Abstract
Retinoic acid-inducible gene I (RIG-I) is a key sensor for viral RNA in the cytosol, and it initiates a signaling cascade that leads to the establishment of an interferon (IFN)-mediated antiviral state. Because of its integral role in immune signaling, RIG-I activity must be precisely controlled. Recent studies have shown that RIG-I CARD-dependent signaling function is regulated by the dynamic balance between phosphorylation and TRIM25-induced K63-linked ubiquitination. While ubiquitination of RIG-I is critical for RIG-I's ability to induce an antiviral IFN response, phosphorylation of RIG-I at S8 or T170 suppresses RIG-I signal-transducing activity under normal conditions. Here, we not only further define the roles of S8 and T170 phosphorylation for controlling RIG-I activity but also identify conventional protein kinase C-α (PKC-α) and PKC-β as important negative regulators of the RIG-I signaling pathway. Mutational analysis indicated that while the phosphorylation of S8 or T170 potently inhibits RIG-I downstream signaling, the dephosphorylation of RIG-I at both residues is necessary for optimal TRIM25 binding and ubiquitination-mediated RIG-I activation. Furthermore, exogenous expression, gene silencing, and specific inhibitor treatment demonstrated that PKC-α/β are the primary kinases responsible for RIG-I S8 and T170 phosphorylation. Coimmunoprecipitation showed that PKC-α/β interact with RIG-I under normal conditions, leading to its phosphorylation, which suppresses TRIM25 binding, RIG-I CARD ubiquitination, and thereby RIG-I-mediated IFN induction. PKC-α/β double-knockdown cells exhibited markedly decreased S8/T170 phosphorylation levels of RIG-I and resistance to infection by vesicular stomatitis virus. Thus, these findings demonstrate that PKC-α/β-induced RIG-I phosphorylation is a critical regulatory mechanism for controlling RIG-I antiviral signal transduction under normal conditions.
INTRODUCTION
The rapid detection of invading viruses by pattern recognition receptors (PRRs) and the subsequent induction of type I interferons (IFN-α/β) is the key to a successful innate immune response to viral infections. For antiviral IFN responses, hosts have evolved at least two main classes of PRRs that sense nucleic acids or other conserved molecular components of viral pathogens: Toll-like receptors (TLRs) and retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) (15, 21). While TLRs play a major role in the detection of incoming virions in specialized immune cells, viral RNA sensing in the cytosol of most cells is carried out by RIG-I and melanoma differentiation-associated gene 5 (MDA5) (1, 29, 31). RIG-I and MDA5 are composed of two N-terminal caspase recruitment domains (CARDs), a central DExD/H box ATPase/helicase, and a C-terminal regulatory domain (RD) (3, 27, 31). The binding of viral RNA to the RD/helicase results in a conformational change that demasks the N-terminal CARDs. The exposed CARDs of RIG-I and MDA5 then interact with the CARD-containing adaptor protein MAVS/VISA/IPS-1/Cardif to trigger downstream signaling, resulting in type I IFN production (16, 20, 28, 30). While RIG-I recognizes the 5′triphosphate-containing RNA of paramyxoviruses, influenza virus, and vesicular stomatitis virus (VSV) as well as that of hepatitis C virus (HCV), MDA5 is a key sensor of picornaviruses (14, 19, 24). In addition, MDA5 was recently shown to play a critical role in the innate immune response to paramyxovirus infections in vivo (9).
The tight regulation of innate immune sensing and the initiation of antiviral signaling is crucial for eliciting an effective immune response. Whereas positive regulatory loops lead to the rapid induction of IFNs and proinflammatory cytokines upon viral infection, multiple negative regulatory checkpoints must be in place to prevent unwanted or excessive cytokine production and autoimmune reactions. A recent series of studies has identified ubiquitination as an important cellular mechanism for regulating or fine-tuning RIG-I signal-transducing ability. RIG-I activity is negatively regulated by K48-linked ubiquitination, leading to its proteasomal degradation (2). Furthermore, the K63-linked ubiquitination of the N-terminal CARDs of RIG-I as well as its C-terminal region is critical for RIG-I's ability to initiate antiviral IFN responses (8, 23). Specifically, the ubiquitination of RIG-I at K172 induced by tripartite motif 25 (TRIM25) is essential for efficient RIG-I-MAVS interaction and for RIG-I's ability to elicit host surveillance against RNA virus infections (8). The necessity of TRIM25-mediated ubiquitination for RIG-I signaling was evidenced by a RIG-I splice variant which is unable to bind TRIM25 and thereby completely loses CARD ubiquitination and antiviral signaling ability (6). Furthermore, a recent study showed that TRIM25 can induce RIG-I signaling in an in vitro-reconstituted cell-free system (32). In addition to ubiquitination, we have recently shown that serine/threonine phosphorylation of the RIG-I CARDs represents an important mechanism to regulate RIG-I antiviral activity (7, 22). The RIG-I CARDs undergo phosphorylation at S8 and T170 under normal conditions, which suppresses RIG-I downstream signaling by inhibiting RIG-I-TRIM25 binding and thereby TRIM25-mediated RIG-I ubiquitination.
Despite the wealth of knowledge resulting from recent studies on RIG-I regulation through ubiquitination, the precise molecular mechanisms by which phosphorylation modulates RIG-I antiviral activity remain elusive. Here, we further characterize the role of S8 and T170 phosphorylation for regulating RIG-I downstream signaling and identify conventional protein kinase C-α (PKC-α) and PKC-β as important negative regulators of the RIG-I-mediated type I IFN response.
MATERIALS AND METHODS
Plasmid construction.
GST-RIG 2CARD, pIRES-RIG-I-2CARD-Flag, pIRES-RIG-I-Δ2CARD-Flag, pIRES-MAVS-CARD-PRD-Flag, and pIRES-TRIM25-V5 were previously described (8). GST-RIG-I 2CARD and RIG-I full-length mutants were generated by PCR using site-directed mutagenesis or overlapping PCR using GST-RIG-I 2CARD or pEF-Bos-Flag-RIG-I (provided by James Chen, University of Texas), respectively, as the template. Introduced mutations were confirmed by DNA sequence analysis. Myc-tagged PKC isozymes and PKG were subcloned into pEF-IRES-Puro encoding a C-terminal Myc tag between AflII and NotI. RIG-I helicase (amino acids [aa] 201 to 734) or RIG-I-RD (aa 735 to 925) was subcloned into pEF-IRES-Puro containing a C-terminal Flag tag. DNA fragments corresponding to the coding sequence of PKC-α and PKC-βII genes were amplified from template DNA by PCR and subcloned into plasmid pEBG between KpnI and NotI. Flag-tagged PKA was provided by Jiuyong Xie, University of Manitoba. The construct for the pcDNA-HA-PKB T308D S473D constitutively active mutant was a gift from Ellen Cahir-McFarland (Harvard). The plasmids encoding green fluorescent protein (GFP)-PKC-α and GFP-PKC-βII were kindly provided by Yusuf Hannun, Medical University of South Carolina. The pSUPER-retro vectors encoding human PKC-α small hairpin RNA (shRNA) or nonsilencing control shRNA were provided by Debra Tonetti (University of Illinois at Chicago) and have been previously described (18).
Cell culture and transfection.
HEK293T, A549, HeLa, Vero, NHLFs (normal human lung fibroblasts), and HPdlFs (human periodontal ligament fibroblasts) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and 1% penicillin-streptomycin (Gibco-BRL). Transient transfections were performed with calcium phosphate (Clontech), FuGENE 6 (Roche), or Lipofectamine 2000 (Invitrogen) according to the manufacturers' instructions. For shRNA transfection, Arrest-In transfection reagent (Open Biosystems) was used. To obtain stable HEK293T cells, cells were transfected with pIRES-puro-vector, pIRES-puro-PKC-α-Myc, pIRES-puro-PKC-βII-Myc, or pIRES-puro-PKC-ζ-Myc, followed by selection using 2 μg/ml of puromycin.
Knockdown of PKC-α and PKC-β using shRNA or siRNA.
For the transient knockdown of endogenous PKC-α and/or PKC-β, HEK293T cells were transfected with retroviral pSM2 encoding human PKC-α-specific shRNA and/or pSM2 encoding PKC-β-specific shRNA (Open Biosystems). As a control, retroviral pSM2 encoding nonsilencing control shRNA was transfected. To generate HEK293T cells in which PKC-α or PKC-β is stably silenced, cells were transfected with retroviral pSM2-PKC-α-specific shRNA or pSM2-PKC-β-specific shRNA, followed by selection using 2 μg/ml of puromycin. To generate PKC-α/PKC-β double-knockdown cells, HEK293T cells were transfected with retroviral pSM2-PKC-β-shRNA (Open Biosystems) together with pSUPER-retro-PKC-α-shRNA (provided by Debra Tonetti, University of Illinois at Chicago), followed by selection using 2 μg/ml puromycin and 400 μg/ml G418. As controls, stable HEK293T cells expressing retroviral pSM2 or pSUPER encoding nonsilencing control shRNA were generated. The transient knockdown of endogenous PKC-α and PKC-β in NHLF cells was achieved by the transfection of siGenome SMARTpool short interfering RNA (siRNA) specific for PKC-α and PKC-β (Dharmacon) with Lipofectamine and Plus reagent (Invitrogen) according to the manufacturer's instructions. A final concentration of 300 nM each PKC-α- and PKC-β-specific SMARTpool siRNA or 600 nM siGenome nontargeting siRNA (Dharmacon) was used.
Viruses.
VSV-enhanced GFP (VSV-eGFP) was provided by Sean Whelan (Harvard). Newcastle disease virus (NDV) expressing GFP (NDV-GFP) and ΔNS1 recombinant A/PR/8/34 (H1N1) influenza virus (ΔNS1) were gifts from Adolfo García-Sastre (Mount Sinai School of Medicine). Sendai virus (SeV; Cantell strain) was obtained from Charles River Laboratories.
Antibodies and reagents.
For immunoblotting, the following primary antibodies were used: anti-Myc (1:2,000) (Covance), anti-V5 (1:5,000) (Invitrogen), anti-Flag (M2; 1:2,000) (Sigma), anti-hemagglutinin (HA) (1:2,000) (clone HA-7; Sigma), anti-glutathione S-transferase (anti-GST) (1:2,000) (Sigma), anti-ubiquitin (P4D1; 1:500) (Santa Cruz), anti-polyubiquitin (Lys63 linkage specific) (1:200) (Biomol), monoclonal anti-TRIM25 (1:2,000) (BD Biosciences), monoclonal anti-RIG-I (Alme-1; 1:1,000) (Alexis), anti-IRF3 (1:1,000) (Santa Cruz), anti-PKC-α (clone M4; 1:1,000) (Millipore), anti-PKC-βI (1:1,000) (Santa Cruz), anti-PKC-βII (1:1,000) (Santa Cruz), anti-PKC-βII (1:200) (Abcam), anti-phospho-S657-PKC-α (1:500) (Millipore), anti-phospho-T500 PKC-β I&II (1:500) (Upstate), anti-PKC-ε (1:1,000) (Millipore), anti-PKC-ζ (1:1,000) (Millipore), anti-β-actin (Abcam), and anti-phosphothreonine (pThr; 1:500) (Cell Signaling Technology). The phospho-specific pS8-RIG-I and pT170-RIG-I antibodies were generated by immunizing rabbits with phosphopeptides and have been previously described (7, 22). Protein kinase C inhibitors bisindolylmaleimide I (BIM I), Gö6976, and Ro-32-0432 were purchased from Calbiochem. Poly(U/UC)-RNA of HCV strain J4L6 was a gift from Lee Gehrke (Harvard/M.I.T.).
GST pulldown assay, immunoprecipitation, and immunoblot analysis.
HEK293T cells were lysed in NP-40 buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1% [vol/vol] NP-40, protease inhibitor cocktail [Roche], and Ser/Thr phosphatase inhibitor cocktail [Sigma]), followed by centrifugation at 13,000 rpm for 20 min. GST pulldown, immunoprecipitation, and Western blot analysis were performed as previously described (7, 8).
VSV-eGFP replication and NDV-GFP bioassay.
Stable HEK293T cells or NHLFs were seeded into 6-well plates and infected with VSV-eGFP at the indicated multiplicity of infection (MOI). At 24 to 48 h postinfection, the culture medium was harvested and the virus yield determined by standard plaque assay on Vero cells. For the NDV-GFP bioassay, supernatants from transfected HEK293T cells were diluted 1:2 in fresh DMEM complete medium and then added to Vero cells. Eighteen hours later, cells were infected with NDV-GFP (MOI of 3). At 24 h postinfection, GFP expression was monitored by epifluorescence, and the percentage of GFP-positive cells was determined by fluorescence-activated cell sorter (FACS) analysis.
IFN-β ELISA.
NHLFs that had been transfected with siGenome nontargeting siRNA or with PKC-α- and PKC-β-specific siRNAs were mock treated or infected with SeV (40 HA units/ml) at 40 h after transfection. Thirty hours later, the supernatants were collected and analyzed for IFN-β production by using enzyme-linked immunosorbent assay (ELISA; PBL Biomedical Laboratories).
RNA pulldown experiments.
Biotinylated 5′-triphosphate containing rabies virus leader RNA (5′pppRVL) was transcribed using the T7 Megashortscript kit (Ambion) and biotin-16-uridine-5′ppp (Roche). For transcription, the annealed oligonucleotides 5′CGCGTAATACGACTCACTATA 3′ and 5′ACA TTT TTG CTT TGC AAT TGA CAA TGT CTG TTT TTT CTT TGA TCT GGT TGT TAA GCG TTA TAG TGA GTC GTA TTA CGC G 3′ were used as the template. The DNA template was removed by DNase I treatment, and RNA was purified using Microspin G-25 Columns (GE Healthcare). For the RNA binding assay, 1 μg of biotinylated RNA was incubated for 1 h at 25°C with 25 μg of cell extract prepared from HEK293T cells transfected with pEF-Bos-Flag-RIG-I plasmids. Following incubation, the mixture was transferred into 400 μl of wash buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40) containing 30 μl streptavidin agarose affinity gel (Sigma) and rocked at 4°C for 2 h. The RNA-protein complexes were collected by centrifugation and washed three times with wash buffer, followed by SDS-PAGE and immunoblotting with anti-Flag antibody.
Protein purification and in vitro phosphorylation assay.
RIG-I 2CARD was cloned into pGEX-4T-1 vector. Escherichia coli XL1-Blue cells were transformed with the plasmid, and RIG-I 2CARD fusion protein was purified using glutathione Sepharose 4B resin (GE Healthcare) according to the manufacturer's instructions. For the in vitro phosphorylation assay, purified GST-RIG-I 2CARD was incubated with 25 ng of purified PKC-α/β/γ (Millipore) in PKC assay dilution buffer II (Millipore) at 30°C for 30 min. The reaction was stopped by adding 2× SDS-Laemmli buffer followed by SDS-PAGE. RIG-I phosphorylation was detected by immunoblotting with phosphospecific pS8- or pT170-RIG-I antibody.
Luciferase reporter assay.
HEK293T cells were seeded into 6-well plates. Twenty-four hours later, the cells were transfected with 500 ng IFN-β luciferase reporter plasmid together with 800 ng constitutive β-galactosidase (β-gal)-expressing pGK-β-gal. In addition, 2 ng of plasmid encoding RIG-I 2CARD-GST fusions was transfected. At 36 to 40 h posttransfection, WCLs were prepared and subjected to a luciferase assay (Promega). Luciferase values were normalized to β-galactosidase to measure the transfection efficiency. To test the effect of PKC inhibitors on SeV-induced IFN-β promoter activation, HEK293T cells transfected with IFN-β luciferase construct and constitutive β-gal-expressing plasmid pGK-β-gal were treated with PKC inhibitors (BIM I, 4 μM; Gö6976, 10 μM; and Ro-32-0432, 12 μM) at 20 h posttransfection. Six hours later, cells were infected with SeV (30 HA units/ml). At 18 to 22 h postinfection, WCLs were prepared and subjected to a luciferase assay (Promega).
Bioinformatic analysis of RIG-I Ser-8 and Thr-170.
Potential protein kinase phosphorylation consensus sites for RIG-I Ser-8 and Thr-170 residues were determined by using EMBL-EBI and Uniprot/GPS2.1 software, using the settings “high threshold” and “low threshold” for Ser-8 and Thr-170, respectively. This identified the motif QRRS8LQ as a putative PKA or PKC consensus site, whereas the motif WPKT170LK was identified as a potential PKC site.
Native PAGE.
Native PAGE was performed as described previously (6).
Confocal immunofluorescence microscopy.
HeLa cells grown on chamber slides were transfected with pBos-Flag-RIG-I and GFP-PKC-α or GFP-PKC-βII. At 20 h posttransfection, cells were fixed with 4% paraformaldehyde for 20 min and permeabilized with 0.2% (vol/vol) Triton. Cell preparation and confocal microscopy analysis were performed as previously described (7, 8). For immunostaining Flag-RIG-I, anti-Flag M2 antibody (Sigma) was used, followed by incubation with anti-mouse Alexa fluor 594 (Invitrogen). Laser-scanning images were taken on a Leica TCS SP5 confocal microscope (Leica Microsystems).
RESULTS
Dephosphorylation of RIG-I at both S8 and T170 is necessary for efficient TRIM25-mediated RIG-I ubiquitination and optimal RIG-I signaling activity.
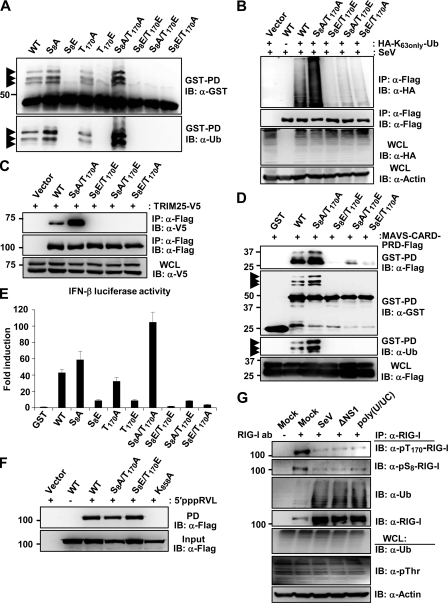
The mutation of either S8 or T170 to phosphomimetic D or E (aspartic/glutamic acid), but not to A (alanine), strongly suppressed TRIM25-mediated RIG-I ubiquitination and antiviral signaling, indicating that RIG-I phosphorylation and TRIM25-mediated RIG-I ubiquitination functionally antagonize each other (7, 22). To define the distinct roles of S8 and T170 phosphorylation in RIG-I regulation, we generated additional point mutants of the RIG-I N-terminal CARDs fused to mammalian glutathione S-transferase (GST-RIG-I 2CARD), as well as of full-length RIG-I: the S8E/T170E and S8A/T170A mutants mimicking constitutive phosphorylation or nonphosphorylation at both residues, and the S8A/T170E and S8E/T170A mutants at which one site is nonphosphorylated while the other mimics constitutive phosphorylation. These RIG-I mutants then were tested for a series of biochemical activities: CARD ubiquitination, TRIM25 and MAVS binding, RNA binding, and downstream signaling activity. Like GST-RIG-I 2CARD S8E and T170E mutants, GST-RIG-I 2CARD S8A/T170E, S8E/T170A and S8E/T170E showed a near-complete loss of ubiquitination compared to that of GST-RIG-I 2CARD wild type (WT) (Fig. 1A). In striking contrast, the GST-RIG-I 2CARD S8A/T170A mutant, mimicking nonphosphorylation at both sites, exhibited a markedly increased level of ubiquitination compared to that of WT GST-RIG-I 2CARD, while T170A and S8A showed a similar or slightly enhanced ubiquitination level compared to that of the WT (Fig. 1A). In line with this, Sendai virus (SeV)-induced K63-linked ubiquitination of the RIG-I S8A/T170A mutant was strongly enhanced compared to that of WT RIG-I, whereas S8A/T170E, S8E/T170A and S8E/T170E RIG-I mutants were minimally ubiquitinated (Fig. 1B). Given that S8E or T170E mutation strongly suppressed RIG-I binding to TRIM25 (7, 22), which apparently abolished RIG-I ubiquitination, we tested Flag-tagged RIG-I S8A/T170E, S8E/T170A, S8A/T170A, and S8E/T170E mutants for their TRIM25 binding abilities (Fig. 1C). RIG-I S8A/T170E, S8E/T170A, and S8E/T170E mutants showed no detectable interaction with V5-TRIM25; in contrast, the S8A/T170A mutant exhibited an increased binding activity for TRIM25 compared to that of WT RIG-I (Fig. 1C). K63-linked ubiquitination of the RIG-I CARDs is essential for efficient RIG-I-MAVS interaction and for RIG-I's ability to induce downstream signal transduction (8). GST-RIG-I 2CARD S8A/T170A, which exhibited elevated ubiquitination levels compared to that of the WT, also showed a stronger binding to the MAVS-CARD-proline-rich domain (PRD) and signal-transducing activity than the GST-RIG-I 2CARD WT (Fig. 1D and E). Furthermore, GST-RIG-I 2CARD S8E/T170E did not bind the MAVS-CARD under the same conditions and did not detectably induce IFN-β promoter activation, whereas S8A/T170E and S8E/T170A mutants showed minimal MAVS binding and IFN-inducing activities (Fig. 1D and E). The inability of the RIG-I S8E/T170E mutant to induce IFN-β promoter activation was not due to an abolished RNA binding activity, since S8E/T170E RIG-I bound in vitro-transcribed 5′triphosphate rabies virus leader RNA (5′pppRVL) with affinity similar to that of the WT or S8A/T170A RIG-I (Fig. 1F). In contrast, a RIG-I mutant in which K858, a critical residue for viral RNA binding to the C-terminal RD (3), is mutated (K858A RIG-I) did not bind 5′pppRVL under the same conditions (Fig. 1F). Finally, we tested the phosphorylation of endogenous RIG-I at S8 and T170 under normal conditions, following viral infection, or upon stimulation with viral RNA by using a phosphospecific pS8- or pT170-RIG-I antibody (Fig. 1G). This showed that RIG-I underwent robust phosphorylation at S8 and T170 in mock-treated cells, and that phosphorylation at both sites declined following infection with SeV or ΔNS1 PR8 influenza virus (ΔNS1), as well as upon stimulation with HCV poly(U/UC) RNA (Fig. 1G). In summary, these results indicate that RIG-I is robustly phosphorylated at S8 and T170 under normal conditions, and that the phosphorylation of either S8 or T170 is sufficient to potently suppress RIG-I ubiquitination by TRIM25, thereby preventing RIG-I downstream signaling. Furthermore, our results indicate that the stimulus-induced dephosphorylation of RIG-I at both residues is necessary for optimal RIG-I-TRIM25 binding, RIG-I CARD ubiquitination, and RIG-I-mediated signal transduction.
Fig 1.
Dephosphorylation of RIG-I at both S8 and T170 is necessary for optimal TRIM25 binding, K63-linked ubiquitination, and signaling activity of RIG-I. (A and B) S8A/T170A mutation enhances RIG-I ubiquitination. (A) HEK293T cells were transfected with GST-RIG-I 2CARD WT or the indicated mutants. Whole-cell lysates (WCLs) were subjected to GST pulldown (GST-PD) followed by immunoblotting (IB) with anti-GST or anti-ubiquitin (Ub) antibody. Arrows indicate the ubiquitinated bands. (B) HEK293T cells transfected with vector, Flag-RIG-I WT, or mutants together with an HA-Ub mutant in which all lysines except K63 are mutated (HA-K63only-Ub) were infected with SeV (50 HA units/ml) for 14 h. WCLs were subjected to IP with anti-Flag, followed by IB with anti-HA or anti-Flag antibody. (C) S8A/T170A mutation enhances RIG-I binding to TRIM25. At 48 h posttransfection with vector, Flag-RIG-I WT, or the indicated mutants together with V5-tagged TRIM25, HEK293T WCLs were used for IP with anti-Flag antibody, followed by IB with anti-V5 antibody. TRIM25 expression was determined by IB with an anti-V5 antibody. (D) S8A/T170A mutation increases RIG-I CARD binding to MAVS. At 48 h posttransfection with GST, GST-RIG-I 2CARD WT, or the indicated mutants together with Flag-tagged MAVS-CARD-PRD, HEK293T WCLs were used for GST-PD followed by IB with anti-Flag, anti-GST, or anti-Ub antibody. MAVS-CARD-PRD expression was determined by IB with an anti-Flag antibody. Arrows indicate the ubiquitinated bands. (E) S8A/T170A mutation increases RIG-I 2CARD-mediated IFN-β promoter activation. GST or GST-RIG-I fusion constructs together with IFN-β luciferase and the constitutive β-gal-expressing plasmid pGK-β-gal were expressed in HEK293T cells. Luciferase and β-galactosidase values were determined as previously described (8). Data represent the means ± standard deviations (SD) (n = 3). (F) RIG-I phosphorylation at S8 and T170 does not affect its RNA binding ability. Cell extracts from HEK293T cells transfected with vector, Flag-RIG-I WT, or the indicated mutants were incubated with in vitro-transcribed, biotinylated 5′-triphosphate containing rabies virus leader RNA (5′pppRVL). RNA-protein complexes were recovered by pulldown assay using streptavidin affinity gel, followed by SDS-PAGE and immunoblotting using an anti-Flag antibody. (G) Viral infection or RIG-I stimulation with viral RNA decreases the S8 and T170 phosphorylation of endogenous RIG-I. HEK293T cells were infected with SeV (25 HA units/ml) or ΔNS1 PR8 virus (MOI, 4) for 10 h or were transfected with HCV poly(U/UC). WCLs were subjected to IP with an anti-RIG-I antibody, followed by IB with anti-pT170-RIG-I, anti-pS8-RIG-I, anti-Ub, or anti-RIG-I antibody.
PKC-α/β phosphorylate the S8 and T170 residues of RIG-I, thereby suppressing RIG-I K63-linked ubiquitination.
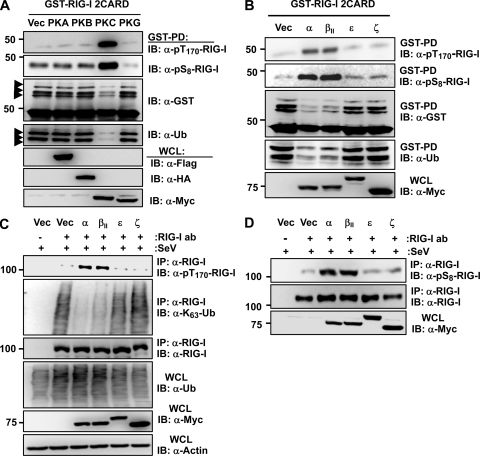
Bioinformatic analysis revealed that the S8 and T170 residues of RIG-I are potential consensus phosphorylation sites for protein kinase A (PKA) and protein kinase C (PKC), respectively. To test the potential role of PKA and PKC in phosphorylation-dependent RIG-I regulation, we examined the effect of the exogenous expression of PKA or PKC on the phosphorylation and ubiquitination of GST-RIG-I 2CARD (Fig. 2A). Protein kinase B (PKB) and protein kinase G (PKG) were included in this assay. PKC expression markedly increased the S8 and T170 phosphorylation of GST-RIG-I 2CARD and suppressed its ubiquitination, whereas PKA, PKB, and PKG showed no effect under the same conditions (Fig. 2A). PKC represents a family of serine/threonine kinases comprising at least 11 isozymes that are classified into three subfamilies based on structural similarities and cofactor dependence: conventional PKCs (α, βI, βII, and γ), novel PKCs (δ, ε, η, and θ), and atypical PKCs (λ and ζ) (25). We thus tested the effect of the ectopic expression of conventional isozyme PKC-α or PKC-βII, novel PKC-ε, or atypical PKC-ζ on the phosphorylation and ubiquitination of GST-RIG-I 2CARD (Fig. 2B). Specifically, the coexpression of PKC-α or PKC-βII enhanced the S8 and T170 phosphorylation of GST-RIG-I 2CARD and suppressed its ubiquitination, whereas PKC-ε and PKC-ζ did not have any effect (Fig. 2B). Accordingly, exogenous PKC-α and PKC-βII, but not PKC-ε or PKC-ζ, strongly increased the phosphorylation of endogenous RIG-I at S8 and T170 and suppressed its K63-linked polyubiquitination (Fig. 2C and D).
Fig 2.
Ectopic PKC-α/β expression enhances RIG-I phosphorylation at S8 and T170, thereby suppressing its K63-linked ubiquitination. (A and B) Exogenous expression of PKC-α or PKC-βII increases the S8 and T170 phosphorylation of RIG-I 2CARD and inhibits its ubiquitination. At 48 h posttransfection with GST-RIG-I 2CARD together with vector (Vec), Flag-tagged PKA, HA-tagged PKB, Myc-tagged PKC-α, or Myc-tagged PKG (A) or with vector, Myc-tagged PKC-α, PKC-βII, PKC-ε, or PKC-ζ (B), WCLs were used for GST-PD followed by IB with anti-pT170-RIG-I, anti-pS8-RIG-I, anti-GST, or anti-Ub antibody (ab). Arrows indicate the ubiquitinated bands. (C) Ectopic expression of PKC-α/β increases the phosphorylation of endogenous RIG-I at T170 and suppresses its K63-linked ubiquitination. HEK293T cells transfected with vector or the indicated Myc-tagged PKC isozymes were infected with SeV (50 HA units/ml) for 6 h. WCLs were used for IP with an anti-RIG-I antibody, followed by IB with anti-pT170-RIG-I, anti-K63-Ub, or anti-RIG-I antibody. (D) Ectopic expression of PKC-α or PKC-β increases the S8 phosphorylation of endogenous RIG-I. HEK293T cells were transfected with vector, Myc-tagged PKC-α, PKC-βII, PKC-ε, or PKC-ζ, followed by infection with SeV (50 HA units/ml) for 6 h. At 48 h posttransfection, WCLs were used for IP with an anti-RIG-I antibody, followed by IB with anti-pS8-RIG-I or anti-RIG-I antibody. The expression of Myc-tagged PKC isozymes was determined in the WCLs by IB with an anti-Myc antibody.
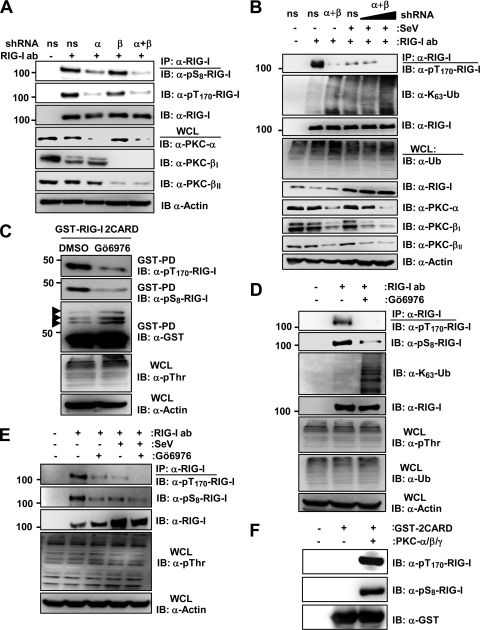
To test the physiological relevance of conventional PKC-α and PKC-β for RIG-I phosphorylation, we examined the phosphorylation of endogenous RIG-I in HEK293T cells, in which PKC-α, PKC-β, or both were stably silenced by using PKC-α- and/or PKC-β-specific small hairpin RNAs (shRNAs) (Fig. 3A). Cells stably expressing a nonsilencing, scrambled shRNA served as the control. This showed that the stable knockdown of PKC-β had little or no effect on endogenous RIG-I S8 and T170 phosphorylation. In contrast, the silencing of PKC-α or both PKC-α and PKC-β markedly reduced the RIG-I phosphorylations compared to those of cells expressing nonsilencing shRNA, with the PKC-α/β double knockdown having the strongest effect (Fig. 3A). Consistent with this finding, the transient knockdown of endogenous PKC-α and PKC-β strongly decreased the T170 phosphorylation of endogenous RIG-I and enhanced its K63-linked ubiquitination in mock-treated cells as well as in SeV-infected cells in a dose-dependent manner (Fig. 3B). Conventional Gö6976 PKC inhibitor treatment also reduced the S8 and T170 phosphorylation of GST-RIG-I 2CARD and increased its ubiquitination (Fig. 3C). In addition, Gö6976 PKC inhibitor treatment profoundly decreased the S8 and T170 phosphorylation of endogenous RIG-I in HEK293T cells as well as primary NHLFs and HPdlFs compared to that of mock-treated cells (Fig. 3D and E and data not shown). In correlation with its decreasing effect on RIG-I phosphorylation, the Gö6976 inhibitor increased the RIG-I K63-linked ubiquitination (Fig. 3D). Finally, conventional PKC robustly phosphorylated bacterially purified GST-RIG-I 2CARD at S8 and T170 in an in vitro phosphorylation assay (Fig. 3F). These results collectively indicate that conventional PKC-α and PKC-β phosphorylate RIG-I at S8 and T170, which inhibits the K63-linked ubiquitination of RIG-I.
Fig 3.
shRNA-mediated depletion or inhibition of PKC-α/β using specific inhibitors decreases RIG-I S8 and T170 phosphorylations and enhances the RIG-I K63-linked ubiquitination. (A) shRNA-mediated knockdown of endogenous PKC-α and PKC-β decreases RIG-I phosphorylation at S8 and T170. WCLs of HEK293T cells stably expressing pSM2-nonsilencing (ns) shRNA, pSM2-PKC-α-shRNA, pSM2-PKC-β-shRNA, or pSUPER-retro-PKC-α-shRNA and pSM2-PKC-β-shRNA were used for IP with an anti-RIG-I antibody, followed by IB with anti-pS8-RIG-I, anti-pT170-RIG-I, or anti-RIG-I antibody. Endogenous PKC-α, PKC-βI, and PKC-βII expression was determined in the WCLs by IB with anti-PKC-α, anti-PKC-βI, or anti-PKC-βII antibody. The loading control was determined by using an anti-actin antibody. (B) shRNA-mediated knockdown of PKC-α and PKC-β decreases RIG-I phosphorylation, thereby enhancing RIG-I K63-linked ubiquitination. At 36 h posttransfection with nonsilencing (ns) shRNA or with PKC-α- and PKC-β-specific shRNAs, HEK293T cells were either mock treated or infected with SeV for 8 h. WCLs were used for IP with anti-RIG-I, followed by IB anti-pT170-RIG-I, anti-K63-Ub, or anti-RIG-I antibody. (C) Conventional PKC inhibitor Gö6976 decreases the S8 and T170 phosphorylation of RIG-I 2CARD. HEK293T cells transfected with GST-RIG-I 2CARD were treated with DMSO or 10 μM Gö6976 for 16 h. WCLs were subjected to GST-PD, followed by IB with anti-pT170-RIG-I, anti-pS8-RIG-I, or anti-GST antibody. Arrows indicate the ubiquitinated bands. (D) Conventional PKC inhibitor Gö6976 decreases the S8 and T170 phosphorylation of endogenous RIG-I and enhances its K63-linked ubiquitination. Primary NHLF cells were treated with DMSO or 10 μM Gö6976 for 16 h. WCLs were used for IP with an anti-RIG-I antibody, followed by IB with anti-pT170-RIG-I, anti-pS8-RIG-I, anti-K63-Ub, or anti-RIG-I antibody. WCLs were further used for immunoblotting with anti-pThr, anti-Ub, or anti-Actin antibody. (E) PKC inhibitor Gö6976 decreases the S8 and T170 phosphorylation of endogenous RIG-I in mock-treated and SeV-infected cells. HEK293T cells were treated with DMSO or 10 μM Gö6976 for 12 h, followed by mock treatment or infection with SeV (50 HA units/ml) for 10 h. WCLs were used for IP with an anti-RIG-I antibody, followed by IB with anti-pT170-RIG-I, anti-pS8-RIG-I, or anti-RIG-I antibody. (F) In vitro phosphorylation of RIG-I 2CARD by conventional PKC. Bacterially purified GST-RIG-I 2CARD was subjected to an in vitro phosphorylation assay using purified PKC-α/β/γ, followed by IB with anti-pT170-RIG-I, anti-pS8-RIG-I, or anti-GST antibody.
PKC-α/β interact with RIG-I.
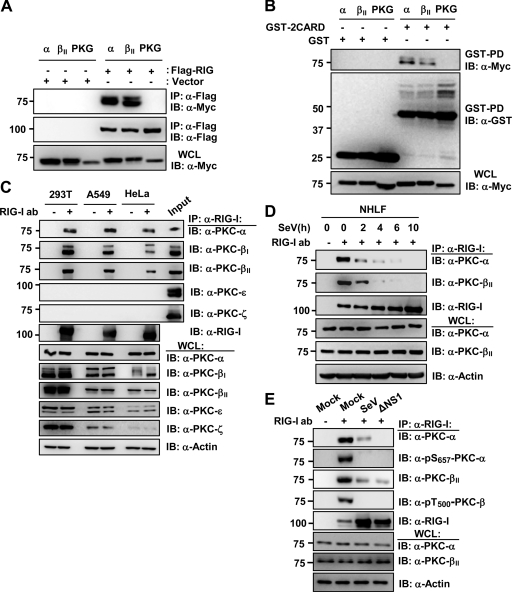
We next tested the potential interaction between Myc-tagged PKC-α or PKC-βII and Flag-tagged RIG-I or GST-RIG-I 2CARD in HEK293T cells (Fig. 4A and B). PKG-Myc was included as a control. Coimmunoprecipitation (co-IP) revealed that PKC-α and PKC-βII, but not PKG, efficiently interacted with Flag-RIG-I and, albeit more weakly, with GST-RIG-I 2CARD (Fig. 4A and B). In addition, we readily observed an interaction of endogenous PKC-α, PKC-βI, and PKC-βII with RIG-I in HEK293T, A549, and HeLa cells (Fig. 4C). In contrast, PKC-ε and PKC-ζ did not bind endogenous RIG-I under the same conditions (Fig. 4C). Confocal microscopy also showed that GFP-PKC-α or GFP-PKC-βII extensively colocalized with Flag-RIG-I in the cytoplasm (data not shown). Furthermore, we examined the interaction of endogenous PKC-α or PKC-βII with RIG-I in primary NHLFs under normal conditions and at different time points after SeV infection (Fig. 4D). Consistent with the results for HEK293T, A549, and HeLa cell lines, PKC-α and PKC-βII efficiently interacted with endogenous RIG-I in mock-infected NHLFs. In contrast, viral infection rapidly led to a strongly decreased binding of PKC-α or PKC-βII to RIG-I (Fig. 4D). In line with this, while PKC-α and PKC-βII strongly bound to endogenous RIG-I in HEK293T cells under normal conditions, only a very weak RIG-I-PKC-α and RIG-I-PKC-βII interaction was observed upon SeV or ΔNS1 influenza virus infection (Fig. 4E). The activation loop phosphorylation of PKC-α at S657 and PKC-β at T500 has been shown to render PKC-α/β catalytically competent (10). As shown in Fig. 4E, endogenous PKC-α and PKC-βII that bound to RIG-I were robustly phosphorylated at S657 and T500, respectively, in noninfected cells; in contrast, no phosphorylation of PKC-α or PKC-βII was detected in cells infected with SeV or ΔNS1 influenza virus (Fig. 4E). In summary, these results indicate that PKC-α and PKC-β efficiently interact with RIG-I under normal conditions, and that they are enzymatically active in the RIG-I-PKC complex.
Fig 4.
PKC-α/β interact with RIG-I. (A and B) Interaction between PKC-α/β and RIG-I or RIG-I 2CARD. At 48 h posttransfection with Myc-tagged PKC-α, PKC-βII, or PKG together with vector or Flag-RIG-I (A) or with GST or GST-RIG-I 2CARD (B), WCLs were used for IP with anti-Flag (A) or GST-PD (B), followed by IB with anti-Myc (A and B), anti-Flag (A), or anti-GST antibody (B). (C) Interaction between endogenous RIG-I and PKC-α/β. WCLs of HEK293T, A549, or HeLa cells were subjected to IP with an anti-RIG-I antibody, followed by IB with anti-PKC-α, anti-PKC-βI, anti-PKC-βII, anti-PKC-ε, anti-PKC-ζ, or anti-RIG-I antibody. The input (2%) for 293T is shown. (D) RIG-I-PKC-α/β interaction in primary NHLFs. NHLFs were mock infected or infected with SeV (50 HA units/ml) for the indicated hours. WCLs were subjected to IP with an anti-RIG-I antibody, followed by IB with anti-PKC-α, anti-PKC-βII, or anti-RIG-I antibody. (E) PKC-α and PKC-β that interact with RIG-I are phosphorylated at S657 or T500, respectively. HEK293T cells were mock treated or infected with SeV (50 HA units/ml) or ΔNS1 PR8 virus (MOI, 2) for 14 h. WCLs were used for IP with an anti-RIG-I antibody, followed by IB with anti-PKC-α, anti-pS657-PKC-α, anti-PKC-βII, or anti-pT500-PKC-β antibody.
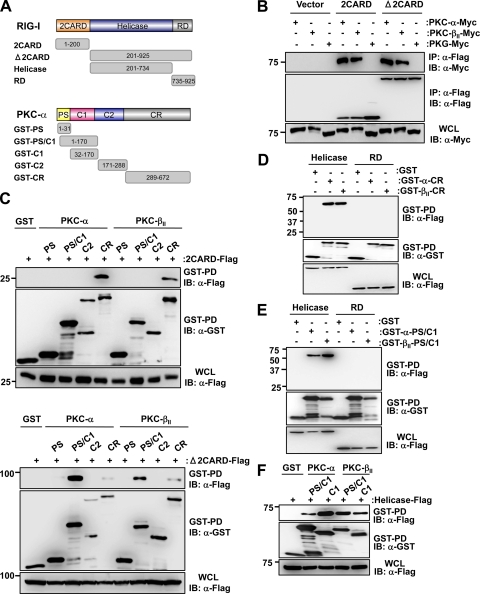
To define the molecular architecture of the RIG-I-PKC complex, we tested Myc-tagged PKC-α and PKC-βII for their ability to interact with Flag-tagged RIG-I 2CARD or with a RIG-I mutant in which the CARDs were deleted (RIG-I Δ2CARD) (Fig. 5B). PKG-Myc was included as a control. PKC-α and PKC-βII strongly interacted with both RIG-I 2CARD and RIG-I Δ2CARD; in contrast, PKG did not bind either under the same conditions (Fig. 5B). As with other PKC family members, PKC-α and PKC-β are comprised of four domains: an N-terminal pseudosubstrate (PS) domain, a central C1 domain, a C2 domain, and a C-terminal catalytic domain (CR) (Fig. 5A). Binding studies using N-terminal GST-fused PKC-α or PKC-βII polypeptides corresponding to PS, PS/C1, C1, C2, and CR domains showed that the C-terminal CR domain of PKC-α/βII bound to Flag-tagged RIG-I 2CARD as well as RIG-I Δ2CARD and the helicase of RIG-I (Fig. 5C and D). Furthermore, the regulatory PS/C1and C1 domain of PKC-α/βII strongly and specifically interacted with RIG-I Δ2CARD and the RIG-I helicase (Fig. 5C, lower, and E and F). These results demonstrate that the N-terminal CARDs of RIG-I interact with the C-terminal CR domain of PKC-α/βII, whereas the helicase domain of RIG-I binds both the CR and regulatory C1 domain of PKC-α/βII.
Fig 5.
Biochemical mapping of the RIG-I-PKC-α/β interaction. (A) Domain structures of RIG-I and PKC-α as well as schematic representation of Flag-tagged RIG-I or GST-fused PKC-α truncation constructs. Numbers indicate amino acids. GST-PKC-βII fusion constructs were constructed corresponding to PKC-α constructs. (B) PKC-α/β interact with the RIG-I 2CARD and RIG-I Δ2CARD. At 48 h posttransfection with Myc-tagged PKC-α, PKC-βII or PKG together with vector, RIG-I 2CARD-Flag or RIG-I Δ2CARD-Flag, WCLs were used for IP with an anti-Flag antibody, followed by IB with anti-Myc or anti-Flag antibody. WCLs were further used for IB with an anti-Myc antibody to determine the expression of PKC-α, PKC-βII, or PKG. (C) The CR and PS/C1 domains of PKC-α/β interact with RIG-I 2CARD and RIG-I Δ2CARD. HEK293T cells were transfected with RIG-I 2CARD-Flag (upper) or RIG-I Δ2CARD-Flag (lower) together with GST or the indicated GST-PKC-α or GST-PKC-βII fusion constructs. WCLs were subjected to GST-PD, followed by IB with anti-Flag or anti-GST antibody. (D and E) The CR and PS/C1 domains of PKC-α/β bind to the helicase of RIG-I. HEK293T cells were transfected with GST, GST-PKC-α-CR, or GST-PKC-βII-CR (D) or with GST, GST-PKC-α-PS/C1, or GST-PKC-βII-PS/C1 (E) together with RIG-I-helicase-Flag or RIG-I-RD-Flag. WCLs were used for GST-PD followed by IB with anti-Flag or anti-GST antibody. The expression of Flag-tagged RIG-I-helicase and RIG-I-RD was determined by IB with an anti-Flag antibody. (F) The C1 domain of PKC-α/β is sufficient for RIG-I-helicase binding. HEK293T cells were transfected with GST, GST-PKC-α-PS/C1, GST-PKC-α-C1, GST-PKC-βII-PS/C1, or GST-PKC-βII-C1 together with Flag-tagged RIG-I-helicase. WCLs were used for GST-PD followed by IB with anti-Flag or anti-GST antibody. The expression of Flag-tagged RIG-I-helicase was determined by IB with an anti-Flag antibody.
PKC-α/β inhibit the RIG-I-TRIM25 interaction, RIG-I-MAVS binding, and RIG-I-mediated antiviral IFN induction.
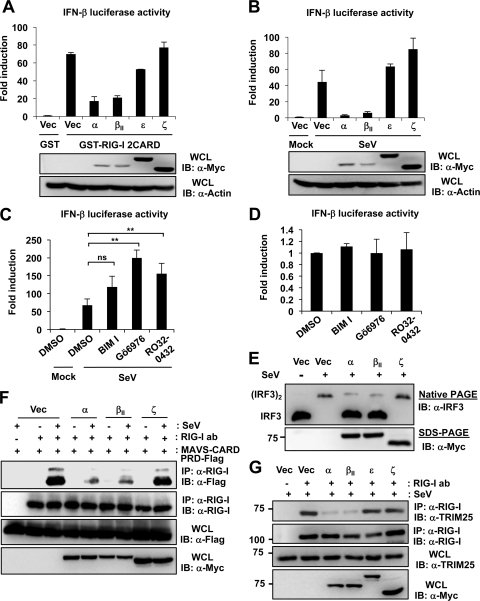
To test the role of PKC-α/β in RIG-I signal transduction, HEK293T cells were transfected with GST-RIG-I 2CARD and IFN-β promoter luciferase together with vector, Myc-tagged PKC-α, PKC-βII, PKC-ε, or PKC-ζ (Fig. 6A). PKC-α or PKC-βII expression markedly inhibited the IFN-β promoter activation induced by GST-RIG-I 2CARD, whereas PKC-ε or PKC-ζ had little or no effect on RIG-I 2CARD-mediated IFN-β promoter activation (Fig. 6A). Furthermore, exogenous PKC-α and PKC-βII but not PKC-ε or PKC-ζ potently suppressed the SeV-induced IFN-β promoter activation (Fig. 6B). Conversely, the treatment of HEK293T with the conventional PKC inhibitor BIM I, Gö6976, or Ro-32-0432 increased the SeV-induced IFN-β promoter activation compared to that of mock treatment (Fig. 6C). However, BIM I, Gö6976, or Ro-32-0432 treatment was not sufficient to detectably induce IFN-β promoter activation in mock-infected cells (Fig. 6D). To further delineate the inhibitory effect of PKC-α/β on the RIG-I-mediated downstream signaling cascade, we examined the virus-induced dimerization of IFN-regulatory factor 3 (IRF3) (Fig. 6E). For this, primary NHLFs were transfected with vector, Myc-tagged PKC-α, PKC-βII, or PKC-ζ, followed by SeV infection. Native PAGE showed that SeV led to an efficient dimerization of endogenous IRF3 in vector- or PKC-ζ-transfected cells; in contrast, IRF3-dimer formation was strongly suppressed in cells expressing exogenous PKC-α or PKC-βII (Fig. 6E). To trigger downstream signaling, RIG-I binding to TRIM25 and to MAVS downstream partner is essential. We thus examined the effect of exogenous PKC-α or PKC-βII expression on the interaction between RIG-I and the MAVS-CARD by co-IP (Fig. 6F). As a control, Myc-tagged PKC-ζ was included. The ectopic expression of PKC-α or PKC-βII potently inhibited the SeV-induced interaction of endogenous RIG-I with the Flag-tagged MAVS-CARD proline-rich domain (PRD); in contrast, PKC-ζ did not have any effect on the binding of RIG-I to the MAVS-CARD (Fig. 6F). Consistent with this, exogenous PKC-α or PKC-βII, but not PKC-ε or PKC-ζ, strongly inhibited the interaction of GST-RIG-I 2CARD and the MAVS-CARD (data not shown). Finally, in correlation with our results indicating that the phosphorylation of RIG-I at S8 and T170 inhibits RIG-I binding to TRIM25 (Fig. 1C), exogenously expressed PKC-α or PKC-βII, but not PKC-ε and PKC-ζ, markedly suppressed the interaction between endogenous RIG-I and TRIM25 (Fig. 6G). Collectively, these results indicate that the PKC-α/β-mediated phosphorylation of RIG-I inhibits the RIG-I-TRIM25 interaction and RIG-I ubiquitination, which subsequently suppresses RIG-I-MAVS binding and, thereby, RIG-I-induced IFN gene expression.
Fig 6.
PKC-α/β inhibit the RIG-I-TRIM25 interaction, RIG-I-MAVS binding, and thereby RIG-I downstream signaling. (A) Ectopic PKC-α/β expression inhibits the RIG-I 2CARD-induced IFN-β promoter activation. HEK293T cells were transfected with GST or GST-RIG-I 2CARD together with vector, Myc-tagged PKC-α, PKC-βII, PKC-ε, or PKC-ζ as well as with IFN-β-luciferase and pGK-β-gal. At 40 h posttransfection, luciferase activity was determined as described in the legend to Fig. 1E. Data represent the means ± SD (n = 3). (B) Exogenous expression of PKC-α/β suppresses SeV-induced IFN-β promoter activation. HEK293T cells were transfected with vector, Myc-tagged PKC-α, PKC-βII, PKC-ε, or PKC-ζ, together with IFN-β-luciferase and pGK-β-gal. At 24 h after transfection, cells were mock treated or infected with SeV. Luciferase activity was determined 18 h later. Data represent the means ± SD (n = 3). (C) Conventional PKC inhibitors enhance SeV-induced IFN-β promoter activation. At 20 h posttransfection with IFN-β-luciferase and pGK-β-gal, HEK293T cells were treated with DMSO or the indicated PKC inhibitors (bisindolylmaleimide I [BIM I], 4 μM; Gö6976, 10 μM; and Ro-32-0432, 12 μM), followed by mock treatment or infection with SeV (30 HA units/ml). Twenty hours later, luciferase and β-galactosidase values were determined and IFN-β luciferase values normalized to β-galactosidase activity for transfection efficiency control. Data represent the means ± SD (n = 3). Statistical analysis was performed by Student's t test. ns, not statistically significant; **, P < 0.01. (D) PKC inhibitor treatment is not sufficient to induce IFN-β promoter activation in mock-infected cells. At 20 h posttransfection with IFN-β-luciferase and pGK-β-gal, HEK293T cells were treated with DMSO or the indicated PKC inhibitors as described for panel C. IFN-β-luciferase and pGK-β-gal values were determined 20 h later. Data represent the means ± SD (n = 3). (E) PKC-α/β inhibit virus-induced IRF3 dimerization in primary NHLFs. At 24 h posttransfection with vector, Myc-tagged PKC-α, PKC-βII, or PKC-ζ, NHLFs were mock treated or infected with SeV (80 HA units/ml) for 22 h. WCLs were used for native PAGE followed by IB with an anti-IRF3 antibody or were subjected to SDS-PAGE followed by IB with an anti-Myc antibody. (F) PKC-α/β suppress the RIG-I-MAVS interaction. HEK293T cells were cotransfected with MAVS-CARD-PRD-Flag together with vector or Myc-tagged PKC isozymes, and subsequently they were either mock treated or infected with SeV (50 HA units/ml) for 5 h. WCLs were subjected to IP with an anti-RIG-I antibody, followed by IB with anti-Flag or anti-RIG-I antibody. WCLs were further used for immunoblotting with anti-Flag or anti-Myc antibody. (G) PKC-α/β inhibit RIG-I binding to TRIM25. At 20 h after transfection with vector, Myc-tagged PKC-α, PKC-βII, PKC-ε, or PKC-ζ, HEK293T cells were infected with SeV (50 HA units/ml) for 8 h. WCLs were subjected to IP with an anti-RIG-I antibody, followed by IB with anti-TRIM25 or anti-RIG-I antibody.
PKC-α/β suppress the IFN-mediated antiviral activity of RIG-I.
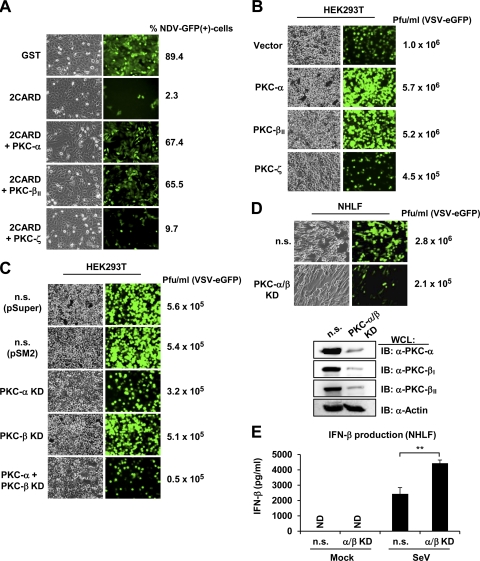
To examine the effect of PKC-α/β on RIG-I-mediated antiviral IFN production, we performed a bioassay using recombinant Newcastle disease virus (NDV) expressing green fluorescent protein (GFP) (NDV-GFP) (Fig. 7A). For this, Vero cells were incubated with supernatants from HEK293T cells that had been transfected with GST, GST-RIG-I 2CARD alone, or GST-RIG-I 2CARD together with PKC-α, PKC-βII, or PKC-ζ, followed by infection with NDV-GFP (Fig. 7A). Supernatants from 293T cells transfected with GST-RIG-I 2CARD alone or GST-RIG-I 2CARD together with PKC-ζ substantially suppressed the replication of NDV-GFP compared to supernatants from cells transfected with GST. In contrast, supernatants from cells transfected with GST-RIG-I 2CARD together with PKC-α or PKC-βII blocked NDV-GFP replication to a far lesser extent than supernatants from transfections with GST-RIG-I 2CARD alone (Fig. 7A). We also performed the same NDV bioassay using the RIG-I S8A/T170A mutant, which cannot be phosphorylated, and observed that supernatants from cells cotransfected with GST-RIG-I S8A/T170A together with PKC-α or PKC-βII suppressed the replication of NDV-GFP to an extent comparable to that of supernatants from cells transfected with GST-RIG-I S8A/T170A alone (data not shown). This corroborates that the inhibitory effect of PKC-α/β on the RIG-I 2CARD-mediated IFN production is due to PKC-α/β-dependent RIG-I phosphorylation at S8 and T170. We next examined the effect of PKC-α/β on the replication of recombinant vesicular stomatitis virus (VSV) expressing enhanced GFP (VSV-eGFP) in HEK293T cells stably expressing PKC-α, PKC-βII, or PKC-ζ (Fig. 7B). This showed that the stable expression of PKC-α or PKC-βII detectably increased the replication of VSV-eGFP compared to that of vector or PKC-ζ expression (Fig. 7B).
Fig 7.
PKC-α/β suppress RIG-I antiviral activity. (A) PKC-α/β inhibit RIG-I 2CARD-mediated antiviral IFN production. Vero cells were incubated for 18 h with supernatants from HEK293T cells that had been transfected with GST or GST-RIG-I 2CARD together with vector, PKC-α, PKC-βII, or PKC-ζ, followed by NDV-GFP infection (MOI, 3). At 24 h after infection, GFP expression was detected by epifluorescence, and GFP-positive cells were determined by FACS analysis. (B) Ectopic PKC-α or PKC-β expression increases VSV-eGFP replication. HEK293T cells stably expressing vector, Myc-tagged PKC-α, PKC-βII, or PKC-ζ were infected with VSV-eGFP at an MOI of 0.2. At 38 h after infection, virus titer and replication were determined by plaque assay and GFP expression, respectively. (C) shRNA-mediated PKC-α/PKC-β double knockdown inhibits VSV-eGFP replication. HEK293T cells stably transfected with nonsilencing (n.s.) shRNA, PKC-α-specific shRNA, PKC-β-specific shRNA, or PKC-α- and PKC-β-specific shRNAs were infected with VSV-eGFP at an MOI of 0.5. Thirty hours after infection, virus titer and GFP expression were determined by plaque assay or microscopy, respectively. (D) Knockdown of endogenous PKC-α and PKC-β in primary normal human lung fibroblasts (NHLF) suppresses VSV-eGFP replication. NHLFs were transiently transfected with nonsilencing control siRNA (n.s.) or with both PKC-α- and PKC-β-specific siRNA. Forty hours after transfection, cells were infected with VSV-eGFP at an MOI of 0.1. Thirty hours after infection, virus titer and GFP expression was determined by plaque assay or microscopy, respectively. The knockdown of endogenous PKC-α, PKC-βI, and PKC-βII was confirmed by immunoblotting using the indicated antibodies. (E) Increased IFN-β production in primary NHLFs in which PKC-α and PKC-β gene expression have been silenced. NHLFs that had been transiently transfected with nonsilencing control siRNA (n.s.) or with both PKC-α- and PKC-β-specific siRNA were either mock treated or infected with SeV (40 HA units/ml). Thirty hours after infection, IFN-β production in the supernatants was determined by ELISA. Statistical analysis was performed by Student's t test. **, P < 0.01. KD, knockdown. ND, not detected.
To determine the physiological role of PKC-α/β in regulating the antiviral activity of RIG-I, we examined the replication of VSV-eGFP in HEK293T cells in which PKC-α, PKC-β, or both have been stably silenced using specific shRNAs (Fig. 7C). While the knockdown of either PKC-α or PKC-β had little or no effect on the replication of VSV-eGFP, the stable silencing of both markedly suppressed the replication of VSV-eGFP compared to the expression of nonsilencing control shRNA (Fig. 7C). To confirm these results in primary human cells, we tested the replication of VSV-eGFP in NHLFs in which endogenous PKC-α and PKC-β have been depleted using specific siRNAs (Fig. 7D). Cells transfected with nonsilencing control siRNA served as the control. In line with our results with HEK293T cells, NHLFs in which both PKC-α and PKC-β were silenced exhibited markedly reduced VSV-eGFP replication; VSV-eGFP titers were ∼10-fold reduced compared to those of cells transfected with control siRNA (Fig. 7D). Finally, we examined the effect of silencing PKC-α and PKC-β on the SeV-induced IFN-β production in NHLFs (Fig. 7E). For this, NHFLs were transiently transfected with nonsilencing control siRNA or with PKC-α- and PKC-β-specific siRNAs, followed by mock treatment or infection with SeV. IFN-β production then was determined by ELISA. This showed that PKC-α/β knockdown cells exhibited detectably increased IFN-β protein levels in the supernatant compared to that of cells transfected with nonsilencing control siRNA (Fig. 7E). In summary, these results indicate that RIG-I phosphorylation by PKC-α/β suppresses RIG-I's ability to induce IFN production and, thereby, its antiviral activity.
DISCUSSION
Dynamic interplay between phosphorylation and ubiquitination is an emerging paradigm for regulating the activities of key signaling molecules (11). Our study reveals that conventional PKC-α/β-induced RIG-I phosphorylation and TRIM25-mediated RIG-I ubiquitination functionally antagonize each other to tightly regulate the RIG-I CARD-mediated antiviral signal transduction. Our results showed that PKC-α and PKC-β interact with RIG-I under normal conditions in various different cell lines, including primary fibroblasts. Furthermore, ectopic expression, gene silencing, and in vitro phosphorylation assays indicated that conventional PKC-α/β phosphorylate RIG-I at S8 and T170, which keeps RIG-I inactive by suppressing RIG-I-TRIM25 binding, RIG-I CARD ubiquitination, and thereby RIG-I-MAVS interaction. Upon viral infection, however, PKC-α/β apparently dissociate from RIG-I, which potentially allows phosphatase-dependent RIG-I dephosphorylation, leading to efficient RIG-I-TRIM25 interaction and RIG-I ubiquitination and ultimately initiating antiviral IFN responses.
Mutational analysis indicated that whereas the phosphorylation of RIG-I at either S8 or T170 prevents TRIM25-mediated RIG-I ubiquitination and RIG-I downstream signaling, the dephosphorylation of RIG-I at both residues is necessary for optimal ubiquitination-mediated RIG-I activation. This kind of dynamic balance between constitutive phosphorylation for cytosolic PRR inactivation and phosphatase-dependent activation represents a novel type of regulation in antiviral innate immunity. Whereas kinases such as IKK-β or TBK-1 are well known to activate innate immune signaling, this study strongly suggests that phosphatases play key roles in the initiation of the RIG-I signal transduction pathway. Thus, future studies directed toward the identification of the phosphatase(s) responsible for RIG-I dephosphorylation promise to decipher the impact of protein dephosphorylation for orchestrating antiviral innate immune responses.
PKC comprises a family of more than 10 isoenzymes divided into conventional, novel, and atypical subgroups (10, 25). Our study showed that specifically conventional PKC-α/β interacted with RIG-I, but neither novel PKC-ε nor atypical PKC-ζ did. Furthermore, the coexpression of PKC-α or PKC-β robustly increased the S8 and T170 phosphorylation of endogenously and exogenously expressed RIG-I, while PKC-ε and PKC-ζ had no effect or only marginally enhanced RIG-I phosphorylation when grossly overexpressed. Furthermore, we employed shRNA-mediated knockdown techniques, specific inhibitor treatment, and a PKC in vitro phosphorylation assay to examine the physiological role of PKC-α/β for phosphorylating RIG-I. This clearly showed that PKC-α and PKC-β are primary kinases responsible for RIG-I S8 and T170 phosphorylation, indicating an important role of conventional PKC-α/β isoforms for regulating RIG-I antiviral activity. Furthermore, whereas the ectopic expression of either PKC-α or PKC-β profoundly inhibited RIG-I downstream signaling, the knockdown of both, but not either of them alone, markedly enhanced RIG-I antiviral activity (Fig. 7C), suggesting a partially redundant function of these two conventional isozymes in regulating RIG-I.
PKC activation typically requires membrane association as well as the binding of cofactors such as phospholipids or Ca2+ (25). In addition, it is well established that the binding of proteins to the regulatory domain of PKC can alter its subcellular localization as well as cofactor dependence and, in some cases, can even bypass the need of allosteric inputs for PKC activation (13). Our detailed interaction studies showed that PKC-α/β bind with their regulatory C1 and catalytic CR domain to the RIG-I CARD and helicase. Confocal microscopy analysis indicated that the RIG-I-PKC complex resides in the cytoplasm (data not shown), and in this complex, PKC-α/β apparently are enzymatically active. However, the molecular details of how PKC-α/β bound to RIG-I are activated and whether cofactors of PKC-α/β play any role for RIG-I phosphorylation remains to be elucidated. Future structural analyses of the RIG-I-PKC complex will not only elucidate the detailed mechanism of RIG-I regulation through phosphorylation but also may provide further insights into the exact mode of PKC activation.
A recent series of studies has suggested the following model of RIG-I activation. Under normal conditions, RIG-I is kept inactive through autorepression by the C-terminal RD (27), which interacts with both CARD and helicase domains of RIG-I. The binding of 5′triphosphate containing viral RNA to the RD and subsequent ATPase activity induces a conformational change in RIG-I, leading to the exposure of the N-terminal CARDs. Our previous study showed that the ubiquitin E3 ligase TRIM25 binds to the first CARD of RIG-I, leading to the K63-linked ubiquitination of the second CARD (6, 8). The ubiquitination of K172 in RIG-I is essential for efficient RIG-I binding to MAVS, thereby initiating downstream signaling. The data presented here reveal that, in addition to RIG-I autoinhibition mediated by the RD, the PKC-α/β-dependent phosphorylation of the CARDs plays a crucial role in keeping RIG-I in an inactive state under normal conditions. The phosphorylation of S8 and T170 did not affect RIG-I binding to viral RNA, but it markedly suppressed RIG-I-TRIM25 binding and thereby RIG-I CARD ubiquitination. This suggests that the negative charge provided by the RIG-I S8 and T170 phosphorylation induces structural alterations within the tandem CARD which interfere with TRIM25 binding. Furthermore, it has been shown recently that the RIG-I CARDs can bind unanchored polyubiquitin chains in an in vitro-reconstituted cell-free system, and that TRIM25 was able to catalyze these free ubiquitin chains for RIG-I binding (32). Thus, future studies will be required to address whether RIG-I S8/T170 phosphorylation by PKC-α/β also affects the free ubiquitin binding capability of the RIG-I CARDs.
Our study further showed that viral infection markedly decreased RIG-I-PKC-α/β binding and RIG-I S8 and T170 phosphorylation, suggesting that, upon viral RNA binding, conformational changes in RIG-I not only cause the dissociation of the RIG-I/PKC complex but also allow the recruitment of a yet-to-be-identified phosphatase for RIG-I S8 and T170 dephosphorylation. In fact, our previous studies demonstrated that RIG-I S8/T170 phosphorylation is robustly enhanced by phosphatase inhibitor treatment (7, 22), suggesting that S8 and T170 phosphorylations are tightly regulated by phosphatase-dependent dephosphorylation. The data presented here further indicate that the dephosphorylation of both S8 and T170 ultimately allows efficient TRIM25 binding, RIG-I CARD ubiquitination, and MAVS-mediated downstream signaling.
During the submission of the manuscript, Zhu et al. reported that PKC-α enhances IRF3-mediated gene induction upon viral infection through the activation of HDAC6 and β-catenin (33). Our study indicates that PKC-α/β act as negative regulators of RIG-I by phosphorylating its N-terminal CARDs, which keeps RIG-I in a latent, inactive state. Consistent with this model, we observed an inhibitory effect of exogenous PKC-α/β expression on RIG-I-CARD-mediated IFN induction and enhanced IFN-β production in primary lung fibroblasts in which PKC-α and PKC-β were silenced using specific siRNAs. Our biochemical analyses also showed that PKC-α/β efficiently bind to RIG-I in noninfected cells and that this interaction leads to RIG-I phosphorylation, which ultimately suppresses the RIG-I signal-transducing activity. It thus is conceivable that conventional PKC has two distinct roles in type I IFN induction: the inhibition of RIG-I downstream signaling by PKC-α/β prior to viral infection and the activation of IRF3-mediated transcription by PKC-α upon viral infection. This also suggests that specific substrate binding and subcellular localization of conventional PKCs determine their functional roles in IFN induction before and after viral infection. Alternatively, it is possible that the different experimental systems used account for the discrepancies between our results and the findings by Zhu et al. (33).
The host has developed numerous checkpoints to regulate viral RNA sensing and IFN signaling initiated by the cytosolic viral RNA receptor RIG-I. Otherwise, the excessive production of type I IFNs or proinflammatory cytokines can be detrimental to the host cell rather than beneficial. RIG-I activity is negatively regulated by K48-linked ubiquitination and the CARD-lacking helicase LGP2 (2, 17, 26). In addition, alternative splicing and deubiquitination by CYLD have been shown to represent effective inhibitory mechanisms of the RIG-I antiviral signaling function (4, 6). Furthermore, it has been shown recently that influenza A virus nonstructural protein 1 (NS1) and cellular HOIL-1L-HOIP linear ubiquitin assembly complex (LUBAC) antagonize TRIM25 activity to ubiquitinate the RIG-I CARDs, thereby leading to RIG-I inhibition (5, 12). In this study, we show that PKC-α/β function as endogenous suppressors to prevent the RIG-I CARD-dependent signal transduction under normal conditions, unveiling PKC-α/β-induced RIG-I phosphorylation as an important mechanism in modulating host IFN-mediated antiviral immune responses.
ACKNOWLEDGMENTS
We greatly thank Jae Jung, Lee Gehrke, Debra Tonetti, Ellen Cahir-McFarland, Jiuyong Xie, Yusuf Hannun, Sean Whelan, and Adolfo García-Sastre for providing reagents.
This work was supported by U.S. Public Health Service grants RO1AI087846 and RR00168 (M.U.G.) and by the German Science Foundation (E.W.).
Footnotes
Published ahead of print 23 November 2011
REFERENCES
- 1. Andrejeva J, et al. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U. S. A. 101:17264–17269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arimoto K, et al. 2007. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl. Acad. Sci. U. S. A. 104:7500–7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cui S, et al. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29:169–179 [DOI] [PubMed] [Google Scholar]
- 4. Friedman CS, et al. 2008. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 9:930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gack MU, et al. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gack MU, et al. 2008. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc. Natl. Acad. Sci. U. S. A. 105:16743–16748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gack MU, Nistal-Villan E, Inn KS, Garcia-Sastre A, Jung JU. 2010. Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J. Virol. 84:3220–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gack MU, et al. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920 [DOI] [PubMed] [Google Scholar]
- 9. Gitlin L, et al. 2010. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 6:e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gould CM, Newton AC. 2008. The life and death of protein kinase C. Curr. Drug Targets 9:614–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hunter T. 2007. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28:730–738 [DOI] [PubMed] [Google Scholar]
- 12. Inn KS, et al. 2011. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol. Cell 41:354–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jaken S, Parker PJ. 2000. Protein kinase C binding partners. Bioessays 22:245–254 [DOI] [PubMed] [Google Scholar]
- 14. Kato H, et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105 [DOI] [PubMed] [Google Scholar]
- 15. Kawai T, Akira S. 2008. Toll-like receptor and RIG-I-like receptor signaling. Ann. N. Y. Acad. Sci. 1143:1–20 [DOI] [PubMed] [Google Scholar]
- 16. Kawai T, et al. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988 [DOI] [PubMed] [Google Scholar]
- 17. Komuro A, Horvath CM. 2006. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J. Virol. 80:12332–12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin X, et al. 2006. Overexpression of PKCalpha is required to impart estradiol inhibition and tamoxifen-resistance in a T47D human breast cancer tumor model. Carcinogenesis 27:1538–1546 [DOI] [PubMed] [Google Scholar]
- 19. Loo YM, et al. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meylan E, et al. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172 [DOI] [PubMed] [Google Scholar]
- 21. Nakhaei P, Genin P, Civas A, Hiscott J. 2009. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin. Immunol. 21:215–222 [DOI] [PubMed] [Google Scholar]
- 22. Nistal-Villán E, et al. 2010. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J. Biol. Chem. 285:20252–20261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. 2009. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J. Biol. Chem. 284:807–817 [DOI] [PubMed] [Google Scholar]
- 24. Pichlmair A, et al. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001 [DOI] [PubMed] [Google Scholar]
- 25. Rosse C, et al. 2010. PKC and the control of localized signal dynamics. Nat. Rev. Mol. Cell Biol. 11:103–112 [DOI] [PubMed] [Google Scholar]
- 26. Rothenfusser S, et al. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 175:5260–5268 [DOI] [PubMed] [Google Scholar]
- 27. Saito T, et al. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. U. S. A. 104:582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682 [DOI] [PubMed] [Google Scholar]
- 29. Uematsu S, Akira S. 2007. Toll-like receptors and type I interferons. J. Biol. Chem. 282:15319–15323 [DOI] [PubMed] [Google Scholar]
- 30. Xu LG, et al. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727–740 [DOI] [PubMed] [Google Scholar]
- 31. Yoneyama M, et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 [DOI] [PubMed] [Google Scholar]
- 32. Zeng W, et al. 2010. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141:315–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu J, Coyne CB, Sarkar SN. 2011. PKC alpha regulates Sendai virus-mediated interferon induction through HDAC6 and beta-catenin. EMBO J. doi:10.1038/emboj.2011.351 [DOI] [PMC free article] [PubMed] [Google Scholar]