Abstract
Cryptococcus neoformans is an opportunistic fungal pathogen that causes meningoencephalitis in immunocompromised patients. Recently, we reported that Toll-like receptor 9 (TLR9) is involved in host defense against C. neoformans: specifically, it detects the pathogen's DNA. In the present study, we aimed to elucidate the mechanisms underlying TLR9-mediated activation of innate immune responses by using the URA5 gene, which encodes a virulent component of this fungal pathogen. A PCR-amplified 345-bp URA5 gene fragment induced interleukin-12 p40 (IL-12p40) production by bone marrow-derived dendritic cells (BM-DCs) in a TLR9-dependent manner. Similar activity was detected in the 5′ 129-bp DNA fragment of URA5 and in a synthesized oligodeoxynucleotide (ODN) with the same sequence. Shorter ODN fragments, which contained GTCGGT or GACGAT but had only 24 or 21 bases, induced IL-12p40 production and CD40 expression by BM-DCs, but this activity vanished when the CG sequence was replaced by GC or when a phosphorothioate modification was introduced. IL-12p40 production caused by active ODN was strikingly enhanced by treatment with DOTAP, a cationic lipid that increases the uptake of DNA by BM-DCs, though DOTAP failed to induce IL-12p40 production by inactive ODN and did not affect the activity of an ODN-containing canonical CpG motif. There was no apparent difference in intracellular trafficking between active and inactive ODNs. Finally, an extremely high dose of inactive ODN suppressed IL-12p40 production by BM-DCs that had been stimulated with active ODN. These results suggest that the C. neoformans URA5 gene activates BM-DCs through a TLR9-mediated signaling pathway, using a mechanism possibly independent of the canonical CpG motif.
INTRODUCTION
Cryptococcus neoformans, an opportunistic fungal pathogen, frequently causes fatal meningoencephalitis in AIDS patients. C. neoformans was recently recognized as an intracellular microorganism that grows in macrophages, and clinical evidence has accumulated showing that this fungal pathogen causes illness following reactivation in immunocompromised patients (4). Host defense against C. neoformans is mediated largely by cellular immunity (15), and CD4+ T cells play a central role in eradicating the infection (7, 21). The outcome of this infection is determined by the balance between Th1 and Th2 immune responses: the polarized synthesis of Th1 cytokines leads to protection, while a biased Th2 immune response means that the host will succumb to the infection (11). This balance is critically regulated by a variety of innate immune cells shortly after the invasion of C. neoformans into lung tissues (10, 16, 18, 19, 27).
The invasion of a microbial pathogen into host tissue results in the development of an inflammatory response, which is initiated by recognition of pathogen-associated molecular patterns (PAMPs) via pattern recognition receptors (PRRs), including the Toll-like receptors (TLRs) (25). The protein MyD88, which is involved in the TLR signaling pathway, plays a critical role in the host immune response to C. neoformans (1, 33), as experiments with mice genetically lacking MyD88 (MyD88KO mice) have shown; this indicates that certain TLRs likewise contribute to the immune response to C. neoformans. In our recent study (22), it was demonstrated that TLR9 contributes to host defense against C. neoformans infection by detecting the DNA of this fungal pathogen. In addition, Zhang and coworkers recently reported that TLR9 signaling plays a critical role in protective immune responses to C. neoformans and in clearance of this fungal pathogen from the lungs (34). Similar findings have been reported regarding other fungal species, including Aspergillus fumigatus, Candida albicans, and Cordyceps sinensis (20, 24, 30).
The unmethylated CpG motif-containing DNA that occurs in prokaryotic microorganisms such as bacteria and viruses is known to trigger host immune responses by interacting with TLR9 (6, 12, 13), which is distributed in the endosomal compartments (8, 14). This interaction causes the activation of a signaling pathway mediated by the adaptor molecule MyD88, leading to the synthesis of proinflammatory cytokines and the expression of costimulatory molecules by macrophages and dendritic cells (DCs) (8, 9). Turning to fungal DNA, Ramirez-Ortiz and coworkers have reported that A. fumigatus DNA contains an unmethylated CpG motif and that it activates mouse bone marrow-derived DCs (BM-DCs) and human plasmacytoid DCs to produce proinflammatory cytokines. This activity was diminished when the fungal DNA was treated with CpG methylase or a CpG-specific endonuclease (24), suggesting that TLR9 senses the unmethylated CpG motif in A. fumigatus DNA.
It remains to be determined whether the immune activation that results when DNA from C. neoformans is introduced into a host is induced by an unmethylated CpG motif in that DNA, like the case with A. fumigatus. In our earlier study (22), treatment with CpG methylase was reported to attenuate the BM-DC-stimulating activity of C. neoformans DNA only partially, suggesting the presence of a certain CpG-independent mechanism. In the present study, therefore, we investigated the mechanism underlying TLR9-mediated immune activation by DNA from C. neoformans by examining the ability of a particular gene known as URA5, which encodes orotidine monophosphate pyrophosphorylase in this fungal pathogen, to induce interleukin-12 p40 (IL-12p40) production and CD40 expression by BM-DCs.
MATERIALS AND METHODS
Mice.
TLR9KO mice were generated as described previously (6). Homozygous mice were backcrossed to C57BL/6 mice for more than 8 generations. Wild-type (WT) C57BL/6 mice were used as controls. Male or female mice at 6 to 10 weeks of age were used for the experiments. All mutant mice were kept under specific-pathogen-free conditions at the Institute for Animal Experimentation, Tohoku University Graduate School of Medicine (Sendai, Japan). The experiments were conducted according to the guidelines of and approved by the ethics committees of Tohoku University.
Preparation of C. neoformans DNA.
An acapsular strain of C. neoformans, designated Cap67 (a kind gift of Stuart M. Levitz, Boston University, Boston, MA), was used. Serotype A encapsulated strains of C. neoformans, designated YC-11 and -13, were established from patients with pulmonary cryptococcosis (32). The yeast cells were cultured on potato dextrose agar (PDA) plates (Eiken, Tokyo, Japan) for 2 to 3 days before use. For preparation of C. neoformans DNA, the yeast cells were lysed with 100 mM Tris-hydrochloride (pH 7.5), 0.5% sodium dodecyl sulfate (SDS), and 30 mM EDTA at 100°C for 15 min. The DNA was purified by extraction with phenol-chloroform-isoamyl alcohol (25:24:1) and isopropanol precipitation. The pellet was washed with 70% ethanol, dried, and resolved with distilled water. The DNA thus obtained was kept at −80°C until use. The ratio of the optical density at 260 nm (OD260) to the OD280 was usually between 1.5 and 2.0. The endotoxin content in the DNA preparations, measured by Limulus amoebocyte lysate assay, was <10 pg/ml.
PCR.
A 345-bp fragment of C. neoformans URA5 DNA was amplified in an automatic DNA thermal cycler (PC320; Astec, Fukuoka, Japan) by using the specific primers 5′-ACG GTG AGG GCG GTA CTA TG-3′ (forward) and 5′-AAG ACC TCT GAA CAC CGT AC-3′ (reverse). The 5′, 3′, and intervening fragments of this 345-bp sequence were amplified using the following specific primers (26): ACG GTG AGG GCG GTA CTA TG (forward) and GCT TCG CTT CAG GGG AGG C (reverse) for the 5′ 129-bp fragment (base numbers 1 to 129), GCC TGT CGA GCC TAT TAT TG (forward) and AAG ACC TCT GAA CAC CGT AC (reverse) for the 3′ 123-bp fragment (base numbers 223 to 345), and GCC TCC CCT GAA GCG AAG C (forward) and CAA TAA TAG GCT CGA CAG GC (reverse) for the intervening 133-bp fragment (base numbers 110 to 242). We added 1.0 μl of C. neoformans DNA solution to 49 μl of a PCR mixture (TaKaRa Ex Taq; TaKaRa, Shiga, Japan) which contained 1.0 μM (each) forward and reverse primers. The mixture was incubated for 36 cycles of 1 min at 94°C, 1 min 30 s at 63°C, and 1 min at 72°C. The PCR products were electrophoresed in 2% agarose gels, stained with 0.5 μg/ml ethidium bromide, and observed with a UV transilluminator. For the stimulation of BM-DCs, PCR products were purified using a QIAquick PCR purification kit (Qiagen, Germantown, MD).
Culture medium and reagents.
RPMI 1640 medium was obtained from Nipro (Osaka, Japan), and fetal calf serum (FCS) was obtained from BioWest (Nuaillé, France). Lipopolysaccharide (LPS) was purchased from Sigma-Aldrich (St. Louis, MO). A prototypic CpG oligodeoxynucleotide (CpG-ODN), CpG1826, and various other ODNs were synthesized and purified by means of high-performance liquid chromatography (HPLC) at Hokkaido System Science (Sapporo, Japan), as shown in Tables 1 and 2. CpG1826 was usually phosphorothioated; in some experiments, ODN112 was phosphorothioated (PS-ODN112) and CpG1826 was in a phosphodiester form (PO-CpG1826). The endotoxin content in these DNA compounds, as measured by Limulus amoebocyte lysate assay, was <10 pg/ml.
Table 1.
Sequences of synthesized ODNs
| ODN (length [bases]) | Sequence (5′–3′) |
|---|---|
| ODN1 (129) | TACGGTGAGGGCGGTACTATGGTCGGTGCGCCTCTCAAGGGACGAATCGTCATCATCGACGATGTTCTCACCTCTGGCAAGGCCATCCGTGAAGCTATTGACATTCTCAAGGCCTCCCCTGAAGCGAAG |
| ODN2 (133) | GCCTCCCCTGAAGCGAAGCTTGTCGGAATTGTCCAGCTTGTCGACAGACAAGAGAAAGGCCAGAGCGGTAGCGGCAAGAGTACCGTACAGGAGGTTGAGGAAGAGTTCGGTGTGCCTGTCGAGCCTATTATTG |
| ODN3 (122) | GCCTGTCGAGCCTATTATTGGTTTGGACGACATTGTGAAGTACTTAGAAAGCTCCGGCAAGTGGGAAAAGGAGCTGCAAGAGGTCAGGAAGTACAGGGCGGAGTACGGTGTTCAGAGGTCTTA |
| ODN11 (53) | ACGGTGAGGGCGGTACTATGGTCGGTGCGCCTCTCAAGGGACGAATCGTCATC |
| ODN12 (43) | AATCGTCATCATCGACGATGTTCTCACCTCTGGCAAGGCCATC |
| ODN13 (53) | CAAGGCCATCCGTGAAGCTATTGACATTCTCAAGGCCTCCCCTGAAGCGAAGC |
| ODN111 (21) | ACGGTGAGGGCGGTACTATGG |
| ODN112 (24) | CTATGGTCGGTGCGCCTCTCAAGG |
| ODN113 (20) | TCAAGGGACGAATCGTCATC |
| ODN121 (18) | AATCGTCATCATCGACGA |
| ODN122 (21) | CGACGATGTTCTCACCTCTGG |
| ODN123 (18) | ACCTCTGGCAAGGCCATC |
Table 2.
Sequences of synthesized ODN variants
| ODN variant | Sequence (5′–3′)a |
|---|---|
| ODN112 | CTATGGTCGGTGCGCCTCTCAAGG |
| ODN112-1 | ---TGGTCGGTGCGCCTCTCAAGG |
| ODN112-2 | CTA---TCGGTGCGCCTCTCAAGG |
| ODN112-3 | CTATGG---GTGCGCCTCTCAAGG |
| ODN112-4 | CTATGGTCG---CGCCTCTCAAGG |
| ODN112-5 | CTATGGTCGGTG---CTCTCAAGG |
| ODN112-6 | CTATGGTCGGTGCGC---TCAAGG |
| ODN112-7 | CTATGGTCGGTGCGCCTC---AGG |
| ODN112-8 | CTATGGTCGGTGCGCCTCTCA--- |
| ODN112-GTCGTT | CTATGGTCGTTGCGCCTCTCAAGG |
| ODN112-GACGTT | CTATGGACGTTGCGCCTCTCAAGG |
| ODN112-GC | CTATGGTGCGTGCGCCTCTCAAGG |
| ODN113 | TCAAGGGACGAATCGTCATC |
| ODN121 | AATCGTCATCATCGACGA |
| ODN122 | CGACGATGTTCTCACCTCTGG |
| ODN122-GC | CGAGCATGTTCTCACCTCTGG |
Underlining shows the portion of nucleic acid deletion.
Preparation and culture of dendritic cells.
BM-DCs were prepared as described by Lutz and coworkers (17), with some modifications. Briefly, BM cells from WT and TLR9KO mice were cultured at 2 × 105/ml in 10 ml RPMI 1640 medium supplemented with 10% FCS, 100 U/ml penicillin G, 100 μg/ml streptomycin, and 50 μM 2-mercaptoethanol and containing 20 ng/ml murine granulocyte-macrophage colony-stimulating factor (GM-CSF; Wako Pure Chemical Industries, Ltd., Osaka, Japan). On day 3, another 10 ml of the same GM-CSF-containing medium was added; on day 6, a half change was performed using the GM-CSF-containing culture medium. On day 8, nonadherent cells were collected and used as BM-DCs. The cells thus obtained were cultured at 1 × 105/ml with various stimulants for 24 h at 37°C in 5% CO2 in an incubator.
DOTAP treatment.
ODNs were treated with N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (DOTAP; Carl Roth, Karlsruhe, Germany) as previously described by Yasuda et al. (31). Briefly, 5 μg of ODN was incubated with 10 μg DOTAP in 100 μl HEPES-buffered saline (20 mM HEPES, 150 mM NaCl, pH 7.4) at room temperature for 15 min.
Cytokine assay.
The concentrations of IL-12p40 in the culture supernatants were measured by an enzyme-linked immunosorbent assay (ELISA) using capture and biotinylated developing antibodies (BD Biosciences, Franklin Lakes, NJ).
Analysis of CD40 expression on BM-DCs.
BM-DCs were preincubated with anti-FcγRII and -III monoclonal antibody (MAb), prepared by use of a protein G column kit (Kirkegaard & Perry Laboratories, Gaithersburg, MD) from the culture supernatants of hybridoma cells (clone 2.4G2), on ice for 15 min in phosphate-buffered saline (PBS) containing 1% FCS and 0.1% sodium azide, stained with fluorescein isothiocyanate (FITC)-conjugated anti-CD11c MAb (clone HL3; BD Biosciences) and phycoerythrin (PE)-conjugated anti-CD40 MAb (clone 1C10; eBioscience, San Diego, CA) for 25 min, and then washed three times in the same buffer. Isotype-matched irrelevant antibodies were used for control staining. The propidium iodide-stained population was excluded as dead cells. The stained cells were analyzed using a FACSCanto II flow cytometer (BD Biosciences). Data were collected from 15,000 to 20,000 individual live cells by using forward scatter/side scatter and FL1 parameters to set a gate on the CD11c+ lymphocyte population.
Analysis of ODN uptake by BM-DCs.
BM-DCs were incubated with rhodamine-labeled ODNs or CpG1826 (prepared by Hokkaido System Science) in the presence or absence of DOTAP for 60 min in a CO2 incubator. The cells were fixed with 4% paraformaldehyde in PBS on ice for 15 min, washed three times with PBS, and then analyzed using a Cytomics FC500 flow cytometer (Beckman Coulter Inc., Fullerton, CA). Data were collected from 15,000 to 20,000 individual live cells by using a forward scatter/side scatter parameter.
Confocal microscopic analysis.
BM-DCs were incubated with rhodamine-labeled ODNs or CpG1826 in microtubes for 2 h. After fixation, the cells were stained with Alexa Fluor 488-conjugated anti-LAMP1 MAb (clone 1D4B; eBioscience) and then cytospun onto glass slides, upon which coverslips were mounted by using a ProLong antifade kit (Molecular Probes, Eugene, OR). Confocal images were acquired using a Carl Zeiss (Oberkochen, Germany) LSM510 microscope. Carl Zeiss LSM Image Browser was used to acquire and process the confocal images.
Statistical analysis.
Analysis was conducted using Statview II software (Abacus Concept, Inc., Berkeley, CA) on a Macintosh computer. Data are expressed as means ± standard deviations (SD). Differences between groups were examined for statistical significance by using one-way analysis of variance (ANOVA) with a post hoc analysis (Fisher's protected least significant difference [PLSD] test). P values of <0.05 were considered significant.
RESULTS
IL-12p40 production by BM-DCs stimulated with C. neoformans URA5.
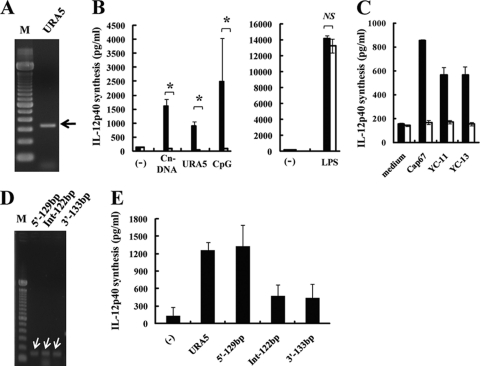
To elucidate the involvement of the C. neoformans URA5 DNA sequence in the activation of immune cells, we examined whether PCR-amplified URA5 DNA from this fungal pathogen induced IL-12p40 production by BM-DCs. As shown in Fig. 1A and B, a PCR-amplified 345-bp DNA fragment from strain Cap67, identified as URA5 by means of nucleic acid sequencing, stimulated BM-DCs to produce IL-12p40 at a level equivalent to that induced by DNA extracted from C. neoformans. IL-12p40 production by URA5 DNA-stimulated BM-DCs was completely abrogated in TLR9KO mice, as was IL-12p40 production induced by C. neoformans DNA and CpG-ODN, whereas IL-12p40 production induced by LPS was not different between WT and TLR9KO mice. Similar results were obtained using PCR-amplified URA5 DNAs from two clinically isolated strains, YC-11 and YC-13 (Fig. 1C). These results indicate that URA5 DNA activates BM-DCs in a TLR9-dependent manner and suggest that a canonical CpG motif, such as GACGTT or GTCGTT, may be contained in this gene. Yet no such motif was found in the PCR-amplified 345-bp DNA fragment, which raises the possibility of involvement of other mechanisms.
Fig 1.
IL-12p40 production by BM-DCs stimulated with URA5 PCR products. PCR-amplified URA5 DNA and its 5′ 129-bp fragment induce IL-12p40 production by BM-DCs. (A) The PCR product of the URA5 gene was amplified from Cap67 and subjected to electrophoresis. The arrow indicates the 345-bp product. (B) BM-DCs from WT (solid columns) or TLR9KO (open columns) mice were cultured with Cap67 DNA (10 μg/ml), the URA5 PCR product from Cap67 (10 μg/ml), CpG1826 (1 μg/ml), or LPS (1 μg/ml) for 24 h, and IL-12p40 concentrations in the culture supernatants were measured by ELISA. (C) BM-DCs from WT (solid columns) or TLR9KO (open columns) mice were cultured with URA5 PCR products from strains Cap67, YC-11, and YC-13 (10 μg/ml for each) for 24 h, and IL-12p40 concentrations in the culture supernatants were measured by ELISA. (D) PCR products of 5′, 3′, and intervening fragments of the URA5 gene were amplified from Cap67 and subjected to electrophoresis. Their sizes were 129, 122, and 133 bp, respectively. Each arrow indicates a PCR product. (E) BM-DCs were cultured with PCR products of the URA5 gene from Cap67 (10 μg/ml) and each fragment (10 μg/ml) for 24 h, and IL-12p40 concentrations in the culture supernatants were measured by ELISA. Data are means ± SD for triplicate cultures. M, 100-bp DNA size marker; 5′-129bp, 5′ PCR product of 129 bp; 3′-133bp, 3′ PCR product of 133 bp; Int-122bp, intervening PCR product of 122 bp; Cn-DNA, Cap67 DNA; CpG, CpG1826. *, P < 0.05; NS, not significant.
IL-12p40 production by BM-DCs stimulated with DNA fragments of the URA5 gene.
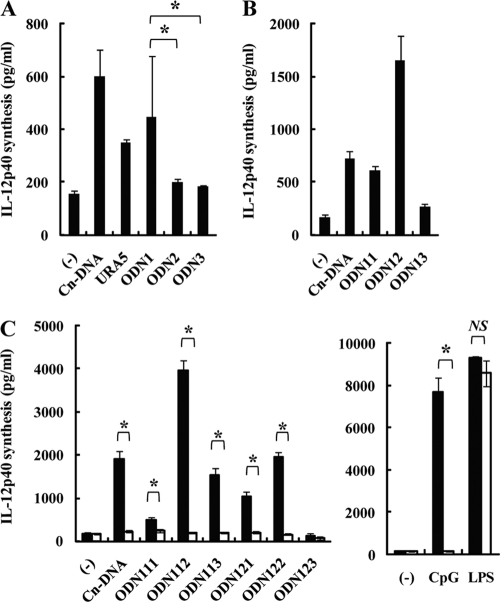
To identify the DNA sequence responsible for activation of BM-DCs, we stimulated BM-DCs with three DNA fragments from within the PCR-amplified 345-bp fragment, namely, the 5′, 3′, and intervening fragments, composed of 129 bp, 122 bp, and 133 bp, respectively, each overlapping the next by 18 to 20 bp (Fig. 1D). As shown in Fig. 1E, the 5′ fragment induced production of IL-12p40 by BM-DCs at a level almost equivalent to that induced by the 345-bp PCR product, whereas the 3′ and intervening fragments induced only weak IL-12p40 production. Additionally, we measured the production levels induced by synthesized ODN fragments with the same sequences as the three DNA fragments (Table 1). As shown in Fig. 2A, as in the case of the PCR products, the ODN with 129 bases (ODN1) induced the production of IL-12p40 by BM-DCs at a considerable level, whereas the levels of IL-12p40 production induced by the other ODN sequences (ODN2 and ODN3, with 133 and 122 bases, respectively) were lower.
Fig 2.
IL-12p40 production by BM-DCs stimulated with synthesized ODNs based on the URA5 gene. There were several ODNs of 18 to 24 bp found in URA5 DNA that induced IL-12p40 production by BM-DCs. (A and B) BM-DCs were cultured with Cap67 DNA (10 μg/ml), the URA5 PCR product from Cap67 (10 μg/ml), various ODNs (10 μg/ml each of ODN1, ODN2, and ODN3 and 30 μg/ml each of ODN11, ODN12, and ODN13), CpG1826 (1 μg/ml), or LPS (1 μg/ml) for 24 h, and IL-12p40 concentrations in the culture supernatants were measured by ELISA. (C) BM-DCs from WT (solid columns) or TLR9KO (open columns) mice were cultured with Cap67 DNA (10 μg/ml), the URA5 PCR product from Cap67 (10 μg/ml), ODN111, -112, -113, -121, -122, or -123 (30 μg/ml), CpG1826 (1 μg/ml), or LPS (1 μg/ml) for 24 h, and IL-12p40 concentrations in the culture supernatants were measured by ELISA. Data are means ± SD for triplicate cultures. Cn-DNA, Cap67 DNA; ODN, synthesized oligodeoxynucleotide; CpG, CpG1826. *, P < 0.05; NS, not significant.
Next, we examined whether even shorter sequences of ODN1 exhibited IL-12p40-inducing activity. BM-DCs were stimulated with three ODN fragments, the 5′, 3′, and intervening fragments of ODN1 (ODN11, ODN13, and ODN12), composed of 53, 53, and 43 bases, respectively (Table 1). As shown in Fig. 2B, ODN11 and ODN12 induced the production of IL-12p40 at a level equivalent to or higher than that caused by C. neoformans DNA, whereas ODN13 induced only weak IL-12p40 production.
Finally, we tested the activity of still shorter sequences of ODN11 and ODN12 (ODN111, ODN112, ODN113, ODN121, ODN122, and ODN123), as shown in Table 1. As Fig. 2C shows, ODN112, ODN113, ODN121, and ODN122 induced IL-12p40 production by BM-DCs, with the highest activity level induced by ODN112, whereas ODN111 and ODN123 induced little or no IL-12p40 production. In addition, IL-12p40 production by these ODN fragments as well as by CpG1826 was completely abrogated when BM-DCs from TLR9KO mice were used, whereas the LPS-induced response was not affected.
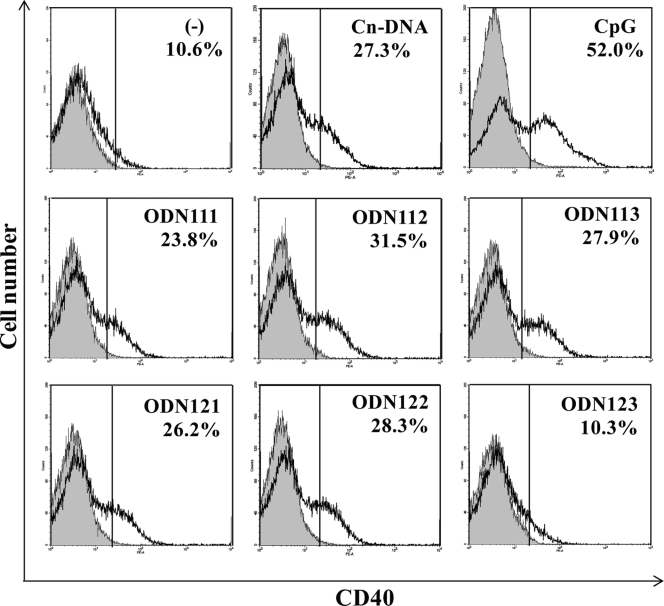
CD40 expression by BM-DCs stimulated with DNA fragments of the URA5 gene.
To determine whether the activity of ODNs was limited to IL-12p40 synthesis, we examined the expression of CD40 by BM-DCs. As shown in Fig. 3, ODN111, -112, -113, -121, and -122 enhanced the expression of CD40, as detected upon stimulation with C. neoformans DNA and CpG1826, whereas ODN123 did not show such an effect.
Fig 3.
CD40 expression by BM-DCs stimulated with synthesized ODNs based on the URA5 gene. ODNs with the ability to induce IL-12p40 production were also active in the expression of CD40 by BM-DCs. BM-DCs were cultured with Cap67 DNA (10 μg/ml), ODN111, -112, -113, -121, -122, or -123 (30 μg/ml), or CpG1826 (1 μg/ml) for 24 h, and CD40 expression by BM-DCs was measured using a flow cytometer. Representative data for three independent experiments are shown. The percentages indicate the proportions of positive cells. Cn-DNA, Cap67; ODN, synthesized oligodeoxynucleotide; CpG, CpG1826.
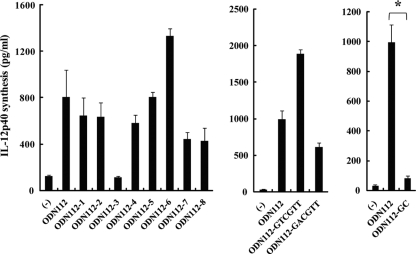
DNA sequence responsible for BM-DC activation.
To determine the DNA sequence responsible for BM-DC activation, BM-DCs were stimulated with modified ODN112 variants from which various sets of three adjacent nucleotides were deleted (Table 2). As shown in Fig. 4, IL-12p40 production by BM-DCs was completely abrogated when ODN112-3 was used and was weakened but not abrogated upon stimulation with ODN112-5, ODN112-7, and ODN112-8. Because a TCG sequence was deleted from ODN112-3, we focused on the six bases in the GTCGGT sequence, which resembles the canonical CpG motifs GACGTT and GTCGTT. Therefore, BM-DCs were stimulated with modified analogues of ODN112 in which one of these two CpG motifs was introduced in place of the targeted six-base sequence (ODN112-GACGTT and ODN112-GTCGTT) (Table 2). Both types of ODN fragment induced IL-12p40 production by BM-DCs, though the activity they induced was not equivalent to that induced by ODN112 (Fig. 4). Next, to address the possible importance of the CG sequence specifically, we tested the activity of a modified ODN112 derivative in which CG was replaced by GC within the six-base sequence (ODN112-GC) (Table 2); this ODN had no capacity to induce IL-12p40 production by BM-DCs (Fig. 4). These results indicate that the CG sequence is essential for the activity of ODN112, though the canonical CpG motif is not.
Fig 4.
IL-12p40 production by BM-DCs stimulated with altered ODNs of the URA5 gene. Some modified ODN derivatives lost the ability to induce IL-12p40 production by BM-DCs. BM-DCs were cultured with various altered ODNs (30 μg/ml) for 24 h, and IL-12p40 concentrations in the culture supernatants were measured by ELISA. Data are means ± SD for triplicate cultures. ODN, synthesized oligodeoxynucleotide. *, P < 0.05.
Effects of phosphorothioate modification and DOTAP treatment.
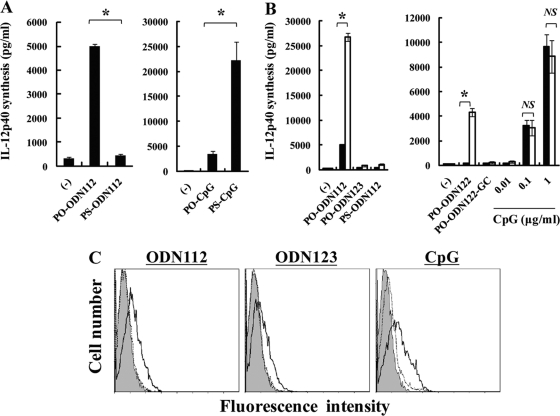
Because natural phosphodiester (PO) DNA causes instability in the presence of endonucleases, phosphorothioate (PS) modification is generally performed on CpG-ODN in order to enable stability in the presence of enzymes, permitting its use in experiments and potentially in the treatment of various diseases (13). In this study, however, all of the synthesized compounds used were PO-DNA. Therefore, we examined the effect of PS modification on the IL-12p40-inducing activity of ODN112. For this purpose, BM-DCs were stimulated with either PO-ODN112 or PS-ODN112 and either PO-CpG1826 or PS-CpG1826. As shown in Fig. 5A, PS-ODN112 completely lacked the capacity to induce IL-12p40 production compared to PO-ODN112, whereas the activity of PO-CpG1826 was much lower than that of PS-CpG1826.
Fig 5.
Effects of phosphorothioate modification and DOTAP on BM-DC production of IL-12p40. ODN112 and CpG1826 were distinct in terms of the effects of PS modification and DOTAP treatment on their ability to induce production of IL-12p40 by BM-DCs, whereas uptake of these ODNs by BM-DCs was equivalent. (A) BM-DCs were cultured with PO-ODN112 (30 μg/ml), PS-ODN112 (30 μg/ml), PO-CpG1826 (1 μg/ml), or PS-CpG1826 (1 μg/ml) for 24 h, and IL-12p40 concentrations in the culture supernatants were measured by ELISA. (B) BM-DCs were cultured with PO-ODN112 (30 μg/ml), PO-ODN123 (30 μg/ml), PS-ODN112 (30 μg/ml), PO-ODN122 (30 μg/ml), PO-ODN122-GC (30 μg/ml), or PS-CpG1826 (0.01, 0.1, or 1 μg/ml) in the presence (open columns) or absence (solid columns) of DOTAP for 24 h, and IL-12p40 concentrations in the culture supernatants were measured by ELISA. Data are means ± SD for triplicate cultures. PO, phosphodiester; PS, phosphorothioate; ODN, synthesized oligodeoxynucleotide DNA; CpG, CpG1826. *, P < 0.05. (C) BM-DCs were incubated with rhodamine-labeled ODN112 (10 μg/ml), ODN123 (10 μg/ml), or CpG1826 (1 μg/ml) in the presence or absence of DOTAP for 60 min, and uptake of the ODNs by these cells was analyzed using a flow cytometer. Data are representative of three independent experiments. Shaded area, no ODN; dotted line, ODN without DOTAP; solid line, ODN with DOTAP. ODN, synthesized oligodeoxynucleotide; CpG, CpG1826.
To exclude the possibility that PS modification may prevent ODNs from entering cells, we examined the effect of DOTAP, a cationic lipid which promotes the entry of DNA into cells, on the production of IL-12p40 by BM-DCs. As shown in Fig. 5B, DOTAP strikingly increased the production of IL-12p40 induced by ODN112 and ODN122 but, interestingly, did not further promote IL-12p40 production induced by CpG1826 and did not enable IL-12p40 production to result from stimulation with the inactive DNA compounds ODN123, ODN112-GC, ODN122-GC, and PS-ODN112. It was considered possible that inactive DNA compounds were not entering the cells even in the presence of DOTAP. To exclude this possibility, we evaluated the uptake of rhodamine-labeled ODN112, ODN123, and CpG1826 into BM-DCs by using flow cytometric analysis. As shown in Fig. 5C, untreated BM-DCs showed marginal or weak uptake of all of these DNA compounds, while DOTAP treatment strikingly increased the uptake not only of ODN112 but also of ODN123 and CpG1826.
Difference in BM-DC activation by active and inactive ODNs.
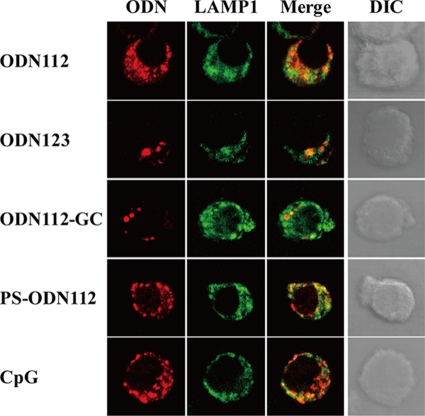
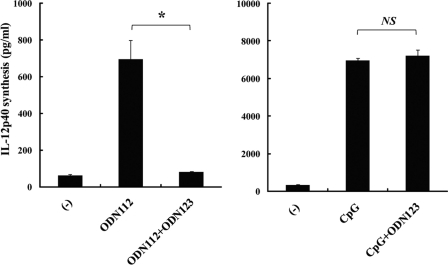
In order to address the mechanism of BM-DC activation in the context of the distinction between active and inactive ODNs, we first examined the intracellular localization of rhodamine-labeled ODNs within BM-DCs by using confocal laser microscopy. As shown in Fig. 6, not only the active ODNs ODN112 and CpG1826 but also the inactive ODNs ODN123, ODN112-GC, and PS-ODN112 were colocalized with LAMP1, a marker of late endosomes. These results indicate that the differences in the activities of the various ODNs could not be ascribed to their different intracellular localizations. Next, we further assessed the intracellular localization of ODNs within BM-DCs by using TLR9; again, no difference could be detected between active and inactive ODNs, although clear colocalization was not easy to observe (data not shown). These results raised the possibility that the interaction of inactive ODNs with TLR9 in late endosomes may not deliver an activation signal, though they may interact with TLR9 at an equivalent level. To address this possibility, we examined whether inactive ODNs prevented active ODN112 from activating BM-DCs. As shown in Fig. 7, IL-12p40 production by ODN112-activated BM-DCs was completely abrogated when 10 times more inactive ODN123 was added to the cultures. In contrast, IL-12p40 production by CpG1826-activated BM-DCs was not affected by the addition of ODN123.
Fig 6.
Colocalization of ODNs and LAMP1. Intracellular trafficking of active ODNs into late endosomes was not very different from that of inactive ODNs. BM-DCs were incubated with various ODNs (30 μg/ml) or CpG1826 (1 μg/ml) labeled with rhodamine for 2 h, and the cells were stained with Alexa Fluor 488-conjugated anti-LAMP1 MAb. Localization of ODNs and LAMP1 was analyzed using confocal laser microscopy. ODN, synthesized oligodeoxynucleotide; PS, phosphorothioate; CpG, CpG1826; Merge, merged image; DIC, differential interference contrast.
Fig 7.
Effect of inactive ODN on BM-DC production of IL-12p40. Excessive doses of ODN123 inhibited IL-12p40 production by BM-DCs stimulated with ODN112 but not by BM-DCs stimulated with CpG1826. BM-DCs were cultured with ODN112 (30 μg/ml) or CpG1826 (1 μg/ml) in the presence or absence of ODN123 (300 μg/ml), and IL-12p40 concentrations in the culture supernatants were measured by ELISA. Data are means ± SD for triplicate cultures. ODN, synthesized oligodeoxynucleotide DNA; CpG, CpG1826. *, P < 0.05; NS, not significant.
DISCUSSION
In the present study, to explore the mechanism underlying TLR9-mediated activation of BM-DCs by DNA from C. neoformans, we examined the ability of a particular gene known as URA5 to induce IL-12p40 production by these cells. The URA5 gene, which encodes orotidine monophosphate pyrophosphorylase, is involved in uracil synthesis (28) and is thought to contribute to the development of C. neoformans infection, as evidenced by the reduced virulence of its mutant strain (3). Our present results have demonstrated that a PCR product of the URA5 gene amplified from C. neoformans DNA, as well as the whole DNA itself, activates BM-DCs to produce IL-12p40 in a TLR9-dependent manner.
In addition, to elucidate whether BM-DC activation was detectable only when cells were stimulated with URA5, we examined the activities of other virulence-associated genes from C. neoformans, namely, CNLAC1 and CAP59 (23). PCR-amplified CNLAC1 and CAP59 DNAs induced the production of IL-12p40 by BM-DCs, as did URA5 DNA (see Fig. S1 in the supplemental material); in addition, the canonical CpG motifs were not found in CNLAC1 and CAP59 DNA fragments, like the case for URA5 DNA. Thus, certain DNA fragments from C. neoformans were shown to induce the activation of BM-DCs.
Ramirez-Ortiz and coworkers recently reported that A. fumigatus DNA contains an unmethylated CpG motif and activates mouse BM-DCs and human plasmacytoid DCs to produce proinflammatory cytokines and that this activity was diminished when the fungal DNA was treated with a CpG methylase or a CpG-specific endonuclease (24). These findings suggest that an unmethylated CpG motif in A. fumigatus DNA is sensed by TLR9, leading to the activation of immune responses. In our study, in contrast, we did not find any canonical CpG motifs in a 345-bp PCR product of the URA5 gene that nevertheless stimulated IL-12p40 production by BM-DCs. Therefore, we attempted to define a putative responsible motif in this gene by dividing the gene into small DNA fragments. Some fragments exhibited the capacity to induce IL-12p40 production and CD40 expression in BM-DCs, while others did not, suggesting the possible involvement of a particular DNA sequence in inducing immune activation. In the highly potent fragment that we called ODN112, a CG dinucleotide similar to that in a prototypic CpG-ODN was observed to be important, because the deletion of either TCG (ODN112-3) or GGT (ODN112-4), but not that of GTC (ODN112-2) or the replacement of TCG with TGC (ODN112-GC), resulted in the complete loss of BM-DC activation. There were some exceptions, however: (i) deletion of a CG dinucleotide at another site in ODN112 (ODN112-5) did not cause a drastic loss of BM-DC-stimulating activity; and (ii) ODN111, another active DNA fragment, showed only marginal activity irrespective of its two CG dinucleotides, located at distinct sites. Thus, the CG dinucleotide does not seem to fully account for the BM-DC-stimulating activity of the highly potent DNA fragment of C. neoformans.
As reported in a previous paper by Casadevall et al. (2), the current 345-bp PCR product of URA5 obtained from strain Cap67, which is identical to that obtained from strain B3501, is located between positions 431 and 775, while ODN112 is composed of 24 bases starting at position 448 and contains GTCGGT, a possible active motif that is conserved among the 5 strains used in this study. In the present study, we found that the PCR products of URA5 from two serotype A clinical strains, YC-11 and YC-13, induced IL-12p40 production by BM-DCs at levels comparable to that caused by URA5 DNA from Cap67 and that the ODN112 sequence from YC-11 and YC-13 was different from that from B3501 only at position 461 (G→A), where it was identical to that from strain ATCC 24064. We showed that this substitution did not affect the activity of ODN112 (Fig. 2C). These results suggest that an active motif of the ODN112 sequence may be conserved in C. neoformans.
In the present study, we used PO-ODN but not PS-ODN for the activation of BM-DCs. PS modification strikingly increases the ability of prototypic CpG-ODNs to activate immune cells by rendering them resistant to breakdown caused by endonucleases (13). When the same modification was introduced to ODN112, in contrast, IL-12p40-inducing activity was completely eliminated. Thus, prototypic CpG-ODNs and active PO-ODNs like ODN112 are likely to exhibit different responses to PS modification, as recently reviewed by Wagner (29).
Recently, it has been considered that the trafficking of DNA toward TLR9 expressed in late endosomes is more important than the particular DNA motif in determining whether immune cells are activated. Moreover, the sugar backbone of DNA, rather than its base sequence, has been identified as a basic element for the activation of DCs (5). In line with these observations, even PO-ODN has a considerable ability to stimulate immune cells when it is added to cell cultures together with DOTAP, which provokes the uptake of DNA into endosomal compartments (31). In the present study, DOTAP enhanced both uptake of ODN112 and IL-12p40 production by BM-DCs, though IL-12p40 production was not changed when CpG1826 was used in place of ODN112. On the other hand, both active and inactive ODNs, such as ODN112 and ODN123, were found in the URA5 gene; the only difference between these lies in their base sequences. These findings suggest that base sequences may still be involved in determining the immunostimulating activity of PO-DNA.
According to our confocal microscopic analysis, there was no apparent difference in the intracellular translocation of the active DNA ODN112 and inactive DNAs such as ODN123, ODN112-GC, and PS-ODN112, as shown by their equivalent uptake by BM-DCs and their overlapping colocalization with LAMP1 and TLR9. These results suggest that the base sequence of DNA, rather than its intracellular trafficking pattern, may be a critical determining factor for the activation of BM-DCs and that inactive DNA may be less effective than active DNA in interacting with TLR9. In agreement with this notion, an extremely high dose of inactive ODN123 completely suppressed the IL-12p40 production otherwise exhibited by BM-DCs stimulated with active ODN112 but not that by BM-DCs stimulated with CpG1826.
Thus, based on the results of the present study, a particular sequence of DNA from C. neoformans is suggested to be involved in activating immune cells through interacting with TLR9 in a manner independent of canonical CpG motifs. However, it remains unclear whether the specific sequence identified in this study is important in the initiation of immune responses when C. neoformans infects the host. In this respect, we found that active ODN112 markedly enhanced IL-12p40 production by BM-DCs stimulated with intact C. neoformans, whereas inactive ODN123 did not (see Fig. S2 in the supplemental material). These results suggest that this specific sequence may have some impact on the immune response during infection and that ODN112 could be used as an immune adjuvant similar to CpG-ODNs. Further investigations will be necessary to define the precise mechanism underlying the initiation of the immune response to C. neoformans.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shizuo Akira (Research Institute for Microbial Diseases, Osaka University, Osaka, Japan) for providing us with TLR9KO mice.
This work was supported in part by a grant (Research on Emerging and Re-Emerging Infectious Diseases [H22-SHINKOU-IPPAN-008]) from the Ministry of Health, Labor and Welfare of Japan and by a Grant-in-Aid for Scientific Research (B) (23390263) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
The authors have no financial conflict of interest.
Footnotes
Published ahead of print 21 November 2011
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Biondo C, et al. 2005. MyD88 and TLR2, but not TLR4, are required for host defense against Cryptococcus neoformans. Eur. J. Immunol. 35:870–878 [DOI] [PubMed] [Google Scholar]
- 2. Casadevall A, Freundlich LF, Marsh L, Scharff MD. 1992. Extensive allelic variation in Cryptococcus neoformans. J. Clin. Microbiol. 30:1080–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Edman JC, Kwon-Chung KJ. 1990. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol. Cell. Biol. 10:4538–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feldmesser M, Tucker S, Casadevall A. 2001. Intracellular parasitism of macrophages by Cryptococcus neoformans. Trends Microbiol. 9:273–278 [DOI] [PubMed] [Google Scholar]
- 5. Haas T, et al. 2008. The DNA sugar backbone 2′ deoxyribose determines Toll-like receptor 9 activation. Immunity 28:315–323 [DOI] [PubMed] [Google Scholar]
- 6. Hemmi H, et al. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740–745 [DOI] [PubMed] [Google Scholar]
- 7. Hill JO, Harmsen AG. 1991. Intrapulmonary growth and dissemination of an avirulent strain of Cryptococcus neoformans in mice depleted of CD4+ or CD8+ T cells. J. Exp. Med. 173:755–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Honda K, et al. 2005. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature 434:1035–1040 [DOI] [PubMed] [Google Scholar]
- 9. Kawai T, et al. 2004. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 5:1061–1068 [DOI] [PubMed] [Google Scholar]
- 10. Kawakami K, et al. 2001. Monocyte chemoattractant protein-1-dependent increase of Vα14 NKT cells in lungs and their roles in Th1 response and host defense in cryptococcal infection. J. Immunol. 167:6525–6532 [DOI] [PubMed] [Google Scholar]
- 11. Koguchi Y, Kawakami K. 2002. Cryptococcal infection and Th1-Th2 cytokine balance. Int. Rev. Immunol. 21:423–438 [DOI] [PubMed] [Google Scholar]
- 12. Krieg AM, et al. 1995. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374:546–549 [DOI] [PubMed] [Google Scholar]
- 13. Krieg AM. 2002. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 20:709–760 [DOI] [PubMed] [Google Scholar]
- 14. Latz E, et al. 2004. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 5:190–198 [DOI] [PubMed] [Google Scholar]
- 15. Lim TS, Murphy JW. 1980. Transfer of immunity to cryptococcosis by T-enriched splenic lymphocytes from Cryptococcus neoformans-sensitized mice. Infect. Immun. 30:5–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lipscomb MF, et al. 1987. Role of natural killer cells in resistance to Cryptococcus neoformans infections in mice. Am. J. Pathol. 128:354–361 [PMC free article] [PubMed] [Google Scholar]
- 17. Lutz MB, et al. 1999. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J. Immunol. Methods 223:77–92 [DOI] [PubMed] [Google Scholar]
- 18. Mansour MK, Latz E, Levitz SM. 2006. Cryptococcus neoformans glycoantigens are captured by multiple lectin receptors and presented by dendritic cells. J. Immunol. 176:3053–3061 [DOI] [PubMed] [Google Scholar]
- 19. Mednick AJ, Feldmesser M, Rivera J, Casadevall A. 2003. Neutropenia alters lung cytokine production in mice and reduces their susceptibility to pulmonary cryptococcosis. Eur. J. Immunol. 33:1744–1753 [DOI] [PubMed] [Google Scholar]
- 20. Miyazato A, et al. 2009. Toll-like receptor 9-dependent activation of myeloid dendritic cells by deoxynucleic acids from Candida albicans. Infect. Immun. 77:3056–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mody CH, Lipscomb MF, Street NE, Toews GB. 1990. Depletion of CD4+ (L3T4+) lymphocytes in vivo impairs murine host defense to Cryptococcus neoformans. J. Immunol. 144:1472–1477 [PubMed] [Google Scholar]
- 22. Nakamura K, et al. 2008. Deoxynucleic acids from Cryptococcus neoformans activate myeloid dendritic cells via a TLR9-dependent pathway. J. Immunol. 180:4067–4074 [DOI] [PubMed] [Google Scholar]
- 23. Petter R, Kang BS, Boekhout T, Davis BJ, Kwon-Chung KJ. 2001. A survey of heterobasidiomycetous yeasts for the presence of the genes homologous to virulence factors of Filobasidiella neoformans, CNLAC1 and CAP59. Microbiology 147:2029–2036 [DOI] [PubMed] [Google Scholar]
- 24. Ramirez-Ortiz ZG, et al. 2008. Toll-like receptor 9-dependent immune activation by unmethylated CpG motifs in Aspergillus fumigatus DNA. Infect. Immun. 76:2123–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takeda K, Kaisho T, Akira S. 2003. Toll-like receptors. Annu. Rev. Immunol. 21:335–376 [DOI] [PubMed] [Google Scholar]
- 26. Tanaka K, et al. 1996. Detection of Cryptococcus neoformans gene in patients with pulmonary cryptococcosis. J. Clin. Microbiol. 34:2826–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Uezu K, et al. 2004. Accumulation of γδ T cells in the lungs and their regulatory roles in Th1 response and host defense against pulmonary infection with Cryptococcus neoformans. J. Immunol. 172:7629–7634 [DOI] [PubMed] [Google Scholar]
- 28. Varma A, Edman JC, Kwon-Chung KJ. 1992. Molecular and genetic analysis of URA5 transformants of Cryptococcus neoformans. Infect. Immun. 60:1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wagner H. 2008. The sweetness of the DNA backbone drives Toll-like receptor 9. Curr. Opin. Immunol. 20:396–400 [DOI] [PubMed] [Google Scholar]
- 30. Xiao G, et al. 2010. Activation of myeloid dendritic cells by deoxynucleic acids from Cordyceps sinensis via a Toll-like receptor 9-dependent pathway. Cell. Immunol. 263:241–250 [DOI] [PubMed] [Google Scholar]
- 31. Yasuda K, Rutz M, Schlatter B. 2006. CpG motif-independent activation of TLR9 upon endosomal translocation of “natural” phosphodiester DNA. Eur. J. Immunol. 36:431–436 [DOI] [PubMed] [Google Scholar]
- 32. Yasuoka A, Kohno S, Yamada H, Kaku M, Koga H. 1994. Influence of molecular sizes of Cryptococcus neoformans capsular polysaccharide on phagocytosis. Microbiol. Immunol. 38:851–856 [DOI] [PubMed] [Google Scholar]
- 33. Yauch LE, Mansour MK, Shoham S, Rottman JB, Levitz SM. 2004. Involvement of CD14, Toll-like receptors 2 and 4, and MyD88 in the host response to the fungal pathogen Cryptococcus neoformans in vivo. Infect. Immun. 72:5373–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y, et al. 2010. TLR9 signaling is required for generation of the adaptive immune protection in Cryptococcus neoformans-infected lungs. Am. J. Pathol. 177:754–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.