Abstract
The human pathogen enterohemorrhagic Escherichia coli (EHEC) O157:H7 has two histidine sensor kinases, QseC and QseE, which respond to the mammalian adrenergic hormones epinephrine and norepinephrine by increasing their autophosphorylation. Although QseC and QseE are present in nonpathogenic strains of E. coli, EHEC exploits these kinases for virulence regulation. To further investigate the full extent of epinephrine and its sensors' impact on EHEC virulence, we performed transcriptomic and phenotypic analyses of single and double deletions of qseC and qseE genes in the absence or presence of epinephrine. We showed that in EHEC, epinephrine sensing seems to occur primarily through QseC and QseE. We also observed that QseC and QseE regulate expression of the locus of enterocyte effacement (LEE) genes positively and negatively, respectively. LEE activation, which is required for the formation of the characteristic attaching and effacing (A/E) lesions by EHEC on epithelial cells, is epinephrine dependent. Regulation of the LEE and the non-LEE-contained virulence factor gene nleA by QseE is indirect, through transcription inhibition of the RcsB response regulator. Finally, we show that coincubation of HeLa cells with epinephrine increases EHEC infectivity in a QseC- and QseE-dependent manner. These results genetically and phenotypically map the contributions of the two adrenergic sensors QseC and QseE to EHEC pathogenesis.
INTRODUCTION
In mammals, the adrenergic hormones epinephrine and norepinephrine are an integral part of the stress response (16). These hormones are recognized by mammalian cells by means of membrane-bound G protein-coupled receptors (GPCRs) to which they bind, initiating a regulatory cascade. In microorganisms, the hormones epinephrine and norepinephrine have been shown to be sensed by a variety of disease-causing organisms, including enterohemorrhagic Escherichia coli (EHEC) O157:H7 (8, 21, 59, 71), enterotoxigenic E. coli (ETEC) (42), Salmonella enterica serovar Typhimurium (1, 3, 4, 49), and Vibrio parahaemolyticus (52), as well as recently in the fish and human pathogen Edwardsiella tarda (79). These enteric pathogens use epinephrine as a signal for differential regulation of virulence factors, including motility (3, 8, 79), invasion (49, 59), and attaching and effacing (A/E) lesion formation, which are typical of EHEC and enteropathogenic E. coli (EPEC) infections (48, 73).
EHEC is an enteric bacterium that causes hemorrhagic colitis (31). In some cases, complications may arise, including hemolytic uremic syndrome (HUS) and thrombotic thrombocytopenic purpura (TTP) (18). In a similar fashion to other pathogens, EHEC controls virulence gene expression aiming for maximal energy efficiency. It senses signals from both the mammalian and intestinal microbial flora, to discern its arrival at its colonization niche, the colon. EHEC has been shown to sense the autoinducer 3 (AI-3) signal produced by the intestinal microbial flora, as well as the aforementioned host-produced hormones epinephrine and norepinephrine (8, 21, 59, 71).
Two histidine sensor kinases have been identified as sensors of epinephrine and norepinephrine in EHEC. The first, histidine kinase QseC, has been reported to increase its autophosphorylation in response to epinephrine, norepinephrine, or AI-3 (8). QseC then transfers its phosphate to, not only its cognate response regulator (RR) QseB, but also two other RRs, QseF and KdpE (21). QseC via QseB regulates flagellar and motility genes through the direct binding of QseB to the promoter region of flhDC, the master regulator of the flagellar regulon (10, 21). Through QseF, QseC activates Shiga toxin production (21, 41, 42).
A/E lesion formation, which is characterized by the attachment of bacteria to colonic epithelial cells followed by an induction of extensive actin rearrangement underneath the bacteria and effacement of surrounding microvilli (29, 36, 48, 73), has been shown to be regulated by QseC through the KdpE RR (21, 59; J. Njoroge et al., submitted for publication). We have shown that KdpE directly binds to the promoter region of ler, which codes for the master regulator of the locus of enterocyte effacement (LEE) genes that are required for A/E lesion formation (44). The LEE genes are mostly organized into five operons (LEE1 to -5), with the first operon encoding the LEE transcriptional activator Ler (13, 44, 45, 69). Most of the genes in the LEE are necessary for A/E lesion formation and include genes that encode the structural components of a type 3 secretion system (TTSS), as well as some effectors that are translocated through this TTSS into the host epithelial cell (26, 44). EspA, a LEE4-encoded secreted protein, forms part of the translocon of the TTSS, providing a structural direct link between the bacteria and the infected host cell (27, 34, 56). The LEE5 gene tir codes for an effector that gets translocated through the TTSS into the host cell, where it serves as a receptor for another LEE5-encoded protein, the adhesin intimin (encoded by the eae gene) (28, 33). The interaction of these two proteins allows for the intimate attachment of EHEC to the host epithelial cell. The TTSS also translocates non-LEE-encoded effectors such as EspFu/TccP (7, 15) and NleA/EspI (19, 50, 51, 63), which mimic mammalian signaling proteins and hijack host cell signal transduction. The NleA effector is an important virulence factor that has been shown to be required for virulence in the Citrobacter rodentium murine model (19, 51). It has been reported to disrupt intestinal tight junctions (74) and to localize to the Golgi apparatus, where it inhibits cellular protein secretion (35). The positive control of the LEE genes, Shiga toxin production, and motility by QseC culminates in the activation of the EHEC virulence repertoire. Deletion of qseC has been shown to attenuate virulence of not only EHEC but also Salmonella enterica Typhimurium, Francisella tularensis, uropathogenic E. coli (UPEC), and Edwarsiella tarda (3, 8, 37, 59, 79) in vitro and in vivo.
The second epinephrine sensor, the histidine kinase QseE, responds to epinephrine, phosphate, and sulfate by increasing its autophosphorylation level and then transfers its phosphate to its cognate RR, QseF (61). Importantly, QseC acts upstream of qseEF, given that QseC activates expression of qseEF (62). The QseEF two-component system has been characterized as being important for espFu transcription (62). The fact that both QseC and QseE increase their phosphorylation in an epinephrine-dependent manner and that QseC has been shown to initiate a signal transduction cascade in response to this hormone posed an interesting question of how this intricate control of epinephrine-dependent pathogenesis is maintained. To answer this question, we performed thorough transcriptional and phenotypical analyses of strains lacking one or both of the genes coding for these kinases, in the absence or presence of the hormone epinephrine. Although the influence of epinephrine on QseC-dependent regulation of the LEE genes, motility, and Shiga toxin production has been previously reported (8, 59), the effect of this hormone on QseE-dependent regulation of downstream genes has not been determined. In this work, we show that the adrenergic kinases QseC and QseE act in an antagonistic manner to regulate both LEE-contained and non-LEE-contained genes in order to control overall virulence of the enteric pathogen EHEC. We also report the role of epinephrine-dependent increase in A/E lesion formation and the important role that these two adrenergic kinases play in the formation of these lesions.
MATERIALS AND METHODS
Strains and growth media.
All bacterial strains used in this study are listed in Table 1. Unless otherwise stated, strains were grown in Luria-Bertani (LB) medium or low-glucose Dulbecco's modified Eagle's medium (DMEM) at 37°C and 250 rpm. Medium was supplemented, when necessary, with 50 μg ml−1 streptomycin, 50 μg ml−1 kanamycin, and 100 μg ml−1 ampicillin. For epinephrine studies, strains were grown under light-protected conditions after addition of epinephrine to a final concentration of 50 μM.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Reference or source | Description |
|---|---|---|
| Strains | ||
| Wild type (wt) | 18 | wt O157:H7 (86-24 clinical isolate) |
| VS138 | 72 | ΔqseC |
| VS179 | 72 | ΔqseC strain complemented with pqseC (pVS155) |
| NR01 | 62 | ΔqseE |
| JN080 | This study | ΔqseE strain complemented with pqseE (pJN62) |
| JN07 | This study | ΔqseC ΔqseE |
| JN071 | This study | ΔqseC ΔqseE strain complemented with pqseC and pqseE |
| JN18 | This study | ΔrcsB |
| JN081 | This study | ΔrcsB strain complemented with prcsB (pJN63) |
| Plasmids | ||
| pKD4 | 11 | λ Red template plasmid |
| pKD46 | 11 | λ Red helper plasmid |
| pCP20 | 11 | λ Red helper plasmid |
| pBAD-myc-hisA | Invitrogen | C-terminal Myc-His tag cloning vector |
| pBAD33 | 20 | cloning vector |
| pRS551 | 68 | lacZ reporter gene fusion vector |
| pVS155 | 72 | qseC in pBADmyc His |
| pJN62 | This study | qseE in pBAD33 |
| pJN63 | This study | rcsB in pBADmyc His |
| pVS175 | 70 | fliC::lacZ in pRS551 |
| pVS182 | 70 | flhD::lacZ in pRS551 |
Recombinant DNA methods.
Methods used for PCR amplification, plasmid purification, restriction enzyme digestion, ligation, and transformation were performed according to standard protocols (64). IDT and Primer Express v1.5 (AB) were used to design the oligonucleotide primers (Table 2) utilized in this work. Construction of the ΔqseC and ΔqseE mutants has been described previously (62, 72). The nonpolar ΔqseC ΔqseE double mutant and the ΔrcsB mutant were constructed using a λ Red-mediated recombination method (11). In brief, using the helper plasmid pKD4 as a template, primer pairs YfhKP1 and YfhKP2 for qseE and JrcsB redF and JrcsB redR for rcsB were used to amplify PCR products that were then gel purified (Qiagen). The ΔqseC and wild-type (wt) strains transformed with the helper plasmid pKD46 were prepared for electroporation and transformed with the qseE and the rcsB PCR products, respectively. The electroporated cells were then recovered in SOC medium (0.5% yeast extract, 2% tryptone, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose) for 6 h at 30°C, plated on kanamycin-supplemented LB plates, and incubated overnight at 42°C. The resultant colonies were screened for ampicillin sensitivity and kanamycin resistance. The kanamycin cassette was then resolved by electroporating deletion candidates with the resolvase plasmid pCP20, heat shocking at 42°C, and then screening the resulting colonies for sensitivity for both ampicillin and kanamycin. Final verification was performed by PCR amplification and sequencing.
Table 2.
Oligonucleotides used in this study
| Oligonucleotide name | Description/use | Oligonucleotide sequence |
|---|---|---|
| yfhKP1 | qseE deletion for JN07 | GGCAAAGCCTGAATGCGCCTTAGCGACCAGGCGGCGCTGGTCAACCGCACCACGCTTATCGATGCCCGGCGCAGCGAAGCAATGACCAACGCGGCGCTGGATGTAGGCTGGAGCTGCTTC |
| yfhKP2 | TTGCCCGCTCTCGTCGACCAGATACAGTTCCCCTTGCATACGGCGAATACAATCCCTGGCAATGCTTAATCCCAGACCGCTGCCCTTCACCGCCCCTTTTATATGAATATCCTCCTTA | |
| JrcsB redF | rcsB deletion for JN18 | CGTGAACGTAATTATTGCCGATGACCATCCGATAGTCTTGTTCGGTATTCGTGTAGGCTGGAGCTGCTTC |
| JrcsB redR | CGCCAGCTTCATCATCGCAGATTTCTTCTGGCTACTGATGGTTTTAATACTGCATATGAATATCCTCCTT | |
| JrcsB checkF | Deletion check for rcsB | CGAACCAGTGACTTTGCTGCGTTAGC |
| JrcsB checkR | CGCTGTTGAAATAATGGGAATCGTAGGACGGA | |
| JqseE bad33F | Complement plasmid pJN62 | GCTCTAGAGGCTATTCGCGTCTGACGAGAGTA |
| JqseE bad33R | CCCAAGCTTTTATTTCGTGTTTTTCGACGACGGTAATTCAATG | |
| JrcsB mycF | Complement plasmid pJN63 | CTCGGTACCAACAATATGAACGTAATTATTGCCGATGACCA |
| JrcsB mycR | CTCGAATTCGTCTTTATCTGCCGGACTTAACGTTACTG | |
| rpoA RTF | rpoA RT-PCR | GCGCTCATCTTCTTCCGAAT |
| rpoA RTR | CGCGGTCGTGGTTATGTG | |
| espA RTF | espA RT-PCR | TCAGAATCGCAGCCTGAAAA |
| espA RTR | CGAAGGATGAGGTGGTTAAGCT | |
| ler RTF | ler RT-PCR | CGACCAGGTCTGCCCTTCT |
| ler RTR | GCGCGGAACTCATCGAAA | |
| tir RTF | tir RT-PCR | CCATGGAGAGCAGACGTAGCT |
| tir RTR | CGGTGATCCTGGATTTAACCTT | |
| eae RTF | eae RT-PCR | GCTGGCCTTGGTTTGATCA |
| eae RTR | GCGGAGATGACTTCAGCACTT | |
| nleArt549F | nleA RT-PCR | AGCCACTACTTCGACGGTAACC |
| nleArt624R | ACGAACCACTTGAGCTGTTAATCC | |
| rcsBF | rcsB RT-PCR | TCTCTCGCCAAAAGAGAGTGAAG |
| rcsBR | CGATCTCGGTCACCAGGAA | |
| qseC RT1 443F | qseC RT-PCR | AATGGGAATACCGTGAAGACATG |
| qseC RT1 505R | CCAACCACGGGATCAATTG | |
| QseE RTF | qseE RT-PCR | CCCTTCACCGCCCCTTT |
| QseE RTR | CGCGCCATGATCTTCGA |
Plasmids for mutant complementation and β-galactosidase assays (see Table 4) were constructed by amplifying the coding regions from the EHEC strain 86-24 using Phusion polymerase, digesting with the appropriate restriction enzymes, and ligating with T4 ligase (NEB) as previously described (72). Briefly, primer pair JqseEbad33F/JqseEbad33R were used to amplify the qseE gene, and the resulting PCR product was ligated into the pBAD33 vector predigested with XbaI and HindIII. The primer pair JrcsBmycF/JrcsBmycR were used to amplify the rcsB gene, and the resulting PCR product was ligated into the pBADmycHis vector predigested with KpnI and EcoRI.
Table 4.
Comparison of effect of epinephrine on wt and mutants
| Comparison and parametera | No. of genes with result: |
|||
|---|---|---|---|---|
| Increased expression | Decreased expression | No change | Total | |
| wt vs wt + epi. | ||||
| MG1655 specific | 727 | 1,011 | 2,332 | 4,070 |
| Pathogen specific | 1,423 | 185 | 4,335 | 5,943 |
| Total | 2,150 | 1,196 | 6,667 | 10,013 |
| ΔqseC mutant vs ΔqseC mutant + epi. | ||||
| MG1655 specific | 5 | 4 | 4,061 | 4,070 |
| Pathogen specific | 12 | 20 | 5,911 | 5,943 |
| Total | 17 | 24 | 9,972 | 10,013 |
| ΔqseE mutant vs ΔqseE mutant + epi. | ||||
| MG1655 specific | 25 | 23 | 4,022 | 4,070 |
| Pathogen specific | 19 | 37 | 5,887 | 5,943 |
| Total | 44 | 60 | 9,909 | 10,013 |
| ΔqseC ΔqseE mutant vs ΔqseC ΔqseE mutant + epi. | ||||
| MG1655 specific | 21 | 83 | 3,966 | 4,070 |
| Pathogen specific | 14 | 76 | 5,853 | 5,943 |
| Total | 35 | 159 | 9,819 | 10,013 |
+ epi., in the presence of epinephrine.
RNA extraction.
Cultures grown overnight aerobically at 37°C in LB were diluted 1:100 into low-glucose DMEM and grown in triplicate to an optical density at 600 nm (OD600) of 1.0. TRIzol (Invitrogen) and the Ribopure bacterial isolation kit (Ambion) were then used to extract RNA from these biological replicates according to the manufacturer's protocols.
qRT-PCR.
Quantitative real-time reverse transcriptase PCR (qRT-PCR) was performed as follows. RNA transcription levels were quantified using the Applied Biosystems ABI 7500 sequence detection system in a one-step reaction as previously described (78). In brief, extracted RNA was added (final dilution of 5 ng/μl) to a mixture containing Sybr green, validated primers (Table 2), RNase inhibitor, and reverse transcriptase (AB). Using ABI Sequence Detection 1.3 software, data were collected and normalized to endogenous rpoA levels. Analysis was performed using the comparative critical threshold cycle (CT) method, and data are presented as fold changes over wt levels. The error bars represent the standard deviations of the ΔΔCT value.
Microarray global analysis.
Microarray (E. coli 2.0 Affymetrix) global analysis was performed on extracted RNA according to the manufacturer's instructions as outlined in the Affymetrix Gene Expression Technical Manual (http://www.affymetrix.com). Briefly, RNA extracted as described above was used as a template for reverse transcription to cDNA. The cDNA was then processed and hybridized to the E. coli Genome GeneChip 2.0. The GeneChips contain over 10,000 probe sets directed toward 20,366 genes from four different strains of E. coli: the K-12 laboratory strain MG1655, the O157:H7 EHEC strain EDL933, the O157:H7 EHEC strain Sakai, and the uropathogenic strain CFT073.
To analyze the results, output from scanning replicates was collected using GCOS v1.4 according to the manufacturer's instructions. The data were then normalized using Robust Multi-array analysis (5, 23) and analyzed for differences in gene expression due to the addition of epinephrine and/or the deletion of qseC and qseE.
Motility assays.
Assays were performed as described previously (21). Briefly, overnight cultures grown shaking at 37°C were used to stab motility agar plates (0.3% agar, 1% tryptone, and 0.25% NaCl). These plates were then incubated at 37°C for 8 h, after which the motility halo diameters were measured and images were taken.
β-Galactosidase assays.
The fliC transcriptional fusion plasmid pVS177 was transformed into appropriate strains, and the resultant transformants were used to perform β-galactosidase assays as previously described (70). Briefly, overnight cultures were grown in LB to the mid-exponential growth phase (OD600, 0.5) and then assayed for β-galactosidase activity using o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate, as described previously (40).
FAS.
To examine pedestal formation, fluorescent actin staining (FAS) assays were performed as previously described (36, 62). HeLa cells were grown on coverslips in plates in DMEM supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and 1% PSG (penicillin-streptomycin-glutamine) antibiotic mixture at 37°C in 5% CO2 overnight to about 80% confluence. The FBS used was dialyzed to remove all molecules with a molecular weight less than 10,000, including any epinephrine that may be present. The wells containing the coverslips were then washed three times with phosphate-buffered saline (PBS) and replaced with fresh medium lacking antibiotics. For epinephrine studies, the drug was added to a final concentration of 50 μM. Overnight static bacterial cultures were then used to infect the washed cells. The plates were light protected and incubated for 6 h at 37°C in 5% CO2. The coverslips were then washed, fixed, and permeabilized. Fluorescein isothiocyanate (FITC)-labeled phalloidin was used to stain actin green, and propidium iodide (PI) was utilized to stain bacteria and HeLa nuclei red. The coverslips were then mounted on slides and visualized with a Zeiss Axiovert microscope. To quantify infected cells, at least 100 cells were counted per coverslip, and the number of bacteria infecting them was counted. Serially diluted samples of the original bacterial cultures were also plated to confirm similar CFU were used for the infection.
Statistical analysis.
To analyze significance of the results obtained from the assays in this work, all experiments were performed at least twice with at least triplicate samples each time. Student's unpaired t test was used to determine statistical significance. A P value of <0.05 was considered significant.
Microarray accession number.
Array data have been deposited in the NCBI GEO database, and the GEO number is GSE33895.
RESULTS
Global assessment of QseC and QseE gene regulation in EHEC.
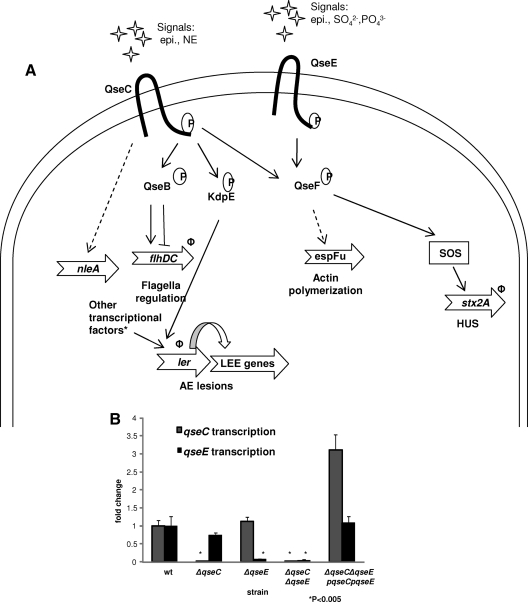
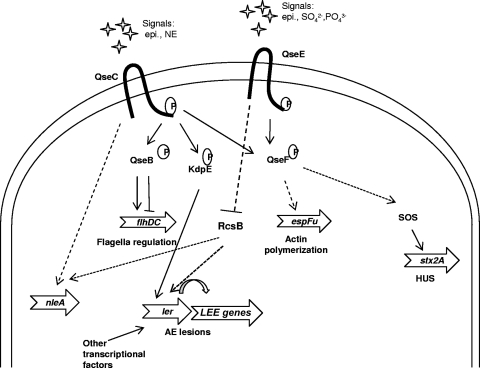
Previous microarray data comparing the single ΔqseC and ΔqseE single mutants to wild-type (wt) EHEC O157 in Dulbecco's modified Eagle's medium (DMEM) have shown divergences in global gene regulation by these two adrenergic receptors (21, 60). We have previously reported the role of QseC in the activation of the expression of genes involved in motility (9, 10), Shiga toxin production (21), and the LEE pathogenicity island (21; Njoroge et al., submitted). We have also reported the role of QseE in the regulation of espFu (62). The regulation of virulence factors by these two sensor kinases as had been identified before this work is summarized in Fig. 1A (8, 21, 61, 62; Njoroge et al., submitted). As the summary indicates, both QseC and QseE have been shown to sense epinephrine (8, 61). Epinephrine-dependent gene expression had only been reported for genes downstream of QseC but not for targets downstream of QseE. Another open question was whether QseC and QseE are the only sensors of epinephrine in EHEC O157. To address these issues, we first needed to define genes that were regulated by both kinases and then test their expression in response to the presence of this adrenergic hormone. Additionally, we hypothesized that if these two kinases were the only epinephrine sensors, deletion of both QseC and QseE would make the resultant double mutant unable to respond to epinephrine. We therefore constructed a nonpolar qseC and qseE gene double deletion mutant (the ΔqseC ΔqseE strain). Using quantitative real-time reverse transcriptase PCR (qRT-PCR), we confirmed the deletion of both genes as well as the efficacy of plasmid encoded QseC and QseE to rescue gene transcription (Fig. 1B).
Fig 1.
Confirmation of nonpolar deletion and rescue of expression of the adrenergic kinase-encoding genes qseC and qseE. (A) Summary of the QseC- and QseE-dependent signaling cascade involved in virulence regulation as reported prior to this work. Genes whose expression had been shown to be affected by epinephrine have Φ next to them. Asterisks indicate that the ler promoter is highly regulated by many transcription factors, including GrlA, Pch, GadE, QseA, and H-NS (2, 6, 25, 30, 67). epi, epinephrine; NE, norepinephrine; AE, attaching and effacing. (B) qRT-PCR analysis examining qseC and qseE expression in the wt and ΔqseC, ΔqseE, and ΔqseC ΔqseE mutant strains and the complemented double mutant strains grown to an OD600 of 1.0 in low-glucose DMEM. The genes' transcript levels were quantified as fold differences normalized to wt gene transcription levels. The samples' rpoA transcript levels were used as internal controls to normalize the output CT values. The data are from at least three independently grown replicates.
Next, using Affymetrix E. coli 2.0 microarrays, we performed a global transcriptomics analysis of the wt, ΔqseC, ΔqseE, and ΔqseC ΔqseE double mutant strains grown in DMEM, which is optimal for the expression of TTSS genes and other EHEC virulence factors. These growth conditions were performed in the presence of AI-3, which is endogenously produced by EHEC O157 and is sensed by QseC to differentially regulate its targets (8, 71, 78). The arrays contain over 10,000 probe sets that cover genes in the genomes of the two sequenced EHEC strains (EDL933 and Sakai), the K-12 strain MG1655, and the UPEC strain CFT073, as well as intergenic regions that can code for small RNAs (sRNAs) or nonannotated small open reading frames (ORFs).
The microarray analysis revealed that although a majority of the probe sets in the double-kinase mutant were unchanged compared to the wt, 510 probe sets showed increased expression, with 47% of these being pathogen specific (Table 3). Additionally, a total of 300 probe sets in the double mutant had decreased expression, with 65% of the genes being pathogen specific. This percentage of pathogen-specific genes that were differentially regulated in the double mutant was similar to that in the ΔqseC strain. The ΔqseC strain had 149 probe sets increased and decreased, with the pathogen-specific ones representing 52% of both the increases and the decreases. On the other hand, the ΔqseE global gene regulation profile revealed more differential expression than is seen in the double mutant, with twice as many probe sets increased in the ΔqseE strain as in the ΔqseC ΔqseE strain (1,282 versus 510). Additionally, more than four times as many probe sets in the ΔqseE strain were decreased than in the ΔqseC ΔqseE mutant (1,294 versus 300). The mostly upregulated probe sets in the double-kinase mutant, as indicated by the microarray, included many hypothetical genes, metabolism genes, and a few (putative) sensor kinase genes, such as yedV and zraS. These genes' expression remained unchanged in the single mutants' profiles, suggesting that QseC's and QseE's regulatory effects on them may be redundant, and only the deletion of both sensors could make a difference in their expression. The most highly downregulated probe sets in the double-kinase mutant, which included pathogen-specific genes such as Z4320, c1516, and c4309, had differential regulation in the single mutants that did not follow a distinct pattern.
Table 3.
Comparison of the effects of deletion of QseC, QseE, or both kinases on global gene expression in EHEC O157
| Comparison and parameter | No. of genes with result: |
|||
|---|---|---|---|---|
| Increased expression | Decreased expression | No change | Total | |
| wt vs ΔqseC mutant | ||||
| MG1655 specific | 71 | 71 | 3,928 | 4,070 |
| Pathogen specific | 78 | 78 | 5,787 | 5,943 |
| Total | 149 | 149 | 9,715 | 10,013 |
| wt vs ΔqseE mutant | ||||
| MG1655 specific | 558 | 871 | 2,641 | 4,070 |
| Pathogen specific | 724 | 423 | 4,796 | 5,943 |
| Total | 1,282 | 1,294 | 7,437 | 10,013 |
| wt vs ΔqseC ΔqseE mutant | ||||
| MG1655 specific | 268 | 104 | 3,698 | 4,070 |
| Pathogen specific | 242 | 196 | 5,505 | 5,943 |
| Total | 510 | 300 | 9,203 | 10,013 |
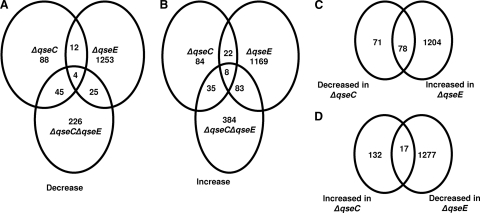
Next we investigated whether there were any commonly regulated genes in the arrays. The ΔqseC ΔqseE mutant has more downregulated genes in common with the ΔqseC mutant (49 genes) than with the ΔqseE mutant (29 genes) (Fig. 2A). Additionally the ΔqseC ΔqseE strain shares more upregulated genes with the ΔqseE strain (91 genes) than with the ΔqseC strain (43 genes) (Fig. 2B). These data suggest that the double-kinase mutant has the plasticity to regulate gene expression to mimic either one of the single mutants, depending on the set of genes being evaluated. Of the 300 genes decreased in the ΔqseC ΔqseE strain, only 4 genes were commonly regulated in the ΔqseC and ΔqseE mutants (Fig. 2A), while of the 510 genes increased in the double-kinase mutant, only 8 genes were commonly regulated in the single-kinase mutants (Fig. 2B). These commonly regulated genes included four that were metabolism related (fruA, rbsD, ais, and srlA) and four that were involved in metal sensing (ygiW, ais, arnF, and basR). The others were hypothetical genes. This leaves a total of 610 genes (226 decreased and 384 increased) that are differentially regulated in the ΔqseC ΔqseE strain that are not shared with the single mutants. This indicates that the double-kinase mutant transcriptome does not fully overlap with the single-kinase mutants, suggesting that deletion of one or both kinases promotes extensive rewiring of downstream signaling.
Fig 2.
Global analysis of QseC's and QseE's effects on EHEC O157 gene transcription. Venn diagrams show the number of overlapping downregulated (A) and upregulated (B) genes between the ΔqseC, ΔqseE, and ΔqseC ΔqseE mutant strains compared to the wt. (C) Venn diagram indicating genes that are decreased in the ΔqseC strain and increased in the ΔqseE strain. (D) Venn diagram indicating genes with expression that is increased in the ΔqseC strain and decreased in the ΔqseE strain. Strains for the microarrays were grown to an OD600 of 1.0 in low-glucose DMEM.
Another possible explanation for the paucity of commonly regulated targets may be that the two kinases conversely regulate similar target genes. Indeed in the single mutant arrays, we identified a total of 95 genes conversely regulated by these two kinases. Expression of 78 genes was decreased in the ΔqseC strain, and increased in the ΔqseE strain, while expression of 17 genes was increased in the ΔqseC strain and decreased in the ΔqseE strain (Fig. 2C and D). These conversely regulated genes included the LEE genes and nleA encoding a non-LEE effector. Altogether, these data indicated that although there may be convergent regulation of some genes by QseC and QseE, other genes may be regulated by only one of these adrenergic kinases.
QseC and QseE conversely regulate transcription of the LEE and nleA.
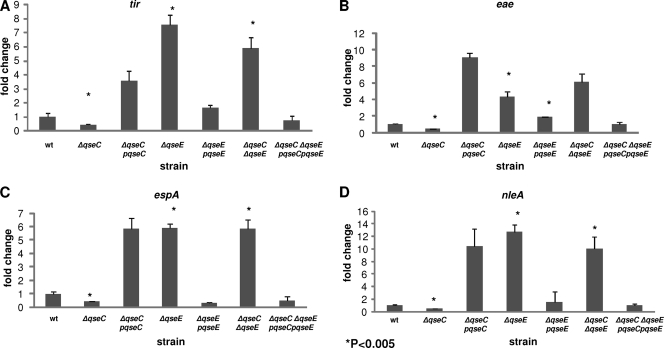
Global transcriptome analysis of the single- and double-kinase mutants indicated that there was differential regulation of some targets (Fig. 2). These included the LEE genes, previously reported to be activated by QseC in DMEM (21, 59), and nleA, which had also been previously reported to be mildly activated by QseC in DMEM (21). However, whether QseE played any role in the regulation of the LEE or nleA was still an open question, as well as if and how QseC and QseE may interface in this regulation. We first performed qRT-PCR to compare the differences in mRNA levels of genes in the LEE4 an LEE5 operons. RNA was extracted from the wt and ΔqseC, ΔqseE, and ΔqseC ΔqseE mutant strains grown in low-glucose DMEM to an OD600 of 1.0 and assessed for differences in transcription of the tir and eae genes (both within LEE5) and the espA gene (LEE4). The mRNA levels of all three of these genes were significantly decreased in the ΔqseC strain compared to the wt (Fig. 3A to C), with tir, eae, and espA transcription decreasing 2-fold for all three. On the other hand, the same genes had a significant increase in transcription in the ΔqseE mutant relative to the wt, with mRNA levels of tir, eae, and espA being augmented 12-fold, 4-fold, and 6-fold, respectively. When the mRNA levels of the three LEE genes in the ΔqseC ΔqseE mutant were evaluated, their levels were comparable to those of the ΔqseE strain (tir up 9-fold and eae and espA up 6-fold). Transcription of all genes was rescued upon complementation.
Fig 3.
Both QseC and QseE regulate the LEE genes and nleA. qRT-PCR analyses of tir (LEE5) (A), eae (LEE5) (B), espA (LEE4) (C), and nleA (D) transcription. The mRNA levels for all of these genes were quantified and normalized to the mRNA levels of the endogenous internal control gene, rpoA. The mRNA levels were graphed as fold changes compared to wt transcript levels. The results are from at least three independent samples.
Next, we evaluated whether this converse gene regulation by QseC and QseE extended beyond those encoded by the LEE pathogenicity island. NleA is a non-LEE-encoded effector translocated by the LEE TTSS into host cells, and it has been shown to play an important role in virulence (19, 35, 74). It has been shown to be mildly activated by QseC in DMEM (21). The microarray data indicated that nleA's expression was decreased in the ΔqseC strain, increased in the ΔqseE strain, and also elevated in the ΔqseC ΔqseE strain. This differential nleA regulation by these two kinases mirrored the LEE regulation. Therefore, we assessed whether nleA transcriptional analysis using the more sensitive qRT-PCR method would also mirror these previous observations. Compared to the wt, nleA mRNA levels were decreased 2-fold in the ΔqseC strain, while we observed over a 10-fold increases in both the ΔqseE and ΔqseC ΔqseE mutants (Fig. 3D). These findings support a positive role and negative role for QseC and QseE, respectively, in the regulation of both LEE genes and the gene encoding the non-LEE effector, NleA. Although both kinases regulated LEE4, LEE5, and nleA (Fig. 4C), QseE is epistatic to QseC, as observed by the fact that the double mutant has a phenotype comparable to that of a mutant with a qseE deletion.
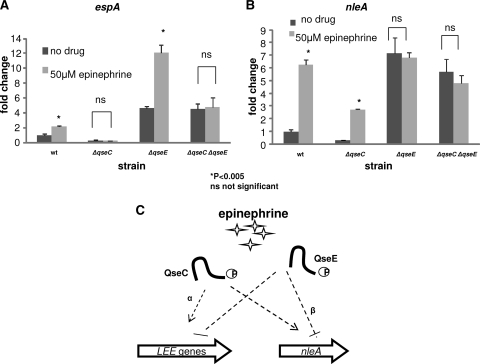
Fig 4.
Effect of epinephrine on QseC- and QseE-dependent regulation of LEE and non-LEE genes. Expression of espA (LEE4) (A) and nleA (B) was evaluated by qRT-PCR in the wt strain and the mutant strains grown to the late exponential phase in the absence and presence of epinephrine (final concentration, 50 μM). The error bars indicate standard deviations of the ΔΔCT values. The levels of endogenous rpoA mRNA were used to normalize the CT values. (C) Representation of the converse regulation of the LEE genes and nleA transcription by the epinephrine-sensing kinases QseC and QseE. Although both kinases regulate the LEE genes and nleA, epinephrine-dependent regulation of the LEE genes is mostly via QseC (dotted arrow with α), while epinephrine-dependent regulation of nleA is mostly via QseE (dotted line with β).
Deletion of both kinases eliminates the epinephrine-dependent regulation of virulence genes.
Previous studies have shown that both QseC and QseE sense the hormone epinephrine (4, 8, 21, 49, 61). Given that both adrenergic kinases regulate the LEE genes as well as nleA (Fig. 2), we next investigated the role that epinephrine plays in this regulation. We grew the wt and the mutants in low-glucose DMEM in the absence or presence of epinephrine (final concentration, 50 μM), extracted RNA, and evaluated nleA and, as a representative of the LEE genes, espA mRNA levels. In the presence of epinephrine, the mRNA levels of both genes were significantly increased in the wt compared to the wt with no drug (Fig. 4), with espA levels increased 2-fold and nleA levels increased 6-fold. Interestingly, the epinephrine effect on transcription in the single-deletion mutants differed depending on the gene evaluated. When espA transcription was compared between nontreatment and treatment with epinephrine, no change was observed in the ΔqseC strain, while there was a 3-fold increase in espA mRNA levels in the epinephrine-treated ΔqseE strain compared to the nontreated ΔqseE strain (Fig. 4A). These results indicate that although both kinases are involved in espA gene regulation, epinephrine-dependent regulation of espA occurs primarily via QseC. In the double mutant, no significant change was observed between nontreatment and treatment with epinephrine. When we evaluated nleA mRNA levels in the absence and presence of epinephrine, we observed a 6-fold increase in wt (Fig. 4B). In the ΔqseC strain in the presence of epinephrine, we observed a 2.5-fold increase in nleA transcription compared to the ΔqseC mutant without epinephrine. However, there was no significant change between the ΔqseE strain in the presence of epinephrine and that without epinephrine. These data suggest that although QseC and QseE both regulate nleA transcription, epinephrine-dependent regulation of nleA occurs primarily via QseE. The ΔqseC ΔqseE double mutant was also blind to the effects of epinephrine. Altogether, these results support our hypothesis that QseC and QseE sense epinephrine to regulate the expression of LEE and non-LEE effectors (Fig. 4C) and that in the absence of these two adrenergic kinases, EHEC is unable to sense this hormone and is consequently unable to differentially regulate these genes.
Global analysis of epinephrine-dependent EHEC gene regulation by the two adrenergic kinases QseC and QseE.
Since transcription of the LEE genes and nleA in the ΔqseC ΔqseE double-kinase mutant is epinephrine independent (Fig. 4), we next investigated the extent of this lack of response to epinephrine. Using Affymetrix E. coli 2.0 microarrays, we performed a global gene analysis of the wt and the single and double mutants grown in low-glucose DMEM in the absence or presence of 50 μM epinephrine. The microarray data indicated that there was more differential regulation when the wt was treated with epinephrine than when the mutants were treated with epinephrine (Table 4). When the wt with epinephrine was compared to wt with no treatment, 21% of the genes were upregulated, while 12% were downregulated, indicating a possible dual role for epinephrine as both an activator and a repressor of its target genes. Altered genes were observed both in the K-12 genes from strain MG1655, which contains the conserved E. coli backbone, and in the pathogen-specific probe sets. It is interesting to note that a higher percentage of the pathogen-specific genes were upregulated than downregulated (24% increased versus 3% decreased). Comparison of the ΔqseC ΔqseE mutant in the presence of epinephrine to the ΔqseC ΔqseE mutant with no treatment indicated very few genes were differentially regulated, with 0.3% being upregulated and 1.4% being downregulated. This indicated to us that deletion of both qseC and qseE left the double mutant strain mostly unable to sense epinephrine, which correlates with the epinephrine unresponsiveness observed by qRT-PCR (Fig. 4). This relative unresponsiveness was also observed in the single mutants. Adding epinephrine to the ΔqseC strain culture only altered the expression of 0.4% of the total genes, while addition of epinephrine to the ΔqseE strain culture led to only 1% of the genes being differentially regulated. The fact that a total of 34% of the genes were differentially regulated when epinephrine was added to the wt culture, while less than 2% of the genes were differentially regulated when epinephrine was added to either the single or the double mutant cultures, indicates that deletion of QseC and QseE results in EHEC being mostly unable to sense epinephrine and that both kinases seem to work in concert toward the proper sensing of this signal.
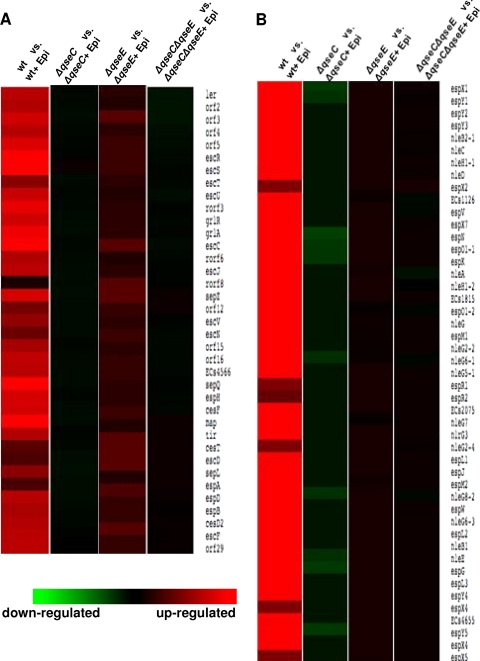
Transcriptome comparison of the four array sets revealed that in the wt, epinephrine increased the regulation of most of the LEE genes (Fig. 5A), as well as most of the genes that encode confirmed and predicted non-LEE EHEC O157 effectors (77) (Fig. 5B). The heat maps comparing the ΔqseC strain with and without epinephrine treatment indicated that in the presence of epinephrine, genes encoding the non-LEE effectors were differentially regulated, while the LEE genes were unaffected. On the other hand, epinephrine increased the LEE genes' expression in the ΔqseE strain but did not affect non-LEE effector gene expression. In the ΔqseC ΔqseE double-kinase mutant, neither set of genes responded to the addition of epinephrine. These heat maps mirrored the qRT-PCR data (Fig. 4), which had suggested that the LEE genes were still responsive to epinephrine in the ΔqseE strain but not in the ΔqseC strain, while non-LEE-encoded effectors such as nleA were still responsive to epinephrine in the ΔqseC strain but not in the ΔqseE strain. These results also confirmed the ΔqseE ΔqseC qRT-PCR data, which indicated that in the double-kinase mutant, the transcription of both the LEE genes and nleA is unaffected by epinephrine.
Fig 5.
Deletion of the two adrenergic kinases QseC and QseE impairs epinephrine-dependent regulation of multiple EHEC virulence factors. Shown are heat maps from microarray analysis representing the effects of epinephrine (Epi) on the wt and ΔqseC, ΔqseE, and ΔqseC ΔqseE mutant strains. The strains treated with epinephrine were compared to the same strains with no treatment. Red indicates upregulation, green indicates downregulation, and black indicates no change. (A) Heat map representing differential regulation of the LEE genes. (B) Heat map showing the differential expression of non-LEE genes.
QseE regulation of the LEE- and non-LEE encoded effectors occurs through RcsB.
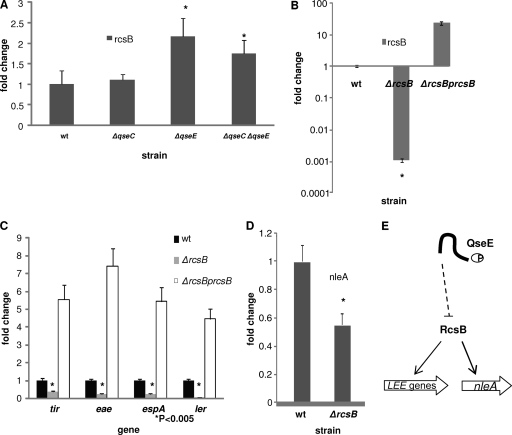
We have shown that QseC and QseE conversely regulate genes both within and outside the LEE pathogenicity island (Fig. 3 and 4). We next explored the mechanism of this differential regulation. We have previously shown that QseC regulation of the LEE occurs through the KdpE RR (21; Njoroge et al., submitted). Unlike QseC, which phosphorylates three RRs (QseB, KdpE, and QseF), QseE phosphorylates only its cognate RR, QseF (80). QseF is a DNA binding transcriptional regulator that binds σ54-dependent promoter regions (62). The transcription of the espA gene's LEE4 operon, as well as the tir and eae LEE5 operon, is σ70 dependent (38, 65). As none of these genes has a σ54-dependent promoter, it is unlikely that QseE's regulation of these genes is through QseF. We have previously shown that QseE regulates expression of several two-component systems at the transcriptional level, including the RcsBC system (60). The response regulator of the system, RcsB, has been shown to be involved in the regulation of the LEE genes in the Sakai strain of EHEC (76). To explore whether RcsB was an intermediate in the QseE regulation of these genes, we assessed rcsB mRNA levels in the wt, ΔqseC, ΔqseE, and ΔqseC ΔqseE mutant. The transcription of rcsB was unaffected in the ΔqseC strain but increased significantly in the ΔqseE and ΔqseC ΔqseE strains (Fig. 6A). These results suggested that the upregulation of the rcsB observed in the ΔqseE mutant and the double mutant may be due to the fact that QseE is an inhibitor of rcsB transcription, which is in agreement with our previous report (60).
Fig 6.
QseE regulates nleA and the LEE genes through its inhibition of rcsB transcription. (A) Transcription (qRT-PCR) of rcsB in the wt and ΔqseC, ΔqseE, and ΔqseC ΔqseE mutant strains. (B) Confirmation by qRT-PCR of the deletion and rescue in expression of rcsBi. (C) Transcriptional LEE gene expression for the wt strain, ΔrcsB mutant, and its complement. (D) qRT-PCR evaluating the transcription of nleA in the wt and ΔrcsB mutant. Error bars indicate the standard deviations of the ΔΔCT values. The mRNA levels of endogenous rpoA were used to normalize the CT values. (E) Representation of how the inhibition of the expression of the LEE genes and nleA by QseE is indirect via RcsB. RcsB, whose transcription is inhibited by QseE, is a transcriptional activator of the LEE genes and nleA.
Next we constructed a nonpolar mutant of rcsB. RNA was then extracted from the wt, the mutant, and the complemented strain, and the absence and rescue of rcsB expression in these strains were confirmed by qRT-PCR (Fig. 6B). We then assessed the impact of RcsB regulation on the expression of the LEE genes tir, eae, and espA. Transcription of all of these genes was significantly decreased in the ΔrcsB strain (2.5-fold for tir and espA and 4-fold for eae), and expression was rescued upon complementation with rcsB on a plasmid (Fig. 6C). It is worth noting that the expression of the genes assessed was much higher in the complement than in the wt, probably due to the fact that the complement overexpressed rcsB. Because the LEE genes are activated by Ler, the master regulator of the LEE pathogenicity island, we assessed the effect of rcsB deletion on ler transcription. We observed a significant downregulation of 5-fold in ler transcription in the mutant. We also observed a 2-fold reduction in the expression of the nleA gene in the rcsB mutant (Fig. 6D). Altogether, these data suggest that QseE repression of the LEE and nleA transcription occurs indirectly via the RcsB RR. The QseEF proteins repress expression of RcsBC, impeding RcsB activation of LEE and nleA expression.
A/E lesion formation.
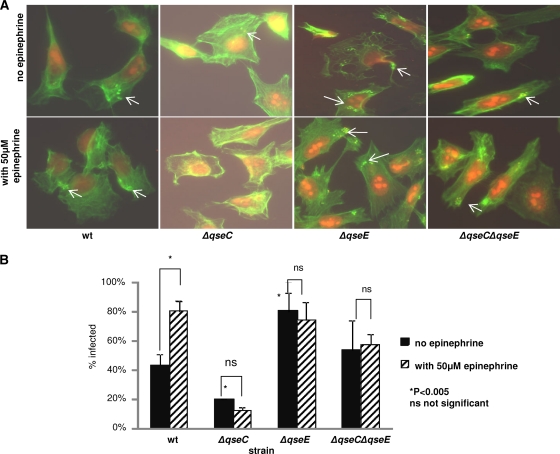
Since the presence of epinephrine and the deletion of qseC, qseE, or both qseC and qseE together affect the expression of nleA as well as the LEE genes, we next used fluorescent actin staining (FAS) to investigate whether this differential regulation affected the formation of A/E lesions. As most commercially available fetal bovine serum (FBS) used to supplement HeLa epithelial cell culture medium contains traces of epinephrine, we used a dialyzed FBS (Gibco, Invitrogen), which has all molecules with a molecular mass less than 10,000 Da removed. HeLa epithelial cells were infected for 6 h with the wt or the mutant strains in the absence or presence of epinephrine to a final concentration of 50 μM. The infected cells were then fixed and stained with FITC-phalloidin (which stains filamentous actin green) and propidium iodide (which stains the HeLa nuclei and bacteria red). The pedestals were visualized as red bacteria cupped by bright green actin (Fig. 7A). To ensure comparable levels of infection by the different strains, an aliquot of the input was also serially diluted and plated to confirm similar bacterial numbers were used for infection. Infection rates were calculated as the number of HeLa cells with bacteria attached as a percentage of the total number of HeLa cells.
Fig 7.
Fluorescent actin staining (FAS) assays. HeLa cells were infected for 6 h in the absence or presence of epinephrine (final concentration, 50 μM). HeLa cell actin was stained green with FITC-phalloidin, while HeLa cell nuclei and bacteria were stained red with propidium iodide. Formation of pedestals was visualized as bright green (actin) cups holding red bacterial cells. The experiments were performed in duplicate at least three times. For every slide, at least 100 cells were evaluated. (A) Visualization of pedestals formed by bacteria on HeLa cells. (B) Representation of the percentage of infected HeLa cells.
Incubation of HeLa cells with wt EHEC O157, in the absence of epinephrine, led to a 40% infection rate (Fig. 7B). When the infection was carried out in the presence of epinephrine, the percentage of cells infected increased a significant 2-fold. Upon ΔqseC mutant incubation with these epithelial cells in the absence of epinephrine, the percentage of infected cells decreased 2-fold compared to that of the wt. Supplementation with epinephrine did not increase infection. These results are consistent with the observation that LEE expression is decreased in the ΔqseC mutant (Fig. 4A) and that addition of epinephrine to the ΔqseC strain did not lead to increased LEE expression. Next, when the FAS assay was performed with the ΔqseE mutant, we observed that in the absence of epinephrine infection, rates were 2-fold higher than that of the wt without epinephrine and comparable to that of the wt in the presence of epinephrine. Addition of epinephrine to the ΔqseE infection assay did not increase infection rates. The infection rate in the ΔqseC ΔqseE strain was comparable to that of the wt but was unaffected by coincubation with epinephrine. These results give further evidence that epinephrine-dependent LEE regulation in EHEC O157 is dependent on only QseC and QseE.
Regulation of motility is dependent on QseC but not QseE.
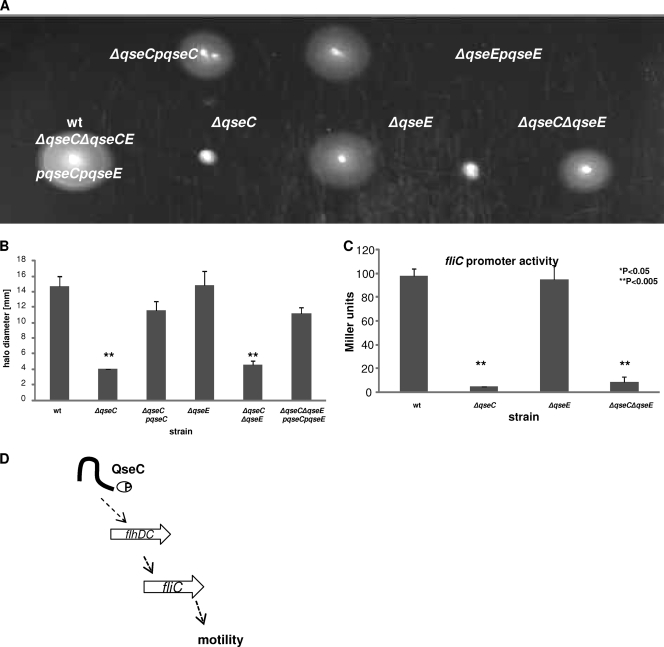
We have previously shown that the regulation of motility in EHEC is QseC dependent (10, 21, 72). Given that LEE gene regulation shows a converse relationship between QseC and QseE, we investigated whether this phenomenon was also observed in motility regulation. We assessed the motility of the wt, ΔqseC, ΔqseE, and ΔqseC ΔqseE strains and their complements in 1% tryptone–agar media. As expected, the motility of the ΔqseC strain compared to the wt was significantly diminished, with the halo diameters of the mutant reduced almost 5-fold (Fig. 8A and B). Deletion of qseE did not affect motility, with halo diameters for the ΔqseE strain comparable to those of the wt. When both qseC and qseE were deleted, the double mutant had a motility defect similar to that of the ΔqseC mutant, and this decrease in swimming could be rescued upon complementation with qseC and qseE in trans.
Fig 8.
Motility regulation is QseC dependent but QseE independent. (A) Tryptone motility plates with the wt strain and the ΔqseC, ΔqseE, ΔqseC ΔqseE mutants and their complemented strains. (B) Representation of the diameter of the bacterial halos. (C) β-Galactosidase assays were performed using plasmid pVS177 with an fliC::lacZ promoter fusion in the wt and ΔqseC, ΔqseE, and ΔqseC ΔqseE mutant strains. (D) Representation indicating the QseC-dependent and QseE-independent activation of motility genes. QseC phosphorylates QseB, which directly binds to the regulatory region of flhDC, encoding the master regulators of flagella, leading to increase fliC expression, production of flagella, and motility (10).
To confirm these motility plate results, we assessed whether the transcription of fliC, which codes for flagellin (Fig. 8C), was affected by deletion of qseC and/or qseE. The strains were transformed with the fliC-lacZ transcription fusions, and β-galactosidase assays were performed. In both the ΔqseC and ΔqseC ΔqseE fliC mutants, transcription was significantly reduced compared to that in the wt. In the ΔqseE strain, transcription of fliC was comparable to that of the wt (Fig. 8C). Altogether these results indicate that regulation of motility is QseE independent but QseC dependent. Also the double mutant data suggest that as far as motility is concerned, qseC is epistatic to qseE.
DISCUSSION
Bacterial populations have evolved the ability to sense their surroundings through chemical signaling (66). In the 1970s, the marine bacteria Vibrio fischeri and Vibrio harveyi were shown to sense increasing concentrations of self-produced compounds (later termed autoinducers), in order to monitor their population density, and at the optimal concentration of these signals, the bacteria activate expression of bioluminescence genes (12, 54, 55). Since then, a multitude of microbes have been shown to communicate within as well as outside their species (22).
Communication between bacterial species has also been reported in EHEC O157, where it has been shown that this enteric pathogen senses the AI-3, which is produced by itself as well as gut resident microbiota (71). As the infectious dose of EHEC O157 is estimated to be ∼50 CFU (31), it is unlikely that the self-produced AI-3 is sufficient to promote gene regulation when this pathogen reaches the intestine. Therefore, it has been proposed that EHEC O157 senses the AI-3 produced by the gut microbial flora to initiate regulation of virulence genes (71). Through the QseC AI-3 sensor, EHEC upregulates motility, which probably allows the bacteria to swim closer to the gut epithelium, where it may be exposed to the host-produced epinephrine and/or norepinephrine hormones (8, 71). This exposure to these human adrenergic hormones is thought to further augment positive regulation of genes important for colonization and formation of A/E lesions.
Here we show that exposure of EHEC O157 to epinephrine increases its ability to infect HeLa cells and form pedestals. This effect is QseC and QseE dependent (Fig. 7). QseC has been previously reported to be an activator of virulence. It has been shown to positively regulate motility in EHEC O157, Salmonella, and UPEC (3, 4, 10, 21, 37, 49, 72), invasion in Salmonella (49), and overall virulence in many other pathogens (47, 57, 59, 79). Here, we have shown that deletion of qseC significantly decreases formation of A/E lesions on HeLa cells and that the qseC mutant's ability to form these lesions is unaffected by epinephrine (Fig. 7). These data are consistent with the observation that the qseC mutant was unable to respond to epinephrine to activate LEE expression (Fig. 4A). However, it is worth noting that with regard to the regulation of nleA transcription, the qseC mutant still appears to sense epinephrine (Fig. 4B). NleA is an important virulence factor, but it is not involved in A/E lesion formation. This would explain why the epinephrine-dependent A/E lesion formation pattern (Fig. 7) mirrored the epinephrine-dependent transcription of the LEE genes (Fig. 4A) and not the epinephrine-dependent transcription of nleA (Fig. 4B). A probable explanation for this may be that although both QseC and QseE regulate nleA transcription, QseE may play a more significant role in this gene's regulation, the result of which would be that in the qseC mutant, the QseE that is present still senses epinephrine and responds to it, consequently altering nleA transcription. We have also shown that the other epinephrine sensor, QseE, inhibits pedestal formation, with the ΔqseE strain forming significantly more pedestals than the wt, and its infection rate is unaffected by epinephrine. Interestingly when espA transcription was assessed, the ΔqseE strain still sensed epinephrine (Fig. 4A). A likely reason for this observation is that in the absence of qseE, qseC is still present, and though both kinases regulate the LEE, QseC is the principal epinephrine-dependent regulator of espA. Therefore, in the qseE mutant, the QseC that is still present senses epinephrine and alters espA transcription. When we tested the ΔqseC ΔqseE double mutant in phenotypic assays with epinephrine, we observed an inability to sense this hormone (Fig. 4A and b, 5, and 7A and B). Transcription of the LEE genes, and consequently A/E lesion formation, was unchanged in the absence and presence of epinephrine, which indicated to us that these two kinases, QseC and QseE, are the only sensors of epinephrine in EHEC O157 involved in the regulation of the LEE. Interestingly, although the ΔqseC ΔqseE strain's regulatory pattern for the LEE genes is similar to that for QseE, the double mutant's pattern for motility regulation is similar to QseC. These data indicate that QseC and QseE have a complex interplay in the regulation of virulence in EHEC.
Bacteria have evolved complex systems to regulate their virulence, with numerous points of control. The first step usually involves the sensing of an environmental signal through a membrane-bound or intracellular sensor (58). The sensor in turn may in a few cases directly alter transcription of target genes or more commonly initiates a regulatory cascade that culminates in gene regulation (14, 17, 46). A multitude of sensors have been shown to be important for bacterial virulence. Enterococcus faecalis, a human enteric pathogen, has been reported to respond to self-produced pheromones through the kinase FsrC in order to differentially regulate virulence (53). The plant pathogen Agrobacterium tumefaciens uses the kinase ChvG to regulate tumorigenesis by directly or indirectly sensing extracellular acidity (39). Other examples include cis-2-dodecenoic acid sensing by Burkholderia cenocepacia BCAM0227 (43) and LAI-1 sensing by Legionella pneumophila LqsS (75).
Here we show that epinephrine sensing is very complex (Fig. 9). QseC senses AI-3, epinephrine, and norepinephrine, and then through the phosphorylation of three RRs (QseB, QseF and KdpE) is able to regulate motility, Shiga toxin production and A/E lesion formation (21). Adding another layer of complexity, QseC also activates expression of the qseEF genes (62). Regulation of motility depends exclusively on QseC, not on QseE (Fig. 8). However, in concert with QseC, QseE play a role in the regulation of the LEE. QseE senses epinephrine, phosphates, and sulfates and subsequently negatively regulates expression of the LEE, and A/E lesion formation (Fig. 3, 4, and 7). This regulation by QseE is indirect through inhibition of rcsB transcription, which is a positive regulator of the LEE and nleA (Fig. 6). Tobe et al. reported that both overexpression and deletion of rcsB led to increased transcription of the LEE in the Sakai strain of EHEC (76). We, however, show by qRT-PCR that in the ΔrcsB strain the transcription of ler, tir, eae, and espA is significantly decreased compared to the wt, and this reduction could be rescued by complementation in trans (Fig. 6). In agreement with Tobe et al., we show that overexpression of rcsB in the complemented strains increased LEE gene expression. It is also important to note that the strain we use in our research, an isolate from an EHEC O157:H7 hemorrhagic colitis outbreak (18), is different from the Sakai strain used by Tobe et al., and this may explain the disparate results. Recent work by Islam et al. and Kendall et al. has also highlighted the occurrence of differential gene regulation among different EHEC strains (24, 32).
Fig 9.
Model of the QseC and QseE regulatory cascade. Solid lines with arrows indicate confirmed positive interactions, while dotted lines indicate indirect or unconfirmed direct interactions. QseC phosphorylates QseB, which directly activates transcription of flhDC to promote expression of flagella. Through phosphorylation of KdpE, QseC activates expression of the LEE genes. QseF is phosphorylated by both QseC and QseE. QseF indirectly activates expression of espFu and Shiga toxin. QseE inhibits rcsB transcription in an as yet undetermined manner. Given that RcsB activates expression of the LEE and nleA, QseE inhibition of rcsB inhibits LEE and nleA expression. How QseC influences nleA expression is unknown. epi, epinephrine; NE, norepinephrine; AE, attaching and effacing; HUS, hemolytic-uremic syndrome.
Here we have shown how EHEC O157 has evolved to use two histidine kinases to sense hormones produced by its host in order to fine-tune the temporal and energy efficient expression of its virulence factors. This control is very complex, and better understanding of the intricacies of this signaling cascade may contribute to the development of future antivirulence therapies.
ACKNOWLEDGMENTS
We would like to acknowledge the UT Southwestern Microarray Core for support with the microarray analysis.
This work was supported by NIH grant AI053067 and by the Burroughs Wellcome Fund.
Footnotes
Published ahead of print 5 December 2011
REFERENCES
- 1. Bailey MT, Karaszewski JW, Lubach GR, Coe CL, Lyte M. 1999. In vivo adaptation of attenuated Salmonella typhimurium results in increased growth upon exposure to norepinephrine. Physiol. Behav. 67:359–364 [DOI] [PubMed] [Google Scholar]
- 2. Barba J, et al. 2005. A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J. Bacteriol. 187:7918–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bearson BL, Bearson SM. 2008. The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb. Pathog. 44:271–278 [DOI] [PubMed] [Google Scholar]
- 4. Bearson BL, Bearson SM, Lee IS, Brunelle BW. 2010. The Salmonella enterica serovar Typhimurium QseB response regulator negatively regulates bacterial motility and swine colonization in the absence of the QseC sensor kinase. Microb. Pathog. 48:214–219 [DOI] [PubMed] [Google Scholar]
- 5. Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193 [DOI] [PubMed] [Google Scholar]
- 6. Bustamante VH, Santana FJ, Calva E, Puente JL. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664–678 [DOI] [PubMed] [Google Scholar]
- 7. Campellone KG, Robbins D, Leong JM. 2004. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev. Cell 7:217–228 [DOI] [PubMed] [Google Scholar]
- 8. Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. 2006. The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U. S. A. 103:10420–10425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clarke MB, Sperandio V. 2005. Transcriptional autoregulation by quorum sensing Escherichia coli regulators B and C (QseBC) in enterohaemorrhagic E. coli (EHEC). Mol. Microbiol. 58:441–455 [DOI] [PubMed] [Google Scholar]
- 10. Clarke MB, Sperandio V. 2005. Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 57:1734–1749 [DOI] [PubMed] [Google Scholar]
- 11. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eberhard A. 1972. Inhibition and activation of bacterial luciferase synthesis. J. Bacteriol. 109:1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elliott SJ, et al. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1–4 [DOI] [PubMed] [Google Scholar]
- 14. Gao R, Stock AM. 2009. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63:133–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garmendia J, et al. 2004. TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell. Microbiol. 6:1167–1183 [DOI] [PubMed] [Google Scholar]
- 16. Gilman AG. 1987. G proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 56:615–649 [DOI] [PubMed] [Google Scholar]
- 17. Gotoh Y, et al. 2010. Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr. Opin. Microbiol. 13:232–239 [DOI] [PubMed] [Google Scholar]
- 18. Griffin PM, et al. 1988. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann. Intern. Med. 109:705–712 [DOI] [PubMed] [Google Scholar]
- 19. Gruenheid S, et al. 2004. Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 51:1233–1249 [DOI] [PubMed] [Google Scholar]
- 20. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hughes DT, Clarke MB, Yamamoto K, Rasko DA, Sperandio V. 2009. The QseC adrenergic signaling cascade in enterohemorrhagic E. coli (EHEC). PLoS Pathog. 5:e1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hughes DT, Sperandio V. 2008. Inter-kingdom signaling: communication between bacteria and host. Nat. Rev. Microbiol. 6:111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Irizarry RA, et al. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264 [DOI] [PubMed] [Google Scholar]
- 24. Islam MS, Bingle LE, Pallen MJ, Busby SJ. 2010. Organization of the LEE1 operon regulatory region of enterohaemorrhagic Escherichia coli O157:H7 and activation by GrlA. Mol. Microbiol. 79:468–483 [DOI] [PubMed] [Google Scholar]
- 25. Iyoda S, Watanabe H. 2004. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157:H7 to HEp-2 cells. Microbiology 150:2357–2571 [DOI] [PubMed] [Google Scholar]
- 26. Jarvis KG, et al. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. U. S. A. 92:7996–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jarvis KG, Kaper JB. 1996. Secretion of extracellular proteins by enterohemorrhagic Escherichia coli via a putative type III secretion system. Infect. Immun. 64:4826–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jerse AE, Kaper JB. 1991. The eae gene of enteropathogenic Escherichia coli encodes a 94-kilodalton membrane protein, the expression of which is influenced by the EAF plasmid. Infect. Immun. 59:4302–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jerse AE, Yu J, Tall BD, Kaper JB. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. U. S. A. 87:7839–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kailasan Vanaja S, Bergholz TM, Whittam TS. 2009. Characterization of the Escherichia coli O157:H7 Sakai GadE regulon. J. Bacteriol. 191:1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 32. Kendall MM, Gruber CC, Rasko DA, Hughes DT, Sperandio V. 2011. Hfq virulence regulation in enterohemorrhagic Escherichia coli O157:H7 strain 86-24. J. Bacteriol. 193:6843–6851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kenny B, et al. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511–520 [DOI] [PubMed] [Google Scholar]
- 34. Kenny B, Lai LC, Finlay BB, Donnenberg MS. 1996. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol. Microbiol. 20:313–323 [DOI] [PubMed] [Google Scholar]
- 35. Kim J, et al. 2007. The bacterial virulence factor NleA inhibits cellular protein secretion by disrupting mammalian COPII function. Cell Host Microbe 2:160–171 [DOI] [PubMed] [Google Scholar]
- 36. Knutton S, Baldwin T, Williams PH, McNeish AS. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kostakioti M, Hadjifrangiskou M, Pinkner JS, Hultgren SJ. 2009. QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli. Mol. Microbiol. 73:1020–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kresse AU, Beltrametti F, Muller A, Ebel F, Guzman CA. 2000. Characterization of SepL of enterohemorrhagic Escherichia coli. J. Bacteriol. 182:6490–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li L, et al. 2002. A global pH sensor: Agrobacterium sensor protein ChvG regulates acid-inducible genes on its two chromosomes and Ti plasmid. Proc. Natl. Acad. Sci. U. S. A. 99:12369–12374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 41. Lyte M, Arulanandam BP, Frank CD. 1996. Production of Shiga-like toxins by Escherichia coli O157:H7 can be influenced by the neuroendocrine hormone norepinephrine. J. Lab. Clin. Med. 128:392–398 [DOI] [PubMed] [Google Scholar]
- 42. Lyte M, et al. 1997. Norepinephrine-induced expression of the K99 pilus adhesin of enterotoxigenic Escherichia coli. Biochem. Biophys. Res. Commun. 232:682–686 [DOI] [PubMed] [Google Scholar]
- 43. McCarthy Y, et al. 2010. A sensor kinase recognizing the cell-cell signal BDSF (cis-2-dodecenoic acid) regulates virulence in Burkholderia cenocepacia. Mol. Microbiol. 77:1220–1236 [DOI] [PubMed] [Google Scholar]
- 44. McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 92:1664–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296–306 [DOI] [PubMed] [Google Scholar]
- 46. Mitrophanov AY, Groisman EA. 2008. Signal integration in bacterial two-component regulatory systems. Genes Dev. 22:2601–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mokrievich AN, et al. 2010. Biological properties and structure of the lipopolysaccharide of a vaccine strain of Francisella tularensis generated by inactivation of a quorum sensing system gene qseC. Biochemistry (Mosc.). 75:443–451 [DOI] [PubMed] [Google Scholar]
- 48. Moon HW, Whipp SC, Argenzio RA, Levine MM, Giannella RA. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moreira CG, Weinshenker D, Sperandio V. 2009. QseC mediates Salmonella enterica serovar Typhimurium virulence in vitro and in vivo. Infect. Immun. 78:914–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mundy R, Jenkins C, Yu J, Smith H, Frankel G. 2004. Distribution of espI among clinical enterohaemorrhagic and enteropathogenic Escherichia coli isolates. J. Med. Microbiol. 53:1145–1149 [DOI] [PubMed] [Google Scholar]
- 51. Mundy R, et al. 2004. Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect. Immun. 72:2288–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nakano M, Takahashi A, Sakai Y, Nakaya Y. 2007. Modulation of pathogenicity with norepinephrine related to the type III secretion system of Vibrio parahaemolyticus. J. Infect. Dis. 195:1353–1360 [DOI] [PubMed] [Google Scholar]
- 53. Nakayama J, et al. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41:145–154 [DOI] [PubMed] [Google Scholar]
- 54. Nealson KH. 1977. Autoinduction of bacterial luciferase. Occurrence, mechanism and significance. Arch. Microbiol. 112:73–79 [DOI] [PubMed] [Google Scholar]
- 55. Nealson KH, Platt T, Hastings JW. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104:313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Neves BC, et al. 1998. Molecular and ultrastructural characterisation of EspA from different enteropathogenic Escherichia coli serotypes. FEMS Microbiol. Lett. 169:73–80 [DOI] [PubMed] [Google Scholar]
- 57. Novak EA, Shao H, Daep CA, Demuth DR. 2010. Autoinducer-2 and QseC control biofilm formation and in vivo virulence of Aggregatibacter actinomycetemcomitans. Infect. Immun. 78:2919–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Parkinson JS, Kofoid EC. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71–112 [DOI] [PubMed] [Google Scholar]
- 59. Rasko DA, et al. 2008. Targeting QseC signaling and virulence for antibiotic development. Science 321:1078–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reading NC, Rasko D, Torres AG, Sperandio V. 2010. A transcriptome study of the QseEF two-component system and the QseG membrane protein in enterohaemorrhagic Escherichia coli O157:H7. Microbiology 156:1167–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reading NC, Rasko DA, Torres AG, Sperandio V. 2009. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 106:5889–5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reading NC, et al. 2007. A novel two-component signaling system that activates transcription of an enterohemorrhagic Escherichia coli effector involved in remodeling of host actin. J. Bacteriol. 189:2468–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Roe AJ, et al. 2007. Analysis of the expression, regulation and export of NleA-E in Escherichia coli O157:H7. Microbiology 153:1350–1360 [DOI] [PubMed] [Google Scholar]
- 64. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 65. Sanchez-SanMartin C, Bustamante VH, Calva E, Puente JL. 2001. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:2823–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shapiro JA. 1998. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52:81–104 [DOI] [PubMed] [Google Scholar]
- 67. Sharp FC, Sperandio V. 2007. QseA directly activates transcription of LEE1 in enterohemorrhagic Escherichia coli. Infect. Immun. 75:2432–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Simons RW, Houman F, Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96 [DOI] [PubMed] [Google Scholar]
- 69. Sperandio V, et al. 2000. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 38:781–793 [DOI] [PubMed] [Google Scholar]
- 70. Sperandio V, Torres AG, Giron JA, Kaper JB. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. U. S. A. 100:8951–8956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sperandio V, Torres AG, Kaper JB. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809–821 [DOI] [PubMed] [Google Scholar]
- 73. Staley TE, Jones EW, Corley LD. 1969. Attachment and penetration of Escherichia coli into intestinal epithelium of the ileum in newborn pigs. Am. J. Pathol. 56:371–392 [PMC free article] [PubMed] [Google Scholar]
- 74. Thanabalasuriar A, et al. 2009. The bacterial virulence factor NleA is required for the disruption of intestinal tight junctions by enteropathogenic Escherichia coli. Cell. Microbiol. 12:31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tiaden A, et al. 2010. The autoinducer synthase LqsA and putative sensor kinase LqsS regulate phagocyte interactions, extracellular filaments and a genomic island of Legionella pneumophila. Environ. Microbiol. 12:1243–1259 [DOI] [PubMed] [Google Scholar]
- 76. Tobe T, et al. 2005. Dual regulatory pathways integrating the RcsC-RcsD-RcsB signalling system control enterohaemorrhagic Escherichia coli pathogenicity. Mol. Microbiol. 58:320–333 [DOI] [PubMed] [Google Scholar]
- 77. Tobe T, et al. 2006. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. U. S. A. 103:14941–14946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Walters M, Sperandio V. 2006. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect. Immun. 74:5445–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang X, et al. 2011. QseBC controls flagellar motility, fimbrial hemagglutination and intracellular virulence in fish pathogen Edwardsiella tarda. Fish Shellfish Immunol. 30:944–953 [DOI] [PubMed] [Google Scholar]
- 80. Yamamoto K, et al. 2005. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 280:1448–1456 [DOI] [PubMed] [Google Scholar]