Abstract
Neisseria meningitidis employs redundant heme acquisition mechanisms, including TonB receptor-dependent and receptor-independent uptakes. The TonB-dependent zinc receptor ZnuD shares significant sequence similarity to HumA, a heme receptor of Moraxella catarrhalis, and contains conserved motifs found in many heme utilization proteins. We present data showing that, when expressed in Escherichia coli, ZnuD allowed heme capture on the cell surface and supported the heme-dependent growth of an E. coli hemA strain. Heme agarose captured ZnuD in enriched outer membrane fractions, and this binding was inhibited by excess free heme, supporting ZnuD's specific interaction with heme. However, no heme utilization defect was detected in the meningococcal znuD mutant, likely due to unknown redundant TonB-independent heme uptake mechanisms. Meningococcal replication within epithelial cells requires a functional TonB, and we found that both the znuD and tonB mutants were defective not only in survival within epithelial cells but also in adherence to and invasion of epithelial cells. Ectopic complementation rescued these phenotypes. Interestingly, while znuD expression was repressed by Zur with zinc as a cofactor, it also was induced by iron in a Zur-independent manner. A specific interaction of meningococcal Fur protein with the znuD promoter was demonstrated by electrophoretic mobility shift assay (EMSA). Thus, the meningococcal ZnuD receptor likely participates in both zinc and heme acquisition, is regulated by both Zur and Fur, and is important for meningococcal interaction with epithelial cells.
INTRODUCTION
Neisseria meningitidis, an exclusive pathogen of humans, is the cause of epidemic bacterial meningitis and sepsis (49). As a result of successful conjugate vaccines in reducing the incidence of meningitis caused by Streptococcus pneumoniae and Haemophilus influenzae infections, N. meningitidis has become a leading cause of bacterial meningitis in children and young adults in the United States (47, 62). Despite the sensitivity of meningococcus to many antibiotics, meningococcal disease still causes substantial mortality and morbidity (5).
Meningococci encounter iron-restricted host environments during colonization and infection, and they are equipped with receptors for many available iron-carrying proteins whose uptake functions depend on the TonB-ExbB-ExbD system to provide energy for active transport (i.e., TonB-dependent receptors) (39). As heme is an abundant potential source of iron from the host and is critical for many important physiological processes, redundant strategies for capturing heme are demonstrated not only for meningococci but also for many other bacterial pathogens (2, 9, 18, 35, 61). The redundancy of heme acquisition systems likely accounts for the efficient capture of diverse heme sources encountered during various stages of infection. The HpuA/B system enables meningococci to use iron from the hemoglobin-haptoglobin (Hb-Hp) complex, while the HmbR receptor is believed to strip heme from hemoglobin and subsequently transport it into the periplasm (52). Subsequently, heme is transported into the cytoplasm via an as-yet-unidentified inner membrane transport system (53), and a cytoplasmic heme oxygenase, HemO, extracts iron from heme and metabolizes the protoporphyrin ring (65). A report of TonB-independent heme utilization in Neisseria gonorrhoeae shows that a point mutation in pilQ, encoding the secretin channel of pilin subunits, enables heme-dependent growth (7). The fact that meningococcal heme-binding outer membrane proteins have been detected but remained uncharacterized (30, 31) suggests that additional TonB-dependent heme receptors are possible. Homology searches using the protein sequence of the HumA heme receptor of Moraxella catarrhalis (15), a strict human respiratory pathogen causing otitis media in infants and children, identified ZnuD with significant sequence similarity. ZnuD was recently reported to be involved in zinc acquisition, and the detection of antibodies specific to ZnuD in human convalescent-phase sera indicates that ZnuD is expressed during infection (55).
Here, we present data that ZnuD also functions in heme utilization. When expressed in E. coli, ZnuD allowed heme capture on the cell surface, and its binding to heme agarose in vitro can be inhibited by the presence of excess heme. The expression of ZnuD in a hemA-deficient E. coli strain enabled growth with heme as the sole iron source. Further, we showed that iron regulated the expression of znuD, and a direct interaction of Fur with the znuD promoter was demonstrated by electrophoretic mobility shift assay (EMSA). Interestingly, a znuD mutation caused defects in meningococcal attachment to, invasion of, and survival within epithelial cells. Thus, ZnuD appears to be capable of interacting with both zinc and heme and is important for the interaction of N. meningitidis with host epithelial cells.
MATERIALS AND METHODS
Bacterial strains and medium.
Strains and plasmids used in this study are listed in Table 1. Meningococcal strains were grown with 5% CO2 at 37°C with GC base agar (Difco) supplemented with 0.4% glucose and 0.68 mM Fe(NO3)3 or in GC broth with the same supplements and 0.043% NaHCO3 or RPMI without phenol red and supplemental with 0.3 mg/ml glutamine. BHI medium (37 g/liter brain heart infusion) with 1.25% fetal bovine serum (FBS) was used when kanamycin selection was required. Antibiotic concentrations (in μg/ml) used for E. coli strains were the following: ampicillin (Amp), 100; kanamycin (Kan), 50; spectinomycin (Spt), 60; tetracycline (Tet), 10; and erythromycin (Ery), 300. For N. meningitidis they were the following: kanamycin, 80; spectinomycin, 60; and erythromycin, 3. E. coli strains DH5α and TOP10 cultured on Luria-Bertani (LB) medium were used for the cloning and propagation of plasmids. Meningococci were transformed by the procedure of Janik et al. (21). E. coli strains were transformed by electroporation with GenePulser (Bio-Rad) or the heat-shock transformation of chemically competent cells according to the manufacturer's protocol.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| N. meningitidis | ||
| IR1072 | Serogroup C, 8013#6 | 51 |
| IR4130 | IR1072, Strr | This study |
| IR3727 | IR4130, hpuB null, hmbR∷ermC | This study |
| 3727RQ | IR3727, znuD∷Ω(Sp), pilQ∷aphA3 | This study |
| 3727BQ | IR3727, tonB∷Ω(Sp), pilQ∷aphA3 | This study |
| IR4130R | IR4130, znuD∷Ω(Sp) | This study |
| IR4130B | IR4130, tonB∷Ω(Sp) | This study |
| IR4130RC | IR4130, znuD∷Ω(Sp), Plac∷znuD in the aspC-lctP locus | This study |
| NMB | B:2b:P1.2,5:L2 (CDC 8201085) (serogroup B) | 50 |
| YT394 | znuD∷lacZ in strain NMB | This study |
| PKT300 | zur∷aphA3 in strain YT394 | This study |
| E. coli | ||
| DH5α | Cloning strain | New England Biolabs |
| TOP10 | Cloning strain | Invitrogen |
| IR1583 | DH5α hemA with cos2 (N. meningitidistonB, exbB, exbD) | 54 |
| Plasmids | ||
| pBAD-TOPO | Arabinose-controlled expression vector | Invitrogen |
| pCR2.1 | Cloning vector | Invitrogen |
| pFlag-CTC | Cloning vector for Flag fusion | Sigma |
| pHP45Ω | Source of Ω(Sp) | 40 |
| pGCC4 | Meningococcal complementation vector | 34 |
| pTA964 | An 1,849-bp fragment of znuD in pCR2.1 | This study |
| pSZ50 | znuD∷Ω(Sp) in pCR2.1 | This study |
| pSZ51 | PBAD∷znuD-V5-His in pBAD | This study |
| pYT387 | Ptrc∷znuD-Flag in pFlag-CTC | This study |
| pYT389 | PBAD∷znuD-V5 in pBAD | This study |
| pYT390 | PBAD∷znuD-Flag in pBAD | This study |
| pYT396 | Fur in pET28a | This study |
| pYT422 | Plac∷znuD-V5-His in pGCC4 | This study |
Construction of ZnuD expression plasmids.
The coding sequence of znuD was obtained by PCR amplification using primers 964-F (5′-TTTGCCATGGGACGACGAGAAGCCAAAATG-3′; the NcoI site is underlined) and 964-R (5′-AAACTTCACGTTCACGCCGCCGGT-3′) and cloned into pBAD-TOPO using the TA cloning kit (Invitrogen). Colonies were screened by colony PCR with primers PBAD-F and 964-R for clones with the correct orientation. The plasmid then was digested with NcoI, purified, and self-ligated to remove the N-terminal leader sequence from the vector, which was confirmed by DNA sequencing. The resulting plasmid, pSZ51, contains the znuD gene with a C-terminal V5-His6 tag under the control of the arabinose-inducible PBAD promoter. To remove the His tag, a PCR product of pSZ51 was obtained with primers B964-F and V5-PmeI (5′-CCGGCGCGCCGTTTAAACTCACGTAGAATCGAGTCCGAG-3′; the PmeI site is underlined). After digestion with NcoI and PmeI, the DNA fragment was cloned into pSZ51, which has been cut with the same enzymes, to replace the original insert. Sequencing analysis of the resulting pYT389 confirmed the introduction of a stop codon at the 3′ end of V5 tag and the absence of His tag sequence. To generate the znuD∷Flag construct, a PCR product of pSZ51 was obtained with primers NMB0964-ERNco (5′-CCGGAATTCCATGGGACGACGAGAAGCCAAAATG-3′; the EcoRI-NcoI the site is underlined) and NMB0964-BglII (5′-GGAAGATCTAAACTTCACGTTCACGCCGCCGGT-3′; the BglII site is underlined) and cloned into pFlag-CTC after digestion with EcoRI and BglII. The resulting plasmid, pYT387, with znuD fused to the Flag tag, was verified by DNA sequencing. The znuD∷Flag fragment was PCR amplified with primers B964-F and Flag-PmeI (5′-CCGGCGCGCCGTTTAAACTCACTTGTCGTCATCGTCCTTG-3′ the PmeI site is underlined) using pYT387 as the template and was used to replace the insert of pSZ51, yielding pYT390.
Heme sedimentation assay.
The heme sedimentation assays were performed as described by Olczak et al. (36). Overnight cultures of E. coli strains expressing recombinant ZnuD proteins were used to inoculate LB broth and grown to mid-log phase. The cultures then were split into two cultures. One culture was induced with 0.4% arabinose, while the other served as a noninduced control. After a 4-h induction, aliquots of cultures were collected, washed once with 1 ml of phosphate-buffered saline (PBS), and adjusted to an optical density at 600 nm (OD600) of 1.0 in 1 ml of PBS. Heme was added to a final concentration of 10 μM, and dimethylsulfoxide (DMSO) (5%) also was added to reduce the self aggregation of heme (36). Triplicates of cells were incubated with heme at room temperature (RT) for 1 h with gentle rocking and then centrifuged to pellet the bacteria. The supernatant was collected and the OD400 determined. A solution containing an identical concentration of heme and DMSO without bacteria was incubated similarly and served as the 100% control. The binding of heme to cells was determined by the decrease of absorbance of the supernatant compared to that of the control.
Outer membrane preparation.
The E. coli strain Top10, carrying pSZ51, was grown in LB broth with 100 μg/ml Amp to an OD600 of ∼0.4, and arabinose was added to induce protein expression. Cells were collected by centrifugation after a 4-h induction, and the enriched outer membrane fractions were prepared as described previously (42). Briefly, cells were resuspended into 0.5 ml of lysis buffer (200 mM Tris, pH 8.0). After diluting the cell suspension with 1 ml of 1 M sucrose in 200 mM Tris, pH 8.0, and 0.1 ml of 10 mM EDTA, freshly prepared lysozyme solution (0.1 ml of 2 mg/ml) was added. Subsequently, water (3.2 ml) was added and the mixture was incubated at RT for 10 min to generate spheroplasts. Outer membranes were extraction from spheroplasts with 5-ml aliquots of extraction buffer (2% Triton X-100, 50 mM Tris, pH 8.0, 10 mM MgCl2) (45, 46, 60). The mixture was further treated with 0.1 ml of DNase (1 μg/μl) and then centrifuged at 35,000 × g for 1 h to pellet the outer membrane fraction. The pellet was washed three times with water and then resuspended in water. Protein concentrations were determined by bicinchoninic acid (BCA) assays (Pierce) with bovine serum albumin (BSA) as the standard.
Heme agarose binding assay.
The heme agarose binding assay was performed as described previously (31). Aliquots of total outer membrane protein preparations (20 μg) in 1 ml of binding buffer (50 mM Tris, pH 8.0, 1 M NaCl) were incubated with 50 μl of heme agarose suspensions (Sigma) that had been washed with 1 ml of binding buffer. After a 1-h incubation at 20°C, EDTA and sarcosyl were added to a final concentration of 10 mM and 0.75%, respectively, and the mixtures were incubated for another hour. The resins were collected by centrifugation at 750 × g for 5 min and washed once for 30 min with 1 ml of buffer A (50 mM Tris, pH 8.0, 1 M NaCl, 5 mM EDTA, 0.5% sarcosyl), followed by one wash with buffer B (50 mM Tris, pH 8.0, 100 mM NaCl). For heme competition, increasing amounts of heme (25 mg/ml in 0.02 N NaOH) were added to the heme agarose prior to the addition of outer membrane proteins, and a sample with the maximal volume of NaOH was included as a control. The bound proteins were dissociated from the resins by boiling in SDS-PAGE buffer, resolved by SDS-PAGE, and then either stained with Coomassie blue or transferred to polyvinylidene difluoride (PVDF) membrane and probed with the anti-His antibody (Invitrogen). The membranes were incubated with the primary antibody at a 1:5,000 dilution for 1 h at RT, followed by incubation with the anti-mouse IgG-horseradish peroxidase (HRP) conjugate (Pierce) at a 1:15,000 dilution for 1 h at RT. The membranes were developed with ECL detection reagents (Pierce).
Heme-dependent growth assay.
The E. coli strain IR1583 carrying either the pSZ51 plasmid or the empty vector pBAD was grown on LB plates supplemented with 5 μg/ml δ-aminolevulinic acid. Bacteria resuspended in PBS were added to 0.5% agarose to an OD600 of 0.1 per ml, and then 3 ml of the cell suspension was immediately poured over an NBD plate (8 g/liter nutrient broth, 5 g/liter NaCl, 250 μM 2,2-dipyridyl) supplemented with appropriate antibiotics and with or without arabinose. Filter paper discs impregnated with 10 μl of test compounds were laid onto the top agar, and growth stimulation around the filter disk was recorded after incubation at 37°C for 48 h. Similar disc diffusion growth assays were performed with N. meningitidis. Bacteria suspended from overnight plate cultures were incorporated into 0.5% soft agar and poured onto GC plates supplemented with 100 μM desferal. Ten μl of an iron source was supplied on discs.
Construction of meningococcal mutations and complementation.
An 1,849-bp coding fragment of znuD was PCR amplified using primers NMB0964-F (5′-CGCATCCGCTCCCGTTATTC-3′) and NMB0964-R (5′-GCCGAGGTTGAGCATATGGTGTC-3′) and cloned into pCR2.1 using the TOPO-TA cloning kit (Invitrogen) to generate pTA964. The Ω(Sp) cassette was released from pHP45Ω by SmaI digestion, gel purified, and ligated with HincII-digested pTA964, which removed 654 bp of the znuD sequence. Sptr and Kanr transformants were selected, and the insertion of the Ω cassette was confirmed by colony PCR. The resulting plasmid, pSZ50, was digested with ScaI to inactivate the gene encoding ampicillin resistance, and the digestion mixture was used to transform meningococcal strains and selected for Sptr colonies. The disruption of znuD with the Ω(Sp) cassette was confirmed by colony PCR using Ω cassette-specific outward primer JS81 (5′-CGAGATCACCAAGGTAGTCGG-3′) and NMB0964-R.
To generate the tonB-exbB-exbD mutation, the chromosomal DNA of a serogroup C strain, FAM18, carrying the ΔtonB-exbB-exbD∷Ω mutation (43), was used for transformation. The mutation was confirmed by colony PCR using primers exbD2 (5′-ATTTTGGTGATTCCTGCC-3′) and SPC1 (5′-GACCAGTTGCGTGAGCGC-3′).
To construct the pilQ∷Ω(Kan) mutation, a PCR product of the pilQ∷Ω(Kan) mutation (J. Folster, unpublished data) was obtained using primers pilQ1 (5′-TCCGGCTTCATCGAGGCTG-3′) and pilQ2 (5′-AGCCGAATGCGCTCGTCTC-3′) and was used to transform meningococcal strains. Kanr transformants were selected and confirmed to carry the pilQ mutation by PCR.
To make the znuD complementation construct, the znuD coding sequence was amplified from pSZ51 using B964-PacI (5′-CCTTAATTAAGAAGGAGATATACATACCCATGGG-3′) and pBAD-R (5′-GATTTAATCTGTATCAGG-3′). The PCR product was digested with PacI and PmeI and then cloned into pGCC4 (34) cut with the same enzymes to yield pYT422.
The zur mutation was generated using the overlapping PCR technique. A 662-bp 5′ fragment and a 644-bp 3′ fragment flanking the zur coding sequence were produced using primer pairs zur-5F (5′-GCTGGTTCTGCCTTATGCGGTTA-3′)/zur-5R (5′-TTCCTCCTAGTTAGTCACCCGCGTGCCTGTTCGATAATTTTCTGT-3′) and zur-3F (5′-CCTGGAGGGAATAATGACCCCTTTGCGCTGAAAGAAGAACACG-3′)/zur-4 (5′-TCCTATTGCGCAATACCCCC-3′), respectively. The aphA3 cassette was obtained using primers aphA3-claI-f3 and apha3-claI-r2 (25) and then cut with SmaI. A mixture of the 5′ fragment and the apha3 cassette were used as the template for the first round of PCR with primers zur-5F and apha3-claI-r2. The resulting PCR product then was digested with SmaI and mixed with the 3′ fragment for the second round of PCR using primers zur-5F and zur-4. The final PCR product was purified and used directly for the transformation of meningococcal strains. Correct transformants were identified by colony PCR using zur-1 (5′-TTCGCCGATGGCGGAATACA-3′) and chromosome-specific primer zur-3R (5′-GTCCGGCAGGGCTTTGAGG-3′).
Cell culture and intracellular survival assay.
The A549 human lung adenocarcinoma cell line (24) was cultured in Dulbecco's modified essential medium (DMEM) supplemented with heat-inactivated fetal bovine serum (FBS) (10%) in T-75 plastic flasks at 37°C and 5% CO2. For the bacterial adherence and invasion assay, cells were seeded at a density of 105 cells per ml in 24-well plates (Corning) 2 days prior to the experiment. To prepare bacterial inocula, meningococcal strains were grown in GC broth to mid-log phase, collected by centrifugation, and resuspended in cell culture media. For intracellular survival, the infection was initiated at a multiplicity of infection (MOI) of 2 without centrifugation and allowed to proceed overnight. After gentamicin killing (time [t] = 0 h), cells were rinsed three times with PBS and then fresh medium was added. Infected cells were incubated for an additional 6 h, and then cells were lysed by incubating with 1% of saponin (Sigma) for 10 min to release intracellular bacteria. Serial dilutions of lysates in PBS were plated on GC plates for survival CFU counts. The direct plating of medium supernatants prior to cell lysis was done to confirm the absence of free extracellular bacteria. Survival results at t = 6 h were normalized to the invasion CFU counts at t = 0 h. For adherence and invasion, bacterial cells were added to cell cultures at an MOI of 100, and serial dilutions of the inoculum were plated to determine the input CFU. Infections were initiated with low-speed centrifugation (80 × g for 5 min) to enhance bacterium-cell contact. After a 2-h infection, the monolayers were washed three times with sterile PBS to remove free bacteria, and the CFU of attached bacteria was determined. Separately infected cells were washed and then incubated in cell culture medium containing 100 μg/ml gentamicin to kill extracellular bacteria, and the recovered bacterial CFU was determined. The levels of attachment were calculated relative to the initial input CFU, while the invasion efficiencies were determined as the gentamicin-resistant counts divided by the adhered counts. The data were expressed as percentages of those of the wild-type strain. Each assay was conducted with two independently infected monolayers and repeated at least two times.
Construction of the znuD∷lacZ reporter and β-galactosidase assays.
A 270-bp znuD promoter fragment was obtained by PCR amplification with primers NMB0964-p-F1 (5′-AGGACGGCGGATACAAATGGACAC-3′) and NMB0964-p-R1 (5′-GACGACGCTGACCGTTTCCAAG-3′) and cloned into pCR2.1. The fragment released by EcoRI digestion subsequently was inserted into the EcoRI site of plasmid pYT328 (58) to yield pYT394. The plasmid construct with a correctly oriented promoter fragment was identified with colony PCR using primers YT168 and a forward primer within the cloned promoter fragment. The plasmid was linearized with NcoI prior to transformation, and Ermr transformants were selected. The integration of znuD∷lacZ at a permissive intergenic chromosomal location (flanked by NMB0428 and NMB0430) was verified by colony PCR. The zur mutation was introduced into the reporter strains using the zur∷aphA3 PCR product with kanamycin selection. The β-galactosidase assays were performed as described previously (58). The reporters were grown in RPMI to mid-log phase (OD550, ∼0.5), divided into 3-ml aliquots, and then treated with various compounds for 1 h.
Primer extension.
Thirty μg of total RNA isolated from mid-log-phase cultures in RPMI with an RNeasy midi kit (Qiagen) was used in primer extension reactions with 32P-labeled primers (znuD-PE1, 5′-GCCCGGCACGCCGTCCAAAGCATC-3′) by following previously described procedure (44). The corresponding DNA sequencing reactions were carried out with the Thermo Sequenase dye primer manual cycle sequencing kit (USB) using the same labeled oligonucleotides and PCR fragments of the promoter regions. The extension products and the sequencing ladders were resolved on an 8% sequencing gel.
Meningococcal Fur protein expression and purification.
The fur coding sequence was amplified by PCR using primers fur-F-Nde (ATTGAATCATATGGAAAAATTCAACAATATTG; the NdeI site is underlined) and fur-R-Bm (CGCGGATCCTTAACGTTTGCCCTTGGCCT; the BamHI site is underlined). The PCR product was cloned into pET28a after the digestion of DNA with NdeI and BamHI, and the ligation was mixture transformed into E. coli strain DH5α. Correct clones were identified by PCR and confirmed by sequencing analysis to yield pYT396. Plasmid pYT396 was transformed into the E. coli strain BL21(DE3) for expression. A 500-ml culture of the BL21(DE3)(pYT396) strain in LB medium was grown to mid-log phase and induced for 3 h with 1 mM IPTG. Cells resuspended in lysis buffer (50 mM Na phosphate, pH 8.0, 300 mM NaCl, 1 mM MnCl2, 10 mM imidazole) were lysed by sonication, and the protein was purified by Ni-nitrilotriacetic acid affinity chromatography under nondenaturing conditions according to the manufacturer's instructions (Qiagen). The fractions were analyzed by SDS-PAGE for purity and then pooled and dialyzed against the lysis buffer without imidazole at 4°C. The protein concentration was determined using the Bradford colorimetric assay (Bio-Rad) with BSA as the standard.
EMSA.
The previously described procedure for EMSA (58) was performed with slight modifications. The probes were prepared by the end phosphorylation of PCR products using T4 kinase. The znuD, fur, and dsbD probes were generated by primer pairs NMB0964-pF1 and NMB0964-pR1, Fur-N-PacI (5′-CAAACCTTAATTAATCACGGAAATGCCTTTCTGTGC-3′) and Fur-qR1 (5′-CTTGCCCGTTTCAAAATGAT-3′), and YT143 and YT144 (25), respectively. The binding buffer is composed of 20 mM HEPES, 60 mM KCl, 5 mM MgCl2, 100 μM MnCl2, 1 mM dithiothreitol (DTT), 0.3 mg/ml BSA, 0.2 mg/ml salmon sperm DNA, and 10% glycerol. The electrophoresis buffer is made of 0.5× Tris-borate-EDTA (TBE) with 100 μM MnCl2.
RESULTS
Heme was captured by E. coli expressing the ZnuD protein.
ZnuD (758 amino acids [aa]) and HumA (818 aa) share 41% identity and 58% similarity across their entire protein sequences (P = 3.7e−167). The alignment of ZnuD with several representative heme receptors revealed the presence of the C-terminal FRAP/NPNL motifs, which have been shown to be characteristic of receptors known to capture heme or hemoproteins (6), and the conserved histidine residue, which has been shown to be critical for the heme utilization of HemR in Yersinia enterocolitica and HmuR in Porphyromonas gingivalis (6, 33) (Fig. 1). Stork et al. (55) reported that a topology model of ZnuD, analogous to that of the HmbR hemoglobin receptor (38), contained a 22-stranded β barrel. Loop 3 of ZnuD was proposed to be involved in zinc binding (55), whereas the FRAP/NPNL motifs are located in extracellular loop 7 of ZnuD. Interestingly, deleting loop 7 of HmbR caused a defect in the removal of heme from Hb, while the deletion of loop 3 of HmbR preserved the use of heme as an iron source (38). The distinct locations of the predicted heme and zinc binding loops implied independent ligand recognition.
Fig 1.
Homology at the C-terminal region between the N. meningitidis ZnuD protein and characterized heme receptors. The conserved motifs, the FRAP and NPNL boxes, and the highly conserved histidine residue are in boldface and marked by black lines and pound signs, respectively. The identical residues are indicated by asterisks. The numbers at either side of the sequences are the residue numbers. Nm, Neisseria meningitidis; Mc, Moraxella catarrhalis; Sd, Shigella dysenteriae; Ye, Yersinia enterocolitica; Pg, Porphyromonas gingivalis; Bp, Bordetella pertussis.
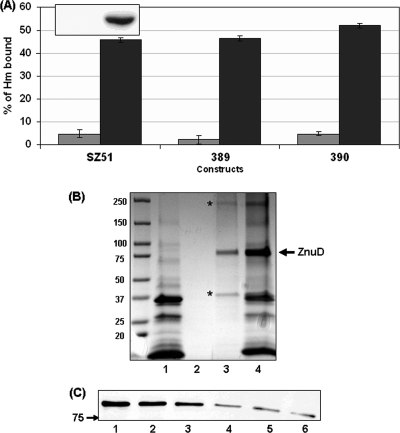
To examine whether ZnuD binds heme, the coding sequence of znuD was cloned into the pBAD vector under the control of arabinose and fused with a C-terminal V5-His6 tag to allow the detection of its expression. A band with the expected molecular size was detected only in the induced cells by anti-His Western blotting (Fig. 2A, inset), confirming the tight regulation by PBAD. Thus, the noninduced sample was used as a control for nonspecific heme absorption. The heme sedimentation experiment using whole bacterial cells was performed as described previously (36). The amount of heme removed from the supernatant by E. coli cells was determined, and the absorbance of a solution similarly incubated without bacteria was treated as 100% for normalization. Figure 2A showed that noninduced cells gave low background binding (∼5%), indicating minimal nonspecific heme trapping by the E. coli cell surface under the experimental conditions. In contrast, E. coli cells expressing ZnuD captured significantly larger amounts of heme (∼ 45%). Two additional recombinant ZnuD constructs with either a V5 (GKPIPNPLLGLDST) or a Flag (DYKDDDDK) tag were generated. Both constructs displayed heme-binding abilities similar to that of the original construct (Fig. 2A), demonstrating that ZnuD, not the tag sequences, enabled heme capture. We also assessed the sensitivity of induced E. coli strains to hydrophobic erythromycin by disc diffusion assays, and no significant difference was seen between the ZnuD-expressing strain and the control strain carrying an empty vector (data not shown), supporting that heme capture by E. coli was not a result of altered outer membrane permeability that allowed the nonspecific diffusion of hydrophobic agents. As ZnuD was reported to interact with zinc (55), we examined whether the presence of zinc would interfere with heme binding. The experiments were repeated in the presence of an equimolar concentration of zinc. Similar levels of heme binding were detected in the presence of zinc (data not shown), thus the binding of heme to ZnuD appeared to be independent of zinc.
Fig 2.
Interaction of ZnuD with heme. (A) Binding of heme to E. coli cells expressing various tagged ZnuD protein constructs under noninduced (gray bars) and induced (black bars) conditions. The binding of free heme to whole cells was calculated by the difference between the OD400 of the cell-free supernatant and the control heme solution without cells (10 μM as 100%). ZnuD constructs were the following: SZ51, ZnuD-V5-His6; 389, ZnuD-V5; and 390, ZnuD-Flag. The data are representative of three independent experiments performed in triplicate. The inset shows the expression level of ZnuD with or without arabinose induction as determined by anti-His Western blotting. Whole-cell lysates from equal numbers of bacteria based on the OD550 readings were loaded. (B) ZnuD binding to heme agarose. Enriched outer membrane fractions prepared from E. coli expressing the ZnuD-V5-His6 construct were used. Lane 1, total outer membrane proteins (12 μg) from noninduced cultures; 2, heme agarose-bound proteins from noninduced samples; 3, heme agarose-bound proteins from induced samples; 4, total outer membrane fraction from induced samples (12 μg). The location of ZnuD is indicated on the right, while the asterisks mark bands that are present only in the induced sample, possibly resulting from incomplete denatured aggregates and degraded proteins during SDS-PAGE sample preparations. The protein molecular size ladders are indicated on the left. (C) Free heme competition. Outer membrane proteins (15 μg) from induced samples were used, and the amounts of ZnuD bound to heme agarose were analyzed by anti-His Western blotting. Samples: 1, no treatment; 2, treatment with 0.02 N NaOH; 3 to 6, treatment with 0.2, 0.5, 1, and 5 μg/μl heme (Hm), respectively.
To further define the interaction of ZnuD with heme, enriched outer membrane fractions were prepared from the E. coli strain expressing His-tagged ZnuD as described previously (42) using Triton X-100 to selectively solubilize and remove inner membrane proteins. A protein band migrated appropriately to the predicted molecular mass (84.1 kDa) was detected by Coomassie staining in the induced samples (Fig. 2B, lane 4) but not in the noninduced (Fig. 2B, lane 1) or empty vector sample (not shown). The in vitro binding of ZnuD to heme was examined using heme agarose suspension (31). Binding was performed in high-ionic-strength conditions to prevent nonspecific ionic interaction with the matrix, and multiple extensive washings in the presence of N-laurylsarcosine were performed to further remove nonspecific interaction prior to the elution of bound proteins. Analogously to other heme receptors demonstrated to interact with heme agarose in vitro (3, 15), ZnuD was retained on heme resins (Fig. 2B), while no nonspecific protein binding to heme agarose was detected in the noninduced sample. To further determine the specificity of ZnuD association with heme agarose, heme competition experiments were conducted and ZnuD binding detected by anti-His Western blotting. As shown in Fig. 2C, the highest concentration of sodium hydroxide present in the heme solution did not affect ZnuD binding (Fig. 2B compare lane 2 to lane 6), while a dose-dependent decrease in ZnuD signals in samples with increasing amounts of free heme was seen, supporting the notion that ZnuD interacted with heme.
ZnuD enabled heme-dependent growth in E. coli.
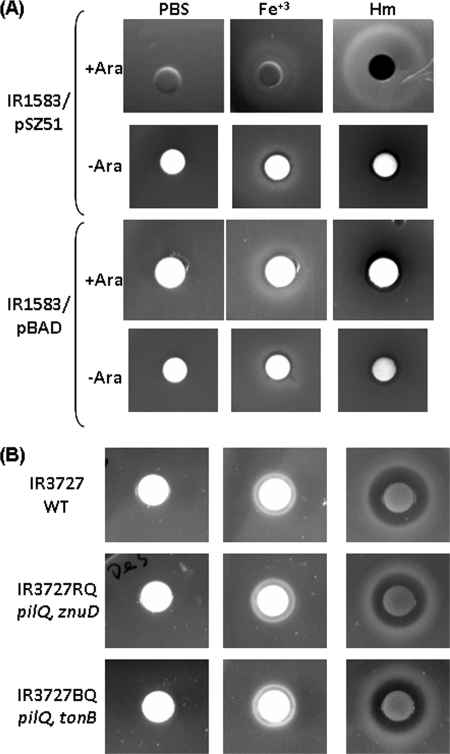
To investigate whether ZnuD supports the heme-dependent growth of E. coli, the znuD-expressing plasmid, pSZ51, was incorporated into E. coli hemA strain IR1583 that contains the meningococcal TonB-ExbB-ExbD system, which has been shown previously to enable the meningococcal hemoglobin receptor HmbR to reconstitute heme utilization in E. coli (54). The growth stimulation assays were performed on NBD plates (4) supplemented with 2,2-dipyridyl to deplete iron. As shown in Fig. 3A, no growth was observed around the negative-control disc, while the addition of free iron restored growth. Supplementing with 5 mM heme yielded a zone of growth around the disc of the pSZ51-carrying strain in the presence of arabinose induction, while no growth was detected without induction, thus supporting that ZnuD enabled E. coli to obtain iron from heme. A control strain carrying the empty vector also was tested under the same conditions, and no heme-dependent growth was observed. The fact that ZnuD expression alone promoted heme-dependent growth suggested that ZnuD likely functioned as a single-component receptor.
Fig 3.
Heme-dependent growth assays. (A) E. coli IR1583 hemA strains carrying either pSZ51 or pBAD were resuspended in soft agar and then poured onto NBD plates with or without arabinose. Discs were placed, and 10-μl aliquots of PBS, 5 mM Fe+3, and 5 mM heme solution was applied. The data are representative of at least three independent growth assays. (B) N. meningitidis strains were resuspended from overnight plate cultures in soft agar and poured onto GC plates supplemented with 100 μM desferal. Aliquots (10 μl) of PBS, 25 mM Fe+3, and 5 mM heme solution then were applied to the discs.
The N. meningitidis znuD mutant retained heme utilization capability.
To examine the contribution of ZnuD in meningococcal heme utilization, an znuD∷Ω mutation was generated in a pilQ mutant of the serogroup C strain IR3727 (mutated in hmbR and hpuB null) that is devoid of all known heme acquisition pathways. When examined by disc diffusion assays, the heme-dependent growth of the znuD mutant (IR3727RQ) was similar to that of the parental strain (Fig. 3B). Thus, an additional heme uptake mechanism exists in N. meningitidis. To determine whether the unknown heme uptake system requires energy from TonB, a tonB pilQ double mutant (IR3727BQ) also was examined, and again this mutant showed no detectable defect in heme-dependent growth. These results indicated that the uncharacterized heme acquisition mechanism(s) is functional in the tonB mutant and thus is TonB independent. Analogously to the mechanism proposed for PilQ, other outer membrane protein channels could allow for the entry of heme. Interestingly, several PilQ- and TonB-independent heme-utilizing gonococcal mutants, designated hgbX (7), have been isolated, but the precise mutations leading to the heme-related phenotypes have not been determined.
ZnuD contributes to meningococcal interaction with epithelial cells.
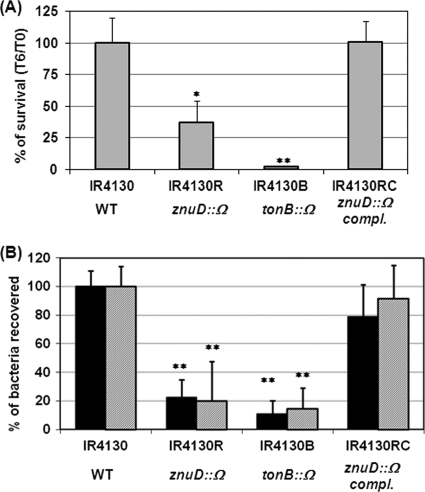
Larson et al. reported that a meningococcal tonB mutant is deficient in intracellular growth, and other uncharacterized TonB-dependent receptors may play a role in meningococcal intracellular growth (28). Analogously, a gonococcal tonB mutant and a strain with mutation in a TonB-dependent receptor, TdfF, also show intracellular growth defect (19). Thus, we examined whether ZnuD contributes to meningococcal interactions with host cells. Because the IR3727 background contains the hmbR∷Erm mutation and prohibited the use of the neisserial intergenic complementation system (nics) at a permissive chromosomal locus (34), we generated the znuD∷Ω mutation (strain IR4130R) in the parental strain IR4130 (hmbR+, hpuB null) (42). HmbR has been shown to have no effect on intracellular survival (28). IR4130R subsequently was complemented with Plac∷znuD integrated as a single copy at a permissive site (strain IR4130RC). A tonB mutant (IR4130B) also was constructed as a control. All strains displayed similar growth rates in standard GC broth and in cell culture media. Initially, the experimental procedure of Larson et al. (28) with an overnight infection was followed using the A549 human lung cell line. After treatment with gentamicin to kill extracellular bacteria, the initial intracellular CFU counts were determined immediately after gentamicin treatment (t = 0 h) and then after an additional 6-h incubation (t = 6 h). When the ratios of t = 6 h versus t = 0 h were normalized to that of the wild-type strain, both the tonB mutant and the znuD mutant yielded reduced survival, and the znuD mutant's defect can be complemented (Fig. 4A). Thus, consistently with the previous report (28), TonB is important for intracellular replication, and ZnuD may contribute to the TonB-dependent function. The gentamicin protection assays also have been performed with the znuD and tonB mutants derived from the IR3727 strain, and similar patterns of survival defects were observed (data not shown).
Fig 4.
Interaction of meningococci with epithelial cells. (A) Intracellular replication of IR4130 (WT), IR4130R (znuD), IR4130B (tonB), and IR4130RC (complemented [compl.] znuD)as determined by gentamicin protection assay after overnight infection at an MOI of 2. The gentamicin-resistant counts at 6 h after gentamicin treatment (t = 6 [T6]) were divided by the resistant counts immediately after gentamicin killing (t = 0 [T0]) and then normalized to that of the wild-type strain. Both the znuD and tonB mutants showed replication defects within cells compared to the wild type. The data represent the means and the standard deviations of two independent experiments. Statistically significant differences as determined by unpaired Student's t test with two-tailed hypotheses are marked with asterisks (∗, P < 0.05; ∗∗, P < 0.001). (B) Adherence and invasion efficiencies. The infection of A549 cells was initiated by low-speed centrifugation at an MOI of 100. The adherence abilities (black bars) relative to the initial bacterial input counts were examined after 2 h of infection. The invasion abilities (hatched bars) were determined immediately after gentamicin treatment, and the values were divided by the adhered bacterial counts. The ratios then were normalized to those of the wild-type strain. The data represent the means and the standard deviations from independent experiments (n ≥ 5). Statistically significant differences as determined by unpaired Student's t test with the two-tailed hypothesis are marked with asterisks (∗∗, P < 0.001).
Further, we noticed that the mutants appeared to have defects in invasion because the gentamicin-resistant counts of the mutants at 0 h (i.e., invasion) were lower than those of the parental strain. To better determine the invasion capability of the mutants and minimize the contribution from intracellular survival differences between the wild type and the mutants, we shortened the overnight infection protocol to 2 h with the incorporation of a low-speed centrifugation step to promote bacterial contact with host cells, and a higher MOI of 100 was used. The invasion efficiencies were calculated as the gentamicin-resistant counts divided by the adhered bacterial counts, while the adherence efficiency was determined by the ratios of attached CFU counts to the initial bacterial inputs. The ratios then were normalized to those for the wild-type strain (Fig. 4B). Surprisingly, statistically significant decreases in attachment and invasion were observed in the znuD mutant. The mutant's defect could be complemented by ectopically located Plac∷znuD (Fig. 4B). The tonB mutant also was impaired in attachment to and invasion of epithelial cells. The initial centrifugation step increased the absolute adhered bacterial numbers, but it did not overcome the adherence defect observed in the mutants. Expression levels of several known adhesins (PilE, PilC, LptA, and Opa) in the wild type and the mutants were compared by Western blotting, and no noticeable difference was detected (data not shown). Taken together, these data indicated that both TonB and ZnuD appeared to contribute in multiple steps during interactions with epithelial cells, and that ZnuD and possibly an additional TonB-dependent receptor contributed to the intracellular survival defect of the tonB mutant.
Analysis of the znuD promoter region.
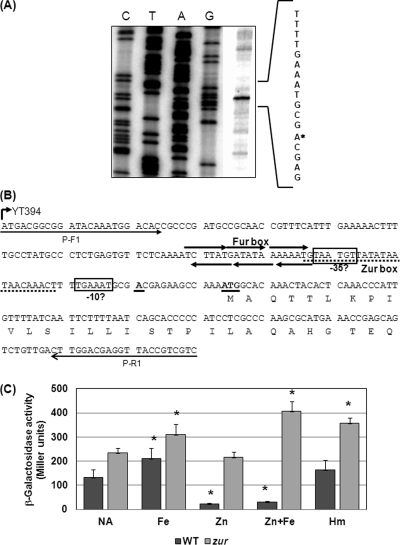
We first determined the transcriptional start site by primer extension using total RNA, and a product was detected corresponding to the −13 adenosine nucleotide relative to the ATG start codon (Fig. 5A). The examination of the sequence upstream of the transcriptional start site revealed no clear σ70 consensus promoter elements, and a possible −10 promoter element (TGAAAT) with a degenerate −35 sequence (TAATGT) separated by 18 bp (Fig. 5B) was the most likely match. As the sequence and spacing of this putative promoter showed poor resemblance to the σ70 consensus, further mutational analyses are required to confirm its function. The putative Zur binding site derived from the Zur binding sequence of E. coli (37), as proposed by Stork et al. (55) (Fig. 5B), overlapped with this putative promoter and, consequently, could repress znuD expression through interference with RNA polymerase binding. As many receptors involved in the acquisition of various iron sources are regulated by Fur (39), we checked for the presence of Fur box sequences in the znuD promoter region using the meningococcal Fur box consensus motif, (natwat)3 (17). A putative Fur box with four mismatches was revealed on both the coding and noncoding strands to locate upstream and adjacent to the putative Zur binding site (Fig. 5B).
Fig 5.
Analysis of the znuD promoter. (A) Primer extension of znuD obtained with the PE1 primer. Lanes C, T, A, and G indicate the dideoxy sequencing reactions. The coding sequence is shown on the right, with an asterisk indicating the transcriptional start site. (B) The nucleotide sequence of the znuD promoter region cloned in the YT394 reporter strain (primers p-F1 and p-R1). The annotated ATG start codon and the transcriptional start site are boldfaced and underlined, while the putative −10 and −35 elements are boxed. The Zur binding site proposed by Stork et al. (55) is marked by a dashed line, while the putative Fur boxes are shown by three consecutive arrows. (C) Iron induction of znuD expression. Strains YT394 (WT, black bars) and PKT300 (zur mutant, gray bars) were grown in RPMI to mid-log phase, divided into aliquots, and then treated for 1 h with 10 μM iron, zinc, or heme as indicated. Cultures grown with no addition (NA) were used as the control. The values are reported as the averages and standard deviations of the β-galactosidase activity in Miller units from at least two independent experiments done in triplicate. Statistically significant differences were determined by unpaired Student's t test between the treated samples and the NA samples in the respective background and are marked with asterisks (∗, P < 0.001).
Transcription of znuD is regulated by Zur and Fur.
To characterize the regulation of znuD in meningococci, we constructed a transcriptional lacZ reporter fusion, YT394, with a 140-bp sequence upstream of the transcriptional start site of znuD (Fig. 5B). The fusions were integrated into the chromosome of the meningococcal serogroup B strain NMB as a single copy (58). The znuD∷lacZ fusion displayed low activities when grown in standard GC broth, while a higher transcriptional activity was detected in cultures grown in RPMI. This pattern correlated with the previous report (55) that detected no ZnuD expression in tryptic soy broth cultures by Western blotting. The degenerate promoter was consistent with the weak expression of znuD, and the lower level of transcription in bacterial growth media indicated that bacterial media likely contain larger amounts of zinc to act as a cofactor of the Zur repressor.
Subsequently, we examined the transcriptional regulatory mechanisms of znuD using the YT394 strain. ZnuD has been shown to be repressed by the Zur homologue (55) (encoded by NMB1266; also termed PerR in N. gonorrhoeae) (63). Thus, to independently examine the possible iron regulation, a zur mutation was created in the reporter to generate strain PKT300. The mid-log-phase cultures grown in RPMI were split and treated with Zn+2, Fe+3, Fe+3 plus Zn+2, or heme at 10 μM for 1 h, and the resulting activities were compared to those of control cultures with no additions. RPMI contains ∼1.7 μM zinc (55), which is insufficient for full repression, since the presence of additional zinc further repressed znuD expression (Fig. 5C). As expected, strain PKT300 was insensitive to the zinc addition and displayed higher znuD expression than the wild type, which is consistent with the repressor role of Zur and zinc (Fig. 5C). Interestingly, the addition of iron enhanced the transcription of znuD in the wild-type reporter, and such iron activation also was observed in the zur mutant, suggesting that iron played a positive role in znuD expression in a Zur-independent manner. However, the iron induction cannot overcome Zur repression, as the presence of an equal concentration of zinc reduced the transcription of znuD in the iron-supplemented wild-type cultures while the zinc addition has no effect on the induction of znuD by iron in the zur mutant. Whether heme can affect znuD expression also was examined, and a statistically significant difference (P < 0.01) was detected only in the zur mutant. Hence, contrary to other receptors that are repressed in the presence of iron, the expression of znuD seemingly was induced by iron. Ducey et al. have found that the gonococcal znuD homolog (NG1205) was significantly induced in cultures grown in iron-supplemented defined media compared to that in the nonsupplemented cultures (14).
Fur directly interacted with the znuD promoter.
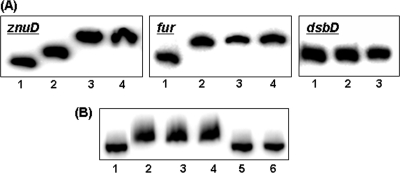
The upstream locations of the putative Fur boxes in the znuD promoter region were consistent with a positive iron regulation and suggested that Fur directly mediates the iron regulation. To investigate whether Fur protein directly binds to the znuD promoter and hence influences znuD transcription, the coding sequence of meningococcal Fur protein was cloned for overexpression in E. coli. A His6 tag was incorporated at the N terminus of the Fur protein to aid purification, since His-tagged Fur proteins have been shown to behave similarly to untagged proteins in several studies (12, 16). The interaction of the purified Fur protein with the znuD promoter DNA fragment was examined by EMSA. Divalent manganese ions were added in the binding buffer and the electrophoresis buffer as described previously (16, 48) to substitute for ferrous iron and promote the interaction of Fur with its target. As shown in Fig. 6A, Fur was able to bind to the znuD promoter in a dose-dependent manner. The fur promoter as a positive control was shifted, while no shift was detected with the dsbD promoter, which is not regulated by Fur. The specificity of the interaction of Fur with the znuD promoter was further demonstrated by competition EMSA. Only the unlabeled znuD probe, but not the same amount of nonspecific DNA competitor, can eliminate the binding (Fig. 6B). Thus, Fur interacted directly with the znuD promoter to mediate iron regulation.
Fig 6.
Interaction of the meningococcal Fur protein with the znuD promoter. (A) Dose-dependent binding of the (His)6-Fur protein to the znuD promoter. The fur and dsbD promoters are included as a positive and a negative control, respectively. Lane 1 in each panel contains free probe, lanes 2 to 4 contain 0.2, 0.4, and 0.6 μg of Fur protein, respectively, for znuD and fur probes, and 0.4 and 1 μg of Fur protein was used in lanes 2 and 3, respectively, for the dsbD probe. (B) Competition EMSA. Lane 1, free probe; lane 2, probe with 1 μg of Fur protein; lanes 3 and 4, probe with 1 μg of Fur protein in the presence of 1 and 0.5 μg nonspecific DNA competitor (a PCR fragment of the misS coding sequence), respectively; lanes 5 and 6, probe with 1 μg of Fur protein in the presence of 1 and 0.5 μg specific DNA competitor, respectively.
DISCUSSION
The diverse repertoire of iron acquisition mechanisms contributes to the success in the colonization and infection of N. meningitidis (53). As heme is an abundant potential source of iron from the host and is critical for several important physiological processes, redundant strategies for capturing heme have been demonstrated for many bacterial pathogens (2, 61). For example, uropathogenic E. coli (18), Vibrio cholerae (35), and Haemophilus influenzae (9) encode two, three, and four TonB-dependent heme receptors, respectively. The redundancy of heme acquisition systems likely accounts for the efficient capture of diverse heme sources encountered during various stages of infection. In this study, we showed that the meningococcal TonB-dependent zinc receptor ZnuD (55) also interacts with heme when expressed in E. coli. ZnuD enables heme capture on E. coli cells and allows the growth of the E. coli hemA strain with heme as the sole iron source. There was, however, no notable in vitro growth defect in the meningococcal znuD mutant lacking all other known heme utilization mechanisms, mostly likely due to the presence of additional as-yet-unidentified heme uptake pathway(s). The possibility of additional TonB-independent uptake mechanism(s) is suggested by the fact that a meningococcal tonB/pilQ double mutant was able to grow with free heme as the only iron source. It is possible that heme enters meningococci through other channel-like protein complexes via passive diffusion. However, the low bioavailability of free heme in vivo suggests that the energy-dependent active transport is more important than the passive process.
In addition to serving as a nutrient iron source, heme also is required as a prosthetic group of many critical bacterial proteins (2). Both heme and zinc acquisitions have been shown to be important for the intracellular growth of bacterial pathogens. The ChuA heme receptor of uropathogenic E. coli is significantly upregulated in intracellular bacteria recovered from urothelial cells, suggesting the need for heme uptake in the intracellular environment (41). The role of exogenous heme uptake in the intracellular survival of Neisseria is not clear. The gonococcal heme auxotrophs fail to survive inside epithelial cells (57), while a meningococcal hemO mutant that cannot degrade heme replicates equally as well as the wild-type strain within epithelial cells (29), suggesting that iron utilization via heme degradation is not required for intracellular meningococci. Indeed, meningococcal invasion has been reported to accelerate ferritin degradation in epithelial cells, thereby yielding a currently unknown iron source that was proposed to support the intracellular replication of N. meningitidis (29). There is no obvious heme-specific regulation of znuD, whereas many heme-specific receptors in other bacteria are highly induced by heme (23, 59). Thus, the primary function of ZnuD may be as a receptor for yet-unknown host-derived ligands, and heme and/or other heme-like compounds are alternative substrates due to their broad ligand specificity. The ZnuABC high-affinity transporter that delivers periplasmic zinc into the cytoplasm also has been shown to be important for the intracellular growth of several pathogens (1, 10, 22, 64). The expression of znuABC is activated in Salmonella enterica serovar Typhimurium recovered from epithelial cells (1), indicating that intracellular zinc is limited for Salmonella. Unlike other bacteria with a dedicated zinc permease, ZnuABC of Neisseria species transports both Zn2+ and Mn2+ with similar metal binding affinities (32) and is named MntABC in N. gonorrhoeae (56). Both gonococcal mntC and mntAB mutants had reduced intracellular survival in a human cervical epithelial cell model (32). Little is known about how zinc passes through the outer membrane. In contrast to the existence of many outer membrane receptors that acquire various iron sources from the environment (39), meningococcal ZnuD is the only outer membrane zinc receptor described to date (55), and whether the free or a complexed form of zinc is the actual ligand is not entirely clear. Curiously, the meningococcal znuD mutant did not have an in vitro growth defect (55 and data not shown), whereas the optimal growth of a gonococcal znuA mutant was reported to require ≥200 μM Zn2+ (8), a concentration that will completely repress znuD expression. This difference thus points to other possible ZnuD-independent zinc uptake processes across the outer membrane, and a TonB-independent zinc uptake has been noted (55).
It is probable that ZnuD plays a more significant role within certain host niches where its expression is desirable. Changes in znuD expression have been detected under three environmental conditions. First, Stork et al. showed that znuD expression is repressed by zinc (55). Second, Dove et al. reported that znuD (NMA1161) in a serogroup A strain is induced ∼20-fold in iron-starved meningococci that subsequently were exposed to 25% normal human serum for 6 h compared to levels for the cultures similarly treated with heat-inactivated serum (13). This observation suggests that certain active complement components upregulate znuD. Although the precise inducing signal in the serum was not defined, Stork et al. hypothesized that the heat-inactivation step denatured serum albumins and increased the free zinc concentration in the sera (55). Third, Ducey et al. have found that, unlike most TonB-dependent iron receptors, the gonococcal znuD homolog (NG1205) was significantly induced in cultures grown in iron-supplemented defined media compared to levels in the nonsupplemented media (14). The sequences of the znuD promoter region are nearly identical between N. meningitidis and N. gonorrhoeae, pointing to potentially related iron-mediated regulatory mechanism(s). Consistently with this notion, we found that the znuD expression increased upon exposure to iron, and the iron-dependent induction of znuD is likely through a direct interaction of the Fur protein with the znuD promoter. The proximity of the predicted Fur and Zur binding sites (Fig. 5B) prompted the investigation of a possible interplay between Fur and Zur regulations. Zur represses znuD, presumably through its binding to the proposed Zur box (55). A possible iron induction mechanism is that in the presence of iron, Fur binds to its operator and thus blocks the binding of Zur to its binding site, leading to the appearance of iron induction. However, the fact that the iron induction persists in the zur mutant (Fig. 5C) suggests that iron induction operates independently of Zur and may further augment ZnuD expression under zinc-limiting conditions. In the presence of sufficient zinc, the iron induction is unable to overcome full Zur repression, which likely explains why znuD has not been identified as an iron-regulated gene in several meningococcal transcriptome studies (11, 17). Thus, znuD is subject to a second level of Fur-iron regulation and is the first example of coordinated regulation by both Fur and Zur.
The deficient heme and/or zinc acquisition capability of the znuD mutant is the most straightforward explanation for its intracellular survival defect. The survival defect of the znuD mutant is not as severe as that of the tonB mutant, suggesting that replication within this environment requires more TonB-dependent receptors to import essential nutrients, or TonB or energy generated from the TonB-ExbB-ExbD system participates in other biological functions important to intracellular survival. For example, gonococcal Mtr efflux pump-mediated inducible resistance requires the TonB-ExbB-ExbD system (43), and a similar TonB requirement in gonococcal intracellular survival has been demonstrated (19). The addition of ferric nitrate supplement during the survival phase completely reversed the intracellular growth defect of the gonococcal tonB mutant (19) but only partially rescued the growth defect of the meningococcal tonB mutant (28), implying possible different intracellular growth requirements between these two closely related pathogens. Indeed, the TdfF receptor, not the gonococcal ZnuD homologue, is responsible for the intracellular growth defect of the gonococcal tonB mutant (19), and a TonB-independent contribution from the gonococcal genetic island, which is absent from meningococci, also has been reported (66).
Consistently with our observation, an ∼2-fold reduction in invasion efficiency has been reported for the gonococcal tonB mutant (19). The tonB mutation likely has a broader physiological consequence on the bacteria than did the znuD mutation. An inability of all TonB-dependent receptors to acquire various iron sources due to the tonB mutation could change the bacterial iron homeostasis, leading to alterations in the overall bacterial surface adhesin/invasin profile, despite the fact that we did not detect changes in several known adhesins. Furthermore, in addition to surface proteins, many nonprotein factors, such as lipooligosaccharide (LOS) and capsule, contribute or interfere with adherence and/or invasion. For example, LptA, a phosphoethanolamine (PEA) transferase, is not an adhesin per se, since its adherence defect is due to changes in the LOS structure caused by the lack of PEA modification of lipid A head groups. Thus, the defects in adherence and invasion caused by the tonB mutation could be a cumulative consequence of multiple indirect effects, and a more detailed analysis of bacterial surface profile is needed to further characterize the cause(s) of the interaction defects. A surprising finding is that ZnuD also contributes to the adherence and invasion capabilities of meningococci to epithelial cells. We currently do not have sufficient data to definitively assign an adhesin/invasin function to the surface-exposed ZnuD, as the adherence and invasion phenotypes presumably also could result from alterations in metal homeostasis, leading to changes on the bacterial surface (20). A normally expressed ZnuD mutant protein with inactivated heme/zinc acquisition function via point mutations will be required to resolve whether these defects are dependent on the ion acquisition functions.
In summary, the ZnuD receptor appears to function as a dual receptor for both zinc (55) and heme uptake. Currently, there is no clear data on the availability of zinc, heme, or Zn-protoporphyrin (26, 27) in various host environments that meningococci encounter during infection. Further studies are needed to define the ZnuD-targeted ligand that significantly contributes to the intracellular replication of N. meningitidis. As antibodies recognizing ZnuD were detected in convalescent-phase sera (55), our data showing ZnuD's involvement in the invasion of and survival within epithelial cells further support the important role of ZnuD in meningococcal pathogenesis.
ACKNOWLEDGMENTS
This work was supported by grant R01 AI061031 to Y.-L.T. from the National Institutes of Health.
We thank Shuming Zhao and Donna Perkins-Balding for technical assistance. We thank J. Folster for providing the pilQ mutational construct and H. Ly for the A549 cell line. We are grateful to Hideyuki Takahashi for the generous gift of antisera.
Footnotes
Published ahead of print 14 November 2011
REFERENCES
- 1. Ammendola S, et al. 2007. High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infect. Immun. 75:5867–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anzaldi LL, Skaar EP. 2010. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect. Immun. 78:4977–4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Battistoni F, et al. 2002. Identification of an iron-regulated, hemin-binding outer membrane protein in Sinorhizobium meliloti. Appl. Environ. Microbiol. 68:5877–5881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bäumler AJ, et al. 1998. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J. Bacteriol. 180:1446–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bilukha OO, Rosenstein N. 2005. Prevention and control of meningococcal disease. Recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm. Rep. 54:1–21 [PubMed] [Google Scholar]
- 6. Bracken CS, Baer MT, Abdur-Rashid A, Helms W, Stojiljkovic I. 1999. Use of heme-protein complexes by the Yersinia enterocolitica HemR receptor: histidine residues are essential for receptor function. J. Bacteriol. 181:6063–6072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen CJ, et al. 2004. A mutant form of the Neisseria gonorrhoeae pilus secretin protein PilQ allows increased entry of heme and antimicrobial compounds. J. Bacteriol. 186:730–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen CY, Morse SA. 2001. Identification and characterization of a high-affinity zinc uptake system in Neisseria gonorrhoeae. FEMS Microbiol. Lett. 202:67–71 [DOI] [PubMed] [Google Scholar]
- 9. Cope LD, et al. 2001. Involvement of HxuC outer membrane protein in utilization of hemoglobin by Haemophilus influenzae. Infect. Immun. 69:2353–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davis LM, Kakuda T, DiRita VJ. 2009. A Campylobacter jejuni znuA orthologue is essential for growth in low-zinc environments and chick colonization. J. Bacteriol. 191:1631–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Delany I, Grifantini R, Bartolini E, Rappuoli R, Scarlato V. 2006. Effect of Neisseria meningitidis fur mutations on global control of gene transcription. J. Bacteriol. 188:2483–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delany I, Spohn G, Rappuoli R, Scarlato V. 2001. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol. Microbiol. 42:1297–1309 [DOI] [PubMed] [Google Scholar]
- 13. Dove JE, Yasukawa K, Tinsley CR, Nassif X. 2003. Production of the signalling molecule, autoinducer-2, by Neisseria meningitidis: lack of evidence for a concerted transcriptional response. Microbiology 149:1859–1869 [DOI] [PubMed] [Google Scholar]
- 14. Ducey TF, Carson MB, Orvis J, Stintzi AP, Dyer DW. 2005. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J. Bacteriol. 187:4865–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Furano K, Campagnari AA. 2004. Identification of a hemin utilization protein of Moraxella catarrhalis (HumA). Infect. Immun. 72:6426–6432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao H, et al. 2008. The iron-responsive Fur regulon in Yersinia pestis. J. Bacteriol. 190:3063–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grifantini R, et al. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. U. S. A. 100:9542–9547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hagan EC, Mobley HL. 2009. Haem acquisition is facilitated by a novel receptor Hma and required by uropathogenic Escherichia coli for kidney infection. Mol. Microbiol. 71:79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hagen TA, Cornelissen CN. 2006. Neisseria gonorrhoeae requires expression of TonB and the putative transporter TdfF to replicate within cervical epithelial cells. Mol. Microbiol. 62:1144–1157 [DOI] [PubMed] [Google Scholar]
- 20. Hiratsuka K, Kiyama-Kishikawa M, Abiko Y. 2010. Hemin-binding protein 35 (HBP35) plays an important role in bacteria-mammalian cells interactions in Porphyromonas gingivalis. Microb. Pathog. 48:116–123 [DOI] [PubMed] [Google Scholar]
- 21. Janik A, Juni E, Heym GA. 1976. Genetic transformation as a tool for detection of Neisseria gonorrhoeae. J. Clin. Microbiol. 4:71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim S, Watanabe K, Shirahata T, Watarai M. 2004. Zinc uptake system (znuA locus) of Brucella abortus is essential for intracellular survival and virulence in mice. J. Vet. Med. Sci. 66:1059–1063 [DOI] [PubMed] [Google Scholar]
- 23. Kirby AE, Metzger DJ, Murphy ER, Connell TD. 2001. Heme utilization in Bordetella avium is regulated by RhuI, a heme-responsive extracytoplasmic function sigma factor. Infect. Immun. 69:6951–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kumar N, Xin ZT, Liang Y, Ly H. 2008. NF-kappaB signaling differentially regulates influenza virus RNA synthesis. J. Virol. 82:9880–9889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumar P, Sannigrahi S, Scoullar J, Kahler CM, Tzeng YL. 2011. Characterization of DsbD in Neisseria meningitidis. Mol. Microbiol. 79:1557–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Labbé RF, Vreman HJ, Stevenson DK. 1999. Zinc protoporphyrin: a metabolite with a mission. Clin. Chem. 45:2060–2072 [PubMed] [Google Scholar]
- 27. Lamola AA, Yamane T. 1974. Zinc protoporphyrin in the erythrocytes of patients with lead intoxication and iron deficiency anemia. Science 186:936–938 [DOI] [PubMed] [Google Scholar]
- 28. Larson JA, Higashi DL, Stojiljkovic I, So M. 2002. Replication of Neisseria meningitidis within epithelial cells requires TonB-dependent acquisition of host cell iron. Infect. Immun. 70:1461–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Larson JA, Howie HL, So M. 2004. Neisseria meningitidis accelerates ferritin degradation in host epithelial cells to yield an essential iron source. Mol. Microbiol. 53:807–820 [DOI] [PubMed] [Google Scholar]
- 30. Lee BC. 1994. Isolation and characterization of the haemin-binding proteins from Neisseria meningitidis. Microbiology 140:1473–1480 [DOI] [PubMed] [Google Scholar]
- 31. Lee BC, Levesque S. 1997. A monoclonal antibody directed against the 97-kilodalton gonococcal hemin-binding protein inhibits hemin utilization by Neisseria gonorrhoeae. Infect. Immun. 65:2970–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lim KH, Jones CE, vanden Hoven RN, et al. 2008. Metal binding specificity of the MntABC permease of Neisseria gonorrhoeae and its influence on bacterial growth and interaction with cervical epithelial cells. Infect. Immun. 76:3569–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu X, Olczak T, Guo HC, Dixon DW, Genco CA. 2006. Identification of amino acid residues involved in heme binding and hemoprotein utilization in the Porphyromonas gingivalis heme receptor HmuR. Infect. Immun. 74:1222–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mehr IJ, Seifert HS. 1998. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol. Microbiol. 30:697–710 [DOI] [PubMed] [Google Scholar]
- 35. Mey AR, Payne SM. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42:835–849 [DOI] [PubMed] [Google Scholar]
- 36. Olczak T, Dixon DW, Genco CA. 2001. Binding specificity of the Porphyromonas gingivalis heme and hemoglobin receptor HmuR, gingipain K, and gingipain R1 for heme, porphyrins, and metalloporphyrins. J. Bacteriol. 183:5599–5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Patzer SI, Hantke K. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275:24321–24332 [DOI] [PubMed] [Google Scholar]
- 38. Perkins-Balding D, Baer MT, Stojiljkovic I. 2003. Identification of functionally important regions of a haemoglobin receptor from Neisseria meningitidis. Microbiology 149:3423–3435 [DOI] [PubMed] [Google Scholar]
- 39. Perkins-Balding D, Ratliff-Griffin M, Stojiljkovic I. 2004. Iron transport systems in Neisseria meningitidis. Microbiol. Mol. Biol. Rev. 68:154–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Prentki P, Krisch HM. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303–313 [DOI] [PubMed] [Google Scholar]
- 41. Reigstad CS, Hultgren SJ, Gordon JI. 2007. Functional genomic studies of uropathogenic Escherichia coli and host urothelial cells when intracellular bacterial communities are assembled. J. Biol. Chem. 282:21259–21267 [DOI] [PubMed] [Google Scholar]
- 42. Richardson AR, Stojiljkovic I. 1999. HmbR, a hemoglobin-binding outer membrane protein of Neisseria meningitidis, undergoes phase variation. J. Bacteriol. 181:2067–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rouquette-Loughlin C, Stojiljkovic I, Hrobowski T, Balthazar JT, Shafer WM. 2002. Inducible, but not constitutive, resistance of gonococci to hydrophobic agents due to the MtrC-MtrD-MtrE efflux pump requires TonB-ExbB-ExbD proteins. Antimicrob. Agents Chemother. 46:561–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sannigrahi S, Zhang X, Tzeng YL. 2009. Regulation of the type I protein secretion system by the MisR/MisS two-component system in Neisseria meningitidis. Microbiology 155:1588–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schnaitman CA. 1971. Effect of ethylenediaminetetraacetic acid, Triton X-100, and lysozyme on the morphology and chemical composition of isolate cell walls of Escherichia coli. J. Bacteriol. 108:553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schnaitman CA. 1971. Solubilization of the cytoplasmic membrane of Escherichia coli by Triton X-100. J. Bacteriol. 108:545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schuchat A, et al. 1997. Bacterial meningitis in the United States in 1995. Active Surveillance Team. N. Engl. J. Med. 337:970–976 [DOI] [PubMed] [Google Scholar]
- 48. Sebastian S, Agarwal S, Murphy JR, Genco CA. 2002. The gonococcal fur regulon: identification of additional genes involved in major catabolic, recombination, and secretory pathways. J. Bacteriol. 184:3965–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stephens DS, Greenwood B, Brandtzaeg P. 2007. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet 369:2196–2210 [DOI] [PubMed] [Google Scholar]
- 50. Stephens DS, Swartley JS, Kathariou S, Morse SA. 1991. Insertion of Tn916 in Neisseria meningitidis resulting in loss of group B capsular polysaccharide. Infect. Immun. 59:4097–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stojiljkovic I, et al. 1995. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol. Microbiol. 15:531–541 [DOI] [PubMed] [Google Scholar]
- 52. Stojiljkovic I, Larson J, Hwa V, Anic S, So M. 1996. HmbR outer membrane receptors of pathogenic Neisseria spp.: iron-regulated, hemoglobin-binding proteins with a high level of primary structure conservation. J. Bacteriol. 178:4670–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stojiljkovic I, Perkins-Balding D. 2002. Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol. 21:281–295 [DOI] [PubMed] [Google Scholar]
- 54. Stojiljkovic I, Srinivasan N. 1997. Neisseria meningitidis tonB, exbB, and exbD genes: Ton-dependent utilization of protein-bound iron in Neisseriae. J. Bacteriol. 179:805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stork M, et al. 2010. An outer membrane receptor of Neisseria meningitidis involved in zinc acquisition with vaccine potential. PLoS Pathog. 6:e1000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tseng HJ, Srikhanta Y, McEwan AG, Jennings MP. 2001. Accumulation of manganese in Neisseria gonorrhoeae correlates with resistance to oxidative killing by superoxide anion and is independent of superoxide dismutase activity. Mol. Microbiol. 40:1175–1186 [DOI] [PubMed] [Google Scholar]
- 57. Turner PC, Thomas CE, Elkins C, Clary S, Sparling PF. 1998. Neisseria gonorrhoeae heme biosynthetic mutants utilize heme and hemoglobin as a heme source but fail to grow within epithelial cells. Infect. Immun. 66:5215–5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tzeng YL, et al. 2006. Autoregulation of the MisR/MisS two-component signal transduction system in Neisseria meningitidis. J. Bacteriol. 188:5055–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vanderpool CK, Armstrong SK. 2003. Heme-responsive transcriptional activation of Bordetella bhu genes. J. Bacteriol. 185:909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Verstreate DR, et al. 1982. Outer membrane proteins of Brucella abortus: isolation and characterization. Infect. Immun. 35:979–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wandersman C, Stojiljkovic I. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215–220 [DOI] [PubMed] [Google Scholar]
- 62. Whitney CG, et al. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348:1737–1746 [DOI] [PubMed] [Google Scholar]
- 63. Wu HJ, et al. 2006. PerR controls Mn-dependent resistance to oxidative stress in Neisseria gonorrhoeae. Mol. Microbiol. 60:401–416 [DOI] [PubMed] [Google Scholar]
- 64. Yang X, Becker T, Walters N, Pascual DW. 2006. Deletion of znuA virulence factor attenuates Brucella abortus and confers protection against wild-type challenge. Infect. Immun. 74:3874–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhu W, Hunt DJ, Richardson AR, Stojiljkovic I. 2000. Use of heme compounds as iron sources by pathogenic Neisseriae requires the product of the hemO gene. J. Bacteriol. 182:439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zola TA, Strange HR, Dominguez NM, Dillard JP, Cornelissen CN. 2010. Type IV secretion machinery promotes ton-independent intracellular survival of Neisseria gonorrhoeae within cervical epithelial cells. Infect. Immun. 78:2429–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]