Abstract
Salmonella enterica serovar Enteritidis causes a systemic, typhoid-like infection in newly hatched poultry and mice. In the present study, a library of 54,000 transposon mutants of S. Enteritidis phage type 4 (PT4) strain P125109 was screened for mutants deficient in the in vivo colonization of the BALB/c mouse model using a microarray-based negative-selection screening. Mutants in genes known to contribute to systemic infection (e.g., Salmonella pathogenicity island 2 [SPI-2], aro, rfa, rfb, phoP, and phoQ) and enteric infection (e.g., SPI-1 and SPI-5) in this and other Salmonella serovars displayed colonization defects in our assay. In addition, a strong attenuation was observed for mutants in genes and genomic islands that are not present in S. Typhimurium or in most other Salmonella serovars. These genes include a type I restriction/modification system (SEN4290 to SEN4292), the peg fimbrial operon (SEN2144A to SEN2145B), a putative pathogenicity island (SEN1970 to SEN1999), and a type VI secretion system remnant SEN1001, encoding a hypothetical protein containing a lysin motif (LysM) domain associated with peptidoglycan binding. Proliferation defects for mutants in these individual genes and in exemplar genes for each of these clusters were confirmed in competitive infections with wild-type S. Enteritidis. A ΔSEN1001 mutant was defective for survival within RAW264.7 murine macrophages in vitro. Complementation assays directly linked the SEN1001 gene to phenotypes observed in vivo and in vitro. The genes identified here may perform novel virulence functions not characterized in previous Salmonella models.
INTRODUCTION
Salmonella is a Gram-negative bacterium belonging to the family Enterobacteriaceae. The genus comprises two species, S. bongori and S. enterica, which include more than 2,500 serovars (17). Salmonella enterica serovar Enteritidis can infect a wide variety of hosts, including birds, rodents (mice), and humans (36). S. Enteritidis is a facultative intracellular pathogen that invades epithelial host cells and uses phagocytic cells to disseminate, causing a systemic, typhoid-like infection in mice and newly hatched poultry. In humans, S. Enteritidis causes a self-limited gastroenteritis characterized by diarrhea, fever, and abdominal cramps (45).
In recent years, S. Enteritidis has emerged as a primary cause of food-borne salmonellosis worldwide. Epidemiological studies suggest that S. Enteritidis filled the ecological niche vacated by the eradication of S. Gallinarum from commercial poultry flocks, increasing human infections progressively due to the consumption of contaminated eggs and egg products from apparently healthy hens (38, 63). Because the infection of adult hens by S. Enteritidis is asymptomatic, the contamination of eggs due to the colonization of the oviduct is hard to detect and prevent (38). To avoid poultry stock infection by this pathogen, some approaches have been addressed to design vaccines for laying hens using live, attenuated mutants (11, 16, 57) or bacteria killed by chemical agents or heat (6, 11, 21). None of these vaccines have been shown to significantly diminish the intestinal colonization or the shedding of the pathogen in the depositions. On the other hand, the emergence of strains resistant to the antibiotics commonly used to treat the disease encourages the identification of new targets for vaccine design.
The mechanisms involved in S. Enteritidis pathogenicity are poorly understood. Most knowledge is based on research conducted on S. Typhimurium, which causes a similar disease in humans and a systemic disease in mice. Although these pathogens share virulence mechanisms central to epithelial cell invasion and survival within phagocytic cells, it is not yet clear how S. Enteritidis is able to cause infection, especially in one of its main reservoirs: mice. Nonetheless, because S. Enteritidis causes a systemic infection in newly hatched poultry and mice, an approach using the in vivo colonization of liver and spleen of BALB/c mice is a suitable model to study the major virulence factors involved in the acute phase of the infection.
Several studies using microarray-based genetic screening have been conducted to identify genes involved in S. Typhimurium pathogenesis in both in vitro and in vivo models (22, 24, 56, 72). However, these approaches have not been used to explore the basis of S. Enteritidis pathogenesis. In the present study, we performed a genome-wide search in order to identify genes and genomic islands of S. Enteritidis that have a role in virulence during the infection of BALB/c mice. To accomplish this, we implemented a microarray-based negative-selection screen for mutants showing an impaired colonization of deep organs in mice. The phenotypes of selected insertion mutants that were less fit during infection in our screen were confirmed by competition assays with individual mutants of some of the genes or genomic islands identified in this study. Furthermore, the colonization defect presented by one of these mutants, the ΔSEN1001 mutant, was directly linked to a defect in intracellular survival within macrophages.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains used in the present study are listed in Table 1. All S. Enteritidis strains are derivatives of the sequenced phage type 4 (PT4) strain P125109 (77). Bacteria were routinely grown in Luria-Bertani (LB) medium (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl) at 37°C with shaking or in E minimal medium (0.2 g/liter MgSO4 · 7H2O, 2 g/liter citric acid · H2O, 13.1 g/liter K2HPO4 · 3H2O, 3.3 g/liter NaNH4HPO4 · 4H2O) with glucose (0.2%). When required, LB medium was supplemented with ampicillin (Amp) (100 mg/liter), chloramphenicol (Cam) (20 mg/liter), kanamycin (Kan) (50 mg/liter), or 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) (50 mg/liter). Media were solidified by the addition of agar (15 g/liter).
Table 1.
Strains used in the present study
| Strain | Genotype or relevant phenotype | Source or reference |
|---|---|---|
| S. Enteritidis | ||
| P125109 | S. Enteritidis PT4 wild type (NCTC13349) | Laboratory collection |
| WT/pNFB9 | attG1::pNFB9 | |
| ΔSEN1001 | ΔSEN1001::FRT | This study |
| ΔSEN1001/pNFB9 | ΔSEN1001::FRT attG1::pNFB9 | This study |
| ΔSEN1001/pCSV1 | ΔSEN1001::FRT attG1::pCSV1 | This study |
| ΔSEN1976 | ΔSEN1976::Cam | This study |
| ΔROD21 | Δ(SEN1976–SEN1999)::Cam | This study |
| Δpeg | Δ(SEN2144A–SEN2145B)::Kan | This study |
| ΔROD40 | Δ(SEN4290–SEN4292)::Kan | This study |
| S. Typhimurium SC203 | 14028 zjg-8103::pir+/F′ proAB lacIq ΔlacZM15 zzf8169::Tn10dKn | Laboratory collection |
| Escherichia coli DH5α | K-12 F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169deoRrecA1endA1hsdR17(rk− mk+) phoAsupE44thi-1gyrA96relA1 λ− | Laboratory collection |
Standard DNA techniques.
Total genomic DNA was obtained by use of the GenElute bacterial genomic DNA kit (Sigma-Aldrich, St. Louis, MO). Plasmid DNA was obtained by use of the QIAprep Spin miniprep kit (Qiagen, Germantown, MD). Digestions using the restriction endonucleases EcoRI, BamHI, and BglII (Fermentas, St. Leon-Rot, Germany) and ligations using T4 DNA ligase (New England BioLabs, Beverly, MA) were conducted as recommended by the manufacturers. DNA samples were routinely analyzed by electrophoresis in 1% agarose gels (0.5× Tris-acetate-EDTA [TAE] buffer) and visualized under UV light after ethidium bromide staining.
Primers for PCR amplification were designed based on the reported sequence of S. Enteritidis PT4 strain P125109 (see Table S1 in the supplemental material), using Vector NTI Advance 9.0 software. PCR amplifications were performed by using GoTaq Flexi DNA polymerase (Promega, Madison, WI). Reaction mixes contained 1× buffer, 2 mM MgCl2, 200 μM each deoxynucleoside triphosphate (dNTP), 200 nM each primer, 0.5 μl of template DNA (50 to 100 ng), and 0.5 U of DNA polymerase. Standard conditions for amplification were as follows: 3 min at 94°C followed by 30 cycles of incubations at 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min, followed by a final extension step at 72°C for 5 min. PCR products were purified by using the QIAquick PCR purification kit (Qiagen, Germantown, MD).
Construction of the transposon library.
S. Enteritidis PT4 strain P125109 was mutagenized with a mini-Tn5 derivative using the EZ-Tn5 <T7/KAN-2> promoter insertion kit (Epicentre Biotechnologies, Madison, WI). Briefly, transposome complexes were assembled in vitro by mixing 2 μl of the EZ-Tn5 <T7/KAN-2> transposon (0.1 pmol/μl), 2 μl of EZ-Tn5 transposase (1 U/μl), and 2 μl of 100% glycerol. The mix was incubated for 30 to 45 min at room temperature. Immediately before mutagenesis, 3 μl of transposome complexes was dialyzed against sterile water for 20 min at room temperature using filters with a 0.025-μm pore size (Millipore). The dialyzed material was recovered, mixed with 1 μl of Type One restriction inhibitor (Epicentre Biotechnologies, Madison, WI), and transformed into fresh electrocompetent P125109 cells. Transformants were allowed to recover in 2 ml of LB medium for 45 min at 37°C. Aliquots of 100 μl from the transformation were plated onto LB supplemented with Kan (LB-Kan) agar plates and incubated overnight at 37°C. The mutagenesis procedure was repeated 4 times. A total of ∼54,000 Kanr mutants generated were scraped from the plates, washed three times in sterile phosphate-buffered saline (PBS), and resuspended in LB medium to assemble the initial transposon library. Glycerol stocks of the library were aliquoted and stored frozen at −80°C.
To confirm transposon insertions, chromosomal DNA obtained from each of 15 randomly selected clones was used as a template for PCR amplification with primers designed to detect the KAN-2 gene (see Table S1 in the supplemental material). All 15 clones contained a transposon insertion (data not shown). The presence of single insertions at different chromosomal locations was confirmed by Southern blotting after DNA digestion with EcoRI and hybridization against a probe generated by the PCR amplification of the KAN-2 gene (data not shown). To select for auxotrophic mutants, 248 colonies were randomly selected and plated onto LB agar plates and onto E medium agar plates supplemented with glucose (0.2%). Of the 248 colonies, 5 were unable to grow in E medium compared to the rich medium. Thus, approximately 2% of auxotrophic mutants were obtained in the library.
Animal studies in BALB/c mice.
A frozen glycerol aliquot of the library was used to inoculate 50 ml of LB-Kan. After overnight growth with agitation at 37°C, the bacteria were pelleted, washed three times, and diluted 1/100 in sterile PBS for inoculation. The exact titer of the inoculum was determined by serial dilution and plating onto LB-Kan agar. A group of 6 BALB/c mice (8- to 10-week-old females) was infected intraperitoneally (i.p.) with ∼106 CFU of the library in 100 μl of PBS. Samples of the inoculated material (input library) were kept frozen at −20°C until further use. Mice were euthanized at 48 h postinfection, and the spleen and liver were removed aseptically and homogenized in 3 ml of ice-cold, sterile PBS. An aliquot of 100 μl from each homogenate was used for serial dilution and determinations of titers on LB-Kan agar plates. The remaining homogenate was inoculated into 100 ml of LB-Kan. After overnight growth with agitation at 37°C, the bacteria (output libraries) were pelleted, washed 3 times in ice-cold sterile PBS, and stored frozen at −20°C until further use.
Individual mutants with an apparent phenotype in our microarray-based negative-selection screen were chosen for confirmation of mixed infections with wild-type S. Enteritidis PT4 strain P125109 to determine the competitive index (CI). Each mutant strain to be tested and the wild-type strain were grown overnight at 37°C with aeration, mixed at a 1:1 ratio, and diluted 1/100 in PBS for inoculation. The exact titer and ratio of strains in the inoculum were determined by serial dilution and plating onto LB agar (total CFU) and LB-Kan or LB-Cam agar (mutant CFU). Groups of 3 to 5 BALB/c mice (8- to 10-week-old females) were inoculated i.p. with ∼106 CFU of the mixture of the mutant and wild-type strains in 100 μl of PBS. Mice were euthanized at 48 h postinfection, and the spleens and livers were removed and homogenized. Each homogenate was serially diluted, and aliquots of the dilutions were plated onto LB and LB-Kan or LB-Cam agar plates to determine the exact ratio of strains recovered. CI values were calculated as the mean ratio of mutant CFU to wild-type CFU, normalized to the input ratio and converted logarithmically. Statistical significance was determined by using a two-tailed Student t test.
Labeling of DNA adjacent to transposon insertions and microarray analysis.
The DNA adjacent to transposon insertions in a mutant library was specifically amplified as described previously (5, 72), with modifications. Genomic DNA was prepared for the input and output libraries by using the GenElute bacterial genomic DNA kit (Sigma-Aldrich, St. Louis, MO). Four micrograms of genomic DNA from each library was then fragmented by sonication using a Branson Sonifier 150 instrument (Branson Ultrasonics). Poly(A) tails were added to the fragmented DNA using terminal transferase (TdT) as follows: 1.5 μg of DNA fragments was incubated for 30 min at 37°C in a total reaction mixture volume of 50 μl containing 40 U TdT (New England BioLabs, Beverly, MA), 1× buffer, 0.25 mM CoCl2, and 0.4 mM dATP. Terminal transferase was subsequently inactivated at 70°C for 10 min, and the tailed product was purified by use of the QIAquick PCR purification kit (Qiagen, Germantown, MD).
A nested PCR strategy was used to amplify only the poly(A)-tailed DNA fragments containing the transposon end carrying PT7 and the genomic DNA flanking the insertion. In the first amplification reaction, 50 ng of purified poly(A)-tailed DNA was used as a template for a PCR using primers DOPR2 and CCT24VN (see Table S1 in the supplemental material). The reaction mixture consisted of 1× buffer, 200 μM each dNTP, 1.5 mM MgCl2, 1.25 U Taq polymerase (Promega, Madison, WI), and 0.2 μM each primer in a total reaction mixture volume of 25 μl. The amplification was carried out under the following conditions: an initial denaturation step at 94°C for 1 min followed by 30 cycles with denaturation at 94°C for 10 s, annealing at 50°C for 10 s, and extension at 72°C for 5 s. The last cycle was followed by a final extension step for 3 min at 72°C. In the second amplification step, a nested PCR was performed by using 1 μl of amplified product from the initial PCR as a template in a total volume of 50 μl. Internal primer KAN2FP1-B and primer CCT24VN (see Table S1 in the supplemental material) were used under cycling conditions identical to those used during the initial PCR.
An aliquot of 5 μl of the nested PCR mixture was used directly as a template for a 20-μl in vitro transcription reaction mixture using the AmpliScribe T7 transcription kit (Epicentre Biotechnologies, Madison, WI) according to the manufacturer's protocol. The RNA was treated with RNase-free DNase (Epicentre Biotechnologies, Madison, WI), purified with the RNeasy minikit (Qiagen, Germantown, MD), and used as a template to synthesize labeled cDNA by the incorporation of Cy5-dCTP or Cy3-dCTP (Amersham Biosciences Piscataway, NJ) using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA). The labeling protocol was repeated with dyes reversed for input and output samples. Finally, the labeled cDNA was purified by using the QIAquick PCR purification kit (Qiagen, Germantown, MD) and hybridized in slides containing a Salmonella microarray based on S. Typhimurium LT2 genes and supplemented with genes specific to S. Enteritidis PT4 strain P125109 (65). The slides containing the microarray were printed in triplicate.
Hybridization and data analysis.
Standard protocols for hybridizations in formamide buffer were applied for prehybridization, hybridization, and posthybridization wash processes (http://catalog2.corning.com/Lifesciences/media/pdf/gaps_ii_manual_protocol_5_02_cls_gaps_005.pdf). Immediately before hybridization, 2 μg of labeled probes from the input sample was combined with 2 μg of labeled probes from one of the output samples, and the volume was adjusted to 40 μl with nuclease-free water. The mixture of probes was combined with 40 μl of 2× hybridization buffer (50% formamide, 10× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.2% SDS) and then denatured at >95°C for 5 min. The probes were hybridized to the Salmonella microarray at 42°C in a water bath. After incubation overnight, the slides were washed and scanned by using the ScanArray 5000 laser scanner (Packard BioChip Technologies, Billerica, MA) with ScanArray 2.1 software. Fluorescence signal intensities were quantified by using the QuantArray 2.0 software package (Packard BioChip Technologies, Billerica, MA). The data were normalized and analyzed by using Webarray (82) and WebarrayDB (http://www.webarraydb.org) (83), with quantile normalization.
Mutant construction and analysis.
Mutant strains with specific deletions and concomitant insertions of a Kan or Cam resistance cassette were constructed by using the lambda Red recombination method (30), with modifications (72). PCR amplifications of an antibiotic resistance cassette flanked by the FRT (Flp recombinase target sequence) sites present in plasmid pCLF2 (Camr) or pCLF4 (Kanr) were carried out under standard conditions using the primers described in Table S1 in the supplemental material. S. Enteritidis PT4 strain P125109 containing plasmid pKD46, which synthesizes the lambda Red recombination system, was grown to an optical density at 600 nm (OD600) of 0.5 at 30°C in LB medium containing Amp and l-arabinose (10 mM) and transformed with approximately 500 ng of each purified PCR product. Transformants were selected on LB agar plates containing Kan or Cam at 37°C. The presence of each mutation was confirmed by PCR amplification and transferred to the wild-type background by generalized transduction using the high-frequency transducing phage P22 HT105/1 int-201.
To generate nonpolar, in-frame deletions of selected genes of interest, the corresponding Kanr or Camr derivatives were transformed with temperature-sensitive plasmid pCP20 that synthesizes the Flp recombinase (25, 30). Transformants were selected at 30°C on LB agar plates containing Amp. A few colonies were streaked two consecutive times at 37°C onto LB agar plates and tested for the loss of the antibiotic resistance cassettes and pCP20 by patching them onto LB agar plates containing Kan or Cam plus Amp. The presence of the mutant allele was confirmed by PCR amplification using primers flanking the sites of substitution.
Cloning of the SEN1001 gene for complementation assays.
Primers SEN1001_out5_BamHI and SEN1001_out3_BamHI (see Table S1 in the supplemental material), including restriction sites for BamHI endonuclease, were used to amplify a 1,181-bp fragment of the S. Enteritidis PT4 strain P125109 genome, including the SEN1001 gene. The PCR product was purified and ligated into plasmid pGEM-T Easy (Promega, Madison, WI). The ligation mix was transformed by electroporation into competent Escherichia coli DH5α cells, and transformants were selected on LB agar plates containing Amp and X-gal. Plasmid DNA was prepared from several independent transformants. The presence of the insert in each plasmid was confirmed by PCR amplification using primers T7 and SP6 (see Table S1 in the supplemental material), flanking the multiple-cloning site in pGEM-T Easy. Subsequently, the cloned fragment was subcloned into plasmid pNFB9, a pir-dependent plasmid carrying the int gene and the attP site of bacteriophage Gifsy-1 (49). To do this, a plasmid with SEN1001 cloned in the same orientation as that of Plac into pGEM-T Easy was digested with BamHI. The DNA fragment including SEN1001 was purified and ligated into plasmid pNFB9 previously digested with BglII. The ligation mix was transformed by electroporation into S. Typhimurium strain SC203, and transformants were selected on LB agar plates containing Amp and X-gal. The presence of the insert in the recombinant plasmids was confirmed by PCR amplification using primers NFB9-MCS_Out5 and NFB9-MCS_Out3 (see Table S1 in the supplemental material). Plasmid pCSV1, carrying SEN1001 cloned into pNFB9 in the same orientation as that of Plac, and plasmid pNFB9 were used to transform the S. Enteritidis P125109 ΔSEN1001 mutant in complementation assays. The chromosomal insertion of pNFB9 derivatives was confirmed by PCR amplification using primers LepA_F and LepA_R (see Table S1 in the supplemental material), flanking the attG1 site.
Cell culture and intracellular survival assay in vitro.
RAW264.7 macrophages were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Monolayers for infection were prepared by seeding ∼105 cells into each well of a 48-well multiwell plate and incubating the cells for 20 h at 37°C in the presence of 5% CO2. Bacteria were grown overnight in LB medium at 37°C under static conditions, washed with PBS, suspended in DMEM-FBS (100 μl), mixed, and added to RAW264.7 macrophages at a multiplicity of infection of ∼50:1. After 45 min of incubation at 37°C, the macrophages were washed three times with PBS and incubated for 2 h or 20 h in DMEM-FBS containing gentamicin (50 μg/ml) to kill extracellular bacteria. Cells were washed three times with PBS and lysed with 0.5% deoxycholate in PBS. Numbers of intracellular CFU were determined by serial dilution and plating. At each time point, the number of viable RAW264.7 cells was assessed by 0.4% trypan blue exclusion and counting. Each experiment was performed on two separate occasions, evaluating samples at least in duplicate. Statistical significance was determined by using the one-way analysis of variance (ANOVA) test and the Dunnett's posttest.
RESULTS AND DISCUSSION
Identification of genes with a role in S. Enteritidis pathogenesis in BALB/c mice by negative selection.
The aim of our study was to identify S. Enteritidis genes that are important for the establishment of a systemic infection in mice. The sequenced S. Enteritidis PT4 strain P125109 (77) colonizes the gastrointestinal tract of streptomycin-pretreated mice when administered orally (76). To evaluate whether this strain is able to colonize systemically during an acute infection of BALB/c mice when administered i.p., 3 mice were inoculated with ∼4.4 × 106 CFU of S. Enteritidis P125109. The mice were euthanized 48 h after the inoculation, and the spleen and liver were aseptically removed. The infected organs looked normal in shape and color, but an enlargement of the infected spleens (splenomegaly) was evident, compared to the organs recovered from a control mouse inoculated with sterile PBS. A small but considerable enlargement of the infected livers was also observed. Bacteria colonizing the spleen and liver of each animal were counted. The number of CFU recovered from spleen ranged from 3.8 × 106 to 4.2 × 106, while the number of CFU recovered from liver ranged from 2.2 × 106 to 6.9 × 107 (see Fig. S1A in the supplemental material).
Next, a library of transposon mutants was generated in S. Enteritidis P125109 using a mini-Tn5 derivative. Approximately 54,000 independent colonies were obtained, representing an average of more than 1 insertion per 100 bases. A group of 6 BALB/c mice was injected i.p. with ∼4.2 × 106 CFU of the transposon library. After 48 h, bacteria were recovered from the spleens and livers of infected mice. The number of CFU recovered from spleen ranged from 2.0 × 106 to 6.6 × 106, while the number of CFU recovered from liver ranged from 2.2 × 106 to 6.9 × 107 (see Fig. S1B in the supplemental material). Mutants carrying insertions in genes contributing to fitness for the colonization of mice, underrepresented in the population recovered from infection, were identified by means of an array-based strategy similar to ABACUS (72), using a microarray designed for the study of several Salmonella serovars which includes S. Enteritidis-specific genes (65).
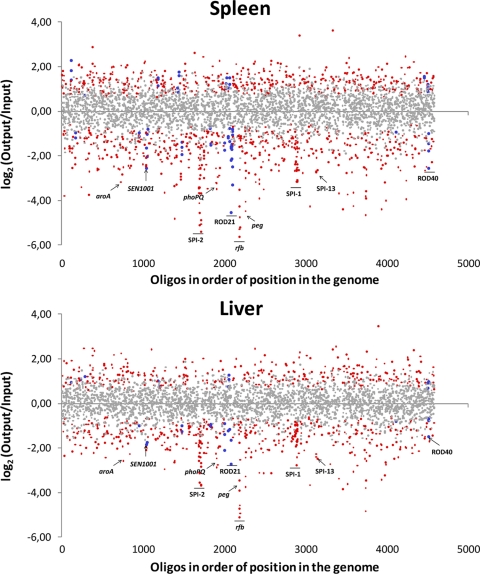
Data for all the genes on the arrays are presented in Table S2 in the supplemental material and are schematized in Fig. 1. Overall, there is an excellent correlation between data from spleen samples and data from liver samples (see Table S2 and Fig. S2 in the supplemental material), indicating that any genes differentially required for the colonization of liver or spleen by S. Enteritidis cannot be detected by this assay. The same observation was reported previously for S. Typhimurium (56). Of the 4,505 “spots” analyzed, representing 4,288 genes of S. Enteritidis (4,231 chromosomal genes and 57 genes carried on the virulence plasmid), 3,700 entries (3,521 genes) with a sufficient signal intensity to be detected were identified. As expected, mutants in genes considered essential for bacterial viability were not detected, since this class of genes does not tolerate insertion mutations. For example, mutants in all 26 genes of the essential ribosomal protein cluster, SEN3244 to SEN3269, remained undetected in the control sample (see Table S2 in the supplemental material). Of the 3,521 genes detected, 306 had a statistically significant log2-fold attenuation score (M) in both organs (M ≤ −0.75 and P ≤ 0.001). The list includes 21 genes absent in the genome of S. Typhimurium (see Table S3 in the supplemental material). Thus, they were considered candidate genes for unique aspects of S. Enteritidis colonization of BALB/c mice by the intraperitoneal route.
Fig 1.
Loss of mutants after in vivo selection by i.p. injection into BALB/c mice and recovery from spleen and liver at 48 h postinfection. Six array hybridizations per organ were performed for a total of six mice. Data with P values of <0.001 (see Materials and Methods) are depicted in red, and those with P values of >0.001 are depicted in gray. Data from S. Enteritidis-specific genes with P values of <0.001 are depicted in blue. The x axis plots the data for each of the probes in the array according to the position of the corresponding gene in the genome of S. Enteritidis strain P125109.
Identification of genes with a role in S. Enteritidis pathogenicity that are shared with other Salmonella serovars.
Analysis of the data in Table S2 in the supplemental material revealed that the most dramatic phenotypes were observed for mutations of genes carried on Salmonella pathogenicity island 2 (SPI-2). These results agree with those obtained previously by using S. Typhimurium in similar infection-based (22, 24, 56, 72) and in vitro (22) studies, consistent with the importance of SPI-2 for the growth within macrophages (26, 47), persistence (56), and pathogenicity (74) of Salmonella.
Mutants lacking the structural genes of the type III secretion system (T3SS) carried on SPI-1, its transcriptional regulators, and secreted effectors were underrepresented in our output libraries. This result agrees with earlier work with S. Typhimurium by Lawley et al. indicating that SPI-1 genes are needed for the colonization of the spleen and liver (56). Even though these results seem to challenge the original model that SPI-1 is needed for the gastrointestinal phase of salmonellosis but not the systemic phase (2, 36, 46, 81), it is conceivable that Salmonella may rapidly occupy additional niches in the mouse (such as the gut) after intraperitoneal infection, and bacteria in these niches may subsequently contribute to the population seen in internal organs. Therefore, mutants in genes that have a role in gastrointestinal infection may be underrepresented in the bacterial population harvested subsequently from spleen and liver (72).
Mutations of the magnesium transport-related genes mgtB and mgtC within SPI-3 were also negatively selected. The mgtC gene is required for growth in low Mg2+ and for the survival in of S. Typhi in macrophages (69) as well as for the survival of S. Typhimurium in macrophages and pathogenicity in mice (12). In contrast, mgtB is dispensable for pathogenicity in BALB/c mice (12). mgtB is located downstream of mgtC in a single transcriptional unit, making it likely that our transposon insertion in mgtC has a polar effect on mgtB.
Mutations of genes encoding a giant nonfimbrial adhesin and the cognate type I secretion system located on SPI-4 (37, 80) were underrepresented in our output libraries. Genes on SPI-4 are needed for the intestinal phase of disease (58) and were associated with the systemic infection of mice by S. Typhimurium and S. Enteritidis when bacteria were administered orally (54).
Three genes located on SPI-5, pipB, pipC and sopB, were also negatively selected in our screen. Genes of SPI-5 contribute to enteric but not to systemic infection by S. Dublin (81). However, this pathogenicity island was described previously to be coregulated with either SPI-1 or SPI-2 genes; thus, it may perform a dual function during infection (55). In fact, PipB is an effector protein translocated by the SPI-2 T3SS, whereas SopB is an effector protein translocated by the SPI-1 T3SS, and PipC is a putative chaperone assisting SopB secretion (55, 67, 81). Hence, it was not surprising to find these genes after negative selection.
Two recent studies evaluated the role of the five major SPIs in the pathogenicity of S. Enteritidis strain 147. One study indicated that bacterial pathogenicity in BALB/c mice was dependent exclusively on the presence of an intact SPI-2 (53), while the other study found that the colonization of chickens' liver and spleen was dependent on the presence of either SPI-1 or SPI-2. The absence of SPI-3, SPI-4, or SPI-5 individually did not influence the pathogenicity of S. Enteritidis for chicken, but the absence of the three islands collectively contributed to the colonization of the spleen (70). Our sensitive screen for the relative fitness of mutants in the presence of wild-type bacteria indicates that, in addition to genes in SPI-1 and SPI-2, some genes in SPI-3, SPI-4, and SPI-5 contribute to the systemic colonization of mice. Thus, infections in mice and chickens may be different with respect to the need for these three SPIs.
Mutations in groups of genes belonging to other pathogenicity islands were also detected in our screen. The S. Enteritidis genes SEN2961 to SEN2164 within SPI-13 and the genes SEN0801 to SEN0804 within SPI-14 were needed for full colonizing abilities. Phenotypes for mutations in genes within SPI-13 and SPI-14 during the systemic infection of mice by S. Typhimurium were also reported previously (22, 43, 72). In addition, both SPI-13 and SPI-14 were identified as being required for S. Gallinarum pathogenicity in a chicken infection model (73). Finally, the genes SEN0535 and SEN0536 within SPI-16 also were negatively selected during S. Enteritidis infection of mice. We have shown previously that the orthologous genes in S. Typhimurium are responsible for the “form variation” of the O12 antigen to generate the 12-2 variant that is critical for the persistence of this serovar in the murine intestine (15).
In addition to the genes belonging to highly conserved SPIs, other well-established pathogenicity-related genes that were negatively selected in our screen included (i) the aromatic amino acid biosynthesis genes aroE, aroD, aroB, and aroP and the purine biosynthesis genes purF, purD, purB, purR, and purA, which were previously identified by Chaudhuri et al. as being needed for the systemic infection of mice by S. Typhimurium (24), in accordance with previous reports showing that auxotrophic mutations of these genes lead to a reduced pathogenicity of S. Typhi, S. Typhimurium, and S. Enteritidis (11, 31, 62); (ii) the anaerobic metabolism genes narH and narI and the oxidative phosphorylation genes cyoC, cyoE, and nuo, which have been shown to contribute to the pathogenicity of S. Typhimurium, S. Dublin, and S. Gallinarum (78); (iii) the lipopolysaccharide (LPS) biosynthesis genes rfaB, rfaG, rfaI, rfaJ, rfaL, rfbF, rfbH, rfbI, rfbK, and rfc, supporting data from previous reports that established an important role of LPS in the invasion of epithelial cells by S. Typhi and S. Typhimurium (50) and in the colonization of the intestine of mice by S. Typhimurium (29, 51, 60) and of chicken by S. Enteritidis (23) and a role for LPS in S. Enteritidis adherence to and survival in eggs (39, 40); and (iv) the membrane stability genes tolA and tolB and the transport system genes sfbB and sfbC (64), SEN0715, SEN0716, and SEN1417 to SEN1420. TolC, a member of the AcrAB-TolC efflux system that was also negatively selected in our screening, was previously shown to be required for the infection of BALB/c mice by S. Enteritidis (75) and for the infection of poultry by S. Typhimurium (18).
Finally, mutants in genes involved in the regulation of responses to external stimuli, such as phoP, phoQ, arcA, arcB, envZ, fimZ, rpoN, crp, and fur, showed colonization deficiencies in our screen, consistent with their global importance in Salmonella pathogenicity (4, 34, 42, 56, 68, 79). In addition, we identified mutants in genes regulated by the PhoP/Q system (pagO, pqaA, and pagD). It is noteworthy that the pagO and pagD genes are dispensable for S. Typhimurium pathogenicity in mice (10, 41), and pqaA mutants of S. Typhi are fully virulent in the mouse mucin virulence assay (7), suggesting that transposon insertion mutants in these genes were identified in our screen due to polar effects on nearby virulence genes. Alternatively, pagO, pqaA, and pagD may play a specific role in S. Enteritidis pathogenicity in mice.
Some S. Enteritidis-specific regions were not sampled in our study either because they were not represented in the array or because the corresponding probes produced weak hybridization signals. For example, the yfg-env locus was reported previously to be needed for chicken colonization by S. Enteritidis after oral inoculation, and a ΔyfgL mutant was less motile than the wild type (3). More recently, it was shown that YfgL is necessary for the assembly and function of T3SSs, including flagella and those encoded in SPI-1 and SPI-2 (35). In addition, Chang et al. described a novel gene involved in the ability to survive or replicate inside the chicken host after subcutaneous delivery: yhbC. This gene encodes a hypothetical protein, and the ΔyhbC mutant had a diminished ability to grow in vitro compared to the wild-type strain (23). We do not yet know the effect of mutations in these genes on mice.
Identification of novel genes with a role in S. Enteritidis pathogenicity.
Thomson et al. described “regions of difference” (RODs) between the genomes of Salmonella serovars Enteritidis, Gallinarum, and Typhimurium (77). The regions reported as being specific to Salmonella serovar Enteritidis include ROD4 (SEN0216) and the prophages ϕSE14 (SEN1378 to SEN1398) and ϕSE20 (SEN1919A to SEN1966). ϕSE14 includes the S. Enteritidis-specific locus lyg, identified previously by Agron and colleagues (1), and correspond to an unstable genetic element that is not required for the systemic colonization of BALB/c mice (71). However, in this study, we found that the genes SEN1921 and SEN1960 in ϕSE20 were needed for the colonization of BALB/c mice. These genes encode a putative tail fiber assembly protein and a conserved hypothetical protein called sb35, respectively. ϕSE20 carries fragments of the sopE and orgA genes previously implicated in Salmonella pathogenicity; however, these were not detected in our screening.
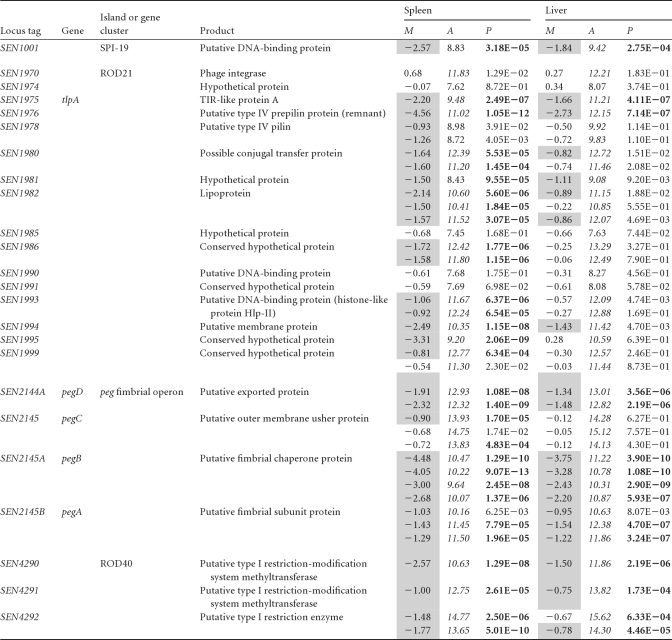
Mutations linked to other genes and genomic islands that have not been previously related to S. Enteritidis pathogenicity were identified as being negatively selected in this study (Table 2). These included SEN1001 and SEN1002, carried on SPI-19 (13); a cluster of genes linked to tRNA-asnT in a putative pathogenicity island (SEN1970 to SEN1999); the peg fimbrial operon (SEN2144A to SEN2145B); and components of a type I restriction-modification system (SEN4290 to SEN4292).
Table 2. New genes and genomic islands related to S. Enteritidis pathogenicitya.
“M” corresponds to the log2-fold attenuation score (M = log2 output − log2 input). M values of ≤−0.75 are shaded. “A” corresponds to the average log2-fold signal intensity [A = (log2 output + log2 input/2)]. A values of ≥9 are in italic type. P values of ≤0.001 are highlighted in boldface type.
SEN1001 and SEN1002 are remnant components of a type VI secretion system (T6SS) carried on SPI-19, a pathogenicity island recently described for Salmonella serovars Enteritidis, Gallinarum, Dublin, Agona, and Weltevreden (13). SEN1002 encodes an Hcp-like protein that appears to be a structural and secreted component of these systems (59). SEN1001 encodes a hypothetical protein of unknown function. The identification of these genes in our screening was unexpected, since S. Enteritidis does not encode all the components described as being essential for T6SS function. It is tempting to speculate that these genes have novel functions unlinked to the T6SS in S. Enteritidis. The T6SS of SPI-19 contributes to the colonization of the gastrointestinal tract and internal organs of chickens by S. Gallinarum (14).
The cluster of genes linked to tRNA-asnT (SEN1970 to SEN1999), officially designated region of difference 21 (ROD21) (77), contains a putative integrase (SEN1970) and is flanked by direct repeats, properties common to genomic islands acquired by lateral gene transfer. This island includes SEN1975, encoding a Toll/interleukin-1 (IL-1) receptor (TIR) domain containing-protein called TlpA, described previously for S. Enteritidis strain LK5 (61). The secretion of proteins with TIR domains was shown previously to interfere with NF-κB induction, allowing the evasion of immune defenses by the pathogen (27, 61). TlpA also induces caspase-1 activation, leading to IL-1β secretion and the induction of apoptosis in macrophages, and is needed for the systemic colonization of mice (61). Interestingly, ROD21 also harbors precursors and structural components of putative type IV pili (SEN1976 to SEN1978), structures linked to motility, adherence, biofilm formation, and bacterial aggregation, and contributes to the invasion of the host cell (44). Our analysis identified these genes as being needed for the colonization of liver and spleen. Other regions of ROD21 identified in our screen include SEN1980 to SEN1982 and SEN1993 to SEN1995, most encoding hypothetical proteins with an unknown function.
Fimbriae play a critical role in host cell adherence (33). According to our screen, two fimbrial operons, peg and sef, were needed for the full pathogenicity of Salmonella serovar Enteritidis in mice. It was previously demonstrated that SEF14 fimbriae are required to avoid uptake by macrophages after intraperitoneal infection of mice and are needed for S. Enteritidis survival in this model (32). Also, the importance of the peg fimbrial operon in the colonization of chicken ceca after oral administration was previously shown (28).
The genes SEN4290 to SEN4292 belong to the region annotated as ROD40 (77), which encodes a putative type I restriction-modification system, systems that protect bacteria from foreign DNA. The type I restriction-modification system consists of a protein complex with three different subunits responsible for the binding to, methylation of, and restriction of DNA (20, 66). Only one previous study conducted with Yersinia pseudotuberculosis linked a type I restriction-modification system with pathogenicity (66). However, the Dam system (type II restriction-modification system) was described previously to play an important role in S. Typhimurium pathogenicity through the epigenetic regulation of pathogenicity-related genes (48).
In conclusion, the microarray-based negative-selection screening developed in this study allowed the identification of novel S. Enteritidis genes playing an important role in the colonization of deep organs in a BALB/c mouse model. These genes have not been described for other serovars of the genus Salmonella.
Validation of negatively selected mutants using competitive infection.
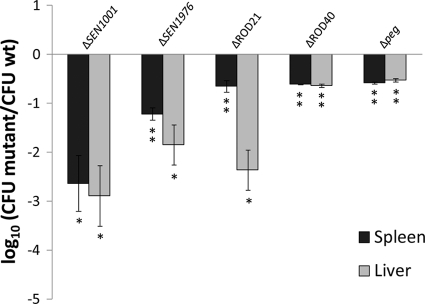
Targeted mutations in selected genes (SEN1001 and SEN1976) and genomic islands (ROD21, ROD40, and the peg operon) of S. Enteritidis identified in our screen were constructed by the Red swap method (30, 72). Competition experiments using groups of BALB/c mice inoculated by the intraperitoneal route with a mixture of the wild-type strain and each mutant were performed. Forty-eight hours after infection, spleens and livers were obtained, and the numbers of wild-type and mutant bacteria were enumerated. Our results indicate that the ΔSEN1001, ΔSEN1976, ΔROD21, ΔROD40, and Δpeg mutants were unable to colonize the internal organs of infected animals as efficiently as the wild-type organism (Fig. 2). These results confirm the predictions that we obtained from our microarray-based screen.
Fig 2.
Competition assays with individual mutants versus the wild-type (wt) strain and recovery from spleen and liver at 48 h postinfection. A total of 106 CFU of each selected mutant was administered at a 1:1 ratio with wild-type strain P125109 by the intraperitoneal route into 5 mice and recovered from spleen and liver at 2 days postinfection. CI values were calculated as the mean ratio of mutant to wild-type CFU, normalized to the input ratio and converted logarithmically. Error bars denote standard errors. Statistical significance was determined by using Student's 2-tailed t test, and asterisks indicate that normalized output ratios were significantly statistically different from the equivalent ratio in the inoculum (∗, P < 0.05; ∗∗, P < 0.001).
To test whether the colonization defect presented by the mutants was linked to an overall fitness defect in a nutrient-limited environment, bacterial growth in E minimal medium was assessed by using both individual cultures and in vitro competition assays with the wild-type strain. Growth curves of individual mutant strains were identical to that of the wild-type parental strain, with the only exception being the ΔSEN1001 mutant, which showed a slightly extended lag phase (data not shown). In vitro competition experiments indicated that each mutant showed a slight but statistically significant defect compared to the parental strain when grown in minimal media (data not shown). However, this effect was tens to hundreds of times less than the defect observed in vivo. These results indicate that the observed large colonization defect presented by the mutants is not attributable to a strong defect in growth in a nutrient-limited environment.
SEN1001 encodes a LysM domain-containing protein needed for systemic infection by S. Enteritidis.
S. Enteritidis harbors a truncated version of SPI-19 that includes an internal deletion compared with the same genomic region in Salmonella serovars Gallinarum, Dublin, Agona, and Weltevreden (13). SEN1001 is one of the genes that remained in the S. Enteritidis chromosome after the deletion of most components of the T6SS.
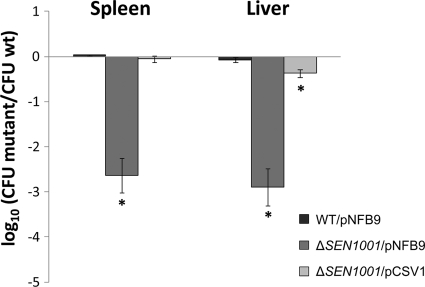
To directly link the defect in systemic infection to the mutation in SEN1001, we complemented our ΔSEN1001 mutant and tested the colonization of the complemented strain during competitive systemic infections. An intact copy of SEN1001 was cloned into pNFB9, and the resultant plasmid (pCSV1) was moved into ΔSEN1001. Since pNFB9 derivatives can integrate at the Gifsy-1 attachment site (attG1) (49), a single copy of the complementing gene was restored in the S. Enteritidis chromosome. The complemented mutant (ΔSEN1001/pCSV1) regained the ability to colonize systemic sites. In contrast, the mutant containing the empty vector (ΔSEN1001/pNFB9) maintained a colonization defect during infection (Fig. 3). The wild-type strain and a derivative harboring an empty pNFB9 vector (WT/pNFB9) displayed similar levels of colonization during competitive systemic infections of mice (Fig. 3), indicating that the presence of the plasmid alone does not interfere with S. Enteritidis pathogenesis in our assay.
Fig 3.
SEN1001 is needed for successful S. Enteritidis colonization in mice. A complementation assay for a ΔSEN1001 mutant was performed. A total of 106 CFU of each strain was administrated at a 1:1 ratio with wild-type strain P125109 by the intraperitoneal route into 5 mice and recovered from spleen and liver at 2 days postinfection. CI values were calculated as a mean ratio of mutant to wild-type CFU, normalized to the input ratio and converted logarithmically. Error bars denote standard errors. Statistical significance was determined by using Student's 2-tailed t test, and asterisks indicate normalized output ratios that were significantly statistically different from the equivalent ratio in the inoculum (∗, P < 0.05).
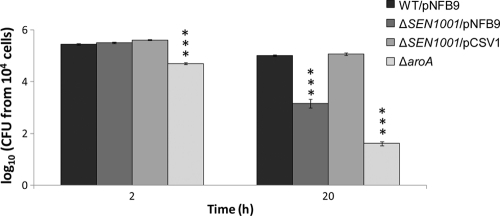
To test whether the defect of our ΔSEN1001 mutant in systemic colonization may be linked to a defect in the intracellular survival within macrophages of the host, the wild-type strain (WT/pNFB9) and the ΔSEN1001/pNFB9 and ΔSEN1001/pCSV1 derivatives were compared for internalization by and survival in RAW264.7 murine macrophages. In these assays, the ΔSEN1001/pNFB9 mutant was defective in intracellular survival after 20 h of infection compared to the wild-type strain. In contrast, the complemented ΔSEN1001/pCSV1 mutant survived as well as the wild-type strain within RAW264.7 macrophages (Fig. 4). As expected, a control ΔaroA mutant included in our assays was impaired for intramacrophage survival from early time points (Fig. 4).
Fig 4.
SEN1001 is needed for intracellular survival of S. Enteritidis within macrophages of the host. Shown are data for intracellular CFU recovered from RAW264.7 macrophages. Each experiment was performed on two separate occasions, evaluating samples at least in duplicate. CFU values were normalized to 104 viable cells and converted logarithmically. Bars represent mean values, and error bars denote standard errors. The statistical significance of differences in the data was determined by using one-way ANOVA and the Dunnett's posttest. Asterisks indicate CFU values that are significantly different from that of the wild-type strain (∗∗∗, P < 0.0001).
For a better understanding of the observed phenotypes, bioinformatic analyses were performed to identify conserved domains and similarities with other proteins present in related species. According to our analyses, SEN1001 includes a lysin motif (LysM) domain (E value = 0.00032) that is thought to bind to peptidoglycan. The LysM domain is 40 to 44 residues long and is found in a variety of enzymes involved in bacterial cell wall degradation (52), similar to those in peptidoglycan hydrolases (19). However, our ΔSEN1001 mutant did not show growth deficiencies, impaired segregation, or membrane stress when cultured in LB medium under standard conditions or in the presence of SDS, EDTA, hydrogen peroxide, or deoxycholate (data not shown). Also, the LysM domain has been found in several proteins considered to be virulence factors, such as intimin from pathogenic E. coli (9) and Slt70 from Shigella flexneri (8), among others.
The LysM domain-containing protein SEN1001 is required for S. Enteritidis pathogenicity in BALB/c mice, and its function enhances survival within macrophages. Thus, positive selection in vivo may explain why SEN1001 was retained in the genome of S. Enteritidis during the degradation of SPI-19 in this serovar. Finally, SEN1001 is a potential new target for the development of live-attenuated vaccines.
Conclusions.
We performed a microarray-based negative-selection screen for S. Enteritidis mutants showing deficiencies in the colonization of spleen and liver in mice. S. Enteritidis PT4 strain P125109 was mutagenized using the mini-Tn5 derivative EZ-Tn5 <T7/KAN-2> to generate a transposon library of ∼54,000 mutants to monitor changes in the library composition after selection during infection using microarrays.
Mutations in genes known to contribute to systemic (e.g., SPI-2, aro, rfa, rfb, phoP, and phoQ) and enteric (e.g., SPI-1 and SPI-5) infections were associated with colonization defects in our assay. The most dramatic phenotypes were observed for mutations in genes required for systemic infection. In addition, colonization defects were observed for mutations in genes and genomic islands restricted to S. Enteritidis or to a limited number of Salmonella serovars. These include genes encoding components of a putative type VI secretion system (SEN1001), a type I restriction-modification system (SEN4290 to SEN4292), the peg fimbrial operon (SEN2144A to SEN2145B), and a cluster of genes linked to tRNA-asnT, forming a putative pathogenicity island (SEN1970 to SEN1999). Conversely, one would also expect a similar or larger number of genes that are present in S. Typhimurium and absent in S. Enteritidis to have a known virulence phenotype in the better-studied S. Typhimurium. Surprisingly, there are few such genes, with possible candidates including STM0274A, STM2599, and STM2231. Thus, not only do these two serovars differ in the details of how they infect mice, but S. Enteritidis might also have a greater number of serovar-specific virulence genes.
Mutants with deletions of individual genes or entire regions presenting novel phenotypes in our screen were generated for confirmation in competition assays with wild-type strain P125109. Targeted mutants with deletions of the genes SEN1001 and SEN1976 and the gene clusters SEN1976 to SEN1999 (ROD21), SEN2144A to SEN2145B (peg operon), and SEN4290 to SEN4292 (ROD40) reiterated the phenotype observed in our microarray-based screening. Furthermore, a ΔSEN1001 mutant was impaired for intracellular survival within RAW264.7 murine macrophages in vitro. The phenotypes observed in vivo and in vitro for this particular mutant could be restored by the complementation of the gene using a plasmid that integrates into the chromosome of S. Enteritidis. With this, SEN1001 is required for the full pathogenicity of S. Enteritidis in BALB/c mice and could be a new target for the development of vaccines. Further experiments to test its use as a live, attenuated vaccine are in progress in our laboratory.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Sang-Ho Choi and Yollande Cheng for technical assistance in microarray hybridizations. We thank Susan Bueno and Lionello Bossi for generous contributions of bacterial strains and plasmids.
This work was supported in part by grants 1100092 and 1110172 from FONDECYT and grant ADI-08/2006 from CONICYT and The World Bank. M.M. and H.L.A.-P. were supported in part by NIH grants R21AI083964, R01AI083646, R56AI077645, and R01AI075093 and AFRI CSREES grant 2009-03579. C.A.S. and C.J.B. were supported by fellowships from CONICYT and from Vicerrectoría de Asuntos Académicos, Departamento de Postgrado y Postítulo, Universidad de Chile.
Footnotes
Published ahead of print 14 November 2011
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Agron PG, et al. 2001. Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar Enteritidis. Appl. Environ. Microbiol. 67:4984–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altier C. 2005. Genetic and environmental control of Salmonella invasion. J. Microbiol. 43:85–92 [PubMed] [Google Scholar]
- 3. Amy M, et al. 2004. Identification of a new Salmonella enterica serovar Enteritidis locus involved in cell invasion and in the colonisation of chicks. Res. Microbiol. 155:543–552 [DOI] [PubMed] [Google Scholar]
- 4. Andrews SC, Robinson AK, Rodriguez-Quinones F. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215–237 [DOI] [PubMed] [Google Scholar]
- 5. Arrach N, et al. 2010. High-throughput screening for Salmonella avirulent mutants that retain targeting of solid tumors. Cancer Res. 70:2165–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Babu U, et al. 2004. Salmonella enteritidis clearance and immune responses in chickens following Salmonella vaccination and challenge. Vet. Immunol. Immunopathol. 101:251–257 [DOI] [PubMed] [Google Scholar]
- 7. Baker SJ, Daniels C, Morona R. 1997. PhoP/Q regulated genes in Salmonella typhi identification of melittin sensitive mutants. Microb. Pathog. 22:165–179 [DOI] [PubMed] [Google Scholar]
- 8. Bartoleschi C, et al. 2002. Selection of Shigella flexneri candidate virulence genes specifically induced in bacteria resident in host cell cytoplasm. Cell. Microbiol. 4:613–626 [DOI] [PubMed] [Google Scholar]
- 9. Bateman A, Bycroft M. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299:1113–1119 [DOI] [PubMed] [Google Scholar]
- 10. Belden WJ, Miller SI. 1994. Further characterization of the PhoP regulon: identification of new PhoP-activated virulence loci. Infect. Immun. 62:5095–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Betancor L, et al. 2005. An attenuated Salmonella Enteritidis strain derivative of the main genotype circulating in Uruguay is an effective vaccine for chickens. Vet. Microbiol. 107:81–89 [DOI] [PubMed] [Google Scholar]
- 12. Blanc-Potard AB, Solomon F, Kayser J, Groisman EA. 1999. The SPI-3 pathogenicity island of Salmonella enterica. J. Bacteriol. 181:998–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blondel CJ, Jimenez JC, Contreras I, Santiviago CA. 2009. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics 10:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blondel CJ, et al. 2010. Contribution of the type VI secretion system encoded in SPI-19 to chicken colonization by Salmonella enterica serotypes Gallinarum and Enteritidis. PLoS One 5:e11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bogomolnaya LM, Santiviago CA, Yang HJ, Baumler AJ, Andrews-Polymenis HL. 2008. ‘Form variation’ of the O12 antigen is critical for persistence of Salmonella Typhimurium in the murine intestine. Mol. Microbiol. 70:1105–1119 [DOI] [PubMed] [Google Scholar]
- 16. Bohez L, Ducatelle R, Pasmans F, Haesebrouck F, Van Immerseel F. 2007. Long-term colonisation-inhibition studies to protect broilers against colonisation with Salmonella Enteritidis, using Salmonella pathogenicity island 1 and 2 mutants. Vaccine 25:4235–4243 [DOI] [PubMed] [Google Scholar]
- 17. Brenner FW, Villar RG, Angulo FJ, Tauxe R, Swaminathan B. 2000. Salmonella nomenclature. J. Clin. Microbiol. 38:2465–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buckley AM, et al. 2006. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell. Microbiol. 8:847–856 [DOI] [PubMed] [Google Scholar]
- 19. Buist G, Steen A, Kok J, Kuipers OP. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68:838–847 [DOI] [PubMed] [Google Scholar]
- 20. Bullas LR, Colson C, Neufeld B. 1980. Deoxyribonucleic acid restriction and modification systems in Salmonella: chromosomally located systems of different serotypes. J. Bacteriol. 141:275–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cerquetti MC, Gherardi MM. 2000. Orally administered attenuated Salmonella enteritidis reduces chicken cecal carriage of virulent Salmonella challenge organisms. Vet. Microbiol. 76:185–192 [DOI] [PubMed] [Google Scholar]
- 22. Chan K, Kim CC, Falkow S. 2005. Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect. Immun. 73:5438–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chang J, Pang E, He H, Kwang J. 2008. Identification of novel attenuated Salmonella Enteritidis mutants. FEMS Immunol. Med. Microbiol. 53:26–34 [DOI] [PubMed] [Google Scholar]
- 24. Chaudhuri RR, et al. 2009. Comprehensive identification of Salmonella enterica serovar Typhimurium genes required for infection of BALB/c mice. PLoS Pathog. 5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 26. Cirillo DM, Valdivia RH, Monack DM, Falkow S. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175–188 [DOI] [PubMed] [Google Scholar]
- 27. Cirl C, et al. 2008. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat. Med. 14:399–406 [DOI] [PubMed] [Google Scholar]
- 28. Clayton DJ, et al. 2008. Analysis of the role of 13 major fimbrial subunits in colonisation of the chicken intestines by Salmonella enterica serovar Enteritidis reveals a role for a novel locus. BMC Microbiol. 8:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Craven SE. 1994. Altered colonizing ability for the ceca of broiler chicks by lipopolysaccharide-deficient mutants of Salmonella typhimurium. Avian Dis. 38:401–408 [PubMed] [Google Scholar]
- 30. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Edwards MF, Stocker BA. 1988. Construction of delta aroA his delta pur strains of Salmonella typhi. J. Bacteriol. 170:3991–3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edwards RA, Matlock BC, Heffernan BJ, Maloy SR. 2001. Genomic analysis and growth-phase-dependent regulation of the SEF14 fimbriae of Salmonella enterica serovar Enteritidis. Microbiology 147:2705–2715 [DOI] [PubMed] [Google Scholar]
- 33. Edwards RA, Schifferli DM, Maloy SR. 2000. A role for Salmonella fimbriae in intraperitoneal infections. Proc. Natl. Acad. Sci. U. S. A. 97:1258–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Escolar L, Perez-Martin J, de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fardini Y, et al. 2007. The YfgL lipoprotein is essential for type III secretion system expression and virulence of Salmonella enterica serovar Enteritidis. Infect. Immun. 75:358–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Galan JE. 1996. Molecular and cellular bases of Salmonella entry into host cells. Curr. Top. Microbiol. Immunol. 209:43–60 [DOI] [PubMed] [Google Scholar]
- 37. Gerlach RG, et al. 2007. Salmonella pathogenicity island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell. Microbiol. 9:1834–1850 [DOI] [PubMed] [Google Scholar]
- 38. Guard-Petter J. 2001. The chicken, the egg and Salmonella enteritidis. Environ. Microbiol. 3:421–430 [DOI] [PubMed] [Google Scholar]
- 39. Guard-Petter J. 1998. Variants of smooth Salmonella enterica serovar Enteritidis that grow to higher cell density than the wild type are more virulent. Appl. Environ. Microbiol. 64:2166–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guard-Petter J, Henzler DJ, Rahman MM, Carlson RW. 1997. On-farm monitoring of mouse-invasive Salmonella enterica serovar Enteritidis and a model for its association with the production of contaminated eggs. Appl. Environ. Microbiol. 63:1588–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gunn JS, Alpuche-Aranda CM, Loomis WP, Belden WJ, Miller SI. 1995. Characterization of the Salmonella typhimurium pagC/pagD chromosomal region. J. Bacteriol. 177:5040–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gunn JS, Miller SI. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857–6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haneda T, Ishii Y, Danbara H, Okada N. 2009. Genome-wide identification of novel genomic islands that contribute to Salmonella virulence in mouse systemic infection. FEMS Microbiol. Lett. 297:241–249 [DOI] [PubMed] [Google Scholar]
- 44. Hansen JK, Forest KT. 2006. Type IV pilin structures: insights on shared architecture, fiber assembly, receptor binding and type II secretion. J. Mol. Microbiol. Biotechnol. 11:192–207 [DOI] [PubMed] [Google Scholar]
- 45. Hennessy TW, et al. 1996. A national outbreak of Salmonella enteritidis infections from ice cream. The Investigation Team. N. Engl. J. Med. 334:1281–1286 [DOI] [PubMed] [Google Scholar]
- 46. Hensel M. 2004. Evolution of pathogenicity islands of Salmonella enterica. Int. J. Med. Microbiol. 294:95–102 [DOI] [PubMed] [Google Scholar]
- 47. Hensel M, et al. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163–174 [DOI] [PubMed] [Google Scholar]
- 48. Heusipp G, Falker S, Schmidt MA. 2007. DNA adenine methylation and bacterial pathogenesis. Int. J. Med. Microbiol. 297:1–7 [DOI] [PubMed] [Google Scholar]
- 49. Ho TD, et al. 2002. Identification of GtgE, a novel virulence factor encoded on the Gifsy-2 bacteriophage of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:5234–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hoare A, et al. 2006. The outer core lipopolysaccharide of Salmonella enterica serovar Typhi is required for bacterial entry into epithelial cells. Infect. Immun. 74:1555–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ilg K, et al. 2009. O-antigen-negative Salmonella enterica serovar Typhimurium is attenuated in intestinal colonization but elicits colitis in streptomycin-treated mice. Infect. Immun. 77:2568–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Joris B, et al. 1992. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol. Lett. 70:257–264 [DOI] [PubMed] [Google Scholar]
- 53. Karasova D, et al. 2010. Influence of 5 major Salmonella pathogenicity islands on NK cell depletion in mice infected with Salmonella enterica serovar Enteritidis. BMC Microbiol. 10:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kiss T, Morgan E, Nagy G. 2007. Contribution of SPI-4 genes to the virulence of Salmonella enterica. FEMS Microbiol. Lett. 275:153–159 [DOI] [PubMed] [Google Scholar]
- 55. Knodler LA, et al. 2002. Salmonella effectors within a single pathogenicity island are differentially expressed and translocated by separate type III secretion systems. Mol. Microbiol. 43:1089–1103 [DOI] [PubMed] [Google Scholar]
- 56. Lawley TD, et al. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Methner U, Barrow PA, Gregorova D, Rychlik I. 2004. Intestinal colonisation-inhibition and virulence of Salmonella phoP, rpoS and ompC deletion mutants in chickens. Vet. Microbiol. 98:37–43 [DOI] [PubMed] [Google Scholar]
- 58. Morgan E, et al. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994–1010 [DOI] [PubMed] [Google Scholar]
- 59. Mougous JD, et al. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nevola JJ, Stocker BA, Laux DC, Cohen PS. 1985. Colonization of the mouse intestine by an avirulent Salmonella typhimurium strain and its lipopolysaccharide-defective mutants. Infect. Immun. 50:152–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Newman RM, Salunkhe P, Godzik A, Reed JC. 2006. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect. Immun. 74:594–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. O'Callaghan D, Maskell D, Liew FY, Easmon CS, Dougan G. 1988. Characterization of aromatic- and purine-dependent Salmonella typhimurium: attention, persistence, and ability to induce protective immunity in BALB/c mice. Infect. Immun. 56:419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Parker CT, Liebana E, Henzler DJ, Guard-Petter J. 2001. Lipopolysaccharide O-chain microheterogeneity of Salmonella serotypes Enteritidis and Typhimurium. Environ. Microbiol. 3:332–342 [DOI] [PubMed] [Google Scholar]
- 64. Pattery T, Hernalsteens JP, De Greve H. 1999. Identification and molecular characterization of a novel Salmonella enteritidis pathogenicity islet encoding an ABC transporter. Mol. Microbiol. 33:791–805 [DOI] [PubMed] [Google Scholar]
- 65. Porwollik S, et al. 2005. Differences in gene content between Salmonella enterica serovar Enteritidis isolates and comparison to closely related serovars Gallinarum and Dublin. J. Bacteriol. 187:6545–6555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pouillot F, Fayolle C, Carniel E. 2007. A putative DNA adenine methyltransferase is involved in Yersinia pseudotuberculosis pathogenicity. Microbiology 153:2426–2434 [DOI] [PubMed] [Google Scholar]
- 67. Raffatellu M, et al. 2005. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype Typhimurium invasion of epithelial cells. Infect. Immun. 73:146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rakeman JL, Miller SI. 1999. Salmonella typhimurium recognition of intestinal environments. Trends Microbiol. 7:221–223 [DOI] [PubMed] [Google Scholar]
- 69. Retamal P, Castillo-Ruiz M, Mora GC. 2009. Characterization of MgtC, a virulence factor of Salmonella enterica serovar Typhi. PLoS One 4:e5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rychlik I, et al. 2009. Virulence potential of five major pathogenicity islands (SPI-1 to SPI-5) of Salmonella enterica serovar Enteritidis for chickens. BMC Microbiol. 9:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Santiviago CA, et al. 2010. Spontaneous excision of the Salmonella enterica serovar Enteritidis-specific defective prophage-like element phiSE14. J. Bacteriol. 192:2246–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Santiviago CA, et al. 2009. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 5:e1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shah DH, et al. 2005. Identification of Salmonella gallinarum virulence genes in a chicken infection model using PCR-based signature-tagged mutagenesis. Microbiology 151:3957–3968 [DOI] [PubMed] [Google Scholar]
- 74. Shea JE, Hensel M, Gleeson C, Holden DW. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 93:2593–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Stone BJ, Miller VL. 1995. Salmonella enteritidis has a homologue of tolC that is required for virulence in BALB/c mice. Mol. Microbiol. 17:701–712 [DOI] [PubMed] [Google Scholar]
- 76. Suar M, et al. 2006. Virulence of broad- and narrow-host-range Salmonella enterica serovars in the streptomycin-pretreated mouse model. Infect. Immun. 74:632–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Thomson NR, et al. 2008. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 18:1624–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Turner AK, et al. 2003. Contribution of proton-translocating proteins to the virulence of Salmonella enterica serovars Typhimurium, Gallinarum, and Dublin in chickens and mice. Infect. Immun. 71:3392–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Unden G, Becker S, Bongaerts J, Schirawski J, Six S. 1994. Oxygen regulated gene expression in facultatively anaerobic bacteria. Antonie Van Leeuwenhoek 66:3–22 [DOI] [PubMed] [Google Scholar]
- 80. Wong KK, et al. 1998. Identification and sequence analysis of a 27-kilobase chromosomal fragment containing a Salmonella pathogenicity island located at 92 minutes on the chromosome map of Salmonella enterica serovar Typhimurium LT2. Infect. Immun. 66:3365–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wood MW, et al. 1998. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol. Microbiol. 29:883–891 [DOI] [PubMed] [Google Scholar]
- 82. Xia X, McClelland M, Wang Y. 2005. WebArray: an online platform for microarray data analysis. BMC Bioinformatics 6:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Xia XQ, et al. 2009. WebArrayDB: cross-platform microarray data analysis and public data repository. Bioinformatics 25:2425–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.