Abstract
Listeria monocytogenes causes a serious food-borne disease due to its ability to spread from the intestine to other organs, a process that is poorly understood. In this study we used 20 signature-tagged wild-type clones of L. monocytogenes in guinea pigs in combination with extensive quantitative data analysis to gain insight into extraintestinal dissemination. We show that L. monocytogenes colonized the liver in all asymptomatic animals. Spread to the liver occurred as early as 4 h after ingestion via a direct pathway from the intestine to the liver. This direct pathway contributed significantly to the bacterial load in the liver and was followed by a second wave of dissemination via the mesenteric lymph nodes (indirect pathway). Furthermore, bacteria were eliminated in the liver, whereas small intestinal villi provided a niche for bacterial replication, indicating organ-specific differences in net bacterial growth. Bacteria were shed back from intestinal villi into the small intestinal lumen and reinfected the Peyer's patches. Together, these results support a novel dissemination model where L. monocytogenes replicates in intestinal villi, is shed into the lumen, and reinfects intestinal immune cells that traffic to liver and mesenteric lymph nodes, a process that occurs even during asymptomatic colonization.
INTRODUCTION
Listeria monocytogenes causes a food-borne disease in humans and other mammals (28, 47). The incidence of confirmed human cases in the United States is ∼1,000 per year (1), with 20% of clinical infections resulting in death (5). Such a high mortality rate is unparalleled by any of the other more common bacterial gastrointestinal pathogens, e.g., Salmonella and Campylobacter (33) and is due to the fact that L. monocytogenes can cross the intestinal barrier and cause invasive disease in predisposed individuals, including pregnant women, newborns, and immunocompromised individuals (13).
Internalin A (InlA), a bacterial cell wall surface protein that binds to E-cadherin (36), induces internalization of L. monocytogenes into enterocytes at the tips of intestinal villi (39), and is important for infection of liver and spleen after oral inoculation (30). Despite the discovery of this molecular mechanism for breaching the intestinal barrier, several key questions about the pathogenesis of listeriosis after oral inoculation remain to be answered. It is unknown how L. monocytogenes disseminates from enterocytes to distant organs, whether dissemination to distant sites occurs during asymptomatic colonization, and whether bacterial replication in the small intestine is important for invasive disease.
Such studies may have been hampered by limitations of the oral inoculation model in the mouse, the predominant animal model for listerial pathogenesis (8). The mouse is not a natural host for L. monocytogenes and often must be infected intravenously or intraperitoneally, routes that are not pertinent to food-borne infections in humans. These limitations are likely due to reduced affinity between InlA and murine E-cadherin (29). Strategies to overcome these limitations by expression of human E-cadherin in transgenic mice (30) or mutation of InlA to improve interaction with murine E-cadherin (52) have improved the oral inoculation model but still require high infectious doses, suggesting that there may be additional differences in the enteral phase of listeriosis between mouse and human. We therefore turned to the guinea pig, which is highly susceptible to oral inoculation with L. monocytogenes (17, 44) and expresses E-cadherin with the same affinity to InlA as human E-cadherin (30).
Bacterial pathogenesis is typically studied by determination of the bacterial load in specific organs and comparison of wild-type and mutant strains. However, this method cannot identify routes of dissemination in mammalian hosts. We have previously used two L. monocytogenes wild-type strains that differ in their antibiotic susceptibility to determine bacterial movement between maternal organs, the placenta, and fetus and to estimate organ specific replication and clearance rates (3). Others have used signature-tagged clones of a given pathogen to explore population dynamics during infection (4, 16). In the present study we used signature-tagged wild-type clones of L. monocytogenes in combination with extensive quantitative data analysis and mathematical modeling. From these studies we gained novel insights into the pathogenesis of food-borne listeriosis. We found that the liver became colonized in all animals, even though they continued to appear healthy. Dissemination to the liver occurred via two routes: a direct pathway from the intestine and an indirect pathway from the intestine via the mesenteric lymph nodes. The bacterial load in the liver was predominantly due to bacterial spread via the direct pathway. Furthermore, intestinal villi provided a niche for bacterial replication from where bacteria continuously shed into the small intestinal lumen and reinfected Peyer's patches.
MATERIALS AND METHODS
Bacterial strains and generation of signature-tagged L. monocytogenes strains.
All L. monocytogenes strains used in the present study were derived from 10403S (6). The Escherichia coli strains XL1-Blue and SM10 were used for cloning. Twenty unique 40-bp signature tags (STs) (Table 1) were inserted into the L. monocytogenes site-specific integration vector pPL2 (27) using QuikChange site-directed mutagenesis (Stratagene) according to the manufacturer's protocol. ST-pPL2 plasmids were transformed into the E. coli strain SM10 using standard techniques generating the plasmid donor strains. The SM10 donor strains were mixed with the L. monocytogenes recipient strain 10403S by streaking equal amounts together on a brain heart infusion (BHI) agar plate. After incubation at 37°C for 2 h, the bacteria were removed from the plate, and dilutions plated onto BHI agar supplemented with chloramphenicol (7.5 μg/ml) and streptomycin (200 μg/ml), followed by incubation at 37°C for 2 days. Individual colonies were picked and screened for plasmid integration within the chromosome as previously described (27).
Table 1.
Sequences of STs and ST-specific primersa
| ST | Sequence (5′-3′) | ST-specific primer sequence (5′-3′) |
|---|---|---|
| T6 | CCATAGCTACCACACGATAGCTCCCCCTAGCCCCCTACAC | CTAGCCCCCTACACGCTAT |
| T11 | CGGGGTGGTGTGTGGGAGGTTTAGAGGTTTATTTCGTGTG | GAGGTTTATTTCGTGTGGAC |
| T58 | CGCAAAATCACTAGCCCTATAGCGACCCCTCTACCCCAAC | GACGTCCGCAAAATCACTAG |
| T102 | AACCATAGCGATATCTACCCCAATCTCTCGCCCCCTCTAC | CGATATCTACCCCAATCTCTC |
| T116 | GGATGGCGGAGATGGAGCTCTTGGGTGCTCTTGCGGTTG | GGAGATGGAGCTCTTGGGT |
| T119 | CGCCCCCTCCCCACATAAAGAGAGCTAAACAAAAACATCA | ATAAAGAGAGCTAAACAAAAACATC |
| T168 | ATATAACAATCCCTAGCCCCCTATCCAGCCATCTCTCCCC | GACGTCATATAACAATCCCTAG |
| T191 | AACTACCGCAAACGCTCGCTAAAAACATCCATATCTAAC | GACGTCAACTACCGCAAACG |
| T205 | CCCCCGCTACCCTGATATCCCCCTTTCCGCTCTCTAGCA | GCTACCCTGATATCCCCCT |
| T209 | TTATGGGTAGAGCGAGATTGTGCTATTGCTAGAGTGATAG | ATTGCTAGAGTGATAGGCTATT |
| T210 | GGAGCGGGATGGAGCGTTGTGGCGGGCTCTAGATCGGGTG | TTGTGGCGGGCTCTAGATC |
| T211 | GGAGAGCGGTCGCTTTATAGCGATTTATATATATCTAGTT | GGTCGCTTTATAGCGATTTATA |
| T219 | CCCTAAAACCCTACAGCAATCACGATATACCGCTCCCGAC | CTACAGCAATCACGATATACC |
| T231 | CTAAAGCACTCTAGCCCGAGAGATCCCAACATCACGCGAC | GACGTCCTAAAGCACTCTTAG |
| T234 | AACCCCCGAACCAGCCAGCACTCCGATATATCTCACCAACC | CAGCCAGCACTCCGATATAT |
| T242 | ATTTCGATGTATATATGTAGCGATAGGGGTGTCGCTTTAG | ATTTCGATGTATATATGTAGCGA |
| T288 | CTATCCAACTAGACCTCTAGCTACATCGCTCACTACCCCC | CTAGACCTCTAGCTACATCG |
| T295 | ACACCTACATCAAACCCTCCCCAGAGAAAGATAAACCTCC | GACGTCACACCTACATCAAAC |
| T296 | GTGGATAGTGAGGGCGGGCGAGAGCTTGCTTGCGCGAGTG | GACGTCGTGGATAGTGAGG |
| T297 | GATCCGTCGCCAAATCGAGTCAGAGGTCTAGCTATCGCGACACTG | CGAGTCAGAGGTCTAGCTAT |
ST, signature tag.
Bacterial growth conditions.
Mixtures of the 20 ST L. monocytogenes strains (clones) were prepared for oral inoculation as follows: overnight cultures of each of the 20 clones were mixed in equal amounts, diluted, and grown in BHI broth at 37°C until mid-log phase was reached. Aliquots were stored at −80°C. The day prior to infection, one aliquot was thawed, diluted in BHI broth, and grown overnight at 30°C without agitation. The following day, the culture was washed once in phosphate-buffered saline (PBS) and resuspended in lactated Ringer's solution to a final concentration of 108 bacteria per 0.5 ml.
Animal studies. (i) Inoculation.
Female Hartley guinea pigs at 3 to 4 weeks of age were purchased from Elm Hill Breeding Labs. Guinea pigs were restricted for food, water allowed, for 2 h prior to inoculation. Oral inoculation of calcium bicarbonate (125 mg in 1 ml of PBS), followed by 20 clones (108 CFU of a ratiometrically equal mixture of all clones in 0.5 ml of lactated Ringer's solution) was performed by slowly dripping the solutions into the mouth of the animal with a 1-ml syringe. Food was returned to the cages following infection. Animals were housed individually after inoculation.
(ii) Dissection of organs and tissues.
At various time points postinoculation (p.i.), the guinea pigs were euthanized. The following organs and tissues were removed: the liver, spleen, mesenteric lymph nodes, villous small intestinal wall, and Peyer's patches. The guinea pig intestine is a large organ; the small intestine, cecum, and colon are approximately 125, 20, and 75 cm in length, respectively (10). In order to determine whether L. monocytogenes preferentially colonizes any part of the guinea pig intestine, we divided the entire intestine in ∼30-cm pieces and cultured the intestinal pieces and their luminal contents. At 4 h and 1 day after inoculation, the small intestine was colonized most heavily (data not shown). We used 30 cm of the small intestine proximal to the ileocecal valve for all subsequent experiments. This section of the small intestine also contains Peyer's patches, which were cultured separately.
(iii) Enumeration of CFU in organs and tissues.
All samples were weighed and homogenized in 0.2% NP-40. The entire homogenate or aliquots were plated on BHI agar plates with 200 μg of streptomycin/ml to enumerate bacterial load per organ. In general, the entire homogenate was plated for the small organs and tissues (spleen, mesenteric lymph nodes, and Peyer's patches), and aliquots were plated for the large organs and tissues (liver and villous small intestinal wall). The remainder of the liver and villous small intestinal wall homogenates was inoculated into BHI broth with 200 μg of streptomycin/ml. After enumeration of the CFU/organ for the spleen, mesenteric lymph nodes, and Peyer's patches, all colonies were scraped off the BHI agar plates and inoculated into BHI broth with 200 μg of streptomycin/ml. Thus, we should have detected each colony in the spleen, mesenteric lymph nodes, and Peyer's patches and identified each clone present in all of the organs unless there were bacteria that were not able to grow on agar plates or in broth culture after organ homogenization and plating. However, loss of bacteria due to such processing should be the same for homogenates from all organs. Broth culture from organ and tissue homogenates were used to prepare genomic DNA for further quantitative real-time PCR (qPCR) analysis. For some organs and tissues, L. monocytogenes only grew in the broth cultures, and we identified the STs by using qPCR. The presence of one ST suggested that at least one CFU of that ST was present in that organ. Therefore, we calculated the minimum CFU/organ by counting the number of distinct STs present in that organ.
(iv) Luminal contents.
We removed 4 g of stool from the rectum as well as the small intestinal contents. To distinguish between L. monocytogenes in the small intestinal lumen versus the wall, we washed the small intestine with PBS four times and then cultured the intestinal wall, the first wash, and the fourth wash separately. Culture from the first wash (small intestinal wash) was used for further analysis. Culture from the fourth wash did not grow L. monocytogenes and was discarded (data not shown) but confirmed that culture from the intestinal villi and Peyer's patches originated from bacteria that were either attached to or had invaded these tissues. The entire sample of stool and intestinal wash was inoculated into broth culture. We determined the presence of clones but were unable to enumerate CFU in stool and wash due to difficulties with contamination. We tried different selection media and antibiotic combinations but were unable to identify a condition that would allow us to enumerate CFU of L. monocytogenes in stool and wash without any doubt. It is important to note that the intestine of guinea pigs differs from mouse. Guinea pigs have a very large cecum, which is full of stool and bacteria. We are inoculating guinea pigs (weight, ∼1 lb) with a relatively small dose of L. monocytogenes (108 CFU/animal). In comparison, most experiments in the murine model of listeriosis use 108 CFU/animal, but the weight of a mouse is only ∼20 g (∼20-fold lower than that of a guinea pig). Obviously, the murine intestine is much smaller than the guinea pig intestine and contains fewer microbes than the guinea pig. Therefore, it is easy to determine CFU/stool in mice because L. monocytogenes takes over but not in guinea pigs.
DNA purification and qPCR.
Chromosomal DNA was purified from bacterial cultures from the inoculum (input pool) and organs, tissues, and luminal contents (output pools) using an Epicentre Gram-positive DNA purification kit (Epicentre Biotechnologies), substituting lysozyme with mutanolysin (5 U/μl; Sigma). Bacterial genomic DNA from the input pool was used for standard curves for each ST in each experiment. All qPCRs were performed in a Stratagene Mx3000P qPCR machine and analyzed using MxPro software (Stratagene). qPCRs were 20 μl in volume and contained 10 μl of Platinum SYBR green 2× reaction mix (Invitrogen), 10 μM ST-specific primer (Table 1), and 10 μM reverse primer (pPL2-359R [CTTAATGAATTACAACAGTACTGC]), 40 ng of chromosomal DNA, and nuclease-free water. Standard curve reactions were identical except for the amount of chromosomal DNA (200, 20, 2, or 0.2 ng per reaction). All cycling conditions were as follows: 50°C for 2 min; 95°C for 2 min; 40 cycles of 95°C for 15 s and 55°C for 1 min; followed by 95°C for 1 min, 55°C for 30 s, and 95°C for 30 s.
Determination of the presence of STs by qPCR.
The qPCR value had to fulfill all of the following conditions for an ST to be determined as present. (i) The qPCR value for each ST had to be above a minimal cutoff, which was calculated based on negative control reactions (see below). (ii) The sum of the qPCR values in each organ of each animal had to be >1 to account for the possibility of additional low levels of nonspecific cross-reactivity of STs against other sources of contaminating DNA from other microbes or the host. (iii) Finally, the stringency (see below) had to be >0.02.
Negative control reactions.
A specific primer was used on a mixture of chromosomal DNA from all clones, excluding the ST clone for which the specific primer was designed. We performed two to five such reactions for each ST and used twice the highest nonspecific value as the minimal cutoff. The minimal cutoff for T205 was calculated based on the qPCR signal for T6 in each reaction because T205 specifically cross-reacted with the sequence of T6 in a concentration-dependent linear manner.
Calculation of CFU/clone and definition of stringency.
CFU/clone was calculated based on the relative qPCR values for each of the STs in an organ and the enumerated total CFU in that organ. Because the total number of clones per organ influenced the proportion of STs, we calculated the “stringency” of the qPCR signal as follows: stringency = the proportion of qPCR signal for an ST × the number of all STs present in this sample. We next plotted the stringency per clone versus the CFU/clone and found that all clones with CFU/clone values <0.25 had stringencies of <0.02, so we used this as our additional cutoff criterion, because it seemed very unlikely that a clone with a CFU of <0.25 was actually present.
Determination of the presence of STs via sequencing.
For some organs individual colonies were picked, and the presence of the ST was confirmed by sequencing. To amplify the ST in the individual colony, we performed colony PCR as follows. Single colonies were sterilely removed from the plate and completely resuspended in 20 μl of sterile water. Then, 1 μl of this resuspension was added to a PCR containing 10 μM concentrations of each primer (pJM1-2374 [TCGCCGACATCACCGATGG] and pPL2-359R [CTTAATGAATTACAACAGTACTGC]), 0.4 mM deoxynucleoside triphosphates, 0.4 μl of Taq polymerase (Qiagen), 2 μl of Taq buffer, and water for a total volume of 20 μl. All cycling conditions were as follows: 94°C for 5 min, followed by 34 cycles of 94°C for 1 min, 55°C for 2 min, and 72°C for 2 min. All PCRs were prepared for sequencing by treatment with ExoSAP-IT (GE Healthcare) according to the manufacturer's protocol. Treated PCRs were used directly for sequencing reactions with the ST sequencing primer pJM1-2901 (GCGCATTGTTAGATTTCATAC). All sequencing was conducted at the University of California DNA Sequencing Facility.
Calculation of the bottleneck for intestinal villi and Peyer's patches.
The total inoculum was 108 CFU. We determined the CFU in the distal 30 cm of the small intestine and found an average CFU of 34 (range, 12.1 to 51.08) at 4 h p.i. The average gastric emptying time in guinea pigs is 2 h (24), and therefore we assumed that the inoculum arrived at the sampled region of the small intestine at 2 h p.i. This leaves only another 2 h during which the bacteria could have replicated, and we therefore assumed that bacterial replication was insignificant during this period. The average length of the entire small intestine is 125 cm (10), and we assumed that bacteria invade the entire length of the small intestine at equal rates. Therefore, the entire small intestine consisted of 4.17 of equal 30-cm sections. From this we could estimate that of 108 bacteria, 34 × 4.17 entered the entire small intestine, giving the result that 108/142 = 7 × 105 or ∼106 bacteria are required for one bacterium to invade any part of the intestinal villi.
Guinea pigs have on average six lymphoid follicles (including Peyer's patches) along the entire length of the small intestine (31). We found an average CFU/three Peyer's patches at 4 h p.i. of only 3.5 (range, 2 to 5). Using the same assumptions about gastric emptying time and replication as described above, we could calculate that 108/(2 × 3.5) = 1.4 × 107 or ∼107 bacteria are required for one bacterium to enter any lymphoid follicle in the small intestine.
Calculation of the contribution of direct and indirect pathway to CFU in the liver.
We used the first-order rate constants for bacterial replication, clearance, and efflux that had been previously estimated for the liver and spleen in the case of intravenous L. monocytogenes infection (3). We assumed that the first-order rate constants remained fixed throughout the first 72 h p.i. and that the influx rate constants from the blood into both, liver and spleen via the indirect pathway were equal. The following equation represents the rate of change in the number of CFU in the spleen (S): dS/dt = ksr× S – ksc × S – kse × S + ksi× B, where ksr and ksc were the rate constants for bacterial replication and clearance in the spleen, and kse was the rate constant for bacterial efflux that left the spleen and entered the bloodstream. The influx of bacteria into the spleen was denoted by the rate constant ksi and was a function of the number of bacteria in the systemically circulating blood (B). Since we used the values for the first three rate constants from Bakardjiev et al. (3), this equation simplified to: dS/dt = −0.13 × S + ksi × B. In the liver the rate of change of CFU that can be attributed to the indirect pathway was dLindirect/dt = −0.02 × Lindirect + kli × B, where ksi × B = kli × B because we are assuming that clones in the blood have equal access to both organs. Therefore, if we knew the value of ksi × B at each time point and the time at which the first bacterium entered the liver, we could calculate the initial dLindirect/dt and thus recalculate Lindirect and dLindirect/dt at each successive time point.
In order to estimate the ksi × B, we first needed to know when, on average, bacteria began entering the spleen. Bacteria were in the spleen at 48 h but not at 24 h p.i. Therefore, infection of the spleen occurred between 24 and 48 h p.i. We assumed that bacteria move from mesenteric lymph nodes into the blood and, once in the blood, circulate throughout the body and are able to seed the spleen immediately. The mesenteric lymph nodes contained one clone with 700 CFU and another clone with 500 CFU at 24 h p.i., neither of which was present in the spleen. This suggested that clones in the mesenteric lymph nodes could disseminate into the spleen once the CFU/clone count in the mesenteric lymph nodes was ≥500. Presumably, the substantial increase in the bacterial load in mesenteric lymph nodes from an average CFU/clone of 131 to 1,188 between 24 and 48 h p.i. occurred in superlinear fashion. Therefore, we estimated that dissemination from mesenteric lymph nodes into the bloodstream occurred approximately three-quarters into the 24 to 48 h p.i. time interval, or at 42 h p.i.
Based on our knowledge of the average CFU/clone in the spleen at 42, 48, and 72 h (0, 5, and 54 CFU/clone, respectively), we calculated the values for ksi × B required to produce the observed rate of change of CFU/clone in the spleen at these time points. Since we do not directly know the behavior of ksi × B over time, we simulated different possible properties for ksi × B: (i) that ksi × B stayed constant, (ii) that ksi × B increased linearly, or (iii) that ksi × B increased exponentially. For all three cases, we calculated ksi × B using estimates for linear or exponential coefficients such that the calculated CFU/clone was similar to the experimentally enumerated CFU/clone in the spleen at 48, and 72 h p.i. (within 3%). Since kli = ksi, we used the same value of ksi × B to calculate Lindirect at 48 and 72 h p.i. Regardless of which of the three cases we used, the resultant estimates for Lindirect were very similar, within a few CFU of each other.
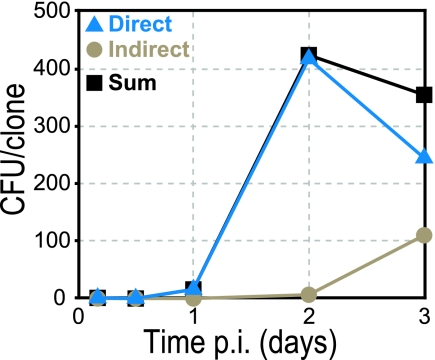
Now we knew the Lindirect at each time point. Clones that were present in both the liver and spleen (LLandS) may have reached these organs via the indirect pathway but also may have traveled to the liver directly (via the hepatic portal). Thus, LLandS is an amalgam of both direct and indirect pathways. On the other hand, the many more clones present in the liver but not the spleen (LLnotS) must have reached the liver via the direct pathway only. Thus, LLnotS + LLandS = Ldirect + Lindirect = Lsum, where Lsum is the sum of the averages of both pathways. We could therefore use the clonal data for LLnotS and LLandS, together with the estimated Lindirect, to calculate Ldirect and Lsum at each time point (see Fig. 5).
Fig 5.
Relative contribution of direct and indirect pathways for liver colonization. The calculated average CFU/clone due to liver colonization via either the direct (blue triangles) or the indirect (beige circles) pathways, and their sums (black squares), at specified time points p.i. (see Materials and Methods) are shown.
Approvals by institutional review boards.
All animals were housed and handled in accordance with federal and institutional guidelines. The animal use committees at the University of California, Berkeley, and the University of California, San Francisco, approved the animal protocol describing our studies.
RESULTS
Oral inoculation of guinea pigs with L. monocytogenes.
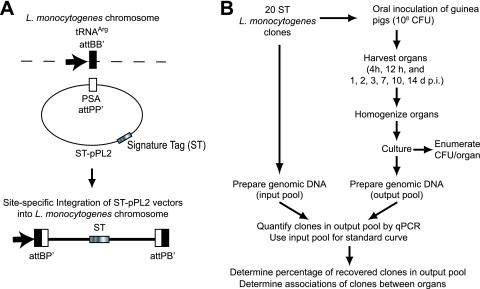
The goal of this study was to characterize the kinetics of colonization, to define the bottlenecks to invasion and spread, and to determine the routes of systemic dissemination after ingestion of L. monocytogenes. For this purpose, we constructed 20 uniquely tagged wild-type L. monocytogenes strains (Fig. 1A; see also Materials and Methods), which were equally virulent in a murine model of listeriosis (data not shown). These clones were distinguished by qPCR detection of their unique signature tag (ST). We orally inoculated guinea pigs, a host that is naturally susceptible to listeriosis (17), with a total of 108 CFU, consisting of a mixture of the 20 signature-tagged clones at equal ratios. All of the animals continued to appear healthy after bacterial inoculation and cleared the infection over the course of 2 weeks. We analyzed the bacterial colonization of liver, spleen, mesenteric lymph nodes, intestinal villi, small intestinal wash, stool, and Peyer's patches at specific time points as early as 4 h and until 14 days postinoculation (p.i.) (Fig. 1B and Table 2). We assessed the following metrics: (i) the percent colonization of each organ, (ii) the total CFU per organ, (iii) the percentage of inoculated clones from all animals at each time point that were recovered from each organ, and (iv) the CFU/clone to determine the absolute size of each individual bacterial population in each organ at any given time point. In addition, we calculated the percentage of clones that were shared between two organs, for example, the number of clones detected in spleen that were also present in mesenteric lymph nodes. These results allowed us to make conclusions about the directionality of bacterial movement between tissues.
Fig 1.
Experimental design. (A) Generation of signature-tagged L. monocytogenes clones (see Materials and Methods). (B) Flowchart of experimental design to determine total CFU/organ and presence of clones by qPCR.
Table 2.
Number of guinea pigs at each time point
| Time p.i.a | No. of animals |
|---|---|
| 4 h | 7 |
| 12 h | 8 |
| 1 day | 7 |
| 2 days | 11 |
| 3 days | 12 |
| 7 days | 6 |
| 10 days | 3 |
| 14 days | 3 |
p.i., postinoculation.
L. monocytogenes disseminates to the liver and spleen.
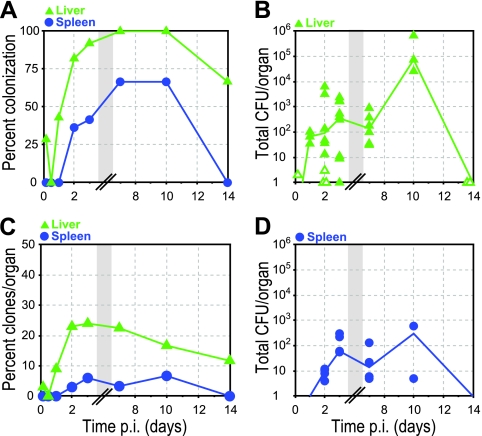
L. monocytogenes is an important human pathogen because it has the ability to cross the intestinal barrier and cause invasive disease (13). In intravenous inoculation models, >90% of bacteria are rapidly taken up by the liver and spleen (2), and therefore the bacterial load in liver and spleen has been used for decades as a representation of bacterial virulence (40). We wanted to determine whether and when liver and spleen become colonized after oral inoculation with L. monocytogenes in guinea pigs. All animals developed infection of the liver while appearing healthy (Fig. 2A). Two animals (29%) had colonization of the liver as early as 4 h p.i. with a small number of bacteria; we estimated a minimum of 2 CFU per liver, because of the number of clones we detected (see Materials and Methods) (Fig. 2B). At 12 h p.i., no animals displayed liver colonization. This may indicate clearing of initial colonization, as observed with Yersinia pseudotuberculosis in mice (4), but the number of guinea pigs in our study was too small for clearance at 12 h to be statistically verifiable. Between 12 h and 2 days p.i. the percentage of infected livers increased sharply (Fig. 2A), the median CFU in the liver rose from 0 to 92 (Fig. 2B), and the bacterial load remained around 100 CFU until day 7 p.i. Between 7 and 10 days p.i. the median CFU in the liver increased from 1.4 × 102 to 7.4 × 104 before it was reduced to less than 4 CFU per liver by day 14 p.i.
Fig 2.
Colonization of the liver and spleen. Guinea pigs were inoculated with 20 signature-tagged L. monocytogenes strains (see Table 2). Liver (green triangles) and spleen (blue circles) were cultured, and the presence of clones was determined by qPCR. (A) Percentage of animals with L. monocytogenes in either liver or spleen at specified times p.i. (B) CFU per liver. Closed symbols represent CFU enumerated on BHI agar plates, and open symbols represent calculated minimum CFU based on qPCR values (see Materials and Methods). (C) Percentage of inoculated clones from all animals at each time point that were recovered from either the liver or spleen at specified times p.i. (D) CFU per spleen. The lines in panels B and D connect median values of enumerated CFU at each time point. Some symbols representing data points may overlap due to similar values. See Table 2 for number of animals at each time point. Gray boxes represent a break in the values on the x axis.
The number of clones recovered from the liver increased from 3 and 0% at 4 and 12 h, respectively, to 23% at 2 days p.i. (Fig. 2C), indicating that bacteria are arriving in the liver over this time period. Between 2 and 7 days p.i., the median CFU and the percentage of recovered clones remained at similar levels, suggesting that bacterial increase due to influx and replication versus bacterial elimination due to clearance and efflux had reached a steady state. During this time period bacterial influx had either ceased or clones already present in the liver were reinfecting this organ. Between 7 and 10 days p.i. the number of clones recovered from the liver decreased from 22.5 to 16.7%, while the median CFU increased ∼1,000-fold (Fig. 2C). Thus, while clones in the liver were eliminated, a subset of clones already present in the liver was able to increase in numbers. It seems unlikely that this increase was due to influx because the bacterial load was decreasing in all other organs that we tested. We noticed that in contrast to other organs and previous time points the liver contained macroscopically visible abscesses on day 10 p.i. It is possible that while infectious foci were walled off and clones were eliminated in these abscesses, some bacteria were also able to replicate temporarily within the abscesses before being cleared by 14 days p.i. (Fig. 2B).
In contrast to the liver, the spleen was the last organ to become colonized at 2 days after ingestion, and not all animals developed spleen infection (Fig. 2A). Once infected, the median bacterial load in infected spleens between 2 and 7 days p.i. was ∼10, which was ∼10-fold lower than the bacterial load in infected livers (compare Fig. 2B and D). Since the weight of the spleen is ∼10-fold lower than the weight of the liver, the detection of CFU/g organ were similar in both organs (data not shown). The spleen contained fewer clones than liver at all time points (Fig. 2C).
Routes of dissemination differ for liver and spleen.
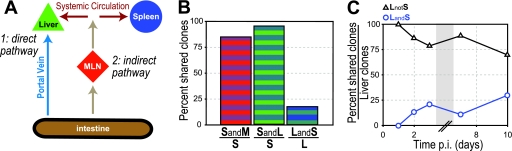
Lower infection rates and fewer clones in the spleen in comparison to the liver could be due to organ-specific differences in bacterial replication, elimination, or influx (3). What are the possible routes L. monocytogenes could take to reach liver and spleen after ingestion? Pathogens are known to disseminate via the lymphatics (mesenteric lymph nodes and thoracic duct) into the systemic circulation and reach the liver and spleen via the hematogenous route (35). However, the liver has an additional circulation system: nutrient-rich blood from the intestine flows directly to the liver via the portal vein. Thus, the liver could be reached by two routes: (i) a direct pathway, by which bacteria can reach the liver from the small intestine via the portal vein, and (ii) an indirect pathway, by which pathogens can gain access to the systemic circulation via the mesenteric lymph nodes (Fig. 3A). In contrast, the spleen can only be colonized via the indirect pathway. Our finding that the liver contains a higher percentage of inoculated clones than the spleen at all time points (Fig. 2C) suggests that the direct pathway contributes significantly to colonization of the liver.
Fig 3.
Routes of bacterial dissemination. (A) Diagram of possible routes of dissemination from the intestine to liver and spleen: bacteria can (pathway 1) traffic from the intestine to the liver via the portal vein (direct pathway) or (pathway 2) traffic via the mesenteric lymph nodes to the bloodstream, followed by hematogenous dissemination via the systemic bloodstream, which leads to colonization of liver and spleen (indirect pathway). (B) Percent clones in spleen of all animals at 2 and 3 days p.i. that were also present in the mesenteric lymph nodes (SandM/S; red/blue) or liver (SandL/S; green/blue) and percent clones in liver of all animals at 2 and 3 days p.i. that were also present in spleen (LandS/L; blue/green). (C) Percent clones in liver of all animals at specified times p.i. that were either also present in the spleen (LandS; blue open circles) or absent from the spleen (LnotS; black open triangles). The gray box represents a break in the values on the x axis.
Consistent with our model that the spleen is preferentially seeded via the indirect pathway, we found a strong correlation between clones present in the spleen and mesenteric lymph nodes: 85% of the clones present in the spleen were also found in mesenteric lymph nodes on days 2 and 3 p.i. (Fig. 3B). A high percentage of clones present in liver was also found in mesenteric lymph nodes (>90% on days 2 and 3 p.i. [data not shown]). Unfortunately, this did not elucidate pathways of dissemination from the intestine to the liver, because once mesenteric lymph nodes were seeded it was no longer possible to differentiate which clones went directly to both liver and mesenteric lymph nodes versus only indirectly to the liver via the mesenteric lymph nodes (Fig. 3A).
Clones that spread beyond mesenteric lymph nodes have to reach the systemic bloodstream and from there settle in the liver and spleen. We have found previously that intravenous injection of L. monocytogenes in guinea pigs leads to seeding of liver and spleen at equal ratios at 30 min p.i (2) and that the rate constants for total bacterial influx (extra- and intracellular bacteria) are the same for the liver and spleen (3). Thus, dissemination via the indirect pathway should lead to seeding of both the liver and spleen. Therefore, we predicted that clones present in the spleen should also be present in the liver, but not vice versa, because the liver can receive clones by an additional direct pathway. Indeed, 95% of the clones present in spleen were also in the liver, but only 18% of clones in the liver were found in the spleen on days 2 and 3 p.i. (Fig. 3B). This alternate route may explain why the liver always contains more clones than the spleen (Fig. 2C). Between days 2 and 10 p.i., when both organs were colonized, >75% of all of the clones present in the liver were absent from the spleen (Fig. 3C). These clones represent influx via the direct pathway. In contrast, <26% of all of the clones that were present in the liver were also present in the spleen. This latter group of clones could have trafficked to the liver via both the direct and the indirect pathways and therefore represents a maximal estimate of the contribution of the indirect pathway to dissemination to the liver. These results are consistent with a major contribution of the direct pathway to liver colonization. Furthermore, if bacteria in the liver could spread back into the systemic circulation and from there into the spleen, we would expect to see the percentage of clones that are present in the liver and also in the spleen increase significantly during the course of infection. Instead, our data suggest that the liver is effective at preventing further bacterial spread.
Mesenteric lymph nodes represent a bottleneck for spread via the indirect pathway.
In order to further determine the role of each pathway for dissemination to the liver, we first characterized the indirect pathway by comparing infection of the mesenteric lymph nodes and spleen. Colonization of the mesenteric lymph nodes was detectable at 12 h p.i., and all of the guinea pigs had infection of mesenteric lymph nodes by day 3 that persisted until at least 10 days after inoculation (Fig. 4A). Peak median total CFU/organ was 6.7 × 103 on day 2 p.i. (Fig. 4B). In contrast, the percentage of spleen infection never reached 100% and was lower than mesenteric lymph node colonization at all time points, suggesting a bottleneck for bacterial trafficking between mesenteric lymph nodes and spleen (compare Fig. 2A and 4A). This was further corroborated by the finding that the spleen had fewer clones than the mesenteric lymph nodes at all time points during infection (compare Fig. 2C and 4C).
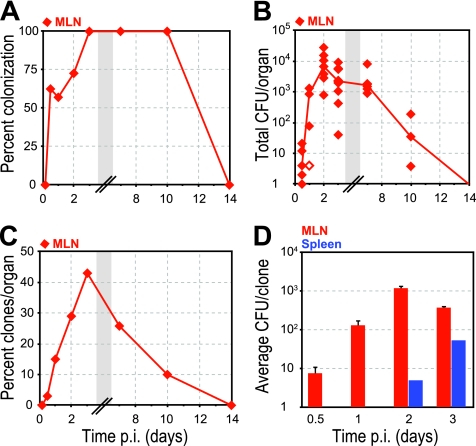
Fig 4.
Indirect pathway. The mesenteric lymph nodes (MLN; red diamonds) were cultured, and the presence of clones was determined by qPCR. (A) Percentage of animals with L. monocytogenes in MLN at specified times p.i. (B) CFU per MLN. Closed symbols represent CFU enumerated on BHI agar plates, and open symbols represent calculated minimum CFU based on qPCR values (see Materials and Methods). A line connects the median values of enumerated CFU at each time point. Some symbols representing data points may overlap due to similar values. See Table 2 for the number of animals at each time point. (C) Percentage of inoculated clones from all animals at each time point that were recovered from MLN at specified times p.i. (D) Average CFU/clone in either MLN (red) or spleen (blue) at specified time points p.i. Error bars indicate the standard errors of the mean (SEM). Gray boxes represent a break in the values on the x axis.
The percentages of clones that were present in mesenteric lymph nodes and also in the spleen were 11.1% (7/63) and 13% (13/100) at 2 and 3 days p.i., respectively. This suggests that only ∼12% of clones from mesenteric lymph nodes reach the spleen. To further quantify the bottleneck between the mesenteric lymph nodes and spleen, we determined the average CFU/clone in these organs (Fig. 4D). The average CFU/clone in mesenteric lymph nodes were ∼102 and ∼103 at 1 and 2 days p.i., respectively. Initial colonization of the spleen occurred during this time period (Fig. 2A), suggesting that the bottleneck between mesenteric lymph nodes and spleen is ∼1 bacterium in 102 to 103.
On days 2 and 3 p.i., 4/11 and 5/13 spleens were infected with a total number of 7 and 13 clones, respectively, translating to and average of 1.75 (range, 1 to 3) and 2.6 (range, 1 to 5) clones per infected spleen (Fig. 2A and C). The average CFU/clone in the spleen were 5 and 54 on days 2 and 3 p.i., respectively (Fig. 4D). Thus, there were only 6 new clones in spleen on day 3 p.i., but 49 new CFU/clone. Based on our previously published model, the spleen has a negative net rate of replication, clearance, and efflux (3). In general, about one in every 7.7 CFU would be eliminated/hour. Therefore, any increase in the bacterial load in the spleen must be due to bacterial influx. This suggests that the clones detected in the spleen were due to multiple infections with the same clone. Between 2 and 3 days p.i. at least 50 CFU of the same clone trafficked from the mesenteric lymph nodes to the spleen since some bacteria that got into the spleen would have been eliminated and other bacteria that got into the blood would have been taken up by the liver at equal rates (2, 3).
The bacterial load in the liver depends largely on bacterial influx via the direct pathway.
As described above, the direct pathway contributes significantly to liver colonization during the entire course of infection, based on the percentage of clones in liver that are absent from spleen (Fig. 3C). However, we wanted to further quantify the relative contribution of each pathway to the bacterial load in the liver. We calculated the CFU/clone in the liver due to influx from direct and indirect pathways. These calculations were based on experimentally determined average CFU/clone in the spleen (indirect pathway), previously determined rate constants for bacterial replication, clearance, and efflux in liver and spleen (3), the resultant calculated influx rates into the spleen, and the assumption that the influx rate constants into liver and spleen via the indirect pathway are equal (see Materials and Methods). We found that the bacterial load in the liver during the first 3 days p.i. was almost entirely due to bacterial influx via the direct pathway (Fig. 5).
As in the spleen, the net bacterial rate for replication, clearance, and efflux in the liver is negative (3). Therefore, any increase in bacterial load in the liver should be due to influx. Influx via the direct and indirect pathways originates in the intestine. Therefore, we decided to analyze infection in the intestine and how it contributes to bacterial dissemination.
Intestinal villi provide a niche for bacterial replication.
L. monocytogenes invades enterocytes of the small intestine at the villous tips via the interaction of InlA with E-cadherin (36, 39), and it has been shown that InlA is important for infection of the liver after oral inoculation (30). Thus, we determined colonization of the distal 30 cm of the villous small intestinal wall, which represents one-fourth of the entire small intestine of the guinea pig (10), and which we refer to as intestinal villi. At 4 h after ingestion, 43% of the animals had evidence of infection of intestinal villi (Fig. 6A). This number increased steadily and reached 100% on day 7 p.i. The median bacterial load in the intestinal villi rose >100-fold between 4 h and 1 day p.i. to 2 × 104 and was followed by a plateau, before the bacterial burden decreased after day 7 p.i., resulting in clearance from intestinal villi by day 10 p.i. (Fig. 6B). The percentage of inoculated clones recovered from intestinal villi was only between 8 and 27.5% between 4 h and 7 days p.i. (Fig. 6C), suggesting a bottleneck between intestinal lumen and colonization of intestinal villi. We calculated that ∼106 bacteria were required for one bacterium to invade any part of the intestinal villi (see Materials and Methods).
Fig 6.
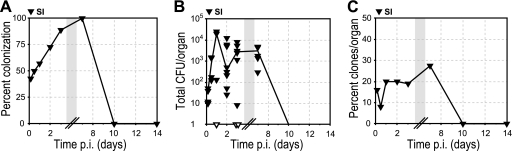
Colonization of intestinal villi. Intestinal villi (SI; black inverted triangles) were cultured, and the presence of clones was determined by qPCR. (A) Percentage of animals with L. monocytogenes in SI at specified times p.i. (B) CFU per SI. Closed symbols represent CFU enumerated on BHI agar plates, and open symbols represent calculated minimum CFU based on qPCR values (see Materials and Methods). A line connects the median values of enumerated CFU at each time point. Some symbols representing data points may overlap due to similar values. See Table 2 for the number of animals at each time point. (C) Percentage of inoculated clones from all animals at each time point that were recovered from SI at specified times p.i. Gray boxes represent a break in the values on the x axis.
Comparison of the bacterial load and the percentage of clones in intestinal villi suggested significant bacterial replication in intestinal villi during the first day after inoculation. The median bacterial load in intestinal villi increased >100-fold between 4 h and 1 day p.i. (Fig. 6B). The bacterial load in each organ is determined by bacterial replication, clearance, influx and efflux. Since the percentage of clones did not change significantly (16% at 4 h and 20% at 1 day p.i.) (Fig. 6C), we concluded that there was no significant influx of additional clones from the initial inoculum into intestinal villi after 4 h p.i., which also suggested that the initial inoculum had been mostly cleared from the small intestinal lumen by 4 h p.i. Therefore, the observed increase in the bacterial load was largely due to bacterial replication of clones that were able to invade intestinal villi by 4 h p.i. This is in contrast to the situation in liver and spleen, where the number of bacteria would decrease without any additional influx.
We observed an increase in the percentage of clones recovered from intestinal villi from 19 to 27.5% between days 3 and 7 p.i. (Fig. 6B). One possible explanation for this observation is spread of clones from the liver back to the intestine. Indeed, it has been reported previously that L. monocytogenes is excreted in the bile of systemically infected mice (7, 20). We could not detect L. monocytogenes in the bile of guinea pigs inoculated with 108 CFU over the entire time course of infection (data not shown). However, we were able to detect L. monocytogenes in the bile of some guinea pigs inoculated orally with a 1,000-fold-higher infectious dose (data not shown). This suggests that excretion of L. monocytogenes into the intestine via the bile is possible in guinea pigs but may occur intermittently, thus making it difficult to detect bacterial colonization of the bile.
Bacteria from intestinal villi are shed into the intestinal lumen.
It is generally accepted that InlA-dependent enterocyte invasion is the first step in crossing the intestinal barrier, but the cellular mechanisms of dissemination from enterocytes to distant organs are unknown. L. monocytogenes could use cell-to-cell spread from infected enterocytes to reach deeper layers of intestinal villi. However, our previous work on infection of human placental organ cultures demonstrates that the basement membrane underlying an epithelial surface constitutes a very good barrier against bacterial cell-to-cell spread (46). We reasoned that it is unlikely that cell-to-cell spread from infected enterocytes to cells in the lamina propria is a significant route of dissemination. What are other possible routes for bacterial dissemination from the small intestine? One possibility is via trafficking inside of infected host cells, such as infected leukocytes from Peyer's patches or the lamina propria (37, 43, 44). Colonization of intestinal villi could therefore serve as an important niche for L. monocytogenes in which bacteria replicate and then are shed into the intestinal lumen and subsequently reinfect susceptible cells that disseminate to local lymphatic and distant organs.
To determine the role of bacterial shedding in the pathogenesis of listeriosis, we analyzed the presence of L. monocytogenes in the intestinal lumen. We determined the number of clones present in the small intestinal wash of the distal 30 cm of small intestine. This is the region that we used for analysis of infection of intestinal villi, and therefore we reasoned that the wash of this section could contain both the clones that had the opportunity to invade enterocytes as well as clones that have been shed from intestinal villi back into the lumen. In addition, we determined the presence of clones in fecal samples from the distal large intestine. These samples should initially include all of the clones from the oral inoculum that have passed through the entire gastrointestinal tract, while later in infection, only clones that have been shed back into the intestinal lumen from the entire small intestinal wall should be present.
The bacterial dynamics in small intestinal wash and stool differed significantly. All infected animals excreted L. monocytogenes in the stool as early as 4 h after ingestion (Fig. 7A), indicating that L. monocytogenes passed rapidly through the gastrointestinal tract. After 12 h p.i., the percentage of animals with fecal excretion of L. monocytogenes decreased steadily. In contrast, the small intestinal wash contained L. monocytogenes in only 29% of animals at 4 h p.i. (Fig. 7A). This percentage increased until day 7 p.i., when 100% of animals contained L. monocytogenes in the small intestinal wash, demonstrating that L. monocytogenes can persist and replicate in the gastrointestinal tract. L. monocytogenes was cleared from both luminal locations by 10 days p.i., correlating strongly with the observed timing of bacterial clearance in intestinal villi (Fig. 6A).
Fig 7.
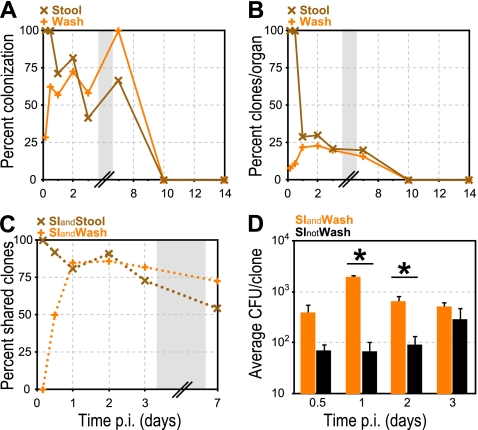
Intestinal lumen colonization. Stool (brown “X” symbol) and small intestinal wash (orange “+” symbol) were cultured, and the presence of clones was determined by qPCR. (A) Percentage of animals with L. monocytogenes in either small intestinal wash or stool at specified times p.i. (B) Percentage of inoculated clones from all animals at each time point that were recovered from either wash or stool at specified times p.i. (C) Percentage of clones in intestinal villi (SI) of all animals at specified time points p.i. that were either also present in the stool (SIandStool) or also in the wash (SIandWash). (D) Average CFU/clone in SI for clones that were either also present (SIandWash; orange) or absent (SInotWash; black) from the wash. Error bars indicate the SEM. The P values were ≤0.0008 and ≤0.02 at 1 and 2 days p.i., respectively. Gray boxes represent a break in the values on the x axis.
The differences in bacterial presence between small intestinal wash and stool were even more pronounced when we compared the percentage of inoculated clones that were recovered. During the first 12 h 98% of clones were detected in the feces but only 21 to 30% past this point (Fig. 7B). In contrast, only 8% of clones were found in the wash at 4 h p.i. (Fig. 7B). This percentage increased and was similar to the percentage of clones recovered from the stool after 12 h p.i. These findings suggest that the clones isolated from the stool at 4 and 12 h p.i. represented the initial inoculum, which was cleared from the colon between 12 h and 1 day p.i. by fecal excretion. We hypothesized that only a subset of clones from the initial inoculum is able to invade enterocytes due to the intestinal bottleneck. After replication in this niche, bacteria are shed continually into the wash, beginning at 12 h p.i. until colonization of intestinal villi has cleared.
If this hypothesis were true, we would expect that clones present in intestinal villi early after infection should strongly correlate with the clones in the stool, because the stool contains all of the clones that were represented in the initial inoculum (Fig. 7B). In contrast, the small intestinal wash should contain none or very few of the clones present in intestinal villi, because the villous intestinal clones need time to replicate before being shed back into the wash. The small percentage of clones in the small intestinal wash early in the course of infection (8 and 11% at 4 and 12 h, respectively [Fig. 7B]) may represent residual clones from the initial inoculum or clones that have already been shed from intestinal villi into the lumen. However, once the initial inoculum has been cleared by excretion into the stool, the clones in wash and stool should all be shed from intestinal villi, and therefore the same clones would be present in all three locations.
Indeed, as expected, at 4 h p.i. 100% of the clones present in the intestinal villi were also in the stool. In contrast, at the same time, none of the clones present in the intestinal villi were in the small intestinal wash (Fig. 7C), suggesting that the 8% of clones in the small intestinal wash at 4 h p.i. were residuals from the initial inoculum. At 12 h p.i., the stool still contained the entire inoculum and, as expected, the percentage of shared clones between intestinal villi and stool was high. The small intestinal wash contained 11% of inoculated clones, and the percentage of shared clones between intestinal villi and wash increased to 50%. On day 1 p.i., after the number of clones in the stool dropped from 98 to 29% (Fig. 7B), the percentage of clones shared among all three intestinal locations increased to >80%, a finding consistent with our hypothesis that once the initial inoculum has cleared, the clones in the intestinal lumen (wash and feces) originate from the intestinal villi. The percentage of shared clones between all three locations remained high (80 to 90%) on days 1 to 3 p.i. On day 7 p.i., the percentage of shared clones between all three compartments declined to 55 to 73%, most likely reflecting different rates of growth and elimination in each location.
Bacterial replication in intestinal villi is important for shedding.
We noted that the average CFU/clone for clones that were present in both the intestinal villi and the wash was significantly higher than the average CFU/clone for clones that were present in intestinal villi but absent from the wash (Fig. 7D). This difference ranged between 2- and 30-fold between 12 h and 3 days p.i. and was statistically significant on days 1 and 2 p.i (Wilcoxon rank-sum P values: 12 h, <0.16; 1 day, <0.0008, 2 days, <0.02; and 3 days, <0.166). We concluded that the intestinal villi represent a niche for bacterial replication, which is important for shedding into the intestinal lumen.
In agreement with the reported InlA dependence of enterocyte invasion (36, 39), we did not find colonization of the intestinal villi, liver, or spleen in guinea pigs inoculated with 108 CFU of InlA-deficient L. monocytogenes at 4, 24, and 72 h p.i. (data not shown). Interestingly, the Peyer's patches were not colonized in these animals either, suggesting that colonization of Peyer's patches does not occur from the initial inoculum or is very rare. Could wild-type L. monocytogenes infection of Peyer's patches occur via reinfection with bacteria that were shed into the intestinal lumen from the intestinal villi?
Bacterial shedding leads to reinfection of Peyer's patches.
The dynamics of bacterial accumulation in Peyer's patches after inoculation with wild-type L. monocytogenes differed from that of intestinal villi (compare Fig. 6B and 8B). Total median CFU/organ in Peyer's patches peaked at 2 days p.i. at 4.6 × 103, followed by a steady decline. Peyer's patch colonization never occurred in 100% of animals, and the percentage of animals with Peyer's patch infection was lower than the percentage of animals with intestinal villus infection at all time points (compare Fig. 6A and 8A). At 4 h p.i., this difference was 1.5-fold, suggesting a lower rate of invasion of Peyer's patches in comparison to villous enterocytes. Consistent with this was the presence of fewer clones in Peyer's patches than intestinal villi throughout the course of infection (compare Fig. 6C and 8C). At 4 h p.i., this difference was 3.2-fold. We determined the bottleneck of Peyer's patch colonization to be ∼107 bacteria to get one bacterium into any lymphoid follicle in the small intestine (see Materials and Methods).
Fig 8.
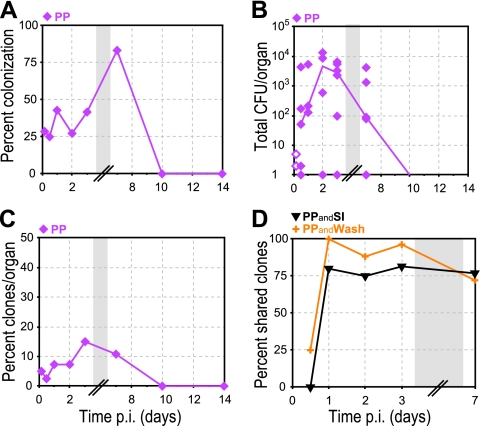
Colonization of Peyer's patches. Peyer's patches (PP; magenta diamonds) were cultured, and the presence of clones was determined by qPCR. (A) Percentage of animals with L. monocytogenes in PP at specified times p.i. (B) CFU per PP. Closed symbols represent CFU enumerated on BHI agar plates, and open symbols represent calculated minimum CFU based on the qPCR values (see Materials and Methods). A line connects the median values of enumerated CFU at each time point. Some symbols representing data points may overlap due to similar values. See Table 2 for number of animals at each time point. (C) Percentage of inoculated clones from all animals at each time point that were recovered from PP at specified times p.i. (D) Percentage of clones in PP of all animals at specified time points p.i. that were either also present in SI (PPandSI; black inverted triangles) or also present in wash (PPandWash; orange “+” symbols). Gray boxes represent a break in the values on the x axis.
We also saw evidence of reinfection of Peyer's patches from intestinal villi. The percentage of clones present in Peyer's patches and also in intestinal villi increased from 0 to 80% between 12 and 24 h p.i. (Fig. 8D). At this time, the initial inoculum had already been cleared from the intestinal lumen. Therefore, infection of Peyer's patches with residual clones from the initial inoculum was unlikely. We reasoned that reinfection of Peyer's patches from the intestinal villi occurred via the small intestinal wash. Indeed, we found an even stronger correlation between clones present in Peyer's patches and wash: on day 1 p.i., 100% of the clones present in Peyer's patches were also found in the wash (Fig. 8D).
DISCUSSION
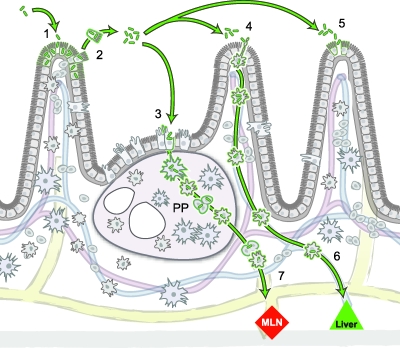
Our results are consistent with a model where L. monocytogenes passes swiftly through the gastrointestinal tract and is rapidly cleared from the stool. However, approximately 1 in 106 bacteria will invade intestinal villi during the first few hours after ingestion. Invasion is dependent on the activity of InlA and allows the bacteria to proliferate in a protected niche. Bacterial replication is important for shedding back into the intestinal lumen and reinfection of Peyer's patches. Bacterial shedding into the small intestinal lumen could also lead to reinfection of other intestinal villi prolonging small intestinal colonization. Bacterial spread beyond the intestine could occur either via the extracellular route or via infected leukocytes (42–44, 49) (Fig. 9).
Fig 9.
Model of bacterial dissemination from intestinal villi to liver and mesenteric lymph nodes. (arrow 1) Ingestion of L. monocytogenes leads to enterocyte invasion at the tip of intestinal villi. The bottleneck for intestinal villus colonization is 1 in 106 bacteria. Once internalized, L. monocytogenes replicates and spreads to neighboring enterocytes via cell-to-cell spread. (arrow 2) The basement membrane under the intestinal epithelium prevents bacterial extension into the lamina propria. Bacteria are shed back from the tips of intestinal villi into the lumen inside extruded enterocytes. Subsequently, bacteria reinfect Peyer's patches (PP) (arrow 3), lamina propria macrophages (arrow 4), and other enterocytes (arrow 5). L. monocytogenes could disseminate in infected leukocytes (dendritic cells and/or macrophages) via the portal vein directly to the liver (arrow 6) and along afferent lymphatic vessels to the mesenteric lymph nodes (MLN) (arrow 7). The mesenteric lymph nodes represent another tissue barrier, since only 1 bacterium out of 102 to 103 bacteria disseminates further via the systemic circulation to liver and spleen.
Lecuit et al. and others have previously shown an important role for InlA-dependent enterocyte invasion and crossing of the intestinal barrier in listeriosis (22, 30, 39). However, how L. monocytogenes spreads from infected enterocytes to cause disseminated disease has remained unclear. L. monocytogenes is able to spread from cell-to-cell without exposure to the extracellular environment (45, 48) and could potentially use this mechanism to spread from enterocytes to underlying cells in the lamina propria where capillaries and lymph vessels are located. However, the basement membrane underlying an epithelial layer provides a barrier for cell-to-cell spread in placental organ cultures (46), and the basement membrane underlying the intestinal epithelium could act in a similar fashion. We have previously determined the bottleneck for bacterial spread from the placenta to the fetus based on placental and fetal infection during the first 3 days after intravenous inoculation and found that it is only one in 104 bacteria (3). Assuming that the bottleneck between placenta and fetus is largely due to the effects of the basement membrane and that the magnitude of the barrier effect is similar in intestine and placenta, then a bacterial load in the enterocytes of 104 CFU for 7 days would only allow 1 to 10 bacteria to spread to deeper layers of the small intestinal wall. Thus, it seems plausible to hypothesize that cell-to-cell spread from infected enterocytes directly to cells in the lamina propria is only a minor route for extra-intestinal dissemination. In contrast, the ability of L. monocytogenes to spread from cell-to-cell may be more important for prolonged enterocyte colonization (39), which in turn can lead to shedding of L. monocytogenes into the intestinal lumen, fecal excretion, contamination of food, and transmission to other mammals. More importantly, we have shown that bacterial shedding can lead to reinfection of Peyer's patches. It is plausible to hypothesize that luminal bacteria can also reinfect other intestinal villi, as well as lamina propria leukocytes, that sample luminal contents and are able to migrate and therefore spread the infection (12, 18, 21, 42, 44).
It has long been known that other gastrointestinal pathogens such as Salmonella and enteropathogenic Yersinia species, are taken up by M cells overlying the Peyer's patches (9, 19, 25), and it has been hypothesized that Peyer's patches are a portal of entry that allows systemic dissemination via mesenteric lymph nodes (35). The role of Peyer's patch colonization in the pathogenesis of listeriosis has been controversial. Several studies have shown that L. monocytogenes is able to colonize murine Peyer's patches (23, 34). Pron et al. found that L. monocytogenes invades villous intestine and Peyer's patches with similar efficiencies but grows much more rapidly in Peyer's patches in a rat ligated ileal loop model (43). The significance of these findings has been unresolved due to the decreased ability of L. monocytogenes to invade enterocytes in these animal species (29, 30). Our results suggest that L. monocytogenes can colonize Peyer's patches if InlA-dependent intestinal villus colonization is established. We found that the bottleneck was approximately 1 bacterium in 106 and 1 bacterium in 107 for intestinal villi and Peyer's patch invasion, respectively. At first sight, it seems that L. monocytogenes is able to invade the villous intestinal wall more easily than the Peyer's patches. However, the surface area of the villous intestinal wall is approximately 100- to 1,000-fold larger than the surface area of the Peyer's patches (51). Thus, taking the differences in surface areas into account, L. monocytogenes was able to invade Peyer's patches approximately 10- to 100-fold more easily than the villous small intestinal wall. If the bottleneck of invasion was overcome, then L. monocytogenes could reach the mesenteric lymph nodes, which represented another tissue barrier because only approximately 1 bacterium in 102 to 103 was able to disseminate further to liver and spleen. These findings show that the intestinal lymphatic tissues could be a portal of entry and route of dissemination for L. monocytogenes, but they also suggest that Peyer's patches and mesenteric lymph nodes represent significant bottlenecks for intestinal invasion and spread to distant organs.
The mesenteric lymph nodes contained the highest percentage of inoculated clones from any of the organs we tested (Fig. 4C). These clones must have come from upstream tissues in the gastrointestinal tract, including Peyer's patches, intestinal villi, and wash. Indeed, ca. 80% of the clones present in mesenteric lymph nodes are also present in these upstream tissues, and only ∼20% of the clones present in mesenteric lymph nodes were not in any of the upstream tissues on day 3 p.i. The most likely explanation for this finding is that the rates of elimination in mesenteric lymph nodes and upstream tissues differ, which resulted in clearance of these 20% of clones in upstream tissues prior to day 3 p.i. Proof of this hypothesis would be possible if we could track clones in the same animal over the entire course of infection. Unfortunately, this is not possible because the animals have to be sacrificed at each time point. Another notable point is that we dissected and analyzed all mesenteric lymph nodes, but we only took one-fourth of the small intestinal villi, the wash from this corresponding small intestinal section, and one-half of the Peyer's patches. Therefore, there may be additional clones in the upstream tissues that we did not detect, although the similar percentage of clones that were present in wash and stool (Fig. 7B and C) suggests that the intestinal sections we used for our analysis were a good representation for the clones in the entire intestinal villi, wash, and Peyer's patches. Nevertheless, it is more likely that we erred on under detecting clones in the small intestinal tract, whereas we should have been able to detect all clones present in the mesenteric lymph nodes.
Another interesting aspect of mesenteric lymph node infection was the observation that only ∼12% of clones were able to spread to systemic organs but that this subset of clones repeatedly infected the spleen. We estimated that at least 50 CFU/clone trafficked from the mesenteric lymph nodes to the spleen. It is unclear why a subset of clones in mesenteric lymph nodes was able to repeatedly seed the spleen. One possibility is that there are different pockets of infection in the MLN and, depending on the exact location of infectious foci, it may be more likely that infection occurs in a cell type that is able to disseminate beyond the mesenteric lymph nodes.
An alternate explanation would be that fewer than 50 CFU, maybe even as little as 1 bacterium/clone, are able to spread from the mesenteric lymph nodes to the bloodstream, where they replicate before infecting spleen and liver. We did not experimentally assess the bacterial load in the blood, because we were previously unable to detect bacteremia in nonpregnant asymptomatic guinea pigs (3). It is more difficult to detect bacteria in the blood in comparison to other organs because only a small portion of the blood is tested compared to entire tissues and the release of bacteria in the blood is likely to be intermittent. However, based on our previously determined rate constants for bacterial replication, clearance, and efflux in the spleen, influx into the spleen from the blood (3), and our observed average CFU/clone in the spleen, we estimated an average CFU/clone in blood of <1 (estimated range, 0.003 to 0.34) at any given time, suggesting that there is no significant continuous pool of bacteria in the blood that could potentially contribute to further infection of the spleen and liver.
On the other hand, L. monocytogenes could also be eliminated in the blood due to microbicidal mechanisms directed toward extracellular and/or intracellular L. monocytogenes. We have found previously that guinea pig serum does not have a bactericidal effect on extracellular L. monocytogenes (data not shown). Furthermore, L. monocytogenes is taken up by liver, spleen, and other organs as early as 30 min after intravenous inoculation (2), and L. monocytogenes cannot be detected in the bloodstream in nonpregnant guinea pigs during the first 3 days after intravenous inoculation. We and others also have shown that L. monocytogenes traffics inside of host cells in the blood of guinea pigs (3) and mice (14), suggesting that if L. monocytogenes is eliminated in the blood it may be due to intracellular killing in phagocytes. However, L. monocytogenes grows well in murine bone marrow-derived macrophages (41), as well as primary human peripheral blood monocytes (data not shown). Taken together, these published and unpublished observations suggest that microbicidal mechanisms in the blood toward L. monocytogenes are negligible.
We have shown that L. monocytogenes can disseminate from the intestine to the liver via a previously unreported direct pathway from the intestine. There is evidence that Salmonella enterica serovar Typhimurium and Y. pseudotuberculosis translocate from the intestine to distant organs via lymphatic tissue-independent routes as well (4, 49). Barnes et al. used a combination of genetically altered mice and signature-tagged Y. pseudotuberculosis clones to show that dissemination to the liver occurs via at least two pathways, one of which is independent of Peyer's patches and the mesenteric lymph nodes. However, the relative contribution of each pathway for spread to distant organs has been unknown. Based on our extensive quantitative data analysis and mathematical modeling, we demonstrated here that the direct pathway contributes substantially more to the bacterial load in the liver than the indirect pathway during listeriosis. It seems likely that L. monocytogenes and other gastrointestinal pathogens are exploiting the portal vein, which carries nutrient rich blood from the intestine to the liver. We and others have previously shown that L. monocytogenes traffics inside of cells to the placenta and brain after infection of the liver and spleen has been established by intravenous inoculation (3, 14). It is plausible to hypothesize that L. monocytogenes exploits host cells for dissemination from the intestine to the liver as well. Consistent with this model is a previous report that S. Typhimurium disseminates via CD18-expressing phagocytes from the intestine to the liver (49). These phagocytes could aid in the dissemination of L. monocytogenes as well.
We have found previously that the net bacterial change over time due to replication, clearance, and efflux in the liver is negative (3). Therefore, any increase in the bacterial load in the liver must be due to influx. During pregnancy the infected placenta provides a niche for bacterial replication and continuous bacterial seeding of the liver (3). Here, we demonstrated that intestinal villi could provide a niche for bacterial replication and subsequent traffic to the liver as well. Consistent with our findings is evidence that Y. pseudotuberculosis disseminates from a replicating pool in the intestine, and oral administration of streptomycin decreased liver colonization in a murine model of yersiniosis (4). This could either be due to the elimination of replicating bacteria in the intestinal lumen or of bacteria that have been shed from intestinal villi back into the lumen. Our results are consistent with the latter possibility. It is important to note that Y. pseudotuberculosis and S. Typhimurium invade enterocytes as well (M. R. Amieva, unpublished data) (15), and more recently it has been shown that epithelial cells that are heavily infected with Salmonella enterica are extruded from an epithelial monolayer (26). Thus, intestinal villi could provide a replicative niche for gastrointestinal pathogens in general, from where pathogens are shed into the lumen and then spread to distant organs.
Why are there differences in bacterial growth in different organs? Lionakis et al. found that organ-specific innate immune responses correlated with the load of the fungal pathogen Candida in liver and kidney (32). An early neutrophil presence is critical for Candida control, and significantly more neutrophils accumulate in the liver, which is able to decrease the fungal burden, than in the kidney, where Candida is able to grow. Another possibility is that host cell-specific differences in intracellular replication and elimination influence the bacterial burden in specific organs (50, 53).
Cytological studies would have added an important dimension to our experiments. However, microscopic examination of the small intestine is usually done in a ligated ileal loop system. Such a system allows instillation of high bacterial numbers directly into a section of the intestine that is only a few centimeters long, e.g., 2 × 107 CFU (39) or 109 CFU (43) per 5-cm ligated ileal loop. Such high bacterial numbers per surface area in a defined section of the intestine are necessary to visualize bacteria in sufficient numbers. In the present study we inoculated guinea pigs with 108 CFU, and the approximate average length of their small intestines is ∼120 cm. Therefore, the CFU/cm of small intestine in our study is significantly lower and distributed over a much larger area than in the ligated ileal loop model, making it extremely unlikely to find bacteria microscopically in our system. Even after the inoculation of guinea pigs with a 1,000-fold-higher dose, 1011 L. monocytogenes via the oral route (data not shown), we were unable to detect sufficient numbers of bacteria in the small intestine via microscopy, making cytological studies in this system very difficult if not impossible.
We found that liver colonization occurred in some animals as early as 4 h p.i. To our knowledge, this is the first study that found infection of the liver within the first few hours of oral inoculation in an animal model that is susceptible to InlA-mediated invasion and therefore relevant to human listeriosis. Early liver colonization has been reported after intragastric inoculation of Salmonella spp. (49) and Y. pseudotuberculosis (4) in mice but is controversial in mice inoculated with L. monocytogenes. For example, liver infection with <10 CFU has been reported after intragastric inoculation of C57BL/6 mice with 107 and 1010 CFU at 4 and 12 h p.i., respectively (11, 52). However, some of these mice succumbed to the infection (11, 52). In contrast, Marco et al. inoculated outbred mice with 1.3 × 109 L. monocytogenes; none of these animals developed infection of the liver or spleen at 6, 12, or 24 h p.i., and none died (34). Similarly, the guinea pigs in our study were outbred, and none of the animals died.
We did not find evidence of further dissemination from the liver to spleen, suggesting that the liver was effective in preventing further systemic hematogenous dissemination under the conditions we used for infection in this study. The bacterial burden in the liver between 1 and 7 days p.i. was only ∼102. It is likely that the bacterial burden correlates with the probability of clearance in the liver, as has been suggested for S. enterica (16). It has also been shown that the spread of S. enterica from the liver to other organs increases under conditions of immune compromise (16). Healthy immunocompetent adults ingest 105 L. monocytogenes approximately four times per year (38), but it is unknown whether L. monocytogenes disseminates to the liver during asymptomatic intestinal colonization. Our results suggest that ingestion of L. monocytogenes by healthy adults may lead to intestinal villi and liver colonization without causing any overt symptoms. However, conditions of immune compromise or pregnancy may promote further dissemination and severe infections such as meningitis and preterm labor.
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health grants K08 AI064951 and R01 AI84928 (to A.I.B.), PO1 A1063302 and R01 AI27655 (to D.A.P.), and F32 AI072988 (to J.A.M.-W.).
We are grateful to Nina Hahn for veterinary assistance. We thank Julie Theriot for helpful discussions and Stephen Gitelman, Byoungkwan Kim, Holly Morrison, and Jennifer Robbins for critical reading of the manuscript.
Footnotes
Published ahead of print 14 November 2011
REFERENCES
- 1. Anonymous 2010. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food: 10 states, 2009. MMWR Morb. Mortal. Wkly. Rep. 59:418–422 [PubMed] [Google Scholar]
- 2. Bakardjiev AI, Stacy BA, Portnoy DA. 2005. Growth of Listeria monocytogenes in the guinea pig placenta and role of cell-to-cell spread in fetal infection. J. Infect. Dis. 191:1889–1897 [DOI] [PubMed] [Google Scholar]
- 3. Bakardjiev AI, Theriot JA, Portnoy DA. 2006. Listeria monocytogenes traffics from maternal organs to the placenta and back. PLoS Pathog. 2:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barnes PD, Bergman MA, Mecsas J, Isberg RR. 2006. Yersinia pseudotuberculosis disseminates directly from a replicating bacterial pool in the intestine. J. Exp. Med. 203:1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bennion JR, Sorvillo F, Wise ME, Krishna S, Mascola L. 2008. Decreasing listeriosis mortality in the United States, 1990–2005. Clin. Infect. Dis. 47:867–874 [DOI] [PubMed] [Google Scholar]
- 6. Bishop DK, Hinrichs DJ. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005–2009 [PubMed] [Google Scholar]
- 7. Briones V, et al. 1992. Biliary excretion as possible origin of Listeria monocytogenes in fecal carriers. Am. J. Vet. Res. 53:191–193 [PubMed] [Google Scholar]
- 8. Busch DH, Vijh S, Pamer EG. 2001. Animal model for infection with Listeria monocytogenes. Curr. Protoc. Immunol. Chapter 19:Unit 19. [DOI] [PubMed] [Google Scholar]
- 9. Clark MA, Hirst BH, Jepson MA. 1998. M-cell surface beta1 integrin expression and invasin-mediated targeting of Yersinia pseudotuberculosis to mouse Peyer's patch M cells. Infect. Immun. 66:1237–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cooper G, Schiller AL. 1975. Anatomy of the guinea pig. Harvard University Press, Cambridge, MA [Google Scholar]
- 11. Czuprynski CJ, Faith NG, Steinberg H. 2003. A/J. mice are susceptible and C57BL/6 mice are resistant to Listeria monocytogenes infection by intragastric inoculation. Infect. Immun. 71:682–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deane HW. 1964. Some electron microscopic observations on the lamina propria of the gut, with comments on the close association of macrophages, plasma cells, and eosinophils. Anat. Rec. 149:453–473 [DOI] [PubMed] [Google Scholar]
- 13. Drevets DA, Bronze MS. 2008. Listeria monocytogenes: epidemiology, human disease, and mechanisms of brain invasion. FEMS Immunol. Med. Microbiol. 53:151–165. [DOI] [PubMed] [Google Scholar]
- 14. Drevets DA, Jelinek TA, Freitag NE. 2001. Listeria monocytogenes-infected phagocytes can initiate central nervous system infection in mice. Infect. Immun. 69:1344–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Francis CL, Starnbach MN, Falkow S. 1992. Morphological and cytoskeletal changes in epithelial cells occur immediately upon interaction with Salmonella typhimurium grown under low-oxygen conditions. Mol. Microbiol. 6:3077–3087 [DOI] [PubMed] [Google Scholar]
- 16. Grant AJ, et al. 2008. Modelling within-host spatiotemporal dynamics of invasive bacterial disease. PLoS Biol. 6:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gray ML, Killinger AH. 1966. Listeria monocytogenes and listeric infections. Bacteriol. Rev. 30:309–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han H, Iwanaga T, Uchiyama Y, Fujita T. 1993. Aggregation of macrophages in the tips of intestinal villi in guinea pigs: their possible role in the phagocytosis of effete epithelial cells. Cell Tissue Res. 271:407–416 [DOI] [PubMed] [Google Scholar]
- 19. Hanski C, et al. 1989. Immunohistochemical and electron microscopic study of interaction of Yersinia enterocolitica serotype O8 with intestinal mucosa during experimental enteritis. Infect. Immun. 57:673–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hardy J, et al. 2004. Extracellular replication of Listeria monocytogenes in the murine gall-bladder. Science 303:851–853 [DOI] [PubMed] [Google Scholar]
- 21. Huang FP, et al. 2000. A discrete subpopulation of dendritic cells transports apoptotic intestinal epithelial cells to T cell areas of mesenteric lymph nodes. J. Exp. Med. 191:435–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jacquet C, et al. 2004. A molecular marker for evaluating the pathogenic potential of food-borne Listeria monocytogenes. J. Infect. Dis. 189:2094–2100 [DOI] [PubMed] [Google Scholar]
- 23. Jensen VB, Harty JT, Jones BD. 1998. Interactions of the invasive pathogens Salmonella typhimurium, Listeria monocytogenes, and Shigella flexneri with M cells and murine Peyer's patches. Infect. Immun. 66:3758–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jilge B. 1980. The gastrointestinal transit time in the guinea-pig. Zentbl. Versuchstierkd. 22:204–210 [PubMed] [Google Scholar]
- 25. Jones BD, Ghori N, Falkow S. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 180:15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Knodler LA, et al. 2010. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc. Natl. Acad. Sci. U. S. A. 107:17733–17738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177–4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lecuit M. 2007. Human listeriosis and animal models. Microbes Infect. 9:1216–1225 [DOI] [PubMed] [Google Scholar]
- 29. Lecuit M, et al. 1999. A single amino acid in E-cadherin responsible for host specificity toward the human pathogen Listeria monocytogenes. EMBO J. 18:3956–3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lecuit M, et al. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722–1725 [DOI] [PubMed] [Google Scholar]
- 31. Levin DM, Rosenstreich DL, Reynolds HY. 1973. Immunologic responses in the gastrointestinal tract of the guinea pig. I. Characterization of Peyer's patch cells. J. Immunol. 111:980–983 [PubMed] [Google Scholar]
- 32. Lionakis MS, Lim JK, Lee CC, Murphy PM. 2011. Organ-specific innate immune responses in a mouse model of invasive candidiasis. J. Innate Immun. 3:180–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lynch M, Painter J, Woodruff R, Braden C. 2006. Surveillance for foodborne-disease outbreaks–United States, 1998–2002. MMWR Surveill. Summ. 55:1–42 [PubMed] [Google Scholar]
- 34. Marco AJ, et al. 1992. A microbiological, histopathological and immunohistological study of the intragastric inoculation of Listeria monocytogenes in mice. J. Comp. Pathol. 107:1–9 [DOI] [PubMed] [Google Scholar]
- 35. Mecsas J, Bilis I, Falkow S. 2001. Identification of attenuated Yersinia pseudotuberculosis strains and characterization of an orogastric infection in BALB/c mice on day 5 postinfection by signature-tagged mutagenesis. Infect. Immun. 69:2779–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mengaud J, Ohayon H, Gounon P, Mege RM, Cossart P. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84:923–932 [DOI] [PubMed] [Google Scholar]
- 37. Milling S, Yrlid U, Cerovic V, MacPherson G. 2010. Subsets of migrating intestinal dendritic cells. Immunol. Rev. 234:259–267 [DOI] [PubMed] [Google Scholar]
- 38. Notermans S, Dufrenne J, Teunis P, Chackraborty T. 1998. Studies on the risk assessment of Listeria monocytogenes. J. Food Prot. 61:244–248 [DOI] [PubMed] [Google Scholar]
- 39. Pentecost M, Otto G, Theriot JA, Amieva MR. 2006. Listeria monocytogenes invades the epithelial junctions at sites of cell extrusion. PLoS Pathog. 2:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Portnoy DA, Auerbuch V, Glomski IJ. 2002. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J. Cell Biol. 158:409–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Portnoy DA, Jacks PS, Hinrichs DJ. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J. Exp. Med. 167:1459–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pron B, et al. 2001. Dendritic cells are early cellular targets of Listeria monocytogenes after intestinal delivery and are involved in bacterial spread in the host. Cell. Microbiol. 3:331–340 [DOI] [PubMed] [Google Scholar]
- 43. Pron B, et al. 1998. Comprehensive study of the intestinal stage of listeriosis in a rat ligated ileal loop system. Infect. Immun. 66:747–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Racz P, Tenner K, Mero E. 1972. Experimental Listeria enteritis. I. An electron microscopic study of the epithelial phase in experimental listeria infection. Lab. Invest. 26:694–700 [PubMed] [Google Scholar]
- 45. Robbins JR, et al. 1999. Listeria monocytogenes exploits normal host cell processes to spread from cell to cell. J. Cell Biol. 146:1333–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Robbins JR, Skrzypczynska KM, Zeldovich VB, Kapidzic M, Bakardjiev AI. 2010. Placental syncytiotrophoblast constitutes a major barrier to vertical transmission of Listeria monocytogenes. PLoS Pathog. 6:e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schlech WF, III, et al. 1983. Epidemic listeriosis: evidence for transmission by food. N. Engl. J. Med. 308:203–206 [DOI] [PubMed] [Google Scholar]
- 48. Theriot JA, Mitchison TJ, Tilney LG, Portnoy DA. 1992. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature 357:257–260 [DOI] [PubMed] [Google Scholar]
- 49. Vazquez-Torres A, et al. 1999. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401:804–808 [DOI] [PubMed] [Google Scholar]
- 50. Westcott MM, Henry CJ, Cook AS, Grant KW, Hiltbold EM. 2007. Differential susceptibility of bone marrow-derived dendritic cells and macrophages to productive infection with Listeria monocytogenes. Cell. Microbiol. 9:1397–1411 [DOI] [PubMed] [Google Scholar]
- 51. Wilson JP. 1967. Surface area of the small intestine in man. Gut 8:618–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wollert T, et al. 2007. Extending the host range of Listeria monocytogenes by rational protein design. Cell 129:891–902 [DOI] [PubMed] [Google Scholar]
- 53. Zeldovich VB, Robbins JR, Kapidzic M, Lauer P, Bakardjiev AI. 2011. Invasive extravillous trophoblasts restrict intracellular growth and spread of Listeria monocytogenes. PLoS Pathog. 7:e1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]