Abstract
Asymptomatic bacteriuria (ABU) is a condition where bacteria stably colonize the urinary tract, in a manner closely resembling commensalism at other mucosal sites. The patients carry >105 CFU/ml for extended periods of time and rarely develop symptoms. Contrasting the properties of ABU strains to those of uropathogenic isolates causing symptomatic infection is therefore highly relevant to understand mechanisms of bacterial adaptation. The prototype ABU strain Escherichia coli 83972 has a smaller genome than uropathogenic E. coli (UPEC) strains with deletions or point mutations in several virulence genes, suggesting that ABU strains undergo a programmed reductive evolution within human hosts. This study addressed if these observations can be generalized. Strains causing ABU in outpatients or hospitalized patients after catheterization or other invasive procedures were compared to commensal E. coli isolates from the intestinal flora of healthy individuals. Notably, clonal complex 73 (CC73) was a prominent phylogenetic lineage dominated by ABU isolates. ABU isolates from outpatients and hospitalized patients had a similar overall virulence gene repertoire, which distinguished them from many commensals, but typical UPEC virulence genes were less frequently attenuated in hospital strains than in outpatient strains or commensals. The decreased virulence potential of outpatient ABU isolates relative to that of ABU strains from hospitalized patients supports the hypothesis that loss of expression or decay of virulence genes facilitates long-term carriage and adaptation to host environments.
INTRODUCTION
Bacteria rapidly adapt to changing environmental conditions, thereby increasing their fitness for new host niches. While it has been generally accepted that bacteria evolve toward virulence, data over the past decade have established that, in addition, bacteria may undergo reductive evolution in human hosts, moving them away from the virulent phenotype (65). Escherichia coli strains recovered from patients with asymptomatic bacteriuria were proposed to have evolved from virulent uropathogenic E. coli strains (UPEC) by genome reduction through systematic inactivation of genes encoding virulence-associated factors either by the accumulation of point mutations or by deletions (29, 66). These observations define urinary tract infections (UTIs) to be a highly interesting and relevant model to study bacterial adaptation to changing host environments. Fully virulent uropathogenic Escherichia coli causes symptomatic UTIs, accompanied by a strong innate immune response and tissue damage resulting from the sequels of acute inflammation. Less virulent strains establish asymptomatic bacteriuria (ABU) accompanied by an innate immune response too weak to cause symptoms. ABU occurs in 2 to 20% of the population, depending on age and gender, and the bacteria may persist in the host for months or years. Epidemiologic studies have established that the severity of UTI reflects the virulence profile of the infecting strain, with tissue-attacking virulence factors being expressed at a higher frequency by UPEC strains than by most strains causing ABU (5, 13), despite the presence of virulence gene sequences in many ABU strains (40). Until recently, the molecular basis for this discrepancy has not been examined, however.
UTIs are the most common health care-associated infections, and E. coli is the most common nosocomial pathogen (59). Prolonged use of indwelling urinary catheters is the critical risk factor for developing a nosocomial UTI (29, 41, 57), as the catheters facilitate bacterial multiplication, which can result in either severe disease or asymptomatic colonization (35). Catheter-associated ABU develops rapidly in patients with urinary catheters (60), but only 10% to 30% develop clinically significant symptomatic UTIs. The host environment and selective forces to which ABU strains are exposed in hospitalized patients may differ greatly from those confronting ABU strains in outpatient settings. Furthermore, the duration of asymptomatic bladder colonization is shorter on average in hospitalized patients than in healthy carriers. Additionally, the hospital environment may preselect certain E. coli variants, e.g., by nosocomial transmission and cross-transmission of strains (58), which may differ in virulence potential and phylogeny from ABU isolates carried by otherwise healthy individuals.
Previous evidence of genome reduction and virulence gene attenuation was obtained for a restricted number of E. coli isolates, including the prototype ABU strain E. coli 83972. This study characterized the virulence gene repertoire and selected virulence phenotypes of E. coli ABU strains isolated from outpatients or hospitalized carriers. Fecal isolates from healthy donors without a history of UTI were used as controls. The results indicate that the loss of functional virulence genes or their reduced expression is a general feature of ABU strains. On the basis of the prevalence of UPEC virulence-associated genes, community-acquired ABU isolates resembled health care-associated ABU isolates, but important virulence-associated genes remained functional in ABU isolates from hospitalized patients. The results suggest that prolonged in vivo growth in the urinary bladder promotes the loss of functional virulence genes, due to mutations or phenotypic attenuation.
MATERIALS AND METHODS
Patient characteristics.
Community-acquired ABU strains (n = 87) were isolated during a screening study in healthy schoolgirls. Bacteriuria was confirmed by repeated urine cultures yielding the same E. coli strain (33). In parallel, fecal E. coli isolates were obtained from healthy children (n = 39) without a history of UTI (12) and used here to represent commensal E. coli isolates without a significant potential to cause UTI. Health care-associated ABU E. coli isolates (n = 25) were obtained from the urine of 23 alert patients treated at the Department of Urology, Hospital St. Elisabeth (Straubing, Germany). Thirteen patients were males, and the mean age was 70 years (range, 37 to 89 years). Ten patients had indwelling urinary catheters (indwelling bladder catheter, n = 8; ureteral catheter, n = 1; percutaneous nephrostomy, n = 1), and their diagnoses included prostate cancer (n = 2), bladder (n = 4) or renal cell (n = 2) carcinoma, benign prostatic hyperplasia (n = 4), stone disease (n = 6), supravesical urinary diversion (n = 3), hypotonic bladder (n = 1), or hydronephrosis (n = 1). The duration of bacteriuria was 1 to 74 days, and 14/23 patients (61%) received antibiotics prior to culturing of the ABU strain. The patients were asymptomatic and had (i) an indwelling urinary catheter within 7 days before one positive culture with ≥105 CFU/ml and no more than two uropathogenic species or (ii) no indwelling urinary catheter within 7 days before the first urine culture and two cultures with ≥105 CFU/ml and no more than two uropathogenic species (24).
Bacterial isolates.
Urine cultures were processed by the respective clinical bacteriology laboratory and maintained as deep agar stabs or frozen glycerol cultures prior to batch evaluation for bacterial characteristics and potential virulence factors. For analysis, E. coli strains were routinely grown in lysogeny broth (LB) or M63B1 glucose medium (52) with or without 1.5% Bacto agar (Difco Laboratories, Detroit, MI).
DNA techniques.
Qiagen products (Hilden, Germany) were used to isolate and purify genomic DNA. Primers were obtained from Operon (Cologne, Germany), while restriction enzymes were purchased from New England BioLabs (Frankfurt am Main, Germany). Primers used for detection of the fim and pap gene clusters are listed in Table S2 in the supplemental material.
Multiplex PCR.
The multiplex PCR assay for detection of virulence-associated genes (hlyA, cnf1, cdtB, iutA, fyuA, fimH, papAH, papEF, papC, papG allele I, papG allele II-papG allele III, sfa-focDE, sfaS, focG, kpsMT K1, kpsMT K5, kpsMT II, kpsMT III, malX, cvaC, ibeA, bmaE, rfc, traT, gafD, afa-draBC, iroN, clbB) of extraintestinal pathogenic E. coli was performed as previously described (27).
Statistical analysis.
Fisher's exact test was used to test for differences in prevalence rates. Average-linkage cluster analysis was performed with Cluster (version 3.0) software (9), based on the prevalence of selected extraintestinal pathogenic E. coli (ExPEC) virulence traits among the three populations, and the result was visualized by a heat map. Similarly, cluster analysis of the PCR screening data was performed with Cluster (version 3.0), based on the presence or absence of genes. The output of the cluster analyses was displayed with the Java TreeView software (51).
MLST.
ABU isolates were allocated to different clonal lineages according to Wirth et al. (63). New sequence types (STs) were submitted to the multilocus sequence typing (MLST) database (http://mlst.ucc.ie/mlst/mlst/dbs/Ecoli). A clonal complex (CC) has been defined to include at least three STs that differ from their nearest neighbor by no more than one of seven alleles. The STs were assigned to one of the four major phylogenetic groups, A, B1, B2, and D, or to recombinant groups A×B1 and ABD using the software Structure (version 2.2.3) (http://pritch.bsd.uchicago.edu/structure_software/) (15, 16, 42, 63). Phylogenetic relationships between distinct sequence types were calculated using the previously described ClonalFrame (version 1.2) software (Xavier Didelot, University of Oxford) (10). This software is based on a model of genetic diversification that takes into account homologous recombination events which occur in E. coli populations. For ClonalFrame analysis, concatenated MLST sequences were used with the default parameters of 50,000 Markov chain Monte Carlo iterations after 50,000 burn-in iterations. The minimum-spanning tree based on the allelic numbers of the MLST loci, which was calculated using SeqSphere software (version 0.9.38 β; Ridom GmbH), reflects the phylogenetic relationships between the sequence types also computed by ClonalFrame.
DNA sequence analysis of papG in health care-associated ABU isolates.
The P-fimbrial adhesin gene papG was amplified by PCR using primers papSQ15 (5′-TGGTTACAGAGTTACAGCAGGTCTG-3′) and papX (5′-CAGAGGCTCACTCTTGCAC-3′), and the PCR product was sequenced by primer walking using an Applied Biosystems 3130 genetic analyzer. Homology searches were performed with the BLAST programs of the National Center for Biotechnology Information (NCBI) (2) (http://www.ncbi.nlm.nih.gov/BLAST/). The relatedness of the PapG variants was inferred using the neighbor-joining method (50). The evolutionary distances were computed using the Poisson correction method (67) in the MEGA5 program (56).
Phenotypic assays.
The expression of d-mannose binding type 1 fimbriae was quantified by agglutination of Saccharomyces cerevisiae cells. Aliquots of bacterial overnight cultures in LB or pooled human urine were incubated with a yeast suspension (10 mg/ml [dry weight]), and agglutination susceptible to inhibition by d-mannose (2%) was scored visually on the basis of the aggregation and precipitation of the cells.
P and S/F1C fimbriae were detected by hemagglutination of defibrinated human and bovine erythrocytes, respectively. Aliquots of bacterial overnight cultures in LB or pooled human urine were incubated with a suspension of human or bovine blood (Elocin Lab, Munich, Germany). Hemagglutination was compared after incubation for some minutes on ice. UPEC strain 536 was used as a positive control, and E. coli strain HB101 was used as a negative control.
Hemolytic activity was detected on sheep blood agar plates (Oxoid), after overnight incubation at 37°C, as the formation of clear halos around the colonies. UPEC strain 536 was used as a positive control, and E. coli strain HB101 was used as a negative control.
The aerobactin siderophore was detected by the aerobactin cross-feeding bioassay (7). Cells (109) of aerobactin-requiring indicator E. coli strain LG1522 were cultured in M9 soft agar containing 200 mM 2′-2′-dipyridyl (Sigma, Deisenhofen, Germany). Aerobactin production by the test strains was indicated by a zone of enhanced growth of E. coli LG1522 around the colonies of the test strains. E. coli ABU strain 83972 was used as a positive control, and E. coli strain HB101 was used as a negative control.
Nucleotide sequence accession numbers.
The nucleotide sequences have been submitted to the EMBL database under accession numbers HE577318 (ABU isolate 1299), HE577319 (ABU isolate 1301), HE577320 (ABU isolate 1302), HE577321 (ABU isolate 1303), HE577322 (ABU isolate 1308), HE577323 (ABU isolate 1312), HE577324 (ABU isolate 1314), HE577325 (ABU isolate 1316), HE577326 (ABU isolate 1321), HE611570 (ABU isolate 2021), HE611571 (ABU isolate 1991), HE611572 (ABU isolate 1973), HE611573 (ABU isolate 256), HE611574 (ABU isolate 2017), HE611575 (ABU isolate 1979), HE611576 (ABU isolate 2035), HE611577 (ABU isolate 2027), HE611578 (ABU isolate 2023), and HE611579 (ABU isolate 2012).
RESULTS
Phylogenetic classification of ABU isolates from healthy and hospitalized carriers.
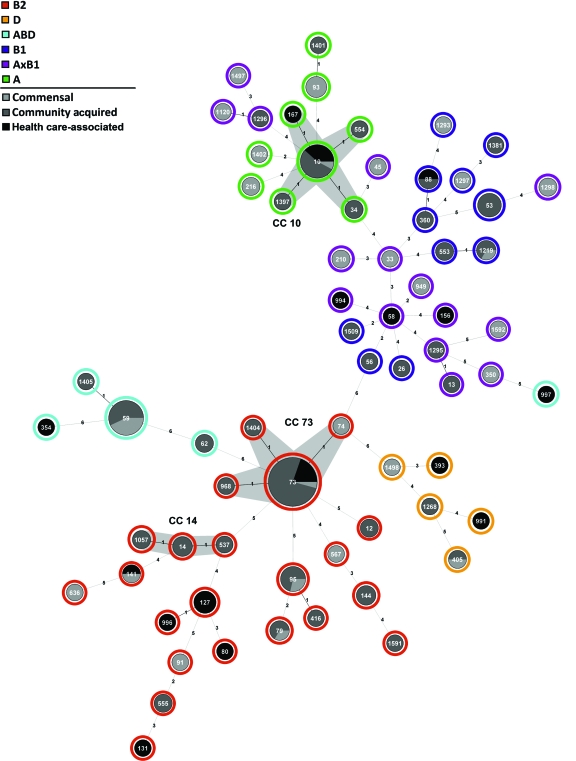
The 151 E. coli strains were allocated to phylogenetic lineages on the basis of MLST and subsequent statistical analyses. The 87 community-acquired ABU isolates and 25 health care-associated ABU isolates showed no significant difference in the distribution to the main E. coli phylogenetic lineages (Table 1), but their distribution differed from that of the 39 commensal E. coli isolates. The phylogenetic lineage B2 was more common among ABU isolates than commensal isolates (community-acquired ABU isolates, P = 0.003), while lineage A×B1 was more common among the commensal strains than among ABU E. coli strains (community-acquired ABU strains, P < 0.0001). Each of the six defined phylogenetic lineages (Fig. 1) included community-acquired ABU isolates, health care-associated ABU isolates, as well as commensal E. coli variants, but the distributions differed. The most prominent lineage was clonal complex 73 (CC73; 25 isolates, 18% of all strains), with a high ratio of community-acquired ABU and health care-associated ABU strains (92%) versus commensal E. coli strains (8%). Within CC73, the vast majority of isolates (23 strains) belonged to sequence type 73 (ST73). Related STs, such as ST74, ST1404, and ST968, were represented by only one isolate each. Other major CCs or STs detected among the strains were CC10 (15 isolates), ST59 (14 isolates), and CC95 (8 isolates). CC10 is common among commensal and diarrheagenic E. coli isolates but is less typical for UPEC variants. In contrast, ST59 and ST95 represent clonal lineages which mainly include UPEC or ExPEC isolates. The MLST analysis therefore demonstrates that ABU strains from healthy carriers and from hospitalized patients can be found in a variety of phylogenetic lineages which are shared among extraintestinal pathogenic and commensal E. coli strains.
Table 1.
Phylogenetic analysis of E. coli isolates from community-acquired ABU or health care-associated patients
| Phylogenetic group | Prevalence (%) relative to total no. of isolates |
Statistical significance of prevalencea |
||||
|---|---|---|---|---|---|---|
| Community-acquired ABU (n = 87) | Health care-associated ABU (n = 25) | Commensal (n = 39) | Community-acquired ABU vs health care-associated ABU | Community-acquired ABU vs commensal | Health care-associated ABU vs commensal | |
| A | 13.79 | 20.00 | 20.51 | — | — | — |
| A×B1 | 3. 45 | 12.00 | 30.77 | — | *** | — |
| B1 | 18.39 | 4.00 | 7.69 | — | — | — |
| ABD | 12.64 | 8.00 | 15.38 | — | — | — |
| D | 2.30 | 8.00 | 5.13 | — | — | — |
| B2 | 49.43 | 48.00 | 20.51 | — | ** | * |
Two-way comparisons were performed for each phylogenetic lineage between the different groups examined using Fisher's exact test. For each comparison, a P value of <0.05 (*), a P value of <0.005 (**), and a P value of <0.001 (***) were considered statistically significant, while a P value of >0.05 (—) was considered not statistically significant.
Fig 1.
Phylogenetic background of ABU E. coli isolates from healthy carriers and hospitalized patients and commensal E. coli isolates. The minimum-spanning tree of the concatenated MLST sequences, calculated by ClonalFrame, has been depicted with the Seqsphere software program (Ridom GmbH). The allocation of the ABU isolates from healthy carriers (community-acquired ABU) or hospitalized patients (health care-associated ABU) as well as of the commensal E. coli isolates is indicated by pie charts. The size of the pie chart mirrors the number of strains allocated to the individual ST. Phylogenetic groups (A, B1, B2, D, A×B1, and ABD), determined by analysis of the MLST sequence data with Structure software, are indicated by the outmost ring of the tree.
We subsequently examined if antibiotic treatment or the presence of long-term indwelling catheters influences the phylogenetic composition of the health care-associated ABU strains. Phylotyping of health care-associated ABU isolates from patients with (n = 10) or without (n = 15) long-term indwelling catheters revealed no significant differences with regard to certain STs or phylogenetic lineages (see Fig. S1A in the supplemental material). Similarly, the phylogenetic composition did not differ between ABU isolates from patients with (n = 10) or without (n = 15) antibiotic treatment (see Fig. S1B in the supplemental material), suggesting that the clinical origins of isolates from hospitalized patients did not disproportionately affect the group of health care-associated ABU isolates as a whole.
Prevalence of ExPEC virulence genes among ABU isolates from healthy or hospitalized carriers relative to commensal E. coli isolates.
The presence of 26 defined virulence genes commonly associated with ExPEC was determined to assess the virulence gene content of the two groups of ABU isolates relative to that of commensal E. coli isolates (Fig. 2; see Table S1 in the supplemental material).
Fig 2.
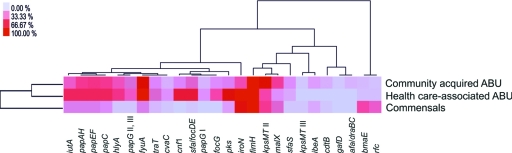
Two-way clustering of gene prevalence results among community-acquired ABU, health care-associated ABU, and commensal E. coli isolates. A heat map was constructed on the basis of the percentage of each gene examined among each of the three groups of isolates. Clustering was performed to illustrate similarities between the prevalence of the genes examined and the groups of isolates.
Based on the prevalence of ExPEC virulence genes, we characterized differences in the genome content of community-acquired and health care-associated ABU isolates relative to commensal E. coli isolates. Generally, the ExPEC virulence gene content was higher in ABU isolates than in commensal isolates. ABU strains isolated from healthy carriers or from hospital patients shared the highest similarity. Cluster analysis for gene correlations showed close overall relationships between genes of the pap operon, the alpha-hemolysin structural gene hlyA, and the high-pathogenicity-island (high-PAI) marker gene fyuA. Similarly, several other PAI-associated genes required for expression of S/F1C fimbriae (sfa, foc), iron uptake (iutA, iroN), group II capsule (kpsMT II), or polyketide colibactin (pks) biosynthesis were frequently present and clustered together in ABU isolates compared to commensals (Fig. 2).
The prevalence of 13 virulence genes differed significantly among the three groups of isolates (see Table S1 in the supplemental material). Eleven virulence genes were significantly more prevalent in ABU isolates than in commensal strains, i.e., those coding for the exotoxins alpha-hemolysin (hlyA) and cytotoxic necrotizing factor (cnf1), the aerobactin and yersiniabactin siderophore receptors iutA and fyuA, P-fimbrial (papAH, papC, papEF, papG alleles II and III) and F1C-fimbrial (sfa-focDE, focG) subunits, and the polyketide colibactin (pks).
In contrast, the blood group M-specific adhesin marker gene bmaE and the serotype O4-specific O-antigen polymerase gene rfc were more frequently detected in commensal isolates than in ABU isolates (Fig. 2; see Table S1 in the supplemental material). Interestingly, five virulence markers were more prevalent among health care-associated ABU isolates than among ABU isolates from healthy carriers, i.e., the genes encoding the cytotoxic necrotizing factor 1 (cnf1), the aerobactin, yersiniabactin, and salmochelin siderophore receptor genes iutA, fyuA, and iroN, as well as the colibactin polyketide determinant pks (Fig. 2; see Table S1 in the supplemental material). Only one marker, indicative of group II capsule gene clusters (kpsMT II), was more common in ABU isolates from healthy carriers.
Two-way clustering of the virulence gene content corroborated the finding that the prevalence of virulence-associated genes was not influenced by long-term catheterization (see Fig. S2A in the supplemental material) or antibiotic treatment (see Fig. S2B in the supplemental material).
Taken together, these results underline the finding that commensal E. coli isolates carry fewer ExPEC virulence genes than ABU isolates. ABU strains from healthy carriers or hospital patients could not be distinguished on the basis of the absence or presence of individual ExPEC virulence genes, but health care-associated ABU strains carried cytotoxin and siderophore determinants more often than community-acquired ABU E. coli strains.
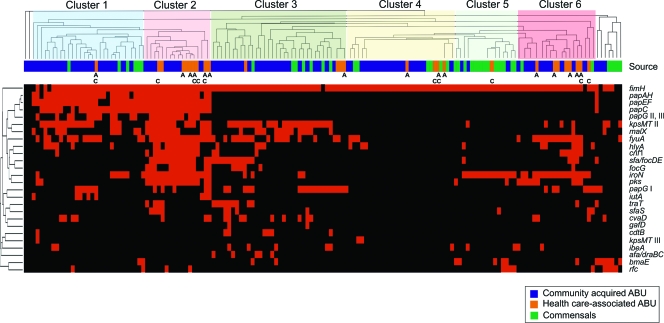
We performed an additional cluster analysis based upon the overall presence or absence of genes, visualizing genetic associations among individual isolates. Six major clusters could be distinguished (Fig. 3), a result which also mirrored the differences in prevalence of ExPEC virulence genes between ABU and commensal isolates indicated in Table S1 in the supplemental material and Fig. 2. Clusters 1 and 2 contain strains with the highest ExPEC virulence gene content. Cluster 1 strains frequently carry the group II capsule determinant, the high-pathogenicity island, as well as a PAI comprising a P-fimbrial operon and the alpha-hemolysin, cytotoxic necrotizing factor 1, and aerobactin determinants. This cluster was mainly populated by community-acquired ABU isolates but included some commensal E. coli isolates as well. Cluster 2 strains possess, in addition to the PAIs frequently present in cluster 1 isolates, other PAIs coding for S/F1C fimbriae and the polyketide colibactin. These strains, which genotypically resemble prototypic UPEC strains, exclusively belong to phylogenetic lineage B2 and represent only ABU isolates, mainly from hospital patients. Many health care-associated ABU isolates were also present in cluster 6, which, together with clusters 3 and 5, included strains with lower-virulence gene profiles. Cluster 4 comprised the isolates with the smallest amount of virulence genes. Whereas commensal isolates predominated in cluster 5, ABU strains were found in all the other clusters (Fig. 3). These clustering results demonstrate that ABU E. coli isolates from healthy or from hospitalized patients are genotypically very heterogeneous with regard to the composition of their ExPEC virulence gene pool. Additionally, these data corroborate the finding that many ABU isolates originate from UPEC but some also genotypically resemble commensal variants without a considerable virulence gene repertoire.
Fig 3.
Cluster analysis based on the presence and absence of ExPEC virulence genes. The source column indicates the origin of an isolate: community-acquired ABU E. coli, health care-associated ABU E. coli, or commensal E. coli. Health care-associated ABU isolates from patients with indwelling catheters (C) or with antibiotic treatment (A) have been labeled. The following columns depict individual PCR results for the presence (red) or absence (black) of typical ExPEC virulence genes.
Comparison of virulence phenotypes of community-acquired ABU, health care-associated ABU, and commensal isolates.
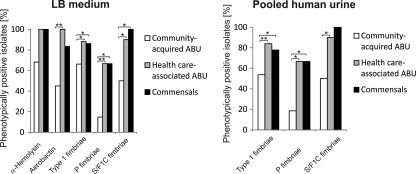
Selected virulence-associated phenotypes were compared after in vitro growth in LB or pooled human urine, such as the expression of alpha-hemolysin, aerobactin, as well as type 1, P-, and S/F1C fimbrial adhesins. Generally, virulence factor expression in LB resembled that in pooled human urine (Fig. 4). The number of genotypically positive strains which expressed the corresponding phenotype was generally lower among community-acquired ABU isolates than among health care-associated ABU or commensal isolates (Fig. 4). In LB, all genotypically hlyA-positive health care-associated ABU and fecal isolates expressed alpha-hemolysin, whereas 68.18% of the community-acquired ABU isolates did so. Interestingly, there were significant differences in the expression of type 1 fimbriae, P fimbriae, S/F1C fimbriae, and the iron uptake system aerobactin in community-acquired isolates relative to health care-associated ABU isolates upon growth in LB. Similarly, fimbrial expression was also reduced in community-acquired ABU isolates relative to health care-associated ABU isolates when the isolates were grown in pooled human urine to mimic conditions of a urinary tract infection (Fig. 4).
Fig 4.
Phenotypic expression of selected virulence factors in community-acquired ABU, health care-associated ABU, or commensal E. coli isolates. The virulence phenotypes were tested upon in vitro cultivation of the isolates in LB or pooled human urine. The percentages of phenotypically positive isolates per group are indicated.
The results show that important UPEC virulence-associated genes are downregulated or inactivated in E. coli isolates which cause ABU in healthy carriers compared to their regulation in isolates from hospitalized patients. In contrast to community-acquired ABU isolates, strains from cases of health care-associated ABU resembled commensal isolates with regard to the phenotypic expression of UPEC virulence factors.
Analysis of type 1 and P-fimbrial adhesin determinants in isolates which are phenotypically negative for type 1 and P fimbriae.
To find out whether the lack of type 1 and P-fimbrial expression results from alterations in the encoding determinants, we screened the integrity of the fimBEAICDFGH and papIBHCDJKEFGX gene clusters in isolates which repeatedly tested negative for phenotypic expression of type 1 or P fimbriae, respectively. In 17 isolates (47.2%) of the 36 type 1 fimbria-negative isolates and in 13 strains (61.9%) of the 21 P fimbria-negative E. coli isolates, multiple genes encoding these determinants could not be amplified by PCR, thus indicating structural variations (see Fig. S4 in the supplemental material). The PCR screenings suggest that large parts of the fim determinant, including the region fimB-fimF (three strains), fimE-fimF (two strains), fimA-fimI (nine strains), or fimI-fimG (three strains), are absent. Structural alterations of the pap determinant included in 1 case the deletion of papJ-papX, in 10 cases the deletion of papGX, and in 2 cases the insertion of an IS3-like sequence between papG and papX.
These results demonstrate that in large fractions of the phenotypically negative strains the absence of type 1 or P-fimbrial expression results from structural differences, including partial deletions of the encoding determinants, thus confirming and extending our previous findings (48, 66).
Sequencing of papG adhesin gene.
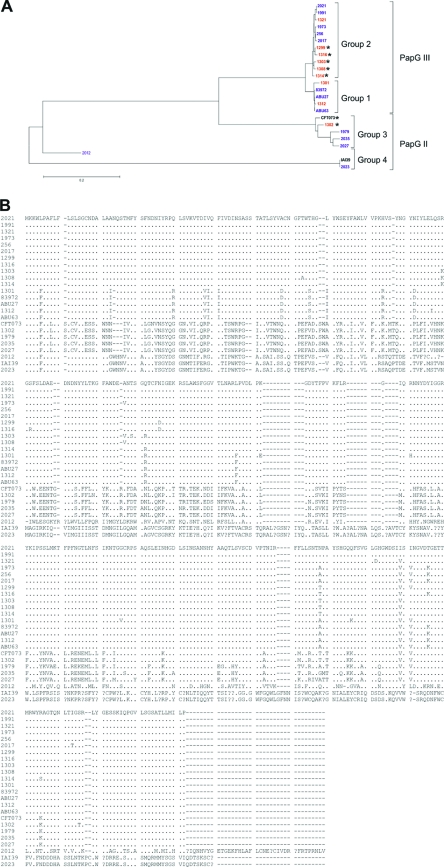
Multiple point mutations in the papG adhesin gene were detected in the prototype ABU strain E. coli 83972 (28, 66). To examine if the reduction in P-fimbrial expression in other ABU strains is also a reflection of such alterations, we sequenced papG in the health care-associated ABU isolates as well as in all genotypically pap-positive but phenotypically Pap-negative community-acquired ABU isolates. Known papG sequences of community-acquired ABU strains as well as P-fimbriated uropathogenic E. coli strains were used as positive controls. On the basis of their length, the different papG sequences were allocated to four groups with 1,005 bp (group 1), 1,008 bp (group 2), 1,011 bp (group 3), and 1,119 bp (group 4), respectively. In health care-associated isolates 2012 and 2023, IS3-like sequences between papG and papX interrupted the pap determinant, and multiple premature stop codons indicated that a functional PapG protein is not expressed in these strains. The structure of the pap determinant in these strains resembles that of an obviously inactivated pap gene cluster in E. coli strain IAI39 (O7:K1) (GenBank accession no. NC_011750), isolated from a patient suffering from acute pyelonephritis.
The PapG amino acid sequences were deduced and compared to known UPEC (CFT073, IAI39) and community-acquired (83972, ABU27, ABU63) ABU sequences. Groups 1 and 2 comprised PapG III variants present in 13 ABU isolates, and group 3 comprised PapG II alleles present in 6 ABU strains. Interestingly, the amino acid sequences of phenotypically Pap-negative isolates were distinct from those of phenotypically Pap-positive strains (Fig. 5). Notably, health care-associated ABU isolates 1301 and 1312 and community-acquired ABU strains 83972, ABU27, and ABU63 carry highly similar nonfunctional PapG III variants.
Fig 5.
Comparison of PapG amino acid sequences in different community-acquired or health care-associated ABU E. coli isolates. The nucleotide sequence of the papG adhesin gene of nine health care-associated (red) and 10 community-acquired (blue) ABU isolates was determined. The deduced amino acid sequence was then compared with the sequences of other phenotypically positive or negative E. coli strains by the ClustalW2 program (31). (A) The phylogram depicts the grouping of the different PapG proteins according to their amino acid similarity. The distances are in units of the number of amino acid substitutions per site. PapG sequences of community-acquired ABU isolates 83972, ABU27, and ABU63, as well as of UPEC strains CFT073 and IAI39, have been used as references. Phenotypically Pap-positive ABU strains have been indicated by an asterisk. (B) Alignment of the deduced amino acid sequences.
These findings demonstrate that virulence gene inactivation due to loss of function mutations is a general and frequent mechanism of phenotypic attenuation in ABU.
DISCUSSION
The in vivo persistence of bacterial pathogens requires different adaptation strategies. In persistent bacterial pathogens such as Pseudomonas aeruginosa or Helicobacter pylori, loss of expression or inactivation of virulence genes during long-term persistence may attenuate highly virulent strains (23, 26, 38, 53). In case of persistent UTI, reductive evolution in UPEC has been explained by gene loss or multiple mutations. UPEC isolates from asymptomatic carriers have been shown to express fewer virulence determinants than strains isolated from patients suffering from symptomatic UTI, due to gene inactivation (18). Similarly, antigenic variation, O-antigen chain reduction or loss, altered capsule or flagella expression, as well as metabolic changes have been observed during the course of chronic UTI or recurrent extraintestinal E. coli infection (6, 39). The present study examined if gene attenuation is a general phenomenon among ABU strains by comparing E. coli strains from healthy children with ABU to those from hospitalized patients. Our results show that these different E. coli isolates share a phylogenetic background and that their virulence gene repertoire distinguishes them from commensal E. coli isolates (Fig. 1 and 2). While similarity to UPEC isolates was reflected by phylogenetic lineages and overall virulence gene content, reduced virulence gene expression and accumulation of mutations were consistent with previous findings in the prototype ABU strain E. coli 83972, suggesting that virulence attenuation may occur broadly among UPEC isolates during long-term carriage in the urinary tract.
ABU is a well-suited model to analyze bacterial adaptation to prolonged in vivo growth in human hosts. Single virulence determinants differ in prevalence among E. coli strains from symptomatic UTI or ABU, reflecting mechanisms of host cell attack and bacterial adherence, such as the expression of fimbrial adhesins, alpha-hemolysin, siderophores, or other significant virulence factors (19, 25). While many virulence factors contribute in the murine UTI model (36, 62), P fimbriae have been confirmed to be an independent virulence factor in the human urinary tract, and the mechanism of tissue attack has been explained, in part, by their glycolipid receptor specificity and the pathogen-specific signaling pathway that they activate in host cells (4, 17, 32). ABU is commonly caused by strains of lower virulence than the UPEC strains with a reduced frequency of virulence factor expression (14, 34, 47, 66). It is interesting to note that inactivating mutations in the papG adhesin gene are shared among E. coli strains causing ABU and that members of the same bacterial clone, e.g., ST73, carry highly similar nonfunctional papG variants. Inactivation of key mechanisms of tissue attack may thus be crucial to avoid overt activation of the host response and pathogen-specific inflammatory pathways that trigger many aspects of symptomatic UTI.
Still, many ABU strains resemble UPEC isolates with regard to genome content and phylogeny. Interestingly, a considerable number of ABU isolates has been allocated to ST73 and its related STs. ST73 mainly comprises UPEC isolates and in particular many ABU isolates. In contrast, other ExPEC-dominated lineages, such as ST95, less frequently include ABU isolates (E. coli MLST database; http://mlst.ucc.ie/mlst/dbs/Ecoli/; as of 18 April 2011). This suggests that ST73 strains may be characterized by particular traits which promote adaptation toward persistence. Comparative genomic analysis of ABU model isolate 83972 indicated that this member of ST73 possesses a large number of UPEC virulence-associated genes (11). This strain's inability to express functional pathogenicity factors is due to deletions or multiple point mutations, resulting in genome reduction as an adaptation to prolonged colonization of the bladder (28, 48, 49, 66).
Furthermore, a limited number of point mutations and small deletions increased in number with prolonged colonization time in ST73 strain 83972 as a molecular adaptation mechanism toward persistence in vivo (65). In the present study, we observed a difference in virulence attenuation between community-acquired isolates and health care-associated strains, which had been carried for shorter periods of time in patients with urological disorders. While it is generally assumed that virulence gene decay in ABU and bacterial adaptation to different infected hosts in general reflect the host response to infection, such host factors remain to be defined. In a recent study, we observed that individual hosts drive rapid genomic changes in bacteria, resulting in host-specific genotypes. The host-specific loss of gene function supports the hypothesis that evolution toward commensalism rather than virulence is favored during asymptomatic bladder colonization. These conclusions were supported by the findings in the present study, as isolates from schoolgirls with long-term ABU were more attenuated than nosocomial isolates from elderly patients (mean age, 70 years) with various comorbidities.
Variation in the host response has also been noted in patients with ABU. Neutrophil numbers in urine vary greatly, and the diagnostic value of pyuria in this patient group has been debated (30). Innate immunity controls the antibacterial defense in the urinary tract, and effector molecules include mucosal cytokines, chemokines, and antibacterial peptides as well as recruited inflammatory cells (1, 8, 22, 43, 45). The Toll-like receptor 4 (TLR4) signaling pathway is critically involved (17, 21), and downstream activation of the transcription factor interferon regulatory factor 3 (IRF3) or NF-κB stimulates the transcription of effector functions of the innate immune response (17). Mice lacking Tlr4 develop an ABU-like state without acute tissue inflammation (22), while mice lacking Irf3, in contrast, develop severe acute pyelonephritis with urosepsis and renal damage (17). In patients with ABU, reduced TLR4 expression (46) and ABU-associated polymorphisms in the TLR4 and IRF3 promoters have been detected, suggesting that the genetic repertoire of the host contributes to the reduced innate immune response in this patient group (17, 44). Recently, the variation in host response to ABU and the influence of the patient TLR4 and IRF3 genotype were documented after inoculation of about 20 patients with the prototype ABU strain E. coli 83972 (20).
The comparison of health care-associated ABU strains from patients (i) with or without long-term indwelling catheter as well as (ii) with or without antibiotic therapy demonstrated that neither of these two factors disproportionally affected the group of health care-associated ABU isolates as a whole, as determined by specific clonal lineages, virulence gene content, or virulence gene expression. As the duration of asymptomatic bladder colonization was longer in healthy carriers (ranging from several months to up to several years [3, 54, 64]) than in the hospitalized patients (ranging from 1 to 74 days), the degree of phenotypic attenuation correlated with prolonged in vivo growth. This suggests that the duration of in vivo colonization is an important determinant of phenotypic attenuation and bacterial adaptation in general. In addition, differences in age, clinical history, and comorbidities may predispose the patients to UTI. The frequency of ABU is increased up to about 20% among elderly individuals (37), possibly due to alterations of their enteric and/or vaginal flora, mucosal surface integrity, and immune status. Ageing is accompanied by a reduction in specific immunity as well as innate defenses and is therefore expected to facilitate the establishment of bacteriuria by strains without specific fitness for the urinary tract. This was not supported by the present study, however, as most of the hospital-acquired strains belonged to the UPEC-associated clonal types and had higher rather than lower virulence factor expression than ABU strains from healthy children.
The findings of our comparison of health care-associated ABU isolates from patients with or without long-term indwelling catheters (see Fig. S1A, S2A, and S3A in the supplemental material) are in accordance with those of a previous study, where ABU E. coli isolates recovered from catheterized and noncatheterized patients from the same hospital have been compared with regard to phylogeny, virulence profiles, as well as virulence phenotypes (61). Generally, the distribution of phylogenetic groups did not differ significantly between these two groups of health care-associated ABU strains, and the majority of them belonged to phylogenetic group B2. Furthermore, these strains possessed similar virulence gene profiles. Significant differences in the ability to functionally express typical UPEC virulence genes could not be observed for alpha-hemolysin, biofilm formation, and type 1 fimbriae. Exceptions were group II capsule marker genes, which had a higher prevalence in catheter-associated ABU (CA-ABU) isolates, and mannose-resistant hemagglutination, which was more prevalent among ABU strains from noncatheterized patients (61). In another study, the genotypic characterization of UPEC and CA-ABU E. coli strains demonstrated that UPEC and CA-ABU E. coli strains predominantly belonged to phylogenetic group B2 and shared virulence determinants. As expected, virulence-associated genes, such as the P-fimbrial determinant (pap), the UPEC pathogenicity island marker malX, as well as iha and ompT, were, however, more common in UPEC isolates than in CA-ABU isolates (55).
The geno- and phenotypic profiles of both ABU E. coli groups and the commensal controls support the hypothesis that bacterial adaptation in vivo is in part driven by positive selection of UPEC variants with a reduced ability to activate the innate immune response (4, 5, 44, 65). On the basis of this attenuation, which may result from virulence gene inactivation and loss of gene expression, such strains achieve long-term carriage in individual hosts. Further studies on the impact of host response and in vivo growth conditions on adaptation of E. coli during colonization of the urinary tract will be necessary to improve our understanding of the microbe-host interaction during symptomatic infection or asymptomatic carriage.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Plaschke (Würzburg, Germany) and O. Mantel (Münster, Germany) for excellent technical assistance, A. Leimbach (Münster, Germany) for software, and S. Weber (Würzburg, Germany) for graphics support.
E.S. received a fellowship from the International Graduate School of Life Sciences of the University of Würzburg. U.D. was supported by the German Research Foundation (DO 789/4-1). A.M. was supported by a grant from the Medical Faculty, University Münster (no. BD9817044). C.S. was supported by the Swedish Medical Research Council, the Royal Physiographic Society, the Medical Faculty, Lund University, and the Söderberg, Österlund, Lundberg, Maggie Stephens, Persson, and Wallenberg Foundations. These studies were carried out within the European Virtual Institute for Functional Genomics of Bacterial Pathogens (CEE LSHB-CT-2005-512061) and the ERA-NET PathoGenoMics II consortium UTI-Interference (Federal Ministry of Education and Research [BMBF] grant no. 0315436A).
Footnotes
Published ahead of print 21 November 2011
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Agace WW, Hedges SR, Ceska M, Svanborg C. 1993. Interleukin-8 and the neutrophil response to mucosal gram-negative infection. J. Clin. Invest. 92:780–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andersson P, et al. 1991. Persistence of Escherichia coli bacteriuria is not determined by bacterial adherence. Infect. Immun. 59:2915–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bergsten G, et al. 2004. PapG-dependent adherence breaks mucosal inertia and triggers the innate host response. J. Infect. Dis. 189:1734–1742 [DOI] [PubMed] [Google Scholar]
- 5. Bergsten G, Wullt B, Svanborg C. 2005. Escherichia coli, fimbriae, bacterial persistence and host response induction in the human urinary tract. Int. J. Med. Microbiol. 295:487–502 [DOI] [PubMed] [Google Scholar]
- 6. Bettelheim KA, Taylor J. 1969. A study of Escherichia coli isolated from chronic urinary infection. J. Med. Microbiol. 2:225–236 [DOI] [PubMed] [Google Scholar]
- 7. Braun V, Gross R, Köster W, Zimmermann L. 1983. Plasmid and chromosomal mutants in the iron(III)-aerobactin transport system of Escherichia coli. Use of streptonigrin for selection. Mol. Gen. Genet. 192:131–139 [DOI] [PubMed] [Google Scholar]
- 8. Chromek M, et al. 2006. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat. Med. 12:636–641 [DOI] [PubMed] [Google Scholar]
- 9. de Hoon MJ, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20:1453–1454 [DOI] [PubMed] [Google Scholar]
- 10. Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dobrindt U, et al. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J. Bacteriol. 185:1831–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edén CS, et al. 1978. Adhesion to normal human uroepithelial cells of Escherichia coli from children with various forms of urinary tract infection. J. Pediatr. 93:398–403 [DOI] [PubMed] [Google Scholar]
- 13. Edén CS, Hanson LA, Jodal U, Lindberg U, Akerlund AS. 1976. Variable adherence to normal human urinary-tract epithelial cells of Escherichia coli strains associated with various forms of urinary-tract infection. Lancet i:490–492 [PubMed] [Google Scholar]
- 14. Edén CS, Janson GL, Lindberg U. 1979. Adhesiveness to urinary tract epithelial cells of fecal and urinary Escherichia coli isolates from patients with symptomatic urinary tract infections or asymptomatic bacteriuria of varying duration. J. Urol. 122:185–188 [DOI] [PubMed] [Google Scholar]
- 15. Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Falush D, et al. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299:1582–1585 [DOI] [PubMed] [Google Scholar]
- 17. Fischer H, et al. 2010. Pathogen specific, IRF3-dependent signaling and innate resistance to human kidney infection. PLoS Pathog. 6:e1001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fünfstück R, et al. 1986. Virulence properties of Escherichia coli strains in patients with chronic pyelonephritis. Infection 14:145–150 [DOI] [PubMed] [Google Scholar]
- 19. Graham JC, et al. 2001. Analysis of Escherichia coli strains causing bacteriuria during pregnancy: selection for strains that do not express type 1 fimbriae. Infect. Immun. 69:794–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grönberg-Hernández J, Sundén F, Connolly J, Svanborg C, Wullt B. 2011. Genetic control of the variable innate immune response to asymptomatic bacteriuria. PLoS One 6:e28289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hagberg L, Briles DE, Eden CS. 1985. Evidence for separate genetic defects in C3H/HeJ and C3HeB/FeJ mice, that affect susceptibility to gram-negative infections. J. Immunol. 134:4118–4122 [PubMed] [Google Scholar]
- 22. Hagberg L, et al. 1984. Difference in susceptibility to gram-negative urinary tract infection between C3H/HeJ and C3H/HeN mice. Infect. Immun. 46:839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hogardt M, Heesemann J. 2010. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int. J. Med. Microbiol. 300:557–562 [DOI] [PubMed] [Google Scholar]
- 24. Horan TC, Andrus M, Dudeck MA. 2008. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am. J. Infect. Control 36:309–332 [DOI] [PubMed] [Google Scholar]
- 25. Hull RA, Rudy DC, Wieser IE, Donovan WH. 1998. Virulence factors of Escherichia coli isolates from patients with symptomatic and asymptomatic bacteriuria and neuropathic bladders due to spinal cord and brain injuries. J. Clin. Microbiol. 36:115–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jelsbak L, et al. 2007. Molecular epidemiology and dynamics of Pseudomonas aeruginosa populations in lungs of cystic fibrosis patients. Infect. Immun. 75:2214–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261–272 [DOI] [PubMed] [Google Scholar]
- 28. Klemm P, Roos V, Ulett GC, Svanborg C, Schembri MA. 2006. Molecular characterization of the Escherichia coli asymptomatic bacteriuria strain 83972: the taming of a pathogen. Infect. Immun. 74:781–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krieger JN, Kaiser DL, Wenzel RP. 1983. Nosocomial urinary tract infections: secular trends, treatment and economics in a university hospital. J. Urol. 130:102–106 [DOI] [PubMed] [Google Scholar]
- 30. Kunin CM. 1997. Urinary tract infections. Detection, prevention and management, 5th ed. The Williams & Wilkins Co., Baltimore, MD [Google Scholar]
- 31. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 32. Leffler H, et al. 1995. Strategies for studying bacterial adhesion in vivo. Methods Enzymol. 253:206–220 [DOI] [PubMed] [Google Scholar]
- 33. Lindberg U, Claesson I, Hanson LA, Jodal U. 1978. Asymptomatic bacteriuria in schoolgirls. VIII. Clinical course during a 3-year follow-up. J. Pediatr. 92:194–199 [DOI] [PubMed] [Google Scholar]
- 34. Mabbett AN, et al. 2009. Virulence properties of asymptomatic bacteriuria Escherichia coli. Int. J. Med. Microbiol. 299:53–63 [DOI] [PubMed] [Google Scholar]
- 35. Nicolle LE, et al. 2005. Infectious Diseases Society of America guidelines for the diagnosis and treatment of asymptomatic bacteriuria in adults. Clin. Infect. Dis. 40:643–654 [DOI] [PubMed] [Google Scholar]
- 36. Nielubowicz GR, Mobley HL. 2010. Host-pathogen interactions in urinary tract infection. Nat. Rev. Urol. 7:430–441 [DOI] [PubMed] [Google Scholar]
- 37. Nordenstam G, Sundh V, Lincoln K, Svanborg A, Eden CS. 1989. Bacteriuria in representative population samples of persons aged 72–79 years. Am. J. Epidemiol. 130:1176–1186 [DOI] [PubMed] [Google Scholar]
- 38. Oh JD, et al. 2006. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc. Natl. Acad. Sci. U. S. A. 103:9999–10004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Olesen B, Kolmos HJ, Orskov F, Orskov I. 1998. Escherichia coli bacteraemia in patients with and without haematological malignancies: a study of strain characters and recurrent episodes. J. Infect. 36:93–100 [DOI] [PubMed] [Google Scholar]
- 40. Plos K, et al. 1995. Intestinal carriage of P fimbriated Escherichia coli and the susceptibility to urinary tract infection in young children. J. Infect. Dis. 171:625–631 [DOI] [PubMed] [Google Scholar]
- 41. Plowman R, et al. 1999. The socio-economic burden of healthcare associated infection. Public Health Laboratory Service, London, United Kingdom [Google Scholar]
- 42. Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ragnarsdottir B, et al. 2008. TLR- and CXCR1-dependent innate immunity: insights into the genetics of urinary tract infections. Eur. J. Clin. Invest. 38(Suppl 2):12–20 [DOI] [PubMed] [Google Scholar]
- 44. Ragnarsdottir B, et al. 2010. Toll-like receptor 4 promoter polymorphisms: common TLR4 variants may protect against severe urinary tract infection. PLoS One 5:e10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ragnarsdottir B, Lutay N, Gronberg-Hernandez J, Koves B, Svanborg C. 2011. Genetics of innate immunity and UTI susceptibility. Nat. Rev. Urol. 8:449–468 [DOI] [PubMed] [Google Scholar]
- 46. Ragnarsdottir B, et al. 2007. Reduced Toll-like receptor 4 expression in children with asymptomatic bacteriuria. J. Infect. Dis. 196:475–484 [DOI] [PubMed] [Google Scholar]
- 47. Raz R. 2003. Asymptomatic bacteriuria. Clinical significance and management. Int. J. Antimicrob. Agents 22(Suppl 2):45–47 [DOI] [PubMed] [Google Scholar]
- 48. Roos V, Nielsen EM, Klemm P. 2006. Asymptomatic bacteriuria Escherichia coli strains: adhesins, growth and competition. FEMS Microbiol. Lett. 262:22–30 [DOI] [PubMed] [Google Scholar]
- 49. Roos V, Schembri MA, Ulett GC, Klemm P. 2006. Asymptomatic bacteriuria Escherichia coli strain 83972 carries mutations in the foc locus and is unable to express F1C fimbriae. Microbiology 152:1799–1806 [DOI] [PubMed] [Google Scholar]
- 50. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 51. Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20:3246–3248 [DOI] [PubMed] [Google Scholar]
- 52. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 53. Smith EE, et al. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sunden F, Hakansson L, Ljunggren E, Wullt B. 2010. Escherichia coli 83972 bacteriuria protects against recurrent lower urinary tract infections in patients with incomplete bladder emptying. J. Urol. 184:179–185 [DOI] [PubMed] [Google Scholar]
- 55. Takahashi A, et al. 2006. Escherichia coli isolates associated with uncomplicated and complicated cystitis and asymptomatic bacteriuria possess similar phylogenies, virulence genes, and O-serogroup profiles. J. Clin. Microbiol. 44:4589–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Turck M, Stamm W. 1981. Nosocomial infection of the urinary tract. Am. J. Med. 70:651–654 [DOI] [PubMed] [Google Scholar]
- 58. Wagenlehner FM, et al. 2002. Epidemiological analysis of the spread of pathogens from a urological ward using genotypic, phenotypic and clinical parameters. Int. J. Antimicrob. Agents 19:583–591 [DOI] [PubMed] [Google Scholar]
- 59. Wagenlehner FM, Niemetz AH, Weidner W, Naber KG. 2008. Spectrum and antibiotic resistance of uropathogens from hospitalised patients with urinary tract infections: 1994-2005. Int. J. Antimicrob. Agents 31(Suppl. 1):S25–S34 [DOI] [PubMed] [Google Scholar]
- 60. Warren JW. 1997. Catheter-associated urinary tract infections. Infect. Dis. Clin. North Am. 11:609–622 [DOI] [PubMed] [Google Scholar]
- 61. Watts RE, et al. 2010. Escherichia coli isolates causing asymptomatic bacteriuria in catheterized and noncatheterized individuals possess similar virulence properties. J. Clin. Microbiol. 48:2449–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wiles TJ, Kulesus RR, Mulvey MA. 2008. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp. Mol. Pathol. 85:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wirth T, et al. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wullt B, et al. 1998. Urodynamic factors influence the duration of Escherichia coli bacteriuria in deliberately colonized cases. J. Urol. 159:2057–2062 [DOI] [PubMed] [Google Scholar]
- 65. Zdziarski J, et al. 2010. Host imprints on bacterial genomes—rapid, divergent evolution in individual patients. PLoS Pathog. 6:e1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zdziarski J, Svanborg C, Wullt B, Hacker J, Dobrindt U. 2008. Molecular basis of commensalism in the urinary tract: low virulence or virulence attenuation? Infect. Immun. 76:695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zuckerkandl E, Pauling L. 1965. Molecules as documents of evolutionary history. J. Theor. Biol. 8:357–366 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.