Abstract
Resistance-associated mutations in the HIV-1 protease modify viral fitness through changes in the catalytic activity and altered binding affinity for substrates and inhibitors. In this report, we examine the effects of 31 mutations at 26 amino acid positions in protease to determine their impact on infectivity and protease inhibitor sensitivity. We found that primary resistance mutations individually decrease fitness and generally increase sensitivity to protease inhibitors, indicating that reduced virion-associated protease activity reduces virion infectivity and the reduced level of per virion protease activity is then more easily titrated by a protease inhibitor. Conversely, mutations at more variable positions (compensatory mutations) confer low-level decreases in sensitivity to all protease inhibitors with little effect on infectivity. We found significant differences in the observed effect on infectivity with a pseudotype virus assay that requires the protease to cleave the cytoplasmic tail of the amphotropic murine leukemia virus (MuLV) Env protein. Additionally, we were able to mimic the fitness loss associated with resistance mutations by directly reducing the level of virion-associated protease activity. Virions containing 50% of a D25A mutant protease were 3- to 5-fold more sensitive to protease inhibitors. This level of reduction in protease activity also resulted in a 2-fold increase in sensitivity to nonnucleoside inhibitors of reverse transcriptase and a similar increase in sensitivity to zidovudine (AZT), indicating a pleiotropic effect associated with reduced protease activity. These results highlight the interplay between enzyme activity, viral fitness, and inhibitor mechanism and sensitivity in the closed system of the viral replication complex.
INTRODUCTION
The addition of protease (PR) inhibitors (PIs) to antiretroviral therapies (ARTs) has led to significant reductions in morbidity and mortality associated with HIV-1 infection (15, 39). Despite these clinical gains, the benefits of ART can be transitory, with some individuals experiencing a rebound of viral load (30). Virologic failure of PI-based ART most often occurs because of characteristic mutations in the HIV-1 protease gene (pro) that reduce the sensitivity of these viruses to one or more inhibitors, and the presence of drug resistance is a significant barrier to achieving long-term viral suppression. Although changes at as many as 45 of the 99 amino acid residues in PR may be associated with selection by a PI, a subset of approximately 26 positions has been identified to be those most commonly involved in PI resistance (8, 9, 24, 37, 59). Mutations at the codons encoding these positions typically accumulate in a stepwise fashion during therapy failure (8, 9, 27, 37, 61), although the order in which they are acquired varies.
PI resistance mutations were originally divided into two groups: primary or active-site mutations and secondary or compensatory mutations. Primary mutations are often selected early in the evolution of PI resistance and are required for high-level PI resistance. These mutations occur at conserved positions that encode amino acids that are clustered around the active site of the enzyme, and these substitutions are generally thought to alter the Ki of the PI-PR interaction (19, 35, 38, 40, 43, 53). While the changes in 50% inhibitory concentrations (IC50s) provided by a single primary mutation are generally small (32, 61), there are examples where significant resistance can be conferred by a single amino acid substitution (7, 32, 41). Concomitantly, the altered enzyme active site is less able to process its normal Gag substrate, resulting in reduced infectivity of these viruses (10, 13, 48, 60). Secondary mutations are generally selected later in PI treatment and occur at codons that encode amino acids outside the enzyme active site. As single mutations, they do not alter drug sensitivity in an appreciable manner. The role of secondary mutations in the evolution of resistance appears to be a compensatory one, as they encode substitutions that recover fitness losses resulting from the incorporation of primary mutations (23, 26, 32–34, 42). In some cases, the amino acid substitutions encoded by secondary mutations have been shown to restore the loss of catalytic activity for mutant proteases (6, 38, 53), which may explain their mechanism of compensation.
In the present study, we have analyzed the effects of 31 common PI resistance-associated mutations on the infectivity of HIV-1 as a means to evaluate their individual contributions to viral fitness and their effects on the sensitivity of HIV-1 to each of seven approved PIs. These data indicate that the classification of resistance-associated mutations in pro as primary or compensatory reflects the biological effects of the substitutions encoded at these positions, as the inclusion of single primary resistance mutations engendered significant fitness losses, while mutations at the more variable/compensatory positions resulted in smaller fitness changes. There were specific examples where a single primary resistance mutation conferred net resistance to a specific inhibitor, but in general, there were small increases in sensitivity to PIs associated with these mutations. Conversely, the compensatory mutations conferred low-level decreases in sensitivity to all PIs, emphasizing the need for multiple mutations to confer high-level resistance to protease inhibitors. When the infectivity values obtained from the specific infectivity assay were compared to the ones from the replication capacity assay in which viruses were pseudotyped with the amphotropic murine leukemia virus (MuLV) Env protein (a modified version of the PhenoSense assay [12, 44]), we found significant differences in the effect of the mutations on viral infectivity. Finally, we were able to mimic the fitness losses associated with primary resistance mutations in the protease by titrating down protease activity in virions by inclusion of a protease active-site mutant with the wild type. The reduction in virion-associated protease activity had pleiotropic effects on sensitivity to reverse transcriptase (RT) inhibitors that highlight the distributive nature of both proteolytic processing and DNA synthesis and the need for multiple enzyme molecules to carry out these two essential steps in viral replication.
MATERIALS AND METHODS
Plasmids, mutagenesis, and cell culture.
The plasmid pARK, containing the ApaI to RsrII fragment of pNL-CH (described below), was used as a template for site-directed mutagenesis of the NL4-3 HIV-1 pro gene. Point mutations were introduced using the QuikChange method (Stratagene) and were confirmed by sequence analysis. The DNA fragment generated by digestion with ApaI and RsrII was used to transfer mutated pro sequences into the infectious molecular clone pNL-CH, which codes for the NL4-3 strain of HIV-1 with a silent T-to-C mutation at nucleotide 2600 (relative to the strain HXB2CG sequence) to introduce an RsrII restriction enzyme site near the 5′ end of pol. Two clones of each mutant were isolated, and the nucleotide sequence of the pro gene was confirmed. The 293 cells and the TZM-bl cells (NIH AIDS Research and Reference Reagent Program) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum in the presence of penicillin and streptomycin at 37°C with 5% CO2. CEMx174 and H9 cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum in the presence of penicillin and streptomycin. Cells of the multinuclear activation of a galactosidase indicator (MAGI) cell line were maintained in DMEM-high glucose (DMEM-H) medium, which consisted of DMEM supplemented with 10% fetal bovine serum, 0.1 mg/ml G418, and 0.1 mg/ml hygromycin B. The TZM-bl cells are HeLa cells which stably express CD4 and CCR5 (57).
Virus stocks.
One day prior to transfection, 3 × 105 293 cells were plated in each well of a 24-well plate. Transfections of 0.5 μg of plasmid DNA were done with each mutant plasmid using Fugene transfection reagent (Roche) according to the manufacturer's instructions. For the phenotypic mixing experiments, a version of pNL-CH with a mutation at the codon encoding the active site of the protease (D25A) was cotransfected with wild-type (WT) DNA at various ratios of the two plasmids while maintaining the total DNA at a fixed mass. After 48 h, virus-containing culture supernatants were collected, filtered through a 0.45-μm-pore-size membrane (Millipore) to remove cell debris, and stored at −80°C.
Real-time RT-PCR.
Cell-free virions harvested from transfected 293 cells were treated with RNase-free DNase (Promega) at 37°C for 1 h to remove any residual plasmid DNA carried over from the transfection. Sindbis virus (Girdwood strain; a gift from Mark Heise) was also treated with RNase-free DNase and used as an internal control for real-time PCR. Viral RNA was extracted using a QIAamp viral RNA kit (Qiagen). The amount of encapsidated genomic viral RNA was detected by quantitative real-time RT-PCR using a TaqMan one-step RT-PCR master mix reagent kit (Applied Biosystems) and an ABI 7000 sequencer detector (Applied Biosystems) and normalized by the amount of Sindbis viral RNA. The sequences of the primers and probe to detect the HIV-1 gag region were previously described (16). Sindbis virus RNA was detected using primers SINRT-F (5′-GCCGCACACGACAATTCAC-3′) plus SINRT-R (5′-GTACCCTCGTACACGGACGAA-3′) and a probe, SINRT-P (5′-FAM-CCGCATCATCTGAATTG-MGBNFQ-3′, where FAM is 6-carboxyfluorescein), which detect the NSP2 region of the genome.
Quantitative determination of CA protein.
The amount of capsid (CA) protein (p24) in the transfection supernatants was measured by an enzyme-linked immunosorbent assay (ELISA) using a set of reagents available from the National Cancer Institute AIDS Vaccine Program by following the manufacturer's instructions. Lysed samples were further diluted at least 50-fold prior to sample binding. All samples were measured in duplicate at two dilutions. Masses were calculated from the standard curve using the average optical density of the duplicate measurements. The dilution-corrected masses were then averaged to determine the final mass of p24 in each transfection supernatant.
Specific infectivity (SpIn) assay.
Virus stocks (i.e., the transfection supernatants) were used to infect TZM-bl cells (14, 57), seeded at 1.3 × 104 cells per well of a 96-well plate 24 h prior to infection. Two dilutions of the transfection supernatant from each mutant, as well as the wild-type NL4-3 transfected in parallel, were used to infect TZM-bl cells in duplicate. After 48 h of incubation, the cells were washed with 150 μl of Dulbecco's phosphate-buffered saline (PBS), lysed in 50 μl of reporter lysis buffer (Promega), and stored at −80°C. After one freeze-thaw cycle, 30 μl of cell lysates was transferred into a 96-well assay plate (Costar) and luciferase (Luc) activity was measured using a luminometer (Promega). Any infections for which the values of the corrected relative light units (RLU) of the two dilutions differed by greater than 30% were discarded. The values of the specific infectivity were normalized either by the amount of p24 determined from the ELISA or by the level of viral RNA determined from the real-time RT-PCR and compared to the value for the parental strain, which was given a value of 1.
RC assay.
Mutant pro sequences were cloned into a previously described replication-incompetent HIV-1 vector that contains a luciferase gene inserted into a deleted portion of the env gene (44). The replication capacity (RC) assay is a modified version of the PhenoSense assay originally designed to measure HIV-1 drug susceptibility (12, 44). Briefly, virus stocks were produced by cotransfecting 293 cells with the HIV-1 luciferase vector encoding the pro sequence of interest and an MuLV amphotropic env-expressing DNA construct. Virus particles were harvested at 48 h after transfection and were used to infect target 293 cells for 48 h. The ratio of luciferase activity in the infected cells to luciferase activity in the transfected cells was calculated to give the single-cycle RC value relative to the wild-type virus, which was given an RC value of 1 (12).
Growth competition assay.
Virus stocks of M46I, G48V, L63P, V82A, I84V, L90M, and wild type (NL-CH) were used to infect 3 × 106 CEMx174 cells in 500 μl. A total of 3 × 103 infectious units of virus, determined using cells of the MAGI cell line, was mixed with the cells (29). Ratios of input infectious units for the two viruses (WT and mutant) in coculture were 1:1 in all cases except for WT/L63P and WT/G48V, where the input ratios were 1:2 and 1:200, respectively. For infection, cells were incubated at 37°C for 2 h, the unbound virus was removed by washing twice with PBS, and the cells were resuspended in a 10 ml of RPMI 1640 medium. The cells were washed to minimize the measurement of input virus in the medium. On each day that the cells were centrifuged, supernatants were collected and fresh medium was added to the culture until the beginning of syncytium formation was detected. Collection was stopped when the first syncytia appeared to ensure that cells were in excess over the virus during the period of analysis.
To quantitate relative viral growth, the culture supernatants (harvested each day, usually days 1 to 3) were treated with RNase-free DNase (Promega) at 37°C for 1 h, viral RNA was extracted using a QIAamp viral RNA kit (Qiagen), and real-time RT-PCR was performed using a TaqMan one-step RT-PCR master mix reagent kit (Applied Biosystems) to document an increase in virus over input. Sindbis virus was used as an internal control to normalize the recovery of viral RNA during the extraction step.
To assess the ratio of the wild-type and mutant viruses in the culture supernatant, a heteroduplex tracking assay (HTA) analysis was performed as described by Resch et al. (47). Briefly, the plasmid pPR-EB was used to generate a labeled probe spanning most of the pro gene. A 247-bp fragment of pro was amplified from viral RNA using primers PRAMPUP (5′-AACTAAAGGAAGCTCTATTAGATACAGGAG-3′) and PRAMPDW (5′-GGAAAATTTAAAGTGCAACCAATCTGA-3′). The labeled probe was annealed to the PCR products representing each day of culture growth, and the labeled heteroduplexes were resolved in a native polyacrylamide gel. The relative abundance of the wild-type and mutant sequences was determined from the intensities of their relative heteroduplexes in the gel, and the magnitude of change in this ratio per day was used to calculate the relative fitness of the mutant compared to the wild type (47).
Silver staining.
A 1-ml aliquot of transfection supernatant was centrifuged at 21,000 × g for 1.5 h in a microultracentrifuge. The pelleted virus particles were lysed in 25 μl of sample lysis buffer (50 mM Tris-HCl, 0.1 M NaCl, 1 mM EDTA, 50 μl of Nonidet P-40, and 1 protease inhibitor tablet [Roche] in 25 ml). Volumes containing equivalent masses of p24, as measured by ELISA, were subjected to denaturing Tricine-SDS-polyacrylamide gel electrophoresis as described previously (52) in a 12% acrylamide gel. Proteins were visualized by silver staining according to the manufacturer's instructions (Bio-Rad).
Western blot analyses.
Viruses pseudotyped with MuLV amphotropic Env protein were harvested from the 293 cells at 48 h after transfection, filtered through a 0.45-μm-pore-size membrane (Millipore) to remove cell debris, and concentrated by ultracentrifugation at 24,000 rpm for 2 h at 4°C using an SW 41 Ti rotor (Beckman). The pelleted virus particles were analyzed to examine viral proteins using polyclonal rabbit anti-MuLV p15E antibody (a kind gift from Alan Rein, NCI-Frederick) or polyclonal rabbit anti-HIV-1 p24 antibody (NIH AIDS Research and Reference Reagent Program). To detect HIV-1 reverse transcriptase (p66/p51), polyclonal rabbit anti-HIV-1 RT antibody (NIH AIDS Research and Reference Reagent Program) was used.
Resistance testing.
Sensitivity to inhibitors was measured using the PhenoSense assay (44). Briefly, virus stocks were prepared as described for the RC assay, but with the addition of serially diluted PI to the cultures at approximately 16 h posttransfection. The addition of RT inhibitors was done at the time of infection. Virus stocks were collected at 48 h posttransfection and used to infect new 293 cell cultures to determine the percent reduction in infectivity, as measured by residual luciferase activity. Inhibition curves were constructed by fitting these data to a sigmoidal function from which the drug concentration at which 50% of viral infectivity was inhibited (50% effective concentration [EC50]) could be calculated. The fold change (FC) in EC50 (FC EC50) value was determined by comparing the EC50 for the protease mutant virus to the EC50 for a wild-type virus. The FC EC50 values obtained from 7 different protease inhibitors (amprenavir [APV], atazanavir [ATV], indinavir [IDV], lopinavir [LPV], nelfinavir [NFV], ritonavir [RTV], and saquinavir [SQV]) were used to calculate the mean FC EC50 values and standard deviations of each mutant for all inhibitors combined.
RESULTS
Specific infectivity (SpIn) measurements of viruses with single mutations associated with protease inhibitor resistance.
Using a single-cycle SpIn assay, we have measured the effects of point mutations in pro on the average per particle infectivity of HIV-1. To conduct the SpIn assay, the relative number of infectious units present in a transfection supernatant was measured using TZM-bl cells, which express Luc upon productive infection by HIV-1. These values were then divided by the mass of p24 CA protein in the transfection supernatant, measured by an antigen-capture ELISA, to give a ratio of the relative number of infectious units per unit mass of p24. Finally, the Luc/p24 ratio for each mutant virus was divided by the Luc/p24 ratio for the parental wild-type virus, which was measured in parallel. While the in vivo fitness of a virus is impacted by many factors (including the presence of an inhibitor), SpIn specifically reflects the changes in the ability of a virus to complete the infection cycle from target cell entry through viral gene expression. Therefore, mutations that result in a reduction in virus release would not score in this assay.
We were concerned that CA in unprocessed or partially processed Gag polyproteins might not be detected with the same sensitivity as the mature, fully processed p24. To assess whether such a bias exists in our data set, we used silver staining to examine SDS-polyacrylamide gels run with virus lysates containing equivalent masses of p24 measured by ELISA (Fig. 1A). Serial dilutions of lysates from the parental NL4-3 virus and a G48V mutant, which is poorly infectious (see below), were compared. Approximately equivalent staining intensity was detected for the bands corresponding to CA and CA-SP1 (p24 and p25) and to MA (matrix, p17) of the wild type (lanes 1 to 3) and the mutant (lanes 4 to 6), indicating that the estimates of protein mass were not seriously discrepant. However, because of concerns about the ability of the ELISA to measure processing intermediates that may constitute small amounts of the total virion protein, we also used a real-time RT-PCR assay to measure the amount of viral RNA in virus particles. When we compared the SpIn values obtained from two different normalization methods, using either p24 or virion RNA, as can be seen in Fig. 1B, there was a good correlation (r2 = 0.55 and P < 0.0001) between these values, with no systematic differences among different classes of protease mutations. This result again confirms that the SpIn value determined by normalization to the value for p24 measured by ELISA did not introduce a systematic error compared to normalization to the value for virion RNA.
Fig 1.
(A) Silver staining showing that p24 ELISA measures equal amounts of Gag products from the wild-type and protease mutant virion. An SDS-polyacrylamide gel was loaded with equal masses of p24, as determined by antigen-capture ELISA. The band signals were compared for the parental NL4-3 virus (lanes 1 to 3) and the poorly infectious G48V mutant (lanes 4 to 6). Threefold dilutions of each mutant were shown (lanes 1 and 4, 150 ng of p24; lanes 2 and 5, 50 ng of p24; lanes 3 and 6, 17 ng of p24). (B) Correlation analysis between SpIn values measured using the p24 ELISA and SpIn values measured using real-time RT PCR. The r2 value was obtained from the statistical analysis using nonparametric (Spearman) correlation.
The SpIn values normalized by either p24 values or viral RNA values for 31 mutant viruses containing a single mutation in protease are shown in Fig. 2A. Mutations were classified on the basis of their amino acid sequence variability in viral pro sequences obtained from PI-treated and untreated subjects (24). Class 1 mutations are found at a similar frequency in both PI-treated and untreated subjects (I15V, E35D, N37D, I64V), class 2/secondary or compensatory mutations are found in untreated subjects, but their frequency increases in treated subjects (L10I, M36I, I62V, L63P, A71V/T, V77I, I93L), and class 3/primary resistance mutations appear only in treated subjects (K20I/R, D30N, V32I, M46I/L, G48V, I54V, G73S, V82A/T, I84V, N88D/S, L90M). We have included the mutations L24I, I47V, I50V, and F53L in the class 3/primary group because they are selected by PIs (for a review, see reference 22) and are not found in the absence of PI selection (24).
Fig 2.
Infectivities of HIV-1 bearing single resistance-associated mutations in pro measured by two separate single-cycle assays. (A) SpIn assay, in which the amount of viruses used for infection was normalized by measuring either p24 mass (ELISA) or viral RNA (real-time RT-PCR). All the values are normalized to that for the parental NL4-3 strain, which is given an infectivity of 1.0. Data from either two or more than two infection values were averaged, and the infectivity data are shown as means ± standard deviations. Standard deviation values for L10I, K20R, and I50V are not determined. (B) RC assay in which the viruses were pseudotyped with the MuLV amphotropic Env protein. Since a luciferase expression cassette is inserted within a deleted region of the HIV-1 env gene, the ratio of luciferase activity in the infected cells to that in the transfected cells was used to normalize viral infectivity. The variability of the RC assay has been validated to be ±0.2 log.
The effects of mutations at class 1 positions on viral fitness are fairly small compared to the wild-type value. These are the most variable positions in the protease whose variability is not affected by the evolution of resistance, suggesting that substitutions at these positions are likely to have little to no effect on viral replication, consistent with the observation that the SpIn values for these mutants remain close to 1 (Fig. 2A). Similar results were obtained from the class 2/secondary mutations, having the SpIn values close to the level of wild type (Fig. 2A). The only exception was the M36I mutant, which was reduced about 2-fold compared to the wild type. As predicted by their absence without selection by a PI, most mutations at class 3/primary resistance positions in PR resulted in a loss in infectivity. Many of the SpIn values for class 3 mutations were well below 0.8, with the lowest value of 0.12 measured for the I50V mutant. In addition to the I50V mutant, D30N, G48V, F53L, and I54V also showed impaired infectivity with values of about 0.5.
Replication capacity values of viruses with single mutations associated with protease inhibitor resistance.
As a complementary comparison to the SpIn assay, we have also used the RC assay (5, 12, 44), which is a related but alternative single-cycle method for examining the fitness effects of mutations in pro. While the SpIn assay measures the effects of mutations on the early steps of infection for infectious HIV-1, the RC assay measures the infectivity of HIV-1 pseudotyped with the MuLV amphotropic Env protein relative to the luciferase expression in the transfected cells that produce virus, thereby incorporating any potential effects of these mutations on virus production into the final RC value. In addition, RC records any effects of mutations in PR on the processing of the C-terminal tail of the MuLV amphotropic Env protein, a step required for activation of the fusion capacity of the MuLV Env protein (46). We have previously observed for a small number of mutants that RC values can cover a wider range of values than those reported by the SpIn assay (48). The RC values for the full set of mutants described in the SpIn assay are shown in Fig. 2B.
Viruses with class 1 mutations had a wider range of RC values than the corresponding SpIn values, especially for E35D (with a 2-fold decrease). For most of the class 2/secondary mutations, the RC values were either higher than (L10I, M36I, I62V, L63P, V77I) or identical to (A71V, A71T) the SpIn values, with only one exception, where the RC was lower (I93L). Several of the class 2/secondary mutations had RC values greater than 1, suggesting enhanced infectivity in this assay.
More dramatic differences between RC and SpIn assay values were seen in comparing the class 3/primary resistance mutations (Fig. 2A and B), such that an overall comparison of RC and SpIn values for all mutants showed no correlation (P = 0.45; data not shown). Only 3 of the 19 mutants in this class had similar RC and SpIn values (the flap mutants M46I, F53L, and I54V, with a difference between RC and SpIn assay values of less than 0.2), and 1 had a higher RC value (I47V). In all other cases, the RC value was much lower than the SpIn value, indicating that the two assays have very different readouts for the effects of these mutations on infectivity. The mean value for the mutants with class 3 mutations tested in the SpIn assay was 0.7 but was only 0.3 in the RC assay.
In an effort to determine which of these assays more accurately reflects the fitness effects of these protease mutations, we also examined a subset of mutants using a growth competition assay. In this analysis, the wild-type virus and the mutant virus are cocultured and the relative ratio of the two viruses is measured daily as they expand through the culture. To determine the relative fitness, the change in abundance of the mutant relative to the wild type is calculated per day, which is assumed to be the generation time of the virus. When we compared the fitness values determined by the growth competition assay for mutants L63P, M46I, G48V, V82A, I84V, and L90M, we found that the values obtained were more similar to those of the SpIn assay than the RC assay, with values in good agreement with earlier studies (Table 1) (23, 32–34, 50).
Table 1.
Relative fitness values of viruses containing a PI resistance-associated mutation compared to wild type determined using a growth competition assay
| Virus | Relative fitness value (mean ± SD) |
|---|---|
| M46I/WT | 1.00 ± 0.03 |
| G48V/WT | 0.53 ± NDa |
| L63P/WT | 1.02 ± 0.12 |
| V82A/WT | 0.85 ± 0.06 |
| I84V/WT | 0.78 ± 0.04 |
| L90M/WT | 0.98 ± 0.04 |
ND, not determined.
Unprocessed p15E MuLV TM protein contributes to the lower RC values for pseudotyped viruses with class 3 mutations.
In the RC assay, the viruses are pseudotyped with the MuLV amphotropic Env protein, which requires proteolytic cleavage of the transmembrane (TM) protein (p15E) by the virally encoded protease for its fusogenic activation (46). This proteolytic processing generates a mature TM protein (p12E) and a 16-amino-acid R peptide (18, 20). Therefore, it is possible that the effects of the impaired catalytic activity of a mutant HIV-1 protease might be amplified by the requirement for cleavage of the amphotropic Env TM subunit. To test this possibility, we selected 11 mutants based on RC values ranging from wild-type to background levels and pseudotyped the virions by adding an expression vector for the MuLV amphotropic env gene used in the RC assay. Phenotypically mixed virions, containing both the HIV-1 Env protein and the MuLV amphotropic Env protein, were analyzed to examine whether the impaired RC values were associated with unprocessed p15E TM protein in virions. As seen in Fig. 3, the mutants with high RC values (L63P and I47V) showed complete processing of p15E protein; however, the amount of the unprocessed p15E protein in the virion was gradually increased with decreasing RC value (Fig. 3). The p12E protein was not detected for the mutants N88S and N88D, which had RC values close to background. Since the amount of processed CA protein detected from each mutant was not greatly affected by the mutations in the class 3 positions, it is likely that the impaired RC values are the result of the lack of processing of the p15E protein in the virion. The only exception among the mutant viruses that we tested was the G48V mutant, which showed more processed p12E protein than unprocessed p15E protein, although the RC value was only 0.04, presumably a reflection of intermediate Env processing plus a reduced fitness value (Fig. 2A).
Fig 3.
Western blot analysis of the selected protease mutant virus particles pseudotyped with the MuLV amphotropic Env protein. Each infectious HIV-1 clone containing the indicated single resistance-associated mutation in the pro region was used along with the MuLV amphotropic Env plasmids to transfect 293 cells. Culture supernatants were harvested at 48 h after transfection and were subjected to ultracentrifugation to concentrate the viral particles. The pelleted viral particles were detected using either anti-MuLV p15E antibody or anti-HIV-1 CA antibody as the primary antibody. For comparison, MuLV particles and HIV-1 protease active-site mutant (D25A) are shown. Unprocessed p15E protein was not detected from the MuLV particles, and in contrast, only unprocessed p15E protein was detected from the D25A mutant.
Effect of single resistance-associated mutations in HIV-1 PR on PI sensitivity.
The PI sensitivities of viruses bearing each of the mutations described above were measured using the PhenoSense assay (44). In this strategy, we took advantage of the increased sensitivity of the pseudotyped virus to measure small changes in drug sensitivity due to limiting amounts of protease activity. The FC EC50, which is the amount of PI required to inhibit 50% of the infectivity of a mutant virus relative to the amount of drug required to inhibit 50% of the wild-type virus infectivity, was measured for each of seven approved PIs (APV, ATV, IDV, LPV, NFV, RTV, and SQV; Fig. 4; see Table S1 in the supplemental material). In this analysis, we have defined a specific interaction between the inhibitor and the mutant protease as having an FC EC50 greater than 2 standard deviations above the mean of all seven FC EC50 values (indicated by open circles in Fig. 4). We pooled the values that did not reach the definition of specific interaction to assess the overall effect of these mutations on the EC50.
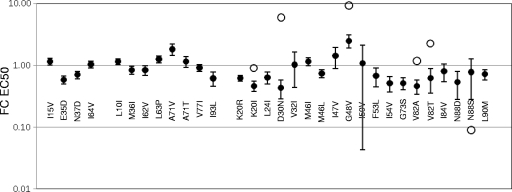
Fig 4.
FC EC50s of mutants to protease inhibitors. The EC50 was measured for each mutant virus with each of the seven approved protease inhibitors (APV, ATV, IDV, LPV, NFV, RTV, and SQV) and compared to the EC50 of the parental virus to calculate the FC EC50. Values are presented on a log scale as the average FC EC50 (filled circles), with error bars indicating the standard deviation of these values. Those FC EC50 values which are greater than 2 standard deviations from the mean of all seven FC EC50s are indicated by open circles (K20I, NFV; D30N, NFV; G48V, SQV; V82A, RTV; V82T, RTV; and N88S, APV), and they are excluded from the calculation of the average FC EC50 and standard deviation.
As was observed for their impact on infectivity, the effects of these mutations on FC EC50 can be generalized by mutation class. The incorporation of either a class 1 or class 2 mutation in pro did not cause a large shift in EC50 (Fig. 4). Furthermore, the FC EC50 responses for these mutants did not vary significantly by drug, as indicated by the small standard deviations of these pooled measurements. Most often, the FC EC50 values for these mutants mirrored their RC values, with mutants that had RC values near 1 showing little change in EC50, while those that had reduced infectivity, such as E35D and I93L, were more sensitive to the PIs. The FC EC50 values for I15V and A71V/T break this trend, as they had higher EC50 values than would have been predicted on the basis of their RC values. Also, the class 2/secondary mutations L10I, L63P, and A71V/T showed a small but largely consistent decrease in drug sensitivity across the protease inhibitors, suggesting that the effects of these mutations were on the protease and not on the interaction between protease and inhibitor.
Most viruses (14 out of 19) bearing mutations at class 3/primary resistance positions exhibited a decrease in FC EC50 (increased PI sensitivity), as shown in Fig. 4 and 5. This suggests that for many mutation/PI combinations there was a sensitization of the protease to inhibition by a PI that outweighed the effect on resistance. There were exceptions to this general trend, with increases in FC EC50 (PI resistance) occurring for expected specific pairings of mutant and PI and with high levels of PI resistance being observed for D30N (which provided a 6-fold increase in EC50 for NFV) and G48V (which provided a 9-fold increase in EC50 for SQV); in addition, G48V conferred decreased sensitivity to all inhibitors, in spite of being one of the less fit proteases. Mutations K20I and V82A/T also conferred significantly higher PI resistance to one PI of the panel (K20I with NFV and V82A/T with RTV). The FC EC50 responses for V32I and I50V with APV approached this cutoff. In contrast, the observed increase in susceptibility for a large number of mutants with class 3 mutations approached or exceeded 2.5-fold (FC EC50, 0.4 or less). At the extreme, the N88S mutant displayed a specific hypersensitive response to APV (open circle in Fig. 4), as reported previously (62). The lower mean values of FC EC50 for the viruses with class 3 mutations compared to the viruses with class 2 mutations are shown in Fig. 5. We interpret the general increase in sensitivity to reflect the fitness loss associated with these mutations. However, the outliers conferring either reduced sensitivity (e.g., D30N, G48V, and V82A/T) or dramatically increased sensitivity (e.g., N88S) reflect direct changes in the interaction between the protease and the inhibitor that confer net resistance (or sensitivity), in spite of the fitness loss. In the absence of a strong specific interaction with the PI, the increase in sensitivity reflects the net loss in catalytic capacity within the virus particle due to the presence of the resistance mutation and the more limited requirement of the PI to titrate down the residual protease activity.
Fig 5.
Comparison of average FC EC50 between class 2 and class 3 mutations. The solid horizontal lines represent the median FC EC50 in each group. The mean FC EC50 values between class 2 and class 3 mutations were compared by statistical analysis using a Mann-Whitney test (nonparametric). The outliers shown as open circles in Fig. 4 are not included in the analysis.
Effect of reduction in protease activity on virion infectivity and inhibitor sensitivity.
We have interpreted the general negative effect of the class 3/primary resistance mutations on virion infectivity as being due to a decrease in total virion-associated protease activity. We modeled this effect by directly reducing the amount of protease activity in the virion. We accomplished this by mixing the infectious molecular clone with an isogenic clone containing an inactivating mutation at the codon encoding the protease active site, D25A. By titrating in an increasing proportion of the protease mutant, we were able to decrease protease activity nonspecifically. Since the protease functions as a homodimer, the fraction of protease dimers that are active is the square of the fraction of the infectious genome compared to the total viral DNA.
The amount of mutant DNA was varied relative to the amount of wild-type DNA, and the infectivity was recorded by both the SpIn assay and the RC assay. As can be seen in Fig. 6A, the two assays had different responses to the inclusion of the mutant DNA. The RC assay showed a linear decrease with increasing amounts of the D25A mutant. In contrast, increasing the amount of the D25A mutant in the SpIn assay resulted in a biphasic reduction in virion infectivity. As shown in Fig. 6D, processing intermediates were apparent in the virion in the presence of 50% of the D25A mutant protease subunits, representing 25% residual protease activity.
Fig 6.
Effect of nonspecific loss of protease activity on viral infectivity and inhibitor sensitivity. (A) Protease activity was reduced in a nonspecific way by transfecting the infectious molecular clone with an isogenic clone containing an inactivating mutation at the protease active site (D25A). Both the fraction of the D25A mutant and the calculated remaining protease activity in each preparation are indicated. The relative infectivity of each mixture was measured using the SpIn (filled circles) and RC (open circles) assays. (B) The effect of nonspecific loss of protease activity on sensitivity to inhibitors of HIV replication was studied by measuring the EC50 for nucleoside reverse transcriptase inhibitors (tenofovir, AZT), nonnucleoside reverse transcriptase inhibitors (EFV), and protease inhibitors (LPV, darunavir [DRV]). Increasing the amount of mutant protease up to 50% (equivalent to 25% residual protease activity) had a systematic effect on the EC50 value for the three classes of antiviral drugs tested. (C) The FC EC50 of 50% D25A relative to the FC EC50 of 0% D25A is shown for all the inhibitors tested. The values of three classes of inhibitors were compared by statistical analysis using a nonparametric Kruskal-Wallis test. The open arrowhead indicates the value from AZT. (D) Western blot analysis showing the extent of processing of p24 and RT in virus particle preparations containing either 100% WT or 50% inactive mutant protease.
We next tested the possibility that the amount of protease activity could affect sensitivity to inhibitors of viral replication by comparing the EC50 for the D25A-containing viruses to the EC50 for the wild-type viruses. As can be seen in Fig. 6B and Table S2 in the supplemental material, increasing the amount of mutant protease up to 50% (equivalent to 25% residual protease activity, the greatest reduction in protease level that could be attained while allowing some residual infectivity in the PhenoSense assay) had a systematic effect on the EC50 value for the three classes of antiviral drugs tested. In the presence of 50% mutant (D25A) protease, the nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs) abacavir (ABC), didanosine (ddI), emtricitabine (FTC), lamivudine (3TC), stavudine (d4T), and tenofovir (TFV) showed only a small effect, with an approximately 20% to 30% decreases in the EC50s. The exceptions to this trend were zalcitabine (ddc), for which there was no change in the EC50, and zidovudine (AZT), for which there was a decrease of nearly 2-fold. The magnitude of change in EC50 for AZT was similar to that seen with the four nonnucleoside RT inhibitors (NNRTIs) delavirdine (DLV), efavirenz (EFV), entecavir (ETV), and nevirapine (NVP), which, again, was in the range of a 2-fold change in the EC50 in the presence of 50% mutant (D25A) protease. An even larger effect was seen among the nine protease inhibitors tested, with decreases in the EC50 ranging from 2.4-fold to 5.3-fold.
These experiments show that by simply decreasing the total amount of protease activity in the virion, we could alter the sensitivity to all three of these classes of inhibitors, with the magnitude of the effect being associated with the inhibitor class and with the protease inhibitors showing the largest effect (Fig. 6C). The pleiotropic effect seen with RT inhibitors is likely the result of the reduced protease activity resulting in a reduced amount of processed RT (Fig. 6D), thus also reducing the amount of total RT activity present in the virion.
DISCUSSION
Previous studies have demonstrated that losses of infectivity are a consequence of resistance-associated mutations in the HIV-1 protease; however, the contributions of individual mutations to changes in fitness have previously been measured for only a subset of these mutations. Therefore, in an effort to acquire a systematic comparison of the effect of HIV-1 PI resistance mutations on viral fitness, in this study the relative fitness and protease inhibitor sensitivity of 31 viruses each bearing a single resistance-associated mutation in the HIV-1 protease were characterized using two different assays, the single-cycle SpIn assay and the RC assay. We found a profound discrepancy in the fitness results between two assays, especially for the mutants with mutations in the class 3 group, where the protease sequence is largely conserved in the absence of a protease inhibitor but becomes variable after selection by a protease inhibitor. Our results demonstrate that the SpIn assay results correlate better with the results of the growth competition assay than the RC assay results do for viruses bearing single PI resistance mutations (Fig. 2 and Table 1). However, in understanding the differences between these assays, we gained insight into the interaction between protease activity, virion infectivity, and inhibitor resistance that highlights the need for multiple enzyme molecules to carry out efficiently the steps of proteolytic processing and DNA synthesis.
The single-cycle SpIn assay is similar to assays described previously (3, 34, 48, 60), in which the infectivity of each virus preparation was measured using a reporter cell line and normalized by the amount of virus used for infection. We used several approaches to show that underprocessing associated with reduced protease activity did not significantly influence our estimation of virion mass in normalizing the different virus preparations. As expected, the fitness values measured by the SpIn assay showed a general trend in the three classes of mutations defining variability in the protease (Fig. 2A). Class 1 mutations, which have similar variability in the sequence database regardless of exposure to protease inhibitor, and class 2 mutations, which are seen in the absence of exposure to protease inhibitors but have increased variability when exposed to these inhibitors, had values close to wild-type level (except for M36I), consistent with these mutations being tolerated within the broad viral population. In contrast, obvious effects on the viral infectivity were seen more frequently with the resistance-associated class 3 mutations, particularly with the flap mutations, G48V, I50V, F53L, and I54V, giving viruses with infectivity values of less than 0.6. Overall, these results are consistent with the fitness values of several mutants reported in the previous studies (32, 50) and collectively provide a much more complete view of the effect of these 31 protease mutations on infectivity. Several studies have reported an increase in fitness associated with some class 3 mutations (32, 50). This can be a feature of a transfection-based approach to generating virus, in that protease may be overproduced in the transfected cells, such that a small reduction in protease activity with either a slightly attenuating mutation, a small amount of a protease inhibitor (data not shown), or a small amount of phenotypic mixing with a mutant with an active-site mutation (Fig. 6A) can give an apparent increase in virion infectivity. We interpret these results as indicating that the SpIn assay slightly overestimates the fitness value in the range of 0.1 or less.
There were differences in the fitness values when the values of the SpIn assay and the RC assay were compared (Fig. 2B). In general, the RC values of the mutants with class 2 mutations were higher than the SpIn values, with mean values of 1.1 and 0.8, respectively, and the RC values of the mutants with class 3 mutations were typically lower than the SpIn values, with mean values of 0.3 and 0.7, respectively. In the RC assay, the viruses are pseudotyped with the MuLV amphotropic Env protein. The cytoplasmic tail of the MuLV TM protein must be cleaved to induce a fusogenic state in the Env protein, a step necessary for MuLV infection. It has been reported that this site is efficiently cleaved by the HIV-1 protease (28), although the cleavage site (VQAL/VLTQ) is suboptimal for HIV-1 protease, with small aliphatic amino acids at P1 and P1′. If cleavage of this heterologous site is actually limiting for infection, then we would predict increases and decreases in infectivity based on both nonspecific changes in protease activity and specific changes in the interaction with the heterologous cleavage site. Consistent with this interpretation, our Western blot analysis demonstrated a strong correlation between the RC values and the level of the processed MuLV TM protein (p15E) in the virion (Fig. 3). The viruses with low RC values contained more unprocessed p15E protein than processed p12E protein and vice versa, suggesting that the infectivity value recorded from the RC assay includes a component that depends on the extent of processing of the MuLV TM protein. Thus, the increase in RC values associated with mutants with class 2 mutations can be viewed as a nonspecific increase in protease activity. Conversely, the low values of the mutants with class 3 mutations represent the reduced activity seen in the SpIn assay. The very low RC values recorded for some mutants likely reflect steric clashes between the protease mutant side chain and the MuLV Env cleavage site, even though some of these mutations (e.g., G73S and N88D) make the PR more MuLV-like (45), indicating the intricate nature of substrate recognition. Conversely, several mutations in the flap, such as M46I and I47V, may enhance the cleavage of this heterologous site. Similarly, the G48V flap mutant generated relatively high levels of processed p12E protein but still had a low RC value, suggesting that the low RC value was the sum of moderate levels of p15E processing and the reduced infectivity associated with this mutation seen in the SpIn assay. Thus, the RC assay represents a complex sum of protease interactions with the heterologous p15E cleavage site and the processing cascade needed for the generation of an infectious HIV-1 core. This suggests that the use of RC data to infer features of viral sequence interactions within the protease on a population level (21) is problematic.
Although the RC assay appears to be limited in its ability to measure absolute fitness changes, the presence of the MuLV cleavage site as a limiting event in virion infectivity can be useful for measuring small changes in protease activity (Fig. 4). When the PI sensitivity of the 31 viruses to 7 different PIs was measured by the PhenoSense assay, mutations at class 1 positions did not provide any PI resistance, as predicted by their lack of positive selection by PI. Likewise, mutations at class 2 positions led to only small changes in EC50s, in agreement with the previous studies (9, 10, 32, 33, 38, 42), with some of the mutants showing a small but consistent decrease in sensitivity across the different inhibitors. This is consistent with the idea that mutants with these mutations have increased protease activity and can partially overcome the effect of the protease inhibitor titrating out protease activity in the virion. In contrast, most of the mutants with class 3 mutations showed increased sensitivity to most of the protease inhibitors, which is consistent with the idea that the reduced activity of these protease mutants makes it easier to titrate out the remaining protease activity by the protease inhibitor to effect a loss of virion infectivity (Fig. 5). The exceptions were the specific/high-level protease inhibitor resistance provided by mutants with class 3 mutations with certain protease inhibitors, in agreement with those reported previously (9, 32, 33). However, it is now clear that the reduced sensitivity to specific PIs is in the background of reduced protease activity that increases sensitivity to PIs in general (Fig. 4). In many cases, the increase in PI susceptibility approached the 2.5-fold (FC EC50 = 0.4) cutoff which has been used to define hypersensitivity (HS) in previous studies (see Table S1 in the supplemental material) (31, 48, 58). The hydrophobic substitutions in flap positions 46, 47, and 48 appear to provide a general resistance to protease inhibitors (albeit at a low level), perhaps by a mechanism that is similar to their ability to enhance interaction with the heterologous p15E substrate (as discussed above). While the analysis of single class 3 mutations reveals their individual impact on protease activity, this approach does not include the contribution that some of these mutations can make in the context of multiple mutations (49) and in the background of class 2 mutations that can compensate for the reduced protease activity to confer high-level resistance to protease inhibitors (34, 38, 53). Our results help bring into focus the dual evolutionary pressures at work that separately select for resistance and fitness.
We were able to model the effect of reduced protease activity by titrating down this activity using a phenotypic mixing strategy with a protease active-site mutation, D25A (Fig. 6). The RC assay was more sensitive to the loss of protease activity than the SpIn assay (Fig. 6A). Consistent with the interpretation that loss in protease activity results in increased sensitivity to protease inhibitors, the virus became increasingly sensitive to these inhibitors with decreasing protease activity (Fig. 6B and C). However, when we tested sensitivity to RT inhibitors, it became apparent that reduced protease activity has pleiotropic effects due to its role in processing the RT and integrase (IN) domains in the Gag-Pro-Pol precursor. The reduced levels of Gag processing that are achieved with reduced protease activity similarly affect the level of processing of RT. Thus, as protease activity is lost, the RT domain is underprocessed, resulting in reduced levels of RT (Fig. 6D). This mechanism could also be at work to further reduce levels of protease; i.e., protease monomers are left in unprocessed or underprocessed Gag-Pro-Pol intermediates.
The effect of reduction of RT activity has previously been studied directly by phenotypic mixing with an RT active-site mutant, and the results obtained (1) were similar to those seen here in the context of reduced protease activity. Specifically, the EC50s for the chain-terminating NRTIs are relatively insensitive to the levels of RT activity (Fig. 6C and D). This is consistent with the idea that while the probability of initiating or completing DNA synthesis is sensitive to the concentration of RT, the probability of incorporating a chain-terminating nucleotide relative to a normal nucleotide is not a function of the RT concentration, which has also been observed for the NRTI inhibitor 3TC (17, 51). The one exception is AZT, for which the EC50 changes with reduced protease/RT activity (Fig. 6C and D) and reduced RT activity (1). This result has previously been seen in the context of protease inhibitor resistance (13). This specific effect with AZT is likely due to the fact that AZT remains in the nucleotide binding site in RT after incorporation and does not translocate to the primer site, in contrast to the other NRTIs (4, 56). In the nucleotide binding site, AZT is susceptible to an excision reaction using ATP which uncaps the growing chain end and allows DNA synthesis to continue (4, 36). The fact that this reaction shows a dependence on RT concentration indicates that the RT molecule that incorporates AZT is not always the same one that excises it; i.e., this can be a distributive action that is dependent on the concentration of active RT. Like the EC50 to AZT, the EC50 to NNRTIs is also sensitive to reduced levels of protease/RT (Fig. 6C and D) and to reduced levels of just RT (1, 25). Since the NNRTIs bind to a pocket in RT to make it enzymatically inactive, the sensitivity of the EC50 to RT concentration is strong evidence that viral DNA synthesis is distributive, involving multiple RT complexes whose availability in the intracellular replication complex is concentration dependent.
The effect of total enzyme activity, either in the number/concentration of enzyme molecules or in the intrinsic activity of each enzyme molecule, displays another feature with respect to interaction with inhibitors. Shen et al. (54) have reported that different classes of inhibitors have different dose-response curves and that at the higher levels of inhibition, protease inhibitors have the most dramatic effect, with NNRTIs showing an intermediate effect and NRTIs the least dramatic effect. This is attributed to the effect of needing multiple enzyme molecules to complete either proteolytic processing or DNA synthesis (55); the ordering of the inhibitors in this analysis is the same as that seen when protease activity is titrated down with an active-site mutant (Fig. 6B and C). Thus, targeting the enzymes in pathways that require multiple enzyme molecules to complete the process (virion assembly/maturation and DNA synthesis) has the added benefit of a dynamic potency, as increasing inhibition reduces the overall probability of completion of the process, thereby enhancing the sensitivity of the residual infectivity to further inhibition. Similarly, the loss of fitness associated with protease inhibitor resistance mutations results in fewer protease molecules (or less protease activity) being available for processing in those virions where processing is successfully completed, again sensitizing the assembly pathway to inhibition by a protease inhibitor (50) (Fig. 6). The pleiotropic effects associated with reduced activity of the viral protease likely account for the persistent benefit that patients can experience even with virologic failure to a protease inhibitor-based regimen (2, 11).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant P01-GM066524. G.J.H. was supported by training grant T32-AI07419 and by fellowship award F30-DA019379. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: U373-MAGI from Michael Emerman and Adam Geballe and TZM-bl from John C. Kappes, Xiaoyun Wu, and Tranzyme Inc.
We thank Dominic Moore for helpful discussions on the statistical analysis of this work. We also thank Jon Condra and Jon Stek for their help in the early stages of this work and Adrienne Swanstrom for assistance in creating some of the mutants.
Footnotes
Published ahead of print 14 November 2011
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Ambrose Z, Julias JG, Boyer PL, Kewalramani VN, Hughes SH. 2006. The level of reverse transcriptase (RT) in human immunodeficiency virus type 1 particles affects susceptibility to nonnucleoside RT inhibitors but not to lamivudine. J. Virol. 80: 2578–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barbour JD, et al. 2002. Evolution of phenotypic drug susceptibility and viral replication capacity during long-term virologic failure of protease inhibitor therapy in human immunodeficiency virus-infected adults. J. Virol. 76: 11104–11112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bleiber G, Munoz M, Ciuffi A, Meylan P, Telenti A. 2001. Individual contributions of mutant protease and reverse transcriptase to viral infectivity, replication, and protein maturation of antiretroviral drug-resistant human immunodeficiency virus type 1. J. Virol. 75: 3291–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boyer PL, Sarafianos SG, Arnold E, Hughes SH. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75: 4832–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Campbell TB, Schneider K, Wrin T, Petropoulos CJ, Connick E. 2003. Relationship between in vitro human immunodeficiency virus type 1 replication rate and virus load in plasma. J. Virol. 77: 12105–12112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Z, et al. 1995. Three-dimensional structure of a mutant HIV-1 protease displaying cross-resistance to all protease inhibitors in clinical trials. J. Biol. Chem. 270: 21433–21436 [DOI] [PubMed] [Google Scholar]
- 7. Colonno R, et al. 2004. Identification of I50L as the signature atazanavir (ATV)-resistance mutation in treatment-naive HIV-1-infected patients receiving ATV-containing regimens. J. Infect. Dis. 189: 1802–1810 [DOI] [PubMed] [Google Scholar]
- 8. Condra JH, et al. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70: 8270–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Condra JH, et al. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374: 569–571 [DOI] [PubMed] [Google Scholar]
- 10. Croteau G, et al. 1997. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J. Virol. 71: 1089–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deeks SG, et al. 2002. CD4+ T cell kinetics and activation in human immunodeficiency virus-infected patients who remain viremic despite long-term treatment with protease inhibitor-based therapy. J. Infect. Dis. 185: 315–323 [DOI] [PubMed] [Google Scholar]
- 12. Deeks SG, et al. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344: 472–480 [DOI] [PubMed] [Google Scholar]
- 13. de la Carriere LC, Paulous S, Clavel F, Mammano F. 1999. Effects of human immunodeficiency virus type 1 resistance to protease inhibitors on reverse transcriptase processing, activity, and drug sensitivity. J. Virol. 73: 3455–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Derdeyn CA, et al. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74: 8358–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Detels R, et al. 1998. Effectiveness of potent antiretroviral therapy on time to AIDS and death in men with known HIV infection duration. Multicenter AIDS Cohort Study Investigators. JAMA 280: 1497–1503 [DOI] [PubMed] [Google Scholar]
- 16. Douek DC, et al. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417: 95–98 [DOI] [PubMed] [Google Scholar]
- 17. Gotte M, Arion D, Parniak MA, Wainberg MA. 2000. The M184V mutation in the reverse transcriptase of human immunodeficiency virus type 1 impairs rescue of chain-terminated DNA synthesis. J. Virol. 74: 3579–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Green N, et al. 1981. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc. Natl. Acad. Sci. U. S. A. 78: 6023–6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gulnik SV, et al. 1995. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry 34: 9282–9287 [DOI] [PubMed] [Google Scholar]
- 20. Henderson LE, Sowder R, Copeland TD, Smythers G, Oroszlan S. 1984. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J. Virol. 52: 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hinkley T, et al. 2011. A systems analysis of mutational effects in HIV-1 protease and reverse transcriptase. Nat. Genet. 43: 487–489 [DOI] [PubMed] [Google Scholar]
- 22. Hirsch MS, et al. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society—USA panel. JAMA 283: 2417–2426 [DOI] [PubMed] [Google Scholar]
- 23. Ho DD, et al. 1994. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J. Virol. 68: 2016–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoffman NG, Schiffer CA, Swanstrom R. 2003. Covariation of amino acid positions in HIV-1 protease. Virology 314: 536–548 [DOI] [PubMed] [Google Scholar]
- 25. Huang W, Gamarnik A, Limoli K, Petropoulos CJ, Whitcomb JM. 2003. Amino acid substitutions at position 190 of human immunodeficiency virus type 1 reverse transcriptase increase susceptibility to delavirdine and impair virus replication. J. Virol. 77: 1512–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaplan AH, et al. 1994. Selection of multiple human immunodeficiency virus type 1 variants that encode viral proteases with decreased sensitivity to an inhibitor of the viral protease. Proc. Natl. Acad. Sci. U. S. A. 91: 5597–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kempf DJ, et al. 2001. Identification of genotypic changes in human immunodeficiency virus protease that correlate with reduced susceptibility to the protease inhibitor lopinavir among viral isolates from protease inhibitor-experienced patients. J. Virol. 75: 7462–7469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kiernan RE, Freed EO. 1998. Cleavage of the murine leukemia virus transmembrane env protein by human immunodeficiency virus type 1 protease: transdominant inhibition by matrix mutations. J. Virol. 72: 9621–9627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kimpton J, Emerman M. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66: 2232–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ledergerber B, et al. 1999. Clinical progression and virological failure on highly active antiretroviral therapy in HIV-1 patients: a prospective cohort study. Swiss HIV Cohort Study. Lancet 353: 863–868 [DOI] [PubMed] [Google Scholar]
- 31. Leigh Brown AJ, et al. 2004. Genetic basis of hypersusceptibility to protease inhibitors and low replicative capacity of human immunodeficiency virus type 1 strains in primary infection. J. Virol. 78: 2242–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mammano F, Trouplin V, Zennou V, Clavel F. 2000. Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug. J. Virol. 74: 8524–8531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Markowitz M, et al. 1995. Selection and analysis of human immunodeficiency virus type 1 variants with increased resistance to ABT-538, a novel protease inhibitor. J. Virol. 69: 701–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martinez-Picado J, Savara AV, Sutton L, D'Aquila RT. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73: 3744–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maschera B, Furfine E, Blair ED. 1995. Analysis of resistance to human immunodeficiency virus type 1 protease inhibitors by using matched bacterial expression and proviral infection vectors. J. Virol. 69: 5431–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meyer PR, Matsuura SE, Mian AM, So AG, Scott WA. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4: 35–43 [DOI] [PubMed] [Google Scholar]
- 37. Molla A, et al. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2: 760–766 [DOI] [PubMed] [Google Scholar]
- 38. Nijhuis M, et al. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13: 2349–2359 [DOI] [PubMed] [Google Scholar]
- 39. Palella FJ, Jr, et al. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N. Engl. J. Med. 338: 853–860 [DOI] [PubMed] [Google Scholar]
- 40. Partaledis JA, et al. 1995. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J. Virol. 69: 5228–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patick AK, et al. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob. Agents Chemother. 42: 2637–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Patick AK, et al. 1996. Antiviral and resistance studies of AG1343, an orally bioavailable inhibitor of human immunodeficiency virus protease. Antimicrob. Agents Chemother. 40: 292–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pazhanisamy S, et al. 1996. Kinetic characterization of human immunodeficiency virus type-1 protease-resistant variants. J. Biol. Chem. 271: 17979–17985 [DOI] [PubMed] [Google Scholar]
- 44. Petropoulos CJ, et al. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44: 920–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rao JK, Erickson JW, Wlodawer A. 1991. Structural and evolutionary relationships between retroviral and eucaryotic aspartic proteinases. Biochemistry 30: 4663–4671 [DOI] [PubMed] [Google Scholar]
- 46. Rein A, Mirro J, Haynes JG, Ernst SM, Nagashima K. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68: 1773–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Resch W, Parkin N, Stuelke EL, Watkins T, Swanstrom R. 2001. A multiple-site-specific heteroduplex tracking assay as a tool for the study of viral population dynamics. Proc. Natl. Acad. Sci. U. S. A. 98: 176–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Resch W, Ziermann R, Parkin N, Gamarnik A, Swanstrom R. 2002. Nelfinavir-resistant, amprenavir-hypersusceptible strains of human immunodeficiency virus type 1 carrying an N88S mutation in protease have reduced infectivity, reduced replication capacity, and reduced fitness and process the Gag polyprotein precursor aberrantly. J. Virol. 76: 8659–8666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rhee SY, et al. 2010. HIV-1 protease mutations and protease inhibitor cross-resistance. Antimicrob. Agents Chemother. 54: 4253–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sampah ME, Shen L, Jilek BL, Siliciano RF. 2011. Dose-response curve slope is a missing dimension in the analysis of HIV-1 drug resistance. Proc. Natl. Acad. Sci. U. S. A. 108: 7613–7618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sarafianos SG, et al. 1999. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc. Natl. Acad. Sci. U. S. A. 96: 10027–10032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schagger H, von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166: 368–379 [DOI] [PubMed] [Google Scholar]
- 53. Schock HB, Garsky VM, Kuo LC. 1996. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. Compensatory modulations of binding and activity. J. Biol. Chem. 271: 31957–31963 [DOI] [PubMed] [Google Scholar]
- 54. Shen L, et al. 2008. Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat. Med. 14: 762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shen L, et al. 2011. A critical subset model provides a conceptual basis for the high antiviral activity of major HIV drugs. Sci. Transl. Med. 3: 91ra63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tong W, et al. 1997. Nucleotide-induced stable complex formation by HIV-1 reverse transcriptase. Biochemistry 36: 5749–5757 [DOI] [PubMed] [Google Scholar]
- 57. Wei X, et al. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46: 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Whitehurst N, Chappey C, Petropoulos C, Parkin N, Gamarnik A. 2003. Polymorphisms in p1–p6/p6* of HIV type 1 can delay protease autoprocessing and increase drug susceptibility. AIDS Res. Hum. Retroviruses 19: 779–784 [DOI] [PubMed] [Google Scholar]
- 59. Wu TD, et al. 2003. Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments. J. Virol. 77: 4836–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zennou V, Mammano F, Paulous S, Mathez D, Clavel F. 1998. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J. Virol. 72: 3300–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang YM, et al. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 71: 6662–6670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ziermann R, et al. 2000. A mutation in human immunodeficiency virus type 1 protease, N88S, that causes in vitro hypersensitivity to amprenavir. J. Virol. 74: 4414–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.