK5 (49) and P. aeruginosa (22). Inactivation of agmR either abolished or reduced the ability of P. putida 11172 and HK5 to grow on ethanol (49, 63). However, in the case of KT2440, inactivation of agmR did not diminish the ability of the strain to grow on ethanol (data not shown), suggesting that an alternative ethanol utilization system might be operating in this strain. Similar results were found by Arias and colleagues (5) when they inactivated an agmR homologous gene, named pedR1, in P. putida U. The analysis of the array assays of the agmR mutant growing with and without chloramphenicol compared to those of the wild-type strain growing in the same conditions indicated that AgmR controls around 15% of all genes whose expression changes in response to chloramphenicol (Table 4). Among the genes regulated by AgmR is a set of 17 genes that are activated by AgmR in response to chloramphenicol and an operon (ORF PP3781/PP3785) that is induced by AgmR regardless of the presence of chloramphenicol. The pqqA gene, the ABC efflux pump, PP2669 through PP2667, and two predicted sensor proteins are induced by AgmR only in the presence of chloramphenicol (Table 4). It seems likely that the sensor proteins are responsible for activation/repression of other genes in response to chloramphenicol, although the nature of the genes under the control of this system is at present unknown.
Does downregulation of genes confer a protective effect against chloramphenicol?
Of the 54 downregulated genes, a subgroup are membrane proteins. Their diminished expression may lead to changes in the bacterial membrane important for defense against this toxic compound. This may be part of a generalized response, since one of the downregulated genes, the major cyclopropane fatty acid synthase, cfaB (PP5365), was also downregulated when cells were grown in the presence of xylene (16). Nonetheless, cfaB mutants are as resistant to chloramphenicol as the parental strain (46), so the question of the importance of cfaB downregulation remains unclear.
Other downregulated membrane proteins include most of the genes of the Cbb3-1 cytochrome oxidase complex, including ccoN-1, ccoQ-1, and ccoP-1 (PP4250, PP4252, and PP4253). This particular cytochrome oxidase complex shows high affinity for O2 and is considered an adaptive trait to microaerobic conditions (12). Small and colleagues (57) found that P. aeruginosa downregulates Cbb3-encoding genes in response to oxidative agents such as sodium hypochlorite, peracetic acid, and hydrogen peroxide, as well as a bacteriostatic antibiotic known as fusidic acid (62). The authors attributed this trait to the decrease in the cellular respiratory function and the loss of metabolic energy, as had been previously found upon exposure to HOCl (4).
In summary, Pseudomonas putida KT2440R can resist chloramphenicol without the presence of specific chloramphenicol-modifying genes. Resistance to chloramphenicol in P. putida KT2440 is mediated by the altered expression of a variety of genes involved in general defense mechanisms that involve efflux pumps, oxidative stress responses, and physiological alterations. This mixed phenomenon may favor P. putida secondary infections in chloramphenicol-treated susceptible patients, but horizontal transfer of chloramphenicol resistance from P. putida to other nosocomial microorganisms is very unlikely.
Supplementary Material
ACKNOWLEDGMENTS
Work in this study was supported by FEDER grants from the EC (BACSIN FP7-KBBE-2007-1) and by ADHERS (BIO2008-04419-E/) from the Pathogenomics program of ERANET. Part of this study is supported by FEDER funds from the Ministry of Science and Innovation, Consolider C (BIO2006-0658), and Plan Nacional (BIO2010-17227).
We thank Ben Pakuts for critical reading of the manuscript.
Footnotes
Published ahead of print 5 December 2011
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Aakra A, et al. 2010. The response of Enterococcus faecalis V583 to chloramphenicol treatment. Int. J. Microbiol. doi:10.1155/2010/483048 [DOI] [PMC free article] [PubMed]
- 2. Abril MA, Michán C, Timmis KN, Ramos JL. 1989. Regulator and enzyme specificities of the TOL plasmid encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J. Bacteriol. 171:6782–6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Albesa I, Becerra MC, Battán PC, Páez PL. 2004. Oxidative stress involved in the antibacterial action of different antibiotics. Biochem. Biophys. Res. Commun. 317:605–609 [DOI] [PubMed] [Google Scholar]
- 4. Albrich JM, Hurst JK. 1982. Oxidative inactivation of Escherichia coli by hypochlorous acid. Rates and differentiation of respiratory from other reaction sites. FEBS Lett. 144:157–161 [DOI] [PubMed] [Google Scholar]
- 5. Arias S, Olivera ER, Arcos M, Naharro G, Luengo JM. 2008. Genetic analyses and molecular characterization of the pathways involved in the conversion of 2-phenylethylamine and 2-phenylethanol into phenylacetic acid in Pseudomonas putida U. Environ. Microbiol. 10:413–432 [DOI] [PubMed] [Google Scholar]
- 6. Ausubel FM, et al. (ed). 2005. Current protocols in molecular biology. John Wiley & Sons Hoboken, NJ. [Google Scholar]
- 7. Bais HP, Prithiviraj B, Jha AK, Ausubel FM, Vivanco JM. 2005. Mediation of pathogen resistance by exudation of antimicrobials from roots. Nature 434:217–221 [DOI] [PubMed] [Google Scholar]
- 8. Bauer A, Kirby WM, Sherris JC, Turck M. 1966. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45:493–496 [PubMed] [Google Scholar]
- 9. Blázquez J, Oliver A, Gómez-Gómez JM. 2002. Mutation and evolution of antibiotic resistance: antibiotics as promoters of antibiotic resistance? Curr. Drug Targets 4:345–349 [DOI] [PubMed] [Google Scholar]
- 10. Burns JL, Hedi LA, Lien DM. 1989. Chloramphenicol resistance in Pseudomonas cepacia because of decreased permeability. Antimicrob. Agents Chemother. 33:136–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cantón R. 2009. Antibiotic resistance genes from the environment: a perspective through newly identified antibiotic resistance mechanisms in the clinical setting. Clin. Microbiol. Infect. 15(Suppl. 1):20–25 [DOI] [PubMed] [Google Scholar]
- 12. Cosseau C, Batut J. 2004. Genomics of the CcoNOQP-encoded cbb3 oxidase complex in bacteria. Arch. Microbiol. 181:89–96 [DOI] [PubMed] [Google Scholar]
- 13. Cytryn EJ, et al. 2007. Transcriptional and physiological responses of Bradyrhizobium japonicum to desiccation-induced stress. J. Bacteriol. 19:6751–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Daniels C, Daddaoua A, Lu D, Zhang X, Ramos JL. 2010. Domain cross-talk during effector binding to the multidrug binding TTGR regulator. J. Biol. Chem. 28:21372–21381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Daniels C, Ramos JL. 2009. Adaptive drug resistance mediated by root-nodulation-cell division efflux pumps. Clin. Microbiol. Infect. 15(Suppl. 1):32–36 [DOI] [PubMed] [Google Scholar]
- 16. Domínguez-Cuevas P, González-Pastor JE, Marqués S, Ramos JL, de Lorenzo V. 2006. Transcriptional tradeoff between metabolic and stress-response programs in Pseudomonas putida KT2440 cells exposed to toluene. J. Biol. Chem. 17:11981–11991 [DOI] [PubMed] [Google Scholar]
- 17. Duque E, et al. 2007. Towards a genome-wide mutant library of Pseudomonas putida strain KT2440, p 227–251, In Ramos JL, Filloux A. (ed), Pseudomonas, vol. V: a model system in biology. Springer, Dordrecht; The Netherlands [Google Scholar]
- 18. Duque E, Segura A, Mosqueda G, Ramos JL. 2001. Global and cognate regulators control the expression of the organic solvent efflux pumps TtgABC and TtgDEF of Pseudomonas putida. Mol. Microbiol. 39:1100–1106 [DOI] [PubMed] [Google Scholar]
- 19. Duque E, et al. 2007. The RpoT regulon of Pseudomonas putida DOT-T1E and its role in stress endurance against solvents. J. Bacteriol. 189:207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Falagas ME, Kopterides P. 2007. Old antibiotics for infections in critically ill patients. Curr. Opin. Crit. Care 5:592–597 [DOI] [PubMed] [Google Scholar]
- 21. Fernández M, et al. 2009. Microbial responses to xenobiotic compounds. Identification of genes that allow Pseudomonas putida KT2440 to cope with 2,4,6-trinitrotoluene. Microb. Biotechnol. 2:287–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gliese N, Khodaverdi V, Schobert M, Görisch H. 2004. AgmR controls transcription of a regulon with several operons essential for ethanol oxidation in Pseudomonas aeruginosa ATCC 17933. Microbiology 150:1851–1857 [DOI] [PubMed] [Google Scholar]
- 23. Godoy P, Molina-Henares AJ, de la Torre J, Duque E, Ramos JL. 2010. Characterization of the RND family of multidrug efflux pumps: in silico to in vivo confirmation of four functionally distinct subgroups. Microb. Biotechnol. 3:691–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goldstein A. 1994. Involvement of the quinoprotein glucose dehydrogenase in the solubilization of exogenous mineral phosphates by gram-negative bacteria, p 197–203. In Torriani-Gorini A, Yagil E, Silver S. (ed), Phosphate in microorganisms: cellular and molecular biology. ASM Press, Washington, DC [Google Scholar]
- 25. Goodwin PM, Anthony C. 1998. The biochemistry, physiology and genetics of PQQ and PQQ-containing enzymes. Adv. Microb. Physiol. 40:1–80 [DOI] [PubMed] [Google Scholar]
- 26. Görisch H. 2003. The ethanol oxidation system and its regulation in Pseudomonas aeruginosa. Biochim. Biophys. Acta 1647:98–102 [DOI] [PubMed] [Google Scholar]
- 27. Guilfoile PG, Hutchinson CR. 1991. A bacterial analog of the mdr gene of mammalian tumor cells is present in Streptomyces peucetius, the producer of daunorubicin and doxorubicin. Proc. Natl. Acad. Sci. U. S. A. 88:8553–8557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Han SH, et al. 2008. Inactivation of pqq genes of Enterobacter intermedium 60-2G reduces antifungal activity and induction of systemic resistance. FEMS Lett. 10:1111–1120 [DOI] [PubMed] [Google Scholar]
- 29. Hopper DJ, Kaderbhai MA. 2003. The quinohaemoprotein lupanine hydroxylase from Pseudomonas putida. Biochim. Biophys. Acta 1647:110–115 [DOI] [PubMed] [Google Scholar]
- 30. Kaur P. 1997. Expression and characterization of DrrA and DrrB proteins of Streptomyces peucetius in Escherichia coli: DrrA is an ATP binding protein. J. Bacteriol. 179:569–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin JT, Connelly MB, Amolo C, Otani S, Yaver DS. 2005. Global transcriptional response of Bacillus subtilis to treatment with subinhibitory concentrations of antibiotics that inhibit protein synthesis. Antimicrob. Agents Chemother. 49:1915–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lombardi G, et al. 2002. Nosocomial infections caused by multidrug-resistant isolates of Pseudomonas putida producing VIM-1 metallo-beta-lactamase. J. Clin. Microbiol. 40:4051–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Magnusson OT, RoseFigura JM, Toyama H, Schwarzenbacher R, Klinman JP. 2007. Pyrroloquinoline quinone biogenesis: characterization of PqqC and its H84N and H84A active site variants. Biochemistry 46:7174–7186 [DOI] [PubMed] [Google Scholar]
- 34. Malla S, Niraula NP, Liou K, Sohng JK. 2009. Self-resistance mechanism in Streptomyces peucetius: overexpression of drrA, drrB and drrC for doxorubicin enhancement. Microbiol. Res. 165:459–467 [DOI] [PubMed] [Google Scholar]
- 35. Matilla MA, Espinosa-Urgel M, Rodríguez-Herva JJ, Ramos JL, Ramos-González MI. 2007. Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol. 9:R179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matilla MA, et al. 2011. Complete genome of the plant growth-promoting rhizobacterium Pseudomonas putida BIRD-1. J. Bacteriol. 193:1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maviglia R, Nestorini R, Pennisi M. 2009. Role of old antibiotics in multidrug resistant bacterial infections. Curr. Drug Targets 10:895–905 [DOI] [PubMed] [Google Scholar]
- 38. Misra HS, et al. 2004. Pyrroloquinoline-quinone: a reactive oxygen species scavenger in bacteria. FEBS Lett. 578:26–30 [DOI] [PubMed] [Google Scholar]
- 39. Montero CI, et al. 2007. Response of wild-type and resistant strains of the hyperthermophilic bacterium Thermotoga maritima to chloramphenicol challenge. Appl. Environ. Microbiol. 73:5058–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. National Committee for Clinical Laboratory Standards 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7–A6. National Committee for Clinical Laboratory Standards, Wayne, PA [Google Scholar]
- 41. Nelson KE, et al. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Microbiology 4:799–808 [DOI] [PubMed] [Google Scholar]
- 42. Ng W-L, Kazmierczak KM, Robertson GT, Gilmour R, Winkler ME. 2003. Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J. Bacteriol. 185:359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nitzan O, et al. 2010. Is chloramphenicol making a comeback? Isr. Med. Assoc. J. 12:371–374 [PubMed] [Google Scholar]
- 44. Parkinson H, et al. 2011. ArrayExpress update—an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 39:D1002–D1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Phoenix P, et al. 2003. Characterization of a new solvent-responsive gene locus in Pseudomonas putida F1 and its functionalization as a versatile biosensor. Environ. Microbiol. 5:1309–2137 [DOI] [PubMed] [Google Scholar]
- 46. Pini CV, Bernal P, Godoy P, Ramos JL, Segura A. 2009. Cyclopropane fatty acids are involved in organic solvent tolerance but not in acid stress resistance in Pseudomonas putida DOT-T1E. Microb. Biotechnol. 2:253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Podlesek Z, et al. 1995. Bacillus licheniformis bacitracin-resistance ABC transporter: relationship to mammalian multidrug resistance. Mol. Microbiol. 16:969–976 [DOI] [PubMed] [Google Scholar]
- 48. Price MN, Huang KH, Alm EJ, Arkin AP. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33:880–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Promden W, et al. 2008. Disruption of quinoprotein ethanol dehydrogenase gene and adjacent genes in Pseudomonas putida HK5. FEMS Microbiol. Lett. 280:203–209 [DOI] [PubMed] [Google Scholar]
- 50. Puehringer S, Metlitzky M, Schwarzenbacher R. 2008. The pyrroloquinoline quinone biosynthesis pathway revisited: a structural approach. BMC Biochem. 27:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Qiu J, et al. 2006. Microarray expression profiling of Yersinia pestis in response to chloramphenicol. FEMS Microbiol. Lett. 263:26–31 [DOI] [PubMed] [Google Scholar]
- 52. Rajpurohit YS, Gopalakrishnan R, Misra HS. 2008. Involvement of a protein kinase activity inducer in DNA double strand break repair and radioresistance of Deinococcus radiodurans. J. Bacteriol. 190:3948–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ramos JL, et al. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56:743–768 [DOI] [PubMed] [Google Scholar]
- 54. Roca A, Rodríguez-Herva JJ, Duque E, Ramos JL. 2008. Physiological responses of Pseudomonas putida to formaldehyde during detoxification. Microb. Biotechnol. 1:158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schlünzen F, et al. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814–821 [DOI] [PubMed] [Google Scholar]
- 56. Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28:519–542 [DOI] [PubMed] [Google Scholar]
- 57. Small DA, Chang W, Toghrol F, Bentley WE. 2007. Comparative global transcription analysis of sodium hypochlorite, peracetic acid, and hydrogen peroxide on Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 76:1093–1105 [DOI] [PubMed] [Google Scholar]
- 58. Terán W, et al. 2003. Antibiotic-dependent induction of Pseudomonas putida DOT-T1E TtgABC efflux pump is mediated by the drug binding repressor TtgR. Antimicrob. Agents Chemother. 47:3067–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Timmis KN. 2002. Pseudomonas putida: a cosmopolitan opportunist par excellence. Environ. Microbiol. 4:779–781 [DOI] [PubMed] [Google Scholar]
- 60. Treviño M, et al. 2010. Nosocomial infection by VIM-2 metallo-beta-lactamase-producing Pseudomonas putida. J. Med. Microbiol. 59:853–855 [DOI] [PubMed] [Google Scholar]
- 61. Tsai TY, Yang CY, Shih HL, Wang AH, Chou SH. 2009. Xanthomonas campestris PqqD in the pyrroloquinoline quinone biosynthesis operon adopts a novel saddle-like fold that possibly serves as a PQQ carrier. Proteins 76:1042–1048 [DOI] [PubMed] [Google Scholar]
- 62. Van Rij ET, Girard G, Lugtenberg BJJ, Bloemberg GV. 2005. Influence of fusaric acid on phenazine-1-carboxamide synthesis and gene expression of Pseudomonas chlororaphis strain PCL1391. Microbiology 151:2805–2814 [DOI] [PubMed] [Google Scholar]
- 63. Vrionis HA, Daugulis AJ, Kropinski AM. 2002. Identification and characterization of the AgmR regulator of Pseudomonas putida: role in alcohol utilization. Appl. Microbiol. Biotechnol. 58:469–475 [DOI] [PubMed] [Google Scholar]
- 64. Walker TS, Bais HP, Halligan KM, Stermitz FR, Vivanco JM. 2003. Metabolic profiling of root exudates of Arabidopsis thaliana. J. Agric. Food Chem. 51:2548–2554 [DOI] [PubMed] [Google Scholar]
- 65. Wu X, et al. 2011. Comparative genomic and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol. Rev. 35:299–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang YH, et al. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yu H, et al. 2011. Complete genome sequence of the nicotine-degrading Pseudomonas putida strain S16. J. Bacteriol. 193:5541–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yun SH, et al. 2006. Proteome analysis of cellular response of Pseudomonas putida KT2440 to tetracycline stress. Curr. Microbiol. 53:95–101 [DOI] [PubMed] [Google Scholar]
- 69. Yuste L, et al. 2006. Growth phase-dependent expression of the Pseudomonas putida KT2440 transcriptional machinery analysed with a genome-wide DNA microarray. Environ. Microbiol. 8:165–177 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.