Abstract
The first stage of human immunodeficiency virus type 1 (HIV-1) infection involves the fusion of viral and host cellular membranes mediated by viral envelope glycoprotein gp120. Inhibitors that specifically target gp120 are gaining increased attention as therapeutics or preventatives to prevent the spread of HIV-1. One promising new group of inhibitors is the peptide triazoles, which bind to gp120 and simultaneously block its interaction with both CD4 and the coreceptor. In this study, we assessed the most potent peptide triazole, HNG-156, for inhibitory breadth, cytotoxicity, and efficacy, both alone and in combination with other antiviral compounds, against HIV-1. HNG-156 inhibited a panel of 16 subtype B and C isolates of HIV-1 in a single-round infection assay. Inhibition of cell infection by replication-competent clinical isolates of HIV-1 was also observed with HNG-156. We found that HNG-156 had a greater than predicted effect when combined with several other entry inhibitors or the reverse transcriptase inhibitor tenofovir. Overall, we find that HNG-156 is noncytotoxic, has a broad inhibition profile, and provides a positive combination with several inhibitors of the HIV-1 life cycle. These results support the pursuit of efficacy and toxicity analyses in more advanced cell and animal models to develop peptide triazole family inhibitors of HIV-1 into antagonists of HIV-1 infection.
INTRODUCTION
The global spread of human immunodeficiency virus type 1 (HIV-1), with an annual incidence of 2.6 million cases in 2009, continues to be a serious public health problem and a daunting challenge for the discovery of interventions that can be effective across all human cultures. Among the populations of greatest occurrence and spread, in Africa and Asia, therapeutic drugs such as reverse transcriptase (RT), protease, and integrase inhibitors represent costly options. Currently, only 50% of those medically eligible have access to effective treatment. A vaccine, which would provide an ideal strategy, is not yet available. In the light of these limitations, novel preventatives, such as a topical microbicide or an oral preexposure prophylactic (PrEP), are an urgent goal (13, 37, 51).
HIV-1 entry into host cells has been proposed as an appealing drug target (50). HIV-1 infects macrophages and T cells by fusion of the viral membrane with the target cell membrane (4, 19). The fusion process is mediated by the viral envelope glycoprotein, which is derived from the proteolytic cleavage of a gp160 precursor into the gp120 surface protein and the gp41 transmembrane protein (26, 33, 34, 38). The initial step of cell entry is initiated by the interaction of gp120 with the T-cell antigen receptor CD4 (2, 15, 44). CD4 induces conformational changes in gp120 that are postulated to promote subsequent steps in cell-virus fusion, such as binding to the chemokine coreceptor CCR5 or CXCR4 and the exposure of heptad repeat 1 on gp41 (48, 49). The latter transitions into a gp41 six-helix bundle, ultimately resulting in membrane fusion (6, 28, 36, 55). Interviral contents, including capsid protein p24 and reverse transcriptase, are released into infected cells after fusion.
Recently, a new group of entry inhibitors that allosterically block gp120 interactions has been developed. One such inhibitor is the small peptide 12p1, which antagonizes gp120 interactions with both CD4 and the coreceptor (5, 17, 23, 24). A peptide triazole derivative of 12p1, HNG-156, contains a ferrocenyl triazole-substituted conjugate and binds to monomeric gp120 with an equilibrium dissociation constant (KD) of 7 nM, in contrast to the 2,600 nM KD value of 12p1 (22, 52). Both enzyme-linked immunosorbent assay (ELISA) and surface plasmon resonance (SPR) analyses revealed that HNG-156 retained the ability to inhibit the interaction of gp120 with both CD4 and the coreceptor and inhibited HIV-1BaL entry with a nanomolar 50% effective concentration (EC50) (22). Additionally, a sequence-minimized form of the peptide was found to retain much of its antiviral potency at a substantially reduced size (52).
In this study, we explored the antiviral breadth and combination potential of the peptide triazoles. We tested HNG-156 and a smaller derivative against a panel of subtype B and C isolates of HIV-1 and found that HNG-156 was able to inhibit the majority of the viruses tested, as well as replication-competent clinical isolates. The smaller peptide was also able to inhibit most of the isolates tested, albeit at higher concentrations. Because the most effective treatment for HIV-1 is the use of a cocktail of multiple drugs targeting the virus, we combined HNG-156 with other entry inhibitors as well as with the RT inhibitor tenofovir. We demonstrated that HNG-156 can be paired with any candidate and that it can be favorably combined with many entry inhibitors at the higher concentrations likely to be used as treatment. Overall, we find that HNG-156 is noncytotoxic and effective and has the potential to be developed as a microbicide candidate to stop the spread of HIV-1.
MATERIALS AND METHODS
Reagents and proteins.
The following reagents were obtained from the NIH AIDS Reference and Reagent Program, Division of AIDS, NIAID: griffithsin (GRFT) from B. O'Keefe and J. McMahon; tenofovir, maraviroc, and the HIV-1 IIIB C34 peptide from the Division of AIDS, NIAID; an anti-HIV-1 gp120 monoclonal antibody (2G12) from H. Katinger; HOS.CD4.CCR5 cells and pNL4-3.Luc.R−E− from N. Landau; TZM-bl cells from John C. Kappes, Xiaoyun Wu, and Tranzyme Inc.; pHEF-VSV-G from Lung-Ji Chang; and CHO-ST4.2 cells from D. Littman. The envelope plasmids for QA013.70I.Env.H1 and QF495.23 M.Env.A3 were obtained from J. Overbaugh. The envelope plasmids for 6535.3, QH0692.42, AC10.0.29, PVO.4, and TRO.11 were from D. Montefiori, F. Gao, and M. Li; plasmid pWITO4160.33 was from B. Hahn and J. F. Salazar-Gonzalez; plasmids DU156.12 and DU172.17 were from D. Montefiori, F. Gao, S. Abdool Karim, and G. Ramjee; plasmid DU422.1 was from D. Montefiori, F. Gao, C. Williamson, and S. Abdool Karim; plasmids ZM53M.PB12, ZM109F.PB4, and ZM135M.PL10a were from E. Hunter and C. Derdeyn; and plasmids CAP45.2.00.G3 and CAP210.2.00.E8 were from L. Morris, K. Mlisana, and D. Montefiori. Plasmid CV-N was a gift from Biosyn, Inc. Envelopes encoding the transmitted founder (T/F) viruses ZM246F.C1G, ZM247F fs, and ZM249M.B10.D4 were a gift from G. Shaw. The plasmid for SIVmac239 was a gift from J. Hoxie. Viruses 92RW009 (A1), 97ZA003 (C5), and Uganda Primary D107 were a gift from Eric Arts at Case Western Reserve University (39).
Peptide synthesis and click conjugation.
Peptides were synthesized by manual solid-phase synthesis using 9-fluoroenylmethoxy carbonyl (Fmoc) chemistry on Rink amide resin at a 0.1-mmol scale. The [3 + 2] cycloaddition reaction of azide and terminal alkynes was carried out by an on-resin method (24). All peptides were purified using reverse-phase high-pressure liquid chromatography (HPLC), and their intended sequences were validated by mass spectrometry. The peptides were cleaved from the resin by using a cocktail mixture of trifluoroacetic acid–1,2-ethanedithiol–water–thioanisole (95:2:2:1). Crude peptides were purified by HPLC (Beckman Coulter) using a C18 column with a water–acetonitrile–trifluoroacetic acid gradient between 95:5:0.1 and 5:95:0.1. Peptide identity was confirmed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry, and purity was confirmed by analytical reverse-phase HPLC.
Cell culture.
Human embryonic kidney 293T cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM; MediaTech) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Clontech), 2 mM l-glutamine (Invitrogen), 25 mM HEPES (Invitrogen), 100 U/ml of penicillin, and 0.1 mg/ml of streptomycin (Invitrogen). Human osteosarcoma (HOS) cells stably expressing human CD4 and CCR5 or CXCR4 receptors were maintained with the complete medium described for the 293 cells supplemented with 1 μg/ml of puromycin (Sigma-Aldrich). TZM-bl cells (41, 42) were grown in DMEM (Sigma-Aldrich; United Kingdom) supplemented with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin and were passaged twice per week with 1× trypsin-EDTA (Sigma-Aldrich; United Kingdom).
sCD4 production.
Soluble CD4 (sCD4) was produced as described previously (52). Briefly, CHO-ST4.2 cells, which secrete the entire extracellular domain of CD4, were grown in a hollow-fiber bioreactor (FiberCell Systems Inc.). The supernatant was fractionated over a sulfopropyl-substituted (SP) ion-exchange column (GE Healthcare) followed by a quaternary ammonium-substituted (Q) ion-exchange column (GE Healthcare) with an AKTA fast protein liquid chromatography (FPLC) instrument (GE Healthcare). The column flowthrough containing purified CD4 was pooled and dialyzed into 1× phosphate-buffered saline (PBS) (pH 7.4) overnight at 4°C. All proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining (Invitrogen) and were found to be >95% homogeneous.
CVN production.
Cyanovirin-N (CVN) was produced as described previously (35). In brief, BL21 CodonPlus (DE3) RIL competent cells (Stratagene) expressing CVN were grown in Superbroth supplemented with 1 mM MgSO4, 0.5% glucose, and 25 μg/ml kanamycin. Protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h at 37°C. The cells were then centrifuged at 3,000 rpm, and protein was isolated from the periplasmic space by osmotic shock. The protein was fractionated over nickel-nitrilotriacetic acid (NTA) agarose beads (Qiagen) by using the manufacturer's protocol. The column flowthrough containing purified CVN was pooled and dialyzed into 1× PBS (pH 7.4) overnight at 4°C. The protein was determined by silver staining to be >95% pure.
Pseudovirus production and inhibition using a single inhibitor.
Envelope-pseudotyped luciferase reporter viruses were produced utilizing envelope expression vectors cotransfected into 293T cells with calcium phosphate precipitation (Profection mammalian transfection system; Promega) or Fugene reagent (Qiagen) together with the envelope-deficient pNL4-3-luc+env− provirus developed by N. Landau (10). Culture supernatants containing pseudotyped particles were collected 48 to 72 h after transfection, clarified by centrifugation, aliquoted, and stored at −80°C until use. The titers of viruses were determined on HOS.CD4.R5 cells, and normalized concentrations were used for all experiments.
For inhibition experiments, the pseudotype stocks were first incubated with the inhibitor at 37°C for 30 to 45 min. The virus-inhibitor mixture was then added to the target cells and was returned to a 37°C incubator with 5% CO2. Assays with the panel of subtype B and C envelopes were performed in the presence of 1.5 μg/ml DEAE-dextran. At 48 to 72 h postinfection, these cells were washed with PBS and were lysed using a combination of Passive Lysis buffer (Promega) and freeze-thaw cycles. The lysate was then combined with luciferase buffer (15 mM KPO4 [pH 7.8], 15 mM MgSO4, 1 mM ATP, 1 mM dithiothreitol [DTT]) and d-luciferin salt. Luciferase activity was detected using a 1450 Microbeta Jet liquid scintillation and luciferase counter (Wallac). Studies using 92UG037, 96ZM65, and 92BR025 envelopes were performed as described above, except that luciferase activity was detected using a Luciferase assay system (Promega).
Inhibition of infection with replication-competent HIV-1.
TZM-bl cells (41, 42) (5 × 104 cells/well), cultured overnight, were pretreated with HNG-156 for 1 h prior to exposure either to HIV-1BaL or to one of three CCR5-utilizing HIV-1 clades designated 92RW009 (A1), 97ZA003 (C5), and D107 (Uganda Primary) in the presence of the compound. After 24 h, cells were washed, lysed, and frozen prior to the determination of luciferase expression (Stratagene, United Kingdom). Viral stocks were quantified by 50% tissue culture infective dose (TCID50) titrations on TZM-bl cells.
Inhibition using inhibitor combinations.
The compounds were combined at a fixed ratio based on their half-maximal effective concentrations (EC50s) and were added to luciferase-expressing HIV-1BaL for 30 min at 37°C. The mixture was then added to HOS cells expressing CD4 and the appropriate coreceptor and was incubated at 37°C under 5% CO2. After 48 to 72 h, the cells were washed with PBS and were lysed with Passive Lysis buffer (Promega) combined with three freeze-thaw cycles. The lysate was combined with luciferase buffer (15 mM KPO4 [pH 7.8], 15 mM MgSO4, 1 mM ATP, 1 mM DTT) and d-luciferin salt. Luciferase activity was detected using a 1450 Microbeta Jet liquid scintillation and luciferase counter (Wallac).
The results of the combination studies were analyzed using CalcuSyn software (Biosoft). This software uses the median-effect equation developed by Chou and Talalay (8, 9) to create dose-effect curves for the drugs alone or in combination. The x intercept and slope of the curves are then used to calculate the combination index (CI) for the inhibitor combinations. We used the following equation for non-mutually nonexclusive drugs: CI = (D1/Dx1) + (D2/Dx2) + [(D1)(D2)/(Dx1)(Dx2)], where D1 is the dose of drug 1 in combination, D2 is the dose of drug 2 in combination, Dx1 is the dose of drug 1 alone, and Dx2 is the dose of drug 2 alone.
A CI value of 0.3 to 0.7 indicates synergism; 0.7 to 0.85 indicates moderate synergism; 0.85 to 0.9 indicates slight synergism; 0.9 to 1.1 indicates additivity; and >1.1 indicates antagonism. The dose reduction index (DRI) was calculated as Dx/D, where D is the dose of the drug in combination and Dx is the dose of the drug alone.
In vitro cytotoxicity.
Cytotoxicity assays were performed under conditions identical to those used for the viral inhibition assay. HOS.CD4.CCR5 cells were seeded 24 h in advance of the addition of the HNG-156 at twofold dilutions starting at 500 µM. The inhibitor solution remained on the cells for 24 h prior to the removal of the inhibitor and the addition of fresh medium. The cells were subsequently washed and assessed for viability using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay of viability (described in reference 29). Concentrations were tested in three independent assays.
RESULTS
Range of activity of the peptide triazoles.
HNG-156 has been shown previously by both single-round and replication-competent virus assays to inhibit infection by HIV-1BaL, a subtype B isolate (22, 52). However, patients in the regions of the world most desperately in need of a preventative against HIV-1, such as sub-Saharan Africa, are infected primarily with subtype C or A strains of the virus. We sought to expand on our previous analysis by including a wider array of strains and subtypes of HIV-1 so as to assess the usefulness of the peptide against more globally relevant viruses. We initially performed a recombinant single-round assay using subtype B (31, 53, 54) and subtype C (12, 32, 56) reference panels from the NIH AIDS Reagent and Reference Repository as well as several other individual subtype A, B, and C envelopes. HNG-156 was able to inhibit subtype B viruses in the range of 0.02 to 33 μM and some subtype C viruses in the range of 11 to 36 μM (Table 1). Other subtype C viruses (CAP45.2, ZM53M, 96ZM651, and 92BR025) were inhibited at higher, although still quantifiable, EC50s. The peptide inhibited subtype A viruses less potently, with EC50s ranging from 45 to 84 μM (Table 1). HNG-156 was unable to inhibit viruses pseudotyped with an envelope from vesicular stomatitis virus glycoprotein (VSV-G) or simian immunodeficiency virus (SIV) (Table 1) (52), indicating that its inhibitory action is specific to the HIV-1 envelope.
Table 1.
Antiviral activities for HNG-156 and UM-24
| Subtype/coreceptor | Virus name | EC50 (μM)a |
|
|---|---|---|---|
| HNG-156 | UM-24 | ||
| A/R5 | 92UG037.8 | 65 ± 3 | ND |
| A/R5 | QF495.23M.Env.A3 | 44.6 ± 1.5 | >100 |
| B/R5 | BaL | 0.58 ± 0.1 | 8.4 ± 0.8 |
| B/R5 | 6535.3 | 0.02 ± 0.008 | 1.1 ± 0.37 |
| B/R5 | QH0692.42 | 0.32 ± 0.09 | 1.0 ± 0.19 |
| B/R5 | WITO4160.33 | 7.7 ± 0.96 | 29.5 ± 7.8 |
| B/R5 | AC10.0.29 | 21.6 ± 3.2 | 53 ± 4 |
| B/R5 | PVO.4 | 33.3 ± 7.4 | 124 ± 9.3 |
| B/R5 | TRO.11 | 29.3 ± 8.6 | 109 ± 9.8 |
| C/R5 | ZM109F.PB4 | 14.1 ± 3.0 | 58.3 ± 9.4 |
| C/R5 | DU172.17 | 11.5 ± 4.3 | 25.7 ± 2.2 |
| C/R5 | CAP210.2.00.E8 | 21.5 ± 1.6 | 60.8 ± 5.3 |
| C/R5 | DU422.1 | 24.3 ± 1.6 | 49.3 ± 2.0 |
| C/R5 | DU156.12 | 36 ± 4.9 | 89.8 ± 5.3 |
| C/R5 | ZM135 M.PL10a | 36 ± 6.2 | 42 ± 2.6 |
| C/R5 | CAP45.2.00.G3 | 58.5 ± 19.4 | >100 |
| C/R5 | 96ZM651.8 | 47.2 ± 2.9 | ND |
| C/R5 | ZM53 M.PB12 | 52 ± 12.0 | >200 |
| C/R5 | 92BR025 | 61.2 ± 14.7 | ND |
| C/R5 | ZM246F.C1Gb | 14.6 ± 1.2 | 60.9 ± 6.8 |
| C/R5 | ZM247F fsb | 5.9 ± 0.9 | 22.7 ± 2.4 |
| C/R5 | ZM249M.B10.D4b | 60.4 ± 3.8 | >100 |
| D/R5 | QA013.70I.Env.H1 | 30.9 ± 2.6 | 24.7 ± 0.6 |
| SIVmac339 | >100 | >100 | |
| VSV-G | >100 | >100 | |
Determined based on the infection of HOS.CD4.CCR5 or HOS.CD4.CXCR4 target cells with recombinant luciferase-expressing HIV-1. All values were calculated using Origin, version 8.0 (OriginLab), by fitting experimental data to a sigmoidal inhibition curve. Experimental deviation was calculated as the standard error of the mean. All assays were repeated a minimum of three independent times in triplicate wells. ND, not determined.
Transmitted or founder virus.
Given these promising results, we next determined whether a truncated version of HNG-156, UM-24, could also inhibit this expanded panel of viruses. This seven-residue peptide was created to elucidate the active core of HNG-156 (52) and inhibits HIV-1BaL with a roughly 15-fold reduction in potency. UM-24 was able to inhibit most subtype B viruses tested (Table 1) with EC50s in the range of 1 to 53 μM. However, it had greatly diminished potency against 2 strains that the larger compound HNG-156 was able to inhibit (PVO and TRO.11) (Table 1). UM-24 was able to inhibit 6 of 8 subtype C strains of HIV-1 tested. While UM-24 was less potent than HNG-156, it also behaved more consistently against the different strains against which it was tested. For example, against comparable subtype C viruses, HNG-156 showed a 19- to 100-fold decrease in efficacy, while UM-24 showed a 2- to 10-fold decrease. Additionally, even in cases where the EC50 of UM-24 was higher than 100 μM, there was still a clear-cut trend toward inhibition of infection.
It has been estimated that infection by a single transmitted or early founder (T/F) virus occurs in 60 to 90% of cases of heterosexual transmission (27, 46, 47). Therefore, we assessed the abilities of both HNG-156 and UM-24 to inhibit viruses pseudotyped with envelopes from the subtype C T/F viruses ZM246F, ZM247F, and ZM249M (Table 1). HNG-156 was able to inhibit all three viruses with EC50s ranging from 6 to 60 μM, while UM-24 was able to inhibit two of the three viruses. These values are comparable to, or slightly better than, the overall efficacy that we observed with the peptide triazoles against subtype C viruses.
Inhibition of replication-competent HIV-1.
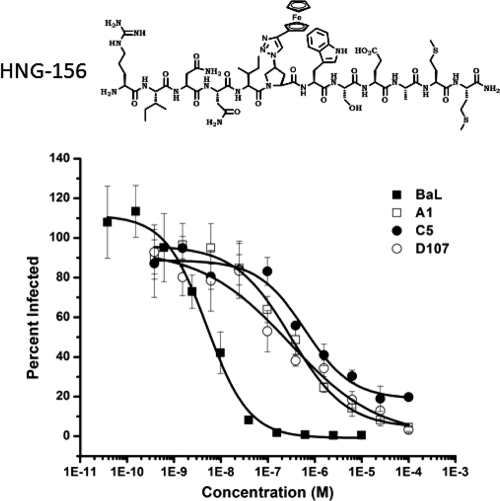
HNG-156 was then tested for its ability to inhibit infection of TZM-bl cells (41, 42) by fully infectious clinical isolates representing subtype A, B, C, or D (39). The peptide was able to inhibit all of the viruses tested (Fig. 1; Table 2), with EC50s in the range of 0.005 to 0.65 μM. It should be noted that HNG-156 was substantially more potent against fully infectious viruses than it was against single-round recombinant viruses, with EC50s for HIV-1BaL of 5 nM and 0.6 μM, respectively. This is consistent with previous reports (22, 52) that demonstrated increased potency against replication-competent viruses. This discrepancy cannot be attributed to differences in cell lines, since the single-round experiment was repeated with the TZM-bl cell line used in the replication-competent virus assay, with similar results (data not shown).
Fig 1.
Inhibition of replication-competent viruses by HNG-156. (Top) Structure of HNG-156 (22). (Bottom) TZM-bl cells were treated with HNG-156 for 1 h prior to the addition of the indicated virus and were then cultured in the presence of the compound. Viral infection was detected by the luciferase activity present in the culture lysates. The data were normalized to the signal from uninhibited virus, which was set at 100%, and were fit to a logistic curve by Origin, version 8.0. The data shown are means ± standard errors of the means for three independent experiments, where each condition was tested in triplicate.
Table 2.
Antiviral potencies of HNG-156 against replication-competent viruses
| Subtype/tropism | Virus name | EC50 (μM)a |
|---|---|---|
| A/R5 | A1 | 0.35 ± 0.11 |
| B/R5 | BaL | 0.005 ± 0.0004 |
| C/R5 | C5 | 0.65 ± 0.09 |
| D/R5 | D107 | 0.17 ± 0.07 |
Determined based on the infection of TZM-bl target cells with replication-competent HIV-1. See Table 1, footnote a, for data treatment. Experimental deviation was calculated as the standard error of the mean. All assays were repeated a minimum of three independent times in triplicate wells.
HNG-156 toxicity studies.
We assessed the toxicity of HNG-156 by using both P4.CCR5 and HOS.CD4.CCR5 cells under the conditions used to determine antiviral potency. No toxicity was observed in P4.CCR5 cells at an HNG-156 concentration of 50 μM for 2 h of continuous exposure or in HOS.CD4.CCR5 cells at concentrations as high as 500 μM for 24 h of continuous exposure (data not shown). These results were subsequently used to estimate an in vitro therapeutic index (TI), which measures the HNG-156 concentration that results in 50% cytotoxicity (CC50) over the concentration needed for 50% efficacy (TI = CC50/EC50). Since the peptide exhibited no toxicity at any doses tested, we can extrapolate a TI value of >600 for P4-CCR5 cells and >850 for HOS.CD4.CCR5 cells.
Combination of HNG-156 with inhibitors that target viral entry.
An effective preventative for HIV-1 will likely contain a combination of inhibitors to improve the efficacy of the treatment and to reduce risk from developing resistance mutations. Ideally, this combination will contain two compounds that cooperatively inhibit HIV-1 infection to produce a potency greater than that expected from either compound alone. All compounds were first assessed separately to determine the dose-response activity profile of each agent in inhibiting the infection of HOS.CD4.CCR5 cells by HIV-1BaL. The EC50s calculated from these results were used to establish a combination molar ratio. Compounds were compared either alone or in combination, and the results were analyzed using CalcuSyn software (8, 9).
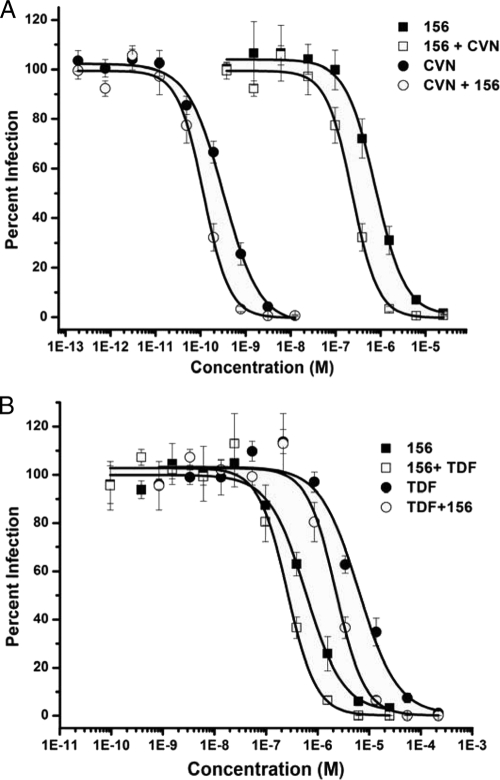
To ensure that HNG-156 could be used with newly emerging entry inhibitors, we combined it with inhibitors that target each stage of the entry process, as well as several carbohydrate binding proteins that target gp120. For entry inhibitors, we used sCD4 (11, 18) and maraviroc (14), which block the interaction of gp120 with CD4 and the coreceptor, respectively. We used C34 (20) as an inhibitor of fusion. Combinations of HNG-156 with sCD4 or C34 were additive at both the EC50 and the EC90 (Table 3), indicating that the use of HNG-156 in combination with either of these inhibitors would have no detrimental effect. When HNG-156 was combined with the coreceptor antagonist maraviroc, it demonstrated a CI value of 0.56 at the EC90, indicative of synergy (Table 3). This corresponds to a 4- to 6-fold dose reduction (Table 4). This result was confirmed with combinations of HNG-156 and TAK-779 (Tables 3 and 4), which also exhibited synergy at the EC90 but a more muted effect at the EC50. It should be noted that both maraviroc and TAK-779 block the interaction of gp120 with the coreceptor by binding directly to CCR5, in contrast to HNG-156, which binds to gp120. Currently, all effective coreceptor antagonists function by targeting the chemokine receptor itself, and therefore, we did not test combinations of HNG-156 with gp120 binding coreceptor antagonists. Finally, combinations of HNG-156 with the lectin cyanovirin-N or griffithsin, or with the high-mannose binding antibody 2G12, were synergistic at both the EC50 and the EC90 (Fig. 2A; Tables 3 and 4). High mannose refers to the type of carbohydrate unit containing 5 to 9 mannose residues and no sialic acid.
Table 3.
CI values for HNG-156 in combination with other inhibitors
| Combinationa | EC50 |
EC90 |
||
|---|---|---|---|---|
| CI value | Conclusionb | CI value | Conclusion | |
| HNG-156 + sCD4 | 1.03 ± 0.24 | Additivity | 0.95 ± 0.17 | Additivity |
| HNG-156 + MVC | 0.83 ± 0.22 | Moderate synergy | 0.56 ± 0.13 | Synergy |
| HNG-156 + TAK | 0.87 ± 0.17 | Slight synergy | 0.56 ± 0.05 | Synergy |
| HNG-156 + C34 | 1.10 ± 0.36 | Additivity | 0.84 ± 0.03 | Moderate synergy |
| HNG-156 + CVN | 0.60 ± 0.10 | Synergy | 0.77 ± 0.14 | Moderate synergy |
| HNG-156 + GRFT | 0.57 ± 0.24 | Synergy | 0.45 ± 0.06 | Synergy |
| HNG-156 + 2G12 | 0.64 ± 0.08 | Synergy | 0.61 ± 0.12 | Synergy |
| HNG-156 + TDF | 1.00 ± 0.40 | Additivity | 0.62 ± 0.10 | Synergy |
MVC, maraviroc; TAK, TAK-779; CVN, cyanovirin-N; GRFT, griffithsin; TDF, tenofovir.
The drug interactions of the combinations were analyzed by the combination index (CI) analysis developed by Chou and Talalay (8, 9) using CalcuSyn software. A CI value of 0.3 to 0.7 indicates synergism; 0.7 to 0.85, moderate synergism; 0.85 to 0.9, slight synergism; 0.9 to 1.1, additivity; and >1.1, antagonism (8, 9). The compounds were combined at fixed ratios based on their EC50s and were mixed with HIV-1BaL for 30 min prior to addition to HOS.T4.R5 cells. All assays were repeated in a minimum of three independent trials in triplicate wells. Experimental deviation was calculated as the standard error of the mean.
Table 4.
Dose reductions for inhibition of HIV-1 by HNG-156 combinations at the EC50 and EC90
| Combinationa | EC50 (nM)b for: |
DRIc at EC50 | EC90 (nM)b for: |
DRI at EC90 | ||
|---|---|---|---|---|---|---|
| Drug alone | Combination | Drug alone | Combination | |||
| HNG-156 + sCD4 | ||||||
| HNG-156 | 1,176 ± 237 | 325 ± 65 | 3.3 ± 0.7 | 5,484 ± 1,735 | 2,155 ± 541 | 3.2 ± 1.1 |
| sCD4 | 10.2 ± 1.4 | 4.2 ± 0.7 | 2.5 ± 0.3 | 41.2 ± 8.8 | 24.0 ± 6.1 | 1.9 ± 0.3 |
| HNG-156 + MVC | ||||||
| HNG-156 | 692 ± 61 | 261 ± 51 | 3.8 ± 1.2 | 4,435 ± 568 | 1,543 ± 320 | 3.9 ± 1.2 |
| MVC | 0.99 ± 0.2 | 0.25 ± 0.05 | 4.8 ± 1.3 | 7.7 ± 1.2 | 1.5 ± 0.3 | 5.9 ± 1.3 |
| HNG-156 + TAK | ||||||
| HNG-156 | 713 ± 131 | 316 ± 103 | 2.8 ± 0.4 | 9,741 ± 3,190 | 2,099 ± 512 | 3.4 ± 2.5 |
| TAK | 16.8 ± 4.0 | 6.0 ± 2.0 | 2.8 ± 0.5 | 101.8 ± 13.5 | 31.5 ± 7.7 | 4.5 ± 0.6 |
| HNG-156 + C34 | ||||||
| HNG-156 | 753 ± 174 | 298 ± 63 | 3.0 ± 0.5 | 3,576 ± 470 | 1,503 ± 155 | 2.4 ± 0.2 |
| C34 | 2.2 ± 0.3 | 0.60 ± 0.13 | 4.6 ± 1.2 | 12.1 ± 1.5 | 3.0 ± 0.3 | 4.3 ± 0.7 |
| HNG-156 + CVN | ||||||
| HNG-156 | 742 ± 90 | 177 ± 37 | 6.9 ± 2.0 | 4,540 ± 498 | 1,069 ± 181 | 5.5 ± 1.0 |
| CVN | 0.44 ± 0.13 | 0.11 ± 0.02 | 7.1 ± 3.1 | 1.8 ± 0.36 | 0.66 ± 0.13 | 3.3 ± 0.7 |
| HNG-156 + GRFT | ||||||
| HNG-156 | 0.61 ± 0.11 | 0.15 ± 0.06 | 3.6 ± 0.3 | 4,516 ± 1,022 | 989 ± 220 | 4.5 ± 1.2 |
| GRFT | 0.06 ± 0.03 | 0.01 ± 0.005 | 4.5 ± 1.5 | 0.66 ± 0.10 | 0.10 ± 0.06 | 10.9 ± 5.0 |
| HNG-156 + 2G12 | ||||||
| HNG-156 | 652 ± 120 | 211 ± 19 | 3.1 ± 0.3 | 3,524 ± 1,193 | 1,147 ± 274 | 2.9 ± 0.3 |
| 2G12 | 1.3 ± 0.22 | 0.28 ± 0.02 | 4.9 ± 1.3 | 11.2 ± 4.4 | 1.5 ± 0.33 | 10.0 ± 6.4 |
| HNG-156 + TDF | ||||||
| HNG-156 | 707 ± 112 | 287 ± 78 | 3.2 ± 1.0 | 7,110 ± 2,191 | 1,192 ± 139 | 6.7 ± 2.8 |
| TDF | 6,824 ± 448 | 2,530 ± 689 | 3.6 ± 1.2 | 43,679 ± 19,239 | 10,489 ± 1,219 | 3.8 ± 1.2 |
MVC, maraviroc; TAK, TAK-79; CVN, cyanovirin-N; GRFT, griffithsin; TDF, tenofovir.
Determined on the basis of infection of HOS.CD4.CCR5 target cells by recombinant luciferase-expressing HIV-1. See Table 1, footnote a, for data treatment.
Dose reduction index (DRI) values for each inhibitor within the combination were calculated using CalcuSyn software.
Fig 2.
Combinations of HNG-156 with CVN (A) or tenofovir (TDF) (B). Serial dilutions of HNG-156, CVN, or TDF, alone or in combination, were added to HIV-1BaL for 30 min prior to addition to HOS.CD4.CCR5 cells. The data were normalized to an uninhibited control, which was set to 100% infected, and thus represent a reduction in infection relative to the control. Data were fit to a logistic model using Origin, version 8.0. Curves representing the combined inhibitors were plotted based on the concentration of HNG-156, CVN, or tenofovir. The data shown are means ± standard errors of the means for three independent experiments, where each condition was tested in triplicate.
Combination of HNG-156 and tenofovir.
Tenofovir has demonstrated some preliminary success as a topical microbicide (1), as part of oral preexposure prophylaxis (PrEP) (25), and as a component of antiretroviral therapy (43). We therefore wanted to determine whether HNG-156 could potentially be used in combination with this compound. The two inhibitors target different viral proteins and different stages of the HIV-1 life cycle, and we hypothesized that they should not exhibit any degree of cross-resistance. We found that HNG-156 was additive with tenofovir at the EC50 and synergistic at the EC90 (Fig. 2B; Tables 3 and 4). This corresponded to dose reductions of 4-fold for tenofovir and 7-fold for HNG-156 at the EC90.
Inhibition of transmitted founder viruses by HNG-156 combinations.
We next wanted to determine whether the favorable combination effect observed against HIV-1BaL could also be seen in other strains of HIV-1, such as the T/F viruses. For this analysis, we chose combinations of HNG-156 with CVN or tenofovir (Table 5). We found that HNG-156 and CVN were synergistic at all doses against strain ZM247F fs, while the effect against ZM246F.C1G was more muted, with moderate synergism to additivity. HNG-156 and tenofovir were moderately synergistic against ZM247F fs and moderately synergistic to additive against ZM246F.C1G.
Table 5.
CI values for 156 combinations against transmitted founder viruses
| Drug combinationa and virus | EC50 |
EC90 |
||
|---|---|---|---|---|
| CI valueb | Conclusion | CI value | Conclusion | |
| HNG + CVN | ||||
| ZM246F.C1G | 0.72 ± 0.14 | Moderate synergy | 0.97 ± 0.09 | Additivity |
| ZM247F fs | 0.65 ± 0.05 | Synergy | 0.57 ± 0.11 | Synergy |
| HNG + TDF | ||||
| ZM246F.C1G | 0.94 ± 0.14 | Additivity | 0.73 ± 0.06 | Moderate synergy |
| ZM247F fs | 0.81 ± 0.03 | Moderate synergy | 0.74 ± 0.10 | Moderate synergy |
CVN, cyanovirin-N; TDF, tenofovir.
The drug interactions of the combinations were analyzed by the combination index (CI) analysis developed by Chou and Talalay (8, 9) using CalcuSyn software. A CI value of 0.3 to 0.7 indicates synergism; 0.7 to 0.85, moderate synergism; 0.85 to 0.9, slight synergism; 0.9 to 1.1, additivity; and >1.1, antagonism. The compounds were combined at fixed ratios based on their EC50s and were mixed with HIV-1ZM246F or HIV-1ZM247F for 30 min prior to addition to HOS.T4.R5 cells. All assays were repeated in a minimum of three independent trials. Experimental deviation was calculated as the standard error of the mean.
DISCUSSION
There is an urgent need for novel compounds that target HIV-1-specific proteins and prevent infection. HNG-156 is a small peptide that binds to gp120 and prevents viral entry by a novel mechanism that includes blocking the interaction of gp120 with both of its receptors. In this study, we explored the potential of HNG-156 by assessing its efficacy, cytotoxicity, and ability to be combined with other inhibitors. We demonstrated that HNG-156 is able to inhibit a broad range of viruses, including the globally relevant subtype C strains. We also showed that HNG-156 can be combined with other antiretrovirals and should be considered for development as a topical microbicide.
An effective preventative against HIV-1 will need to inhibit subtypes of the virus that are prevalent in the regions of the world, such as sub-Saharan Africa, that have the greatest incidence of infection. We therefore tested HNG-156 and its smaller derivative UM-24 against a panel of subtype C isolates. HNG-156 was able to inhibit all of the strains tested, including transmitted or early founder viruses. The significantly smaller compound UM-24 was able to inhibit 6 of 8 strains over the dose range tested, and the dose inhibition curves for the two outliers suggested that infection with these viruses would have been inhibited over a broader dose range. Interestingly, while UM-24 is less potent than the larger compound HNG-156, it exhibited less variation in the extent of inhibition of viruses pseudotyped with different envelopes. One possible explanation for this is that the binding site for the larger peptide triazoles could be partially occluded or disrupted in some of the subtype C isolates. The substantially smaller compound UM-24 may be able to access its binding site more easily than HNG-156. This result is consistent with the possibility that smaller peptidomimetics of peptide triazoles could viably access a common, localized neutralization site within the peptide triazole binding region.
Given the promising results obtained using pseudotyped virus, we expanded our study to include replication-competent clinical isolates of HIV-1. We found that HNG-156 was able to inhibit all of the viral isolates tested. Interestingly, HNG-156 was significantly better at inhibiting replication-competent virus than pseudotyped viruses, as exemplified by the nearly 100-fold reduction in the level of HIV-1BaL in this study. The results obtained from the replication-competent virus studies closely mirror the calculated KD for HNG-156 (22, 52), 5 nM versus 7 nM, respectively. We confirmed that this result was not due to the TZM-bl cell line used in the replication-competent virus experiment (data not shown). These results were also observed in previous work (22, 52), which demonstrated that HNG-156 is more potent against replication-competent viruses. We speculate that this could be due to a higher number of envelope spikes on the single-round virus than on fully infectious virus. The number of envelope spikes on a replication-competent virus is predicted to be 7 to 14 (7, 30, 57), whereas we believe that the pseudoviral preparations used in this study have significantly greater concentrations of both free and virally incorporated envelope. The differences between the plasmid types and backbone-to-envelope ratios used in different laboratories make it difficult to draw a global conclusion about how much envelope will be incorporated into a given pseudoviral preparation. Additionally, the preparations used in this study were not purified and likely contain an elevated concentration of unincorporated envelope. In support of this, when we ran our pseudoviral preparations over a density gradient, we observed substantial concentrations of both free and virally incorporated gp120.
HNG-156 is believed to bind to a location close to, but not overlapping, the CD4 binding site. Current research on HNG-156 suggests that strong resistance is conferred by D474A and T257A substitutions on gp120. This result was demonstrated by both single-round inhibition experiments and the loss of direct binding determined by surface plasmon resonance (SPR) (F. Tuzer, unpublished data). While T257 is conserved among the envelopes tested, a natural substitution of D474N is commonly found in many of the envelopes used in this study. There appeared to be no correlation between the concentration of peptide needed for 50% inhibition and the presence of either asparagine or aspartic acid at residue 474. Alignment of the amino acid sequences of the tier 2 subtype B or C panel did not reveal any specific variation that corresponded to a loss of antiviral potency.
A successful antiretroviral formulation will likely contain a combination of several inhibitors. We therefore wanted to ensure that the peptide triazole could be combined with other inhibitors. Several methods can be used to analyze drug combinations. For our studies, we used the method of Chou and Talalay, based on the median-effect principle, using the formula for nonexclusive drug combinations (8, 9). The results obtained by this method do not always correlate with those of other methods used to calculate drug combinations (40). We therefore also represented the dose reduction seen with our compounds in combination by comparison with the effect of the individual compound alone. Since drug combinations will likely be made with the maximal amount of soluble compound and not at the EC50, we put more weight on results from the EC90 analysis. Although this value does not represent the concentrations likely to be used in formulations, it provides an easily quantifiable starting point for the assessment of synergy.
HNG-156 was combined with a number of inhibitors that target distinct stages of the entry process. HNG-156 demonstrated the predicted inhibition profile with both sCD4 and C34 but showed greater than predicted inhibition when combined with the coreceptor inhibitor maraviroc or TAK-779. The CI values in the latter cases were 0.83 to 0.87 at the EC50 and 0.56 at the EC90, representing a modest but consistent improvement in efficacy. A previous investigation looking at combinations of the anti-CCR5 monoclonal antibody PA14 and the small-molecule inhibitor TAK-779 or SCH-C found that the combinations were synergistic at both the EC50 and the EC90 (45), with CI values of 0.25 or 0.06 at the EC50 and 0.57 or 0.17 at the EC90, respectively. The authors speculated that this was due to inhibition of temporally distinct phases of coreceptor binding. HNG-156 appears to inhibit the formation of the coreceptor binding site on gp120 (22), while maraviroc blocks the formation of the binding site on CCR5 (21). The favorable efficacy of HNG-156 in combination with maraviroc or TAK-779 may be due to inhibition of spatially and temporally distinct phases of coreceptor binding.
Compounds that target carbohydrate residues on gp120 are potent and broadly neutralizing and have been proposed as potential entry inhibitors for the prevention of HIV-1 infection. We knew from past studies that 12p1, the parent peptide of HNG-156, synergized with CVN, a small protein lectin that binds high mannose residues on gp120. We expanded on this study to look at combinations of HNG-156 with CVN; with griffithsin (GRFT), a lectin that is able to potently inhibit diverse isolates of HIV-1 without the toxicity concerns of CVN; and with the high-mannose binding antibody 2G12. We found that HNG-156 was able to synergize with all three lectins. A number of newly emerging carbohydrate binding agents are being proposed as potential drug leads (3). HNG-156 may be a suitable partner for any of these compounds, and an expanded analysis should be considered.
We next tested the effect of HNG-156 with the RT inhibitor tenofovir. Since the two inhibitors target different proteins that act at different stages of the entry process, we hypothesized that they would not interfere with each other. When we combined HNG-156 and tenofovir, we were surprised to see a modest level of synergy between the two compounds against HIV-1BaL (CI at the EC90, 0.62). The mechanism behind the favorable effect of the combination of tenofovir and the peptide is difficult to explain. However, these findings are not without precedent, since a recent study demonstrated that tenofovir was able to synergize with carbohydrate binding agents such as 2G12 (CI at the EC90, 0.18) and the coreceptor antagonist maraviroc (CI at the EC90, 0.53) against HIV-1BaL infection of peripheral blood mononuclear cells (PBMCs) (16).
Overall, this study demonstrated that the peptide triazole HNG-156 has broad anti-HIV-1 activity and is noncytotoxic. HNG-156 combined well with all of the inhibitors with which it was tested. While the synergy seen with HNG-156 was generally modest, it was also consistent. These results argue in favor of future efforts to develop the peptide triazole family of gp120 antagonists as effective inhibitors of HIV-1 transmission.
ACKNOWLEDGMENTS
This work was supported by grants NIH P01 GM56550 and IPM/USAID GPO-A-00-05-00041-00 (to I.C.) and by NIH/NINDS grant NS065727 (to J.M.-G.).
Footnotes
Published ahead of print 14 November 2011
REFERENCES
- 1. Abdool Karim Q, et al. 2010. Effectiveness and safety of tenofovir gel, an antiretroviral microbicide, for the prevention of HIV infection in women. Science 329: 1168–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allan JS, et al. 1985. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science 228: 1091–1094 [DOI] [PubMed] [Google Scholar]
- 3. Balzarini J. 2007. Targeting the glycans of glycoproteins: a novel paradigm for antiviral therapy. Nat. Rev. Microbiol. 5: 583–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barre-Sinoussi F, et al. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220: 868–871 [DOI] [PubMed] [Google Scholar]
- 5. Biorn AC, et al. 2004. Mode of action for linear peptide inhibitors of HIV-1 gp120 interactions. Biochemistry 43: 1928–1938 [DOI] [PubMed] [Google Scholar]
- 6. Bullough PA, Hughson FM, Skehel JJ, Wiley DC. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371: 37–43 [DOI] [PubMed] [Google Scholar]
- 7. Chertova E, et al. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 76: 5315–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chou TC. 1976. Derivation and properties of Michaelis-Menten type and Hill type equations for reference ligands. J. Theor. Biol. 59: 253–276 [DOI] [PubMed] [Google Scholar]
- 9. Chou TC, Talalay P. 1981. Generalized equations for the analysis of inhibitions of Michaelis-Menten and higher-order kinetic systems with two or more mutually exclusive and nonexclusive inhibitors. Eur. J. Biochem. 115: 207–216 [DOI] [PubMed] [Google Scholar]
- 10. Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206: 935–944 [DOI] [PubMed] [Google Scholar]
- 11. Daar ES, Li XL, Moudgil T, Ho DD. 1990. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc. Natl. Acad. Sci. U. S. A. 87: 6574–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Derdeyn CA, et al. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303: 2019–2022 [DOI] [PubMed] [Google Scholar]
- 13. Doncel G, Mauck C. 2004. Vaginal microbicides: a novel approach to preventing sexual transmission of HIV. Curr. HIV/AIDS Rep. 1: 25–32 [DOI] [PubMed] [Google Scholar]
- 14. Dorr P, et al. 2005. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 49: 4721–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Earl PL, Koenig S, Moss B. 1991. Biological and immunological properties of human immunodeficiency virus type 1 envelope glycoprotein: analysis of proteins with truncations and deletions expressed by recombinant vaccinia viruses. J. Virol. 65: 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferir G, et al. 2011. Synergistic in vitro anti-HIV type 1 activity of tenofovir with carbohydrate-binding agents (CBAs). Antiviral Res. 90: 200–204 [DOI] [PubMed] [Google Scholar]
- 17. Ferrer M, Harrison SC. 1999. Peptide ligands to human immunodeficiency virus type 1 gp120 identified from phage display libraries. J. Virol. 73: 5795–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fisher RA, et al. 1988. HIV infection is blocked in vitro by recombinant soluble CD4. Nature 331: 76–78 [DOI] [PubMed] [Google Scholar]
- 19. Gallo RC, et al. 1984. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 224: 500–503 [DOI] [PubMed] [Google Scholar]
- 20. Gallo SA, Sackett K, Rawat SS, Shai Y, Blumenthal R. 2004. The stability of the intact envelope glycoproteins is a major determinant of sensitivity of HIV/SIV to peptidic fusion inhibitors. J. Mol. Biol. 340: 9–14 [DOI] [PubMed] [Google Scholar]
- 21. Garcia-Perez J, et al. 2011. New insights into the mechanisms whereby low molecular weight CCR5 ligands inhibit HIV-1 infection. J. Biol. Chem. 286: 4978–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gopi H, et al. 2009. Introducing metallocene into a triazole peptide conjugate reduces its off-rate and enhances its affinity and antiviral potency for HIV-1 gp120. J. Mol. Recognit. 22: 169–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gopi H, et al. 2008. Structural determinants for affinity enhancement of a dual antagonist peptide entry inhibitor of human immunodeficiency virus type-1. J. Med. Chem. 51: 2638–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gopi HN, Tirupula KC, Baxter S, Ajith S, Chaiken IM. 2006. Click chemistry on azidoproline: high-affinity dual antagonist for HIV-1 envelope glycoprotein gp120. Chem.MedChem 1: 54–57 [DOI] [PubMed] [Google Scholar]
- 25. Grant RM, et al. 2010. Preexposure chemoprophylaxis for HIV prevention in men who have sex with men. N. Engl. J. Med. 363: 2587–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Helseth E, Olshevsky U, Furman C, Sodroski J. 1991. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J. Virol. 65: 2119–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keele BF, et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105: 7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kessler JA, II, et al. 1997. Recombinant human monoclonal antibody IgG1b12 neutralizes diverse human immunodeficiency virus type 1 primary isolates. AIDS Res. Hum. Retroviruses 13: 575–582 [DOI] [PubMed] [Google Scholar]
- 29. Krebs FC, Miller SR, Malamud D, Howett MK, Wigdahl B. 1999. Inactivation of human immunodeficiency virus type 1 by nonoxynol-9, C31G, or an alkyl sulfate, sodium dodecyl sulfate. Antiviral Res. 43: 157–173 [DOI] [PubMed] [Google Scholar]
- 30. Layne SP, et al. 1992. Factors underlying spontaneous inactivation and susceptibility to neutralization of human immunodeficiency virus. Virology 189: 695–714 [DOI] [PubMed] [Google Scholar]
- 31. Li M, et al. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79: 10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li M, et al. 2006. Genetic and neutralization properties of acute and early subtype C human immunodeficiency virus type 1 molecular env clones from heterosexually acquired infections in Southern Africa. J. Virol. 80: 11776–11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCune JM, et al. 1988. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell 53: 55–67 [DOI] [PubMed] [Google Scholar]
- 34. McDougal JS, et al. 1986. Binding of HTLV-III/LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecule. Science 231: 382–385 [DOI] [PubMed] [Google Scholar]
- 35. McFadden K, et al. 2007. A recombinant allosteric lectin antagonist of HIV-1 envelope gp120 interactions. Proteins 67: 617–629 [DOI] [PubMed] [Google Scholar]
- 36. Melikyan GB, et al. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151: 413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mertenskoetter T, Kaptur PE. 2011. Update on microbicide research and development—seeking new HIV prevention tools for women. Eur. J. Med. Res. 16: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moore JP, Sattentau QJ, Wyatt R, Sodroski J. 1994. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J. Virol. 68: 469–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. O'Keefe BR, et al. 2000. Analysis of the interaction between the HIV-inactivating protein cyanovirin-N and soluble forms of the envelope glycoproteins gp120 and gp41. Mol. Pharmacol. 58: 982–992 [DOI] [PubMed] [Google Scholar]
- 40. Pirrone V, Thakkar-Rivera N, Jacobson JM, Wigdahl B, Krebs FC. 2011. Combinatorial approaches to the prevention and treatment of HIV-1 infection. Antimicrob. Agents Chemother. 55: 1831–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. 2009. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J. Virol. 83: 8289–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72: 2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pozniak A. 2008. Tenofovir: what have over 1 million years of patient experience taught us? Int. J. Clin. Pract. 62: 1285–1293 [DOI] [PubMed] [Google Scholar]
- 44. Robey WG, et al. 1985. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science 228: 593–595 [DOI] [PubMed] [Google Scholar]
- 45. Safarian D, Carnec X, Tsamis F, Kajumo F, Dragic T. 2006. An anti-CCR5 monoclonal antibody and small molecule CCR5 antagonists synergize by inhibiting different stages of human immunodeficiency virus type 1 entry. Virology 352: 477–484 [DOI] [PubMed] [Google Scholar]
- 46. Salazar-Gonzalez JF, et al. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82: 3952–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Salazar-Gonzalez JF, et al. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206: 1273–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sattentau QJ, Moore JP. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174: 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sattentau QJ, Zolla-Pazner S, Poignard P. 1995. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology 206: 713–717 [DOI] [PubMed] [Google Scholar]
- 50. Shattock RJ, Moore JP. 2003. Inhibiting sexual transmission of HIV-1 infection. Nat. Rev. Microbiol. 1: 25–34 [DOI] [PubMed] [Google Scholar]
- 51. Turpin JA. 2002. Considerations and development of topical microbicides to inhibit the sexual transmission of HIV. Expert Opin. Invest. Drugs 11: 1077–1097 [DOI] [PubMed] [Google Scholar]
- 52. Umashankara M, et al. 2010. The active core in a triazole peptide dual-site antagonist of HIV-1 gp120. ChemMedChem 5: 1871–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wei X, et al. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46: 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wei X, et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422: 307–312 [DOI] [PubMed] [Google Scholar]
- 55. Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387: 426–430 [DOI] [PubMed] [Google Scholar]
- 56. Williamson C, et al. 2003. Characterization and selection of HIV-1 subtype C isolates for use in vaccine development. AIDS Res. Hum. Retroviruses 19: 133–144 [DOI] [PubMed] [Google Scholar]
- 57. Zhu P, et al. 2003. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl. Acad. Sci. U. S. A. 100: 15812–15817 [DOI] [PMC free article] [PubMed] [Google Scholar]