Abstract
As the receptor-binding protein of herpes simplex virus (HSV), gD plays an essential role in virus entry. In its native state, the last 56 amino acids of the ectodomain C terminus (C-term) occlude binding to its receptors, herpesvirus entry mediator (HVEM) and nectin-1. Although it is clear that movement of the C-term must occur to permit receptor binding, we believe that this conformational change is also a key event for triggering later steps leading to fusion. Specifically, gD mutants containing disulfide bonds that constrain the C-term are deficient in their ability to trigger fusion following receptor binding. In this report, we show that two newly made monoclonal antibodies (MAbs), MC2 and MC5, have virus-neutralizing activity but do not block binding of gD to either receptor. In contrast, all previously characterized neutralizing anti-gD MAbs block binding of gD to a receptor(s). Interestingly, instead of blocking receptor binding, MC2 significantly enhances the affinity of gD for both receptors. Several nonneutralizing MAbs (MC4, MC10, and MC14) also enhanced gD-receptor binding. While MC2 and MC5 recognized different epitopes on the core of gD, these nonneutralizing MAbs recognized the gD C-term. Both the neutralizing capacity and rate of neutralization of virus by MC2 are uniquely enhanced when MC2 is combined with MAb MC4, MC10, or MC14. We suggest that MC2 and MC5 prevent gD from performing a function that triggers later steps leading to fusion and that the epitope for MC2 is normally occluded by the C-term of the gD ectodomain.
INTRODUCTION
Herpes simplex virus (HSV) is an important human pathogen that infects epithelial cells before spreading to the peripheral nervous system, where it establishes a lifelong latent infection. Four virion envelope glycoproteins, gD, gB, and gH plus gL (gH/gL), are essential for HSV entry into all relevant cell types (19). Two surface proteins, nectin-1 and herpesvirus entry mediator (HVEM), can serve as gD receptors. Nectin-1 is an immunoglobulin (Ig) superfamily member, while HVEM is a tumor necrosis factor receptor family member (50). A combination of crystal structure, mutagenesis, and monoclonal antibody (MAb)-binding studies has shown that the sites for HVEM and nectin-1 binding are largely distinct (19, 30, 51).
Crystallography studies have also shown that the C terminus of the gD ectodomain (C-term) normally occludes the binding site for nectin-1 and prevents formation of the N-terminal loop needed for HVEM binding (19, 30). Thus, for either receptor to bind to gD, the C-term residues must be displaced. Notably, a gD mutant engineered to contain an additional disulfide bond that constrained the motion of the C-term was able to bind both HVEM and nectin-1 normally. However, this mutant failed to trigger cell-cell fusion and did not complement a gD-null virus (31). Thus, the phenotype of this mutant dissociates receptor binding from downstream post-receptor-binding effects mediated by gD. This led us to hypothesize that a common conformational change is responsible for triggering the downstream events involved in virus-cell fusion.
The recent resolution of the structures of gB and gH/gL for both HSV and Epstein-Barr virus (EBV) (4, 12, 15, 20, 33) revealed that, while gB is a class III fusion protein, the structure of gH/gL does not resemble any known viral fusogen. Thus, the function of gH/gL as part of the core-fusion machinery is still unclear. Some have suggested that the highly conserved and highly hydrophobic C-terminal regions of the gH ectodomain may play a direct role in fusion (15, 32, 33). However, even this suggestion leaves many questions unanswered, since this region does not contain a readily recognizable fusion loop or peptide such as is found in fusion proteins of known structure (18).
Another hypothesis is that gH/gL plays a regulatory role in promoting the fusion activity of gB (12). In support of this concept, it was recently discovered that gH/gL does not have to be in the same cell as gB in order for cell-cell fusion to occur (55). In fact, our data suggest that the gH/gL ectodomain can function without being membrane bound at all (2). We found that when nectin-1-bearing cells (called C10 cells) express gB, they can be triggered to fuse by the addition of a combination of soluble forms (ectodomains) of gD and gH/gL (2). In addition, we found that brief exposure of C10 cells bearing gH/gL to soluble gD was sufficient to make them fusion competent when cocultured with cells expressing gB. Importantly, the converse did not happen, i.e., cells expressing gB and a gD receptor that were first exposed to soluble gD could not fuse with cells expressing gH/gL (2, 31). These observations led us to propose that HSV-induced fusion (and possibly virus entry) consists of several sequential steps: (i) binding of gD to an appropriate receptor, followed by (ii) a conformational change in gD that allows it to activate gH/gL, leading to (ii) activation of gB into a fusogenic state.
To test this hypothesis, we first examined the mechanism by which virus-neutralizing antibodies work. Here, we focused on MAbs to gD. In prior studies, we and others showed that virus-neutralizing antibodies can be directed at HSV gD, gH/gL, or gB (6, 8, 36, 39). Among those directed at gD, some blocked binding of gD to HVEM but not nectin-1 and others blocked gD binding to both receptors (28, 40). Here we report the identification of two separate monoclonal antibodies, MC2 and MC5, that bind to gD in the presence of either HVEM or nectin-1 and do not block receptor binding to gD but effectively neutralize HSV infection.
Competition studies showed that MC2 and MC5 bind to distinct gD epitopes, suggesting that they may function in different ways. Surprisingly, we found that MC2 significantly enhances the affinity of gD for its receptors. We propose that this increased affinity reflects the stable displacement of the gD C-term upon MC2 binding. At the same time, binding of MC2 to its epitope likely interferes with the second function of gD (which we propose is the activation of gH/gL). Moreover, both the neutralizing capacity and rate of neutralization of virus by MC2 were enhanced when it was combined with nonneutralizing MAb MC4, MC10, or MC14, all of which map to the same linear epitope (amino acids 262 to 272). In contrast, MC5 did not enhance receptor binding to gD, and its neutralizing activity was not enhanced by combining it with MC14. We suggest that MAbs MC2 and MC5 neutralize virus by interfering with the ability of gD to activate gH/gL, thereby preventing activation of gB into its fusogenically active state. As such, these antibodies can be utilized to delineate essential information about the step that occurs just after receptor binding. Additionally, these MAb studies may supply insights into the efficacy of gD as a vaccine component (14, 52) by revealing whether responses to the vaccine include antibodies that target both functions of gD.
MATERIALS AND METHODS
Cells and viruses.
293T cells were grown in Dulbecco's minimal essential medium (DMEM) supplemented with 10% fetal calf serum (FCS). Sf9 (Spodoptera frugiperda) cells were grown in Sf900II serum-free medium (Gibco). Vero cells were grown in DMEM supplemented with 5% FCS. Gradient-purified HSV-2 strain 333 was propagated in Vero cells and the titer determined. Hybridoma cells were cultured in supplemented Kennett's Hy medium.
Production and purification of recombinant baculovirus-produced proteins.
The methods for production and DL6 affinity purification of gD1(306t), gD2(306t), and gD2(285t) have been described previously (29, 41, 48). Production of HveA(200t) (HVEM) and HveC(346t) (nectin-1) by nickel-agarose purification has also been described previously (28, 59). Recombinant baculovirus expressing gD2(s322) was obtained by first cloning the 0.3-kbp ApaI (blunted with mung bean nuclease)-BamHI fragment from plasmid D2-H20 (11) (which contains an insertion of charged amino acids into the middle of the transmembrane region) into plasmid pAN243 (41) (which had been digested with EcoRI and blunted with Klenow fragment) and BamHI. The resulting baculovirus transfer vector (pCW259), which contained full-length gD2 with the insertion at amino 322, was then cotransfected with BaculoGold baculovirus DNA (BD Pharmingen) to generate a recombinant baculovirus expressing this form of gD2. Recombinant gD2(s322) was purified via DL6 affinity purification as described above.
Production of MAbs against HSV2 gD.
Mice were immunized with gD2(s322) until suitable serum antibody titers were achieved. Hybridoma production was performed using standard procedures, and stable hybridomas secreting IgG reactive with HSV-2 gD by an enzyme-linked immunosorbent assay (ELISA) were subcloned twice. IgG was purified from mouse ascitic fluid by protein G chromatography (HiTrap; GE Healthcare) according to the manufacturer's instructions. Following purification, MAbs were dialyzed against phosphate-buffered saline (PBS).
Other antibodies used.
Rabbit polyclonal antibody (PAb) R8 was raised against full-length HSV-2 gD and cross-reacted with HSV-1 gD (22). Anti-gD MAbs used in this study included BD66 and BD78 (kindly supplied by Becton Dickinson Co.) and DL6, which recognize linear epitopes within residues 272 to 279, 284 to 301, and 264 to 275, respectively (22). We also used MAbs DL11 (13, 36), HD1 (42, 47), H128 (46), and LP2 (35), the latter kindly provided by A. Minson and H. Browne.
SDS-PAGE and Western blotting.
Purified glycoproteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under “native” conditions (0.1% SDS, no reducing agent, no boiling) (13) or denaturing conditions (samples boiled for 5 min in sample buffer containing dithiothreitol [DTT]) in precast 10% Tris-glycine gels (Invitrogen). Following SDS-PAGE, separated proteins were transferred to nitrocellulose, probed with antibodies, and visualized by enhanced chemiluminescence (Amersham Pharmacia).
Coimmunoprecipitation of gD-receptor complexes with gD MAbs.
gD2(285t) (300 ng) was combined with 1.5 μg of nectin-1(346t) or HVEM(200t) in 100 μl of binding buffer (60) and incubated for 1 h at 4°C. Next, 500 ng of the indicated MAb IgG was added and samples were incubated for an additional hour at 4°C. Finally, protein A-Sepharose (ProA-Sepharose) was added to each sample and incubated for 1 h at 4°C. The resulting immune complexes were pelleted, washed, and subjected to Western blot analysis. Blots were probed with rabbit polyclonal antisera against HSV gD (R7) and human nectin-1 (R154) or human HVEM (R140).
Blocking of receptor binding by MAbs.
ELISA plates were coated overnight with soluble receptors [5 μg/ml of HVEM(200t), 10 μg/ml of nectin-1(346t)] and then blocked with 5% milk–0.1% Tween–PBS for 1 h. Concurrently, a constant amount of gD2(306t) was incubated with dilutions of each MAb (whole IgG or Fab) for 1 h at room temperature (RT) in 5% milk–0.1% Tween–PBS. The MAb-gD mixtures were then added to receptor-coated plates for 1 h at RT. Bound gD was detected with anti-gD PAb R8 followed by horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody. This assay was repeated a minimum of three times for each MAb.
Enhancement of receptor binding by MAbs.
ELISA plates were coated overnight with soluble receptors [5 μg/ml of HVEM(200t), 10 μg/ml of nectin-1(346t)] (28, 59) and then blocked with 5% milk–0.1% Tween–PBS for 1 h. Concurrently, 0.5 μM each MAb was incubated with dilutions of gD2(306t) or gD2(285t) starting at 0.5 μM for 1 h at RT in 5% milk–0.1% Tween–PBS. The MAb-gD mixture was then added to the receptor-coated plates for 1 h at RT. Bound gD was detected with anti-gD PAb R8 followed by HRP-conjugated anti-rabbit secondary antibody. This assay was carried out a minimum of three times for each MAb.
Map competition by surface plasmon resonance.
Experiments were carried out on a BIACORE 3000 optical biosensor (Biacore AB) at 25°C as previously described (1, 27, 60). The running buffer for the experiments was HBS-EP (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% polysorbate 20). An anti-His MAb (Qiagen Inc.) was directly coupled to a CM5 Biacore chip for competition studies. At the beginning of each cycle, gD2(306t) (∼600 response units [RU]) was captured via its C-terminal 6-His tag on flow cell 1 (Fc1) and Fc2. The first competing antibody was injected at a concentration of 20 μg/ml (2 min at 5 μl/min) across Fc1. Finally, the second competing MAb (20 μg/ml) was injected across both Fc1 and Fc2 for 2 min to monitor its binding to gD2(306t). To regenerate the anti-His Ig chip surface, brief pulses of 0.2 M Na2CO3–1 M NaCl (pH 10.5) were injected until the response signal returned to baseline.
Virus neutralization assay.
Serial dilutions of IgG of each MAb were mixed with sucrose gradient-purified HSV-2, and the mixtures were incubated at 37°C for 1 h. The virus-MAb mixtures were then added to Vero cell monolayers grown in 48-well plates and incubated for 18 to 24 h at 37°C. Cells were fixed with methanol:acetone (2:1), and plaques were visualized via immunoperoxidase staining (black plaque) as described elsewhere (6).
Virus neutralization with MAb combinations.
To assess whether combinations of antibodies would neutralize to a greater extent than the sum of the results obtained with each of the two antibodies alone, we carried out two types of experiments. In the first, MAbs were diluted serially and then added to media with or without a constant amount of a second antibody. Virus and the combination of MAbs were incubated for 1 h at 37°C and then plated on Vero cell monolayers for plaque assays. The second assay examined the kinetics of virus neutralization (26, 46). Here, we used fixed concentrations of MAbs and adjusted the time during which virus and MAbs were incubated at 37°C (0 to 60 min). At each time point, the virus-antibody mixture was rapidly diluted in growth medium and added to cells to measure the virus neutralization over time. The results were plotted as V/V0, where V is the number of plaques remaining at each time point and V0 is the number of plaques at time zero. This number was plotted against time. A straight-line relationship between V/V0 and time represents first-order kinetics, and the slope of the line represents the rate of virus neutralization. This assay was carried out a minimum of 5 times for each MAb combination.
RESULTS
A new panel of anti-gD MAbs revealed two MAbs that neutralize HSV independently of receptor binding.
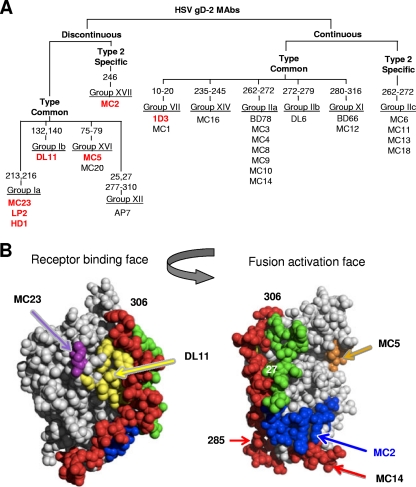
In previous studies of gD, we characterized gD-specific MAbs, studied their properties, and arranged them into antigenic groups, a small subset of which is shown in Fig. 1A. Of particular interest were MAbs that targeted a biologic activity, e.g., the ability to block the interaction between gD and at least one receptor (Fig. 1A; MAbs shown in red) as well as the ability to neutralize HSV (Fig. 1A and Table 1). For example, group Ib-neutralizing MAbs, exemplified by DL11, are type common (bind both HSV-1 and -2), react with discontinuous (38) epitopes, and block binding of gD to both receptors, HVEM and nectin-1 (28, 38, 40, 58). Others, such as those in group VII, have virus-neutralizing activity but block binding to only one of the two receptors (Fig. 1A and Table 1) (28, 36, 40).
Fig 1.
Grouping of MC series of MAbs to gD and their neutralization properties. (A) Tree-like diagram showing how the MC MAbs relate to each other and to previously characterized anti-gD MAbs (22, 37). MC MAbs were grouped according to several criteria, starting with whether they react with any of a series of overlapping peptides to gD2. Additionally, we used truncated forms of gD to delineate the C-terminal end of the discontinuous epitopes. In some cases, the numbers refer to the amino acid changed in monoclonal antibody-resistant (mar) mutants as well as to the position of linker insertion mutations found to affect MAb binding to gD (11). Western blotting of gD proteins was used to determine MC MAb type specificity. They were compared to monoclonal antibodies that had previously been characterized and grouped using biosensor competition as well as being grouped according to which peptides they recognized. Previous groups (group I to group XI) were maintained, and new groups (XII to XVI) were added to accommodate MAbs that did not fit into known groups (37). Those shown in bold and in red have virus-neutralizing activity. MC12 recognizes the same amino acids as the previously mapped MAb BD66 and was placed in group XI. MC1 recognized synthetic peptide 1-20 and was placed in group VII. Eleven type-common MC MAbs were added to group IIa (previously defined only by BD78). Five new groups (IIc, XIV, XV, XVI, and XVII) accounted for the remaining MC MAbs. (B) Structures of gD showing the receptor-binding face and the fusion activation face.
Table 1.
Virus neutralization by anti-gD MAbsa
| MAb | IgG (μg /ml) needed for 50% neutralization of: |
|
|---|---|---|
| HSV-1 (KOS) | HSV-2 (333) | |
| MC2 | >25 | 0.781 |
| MC5 | 3.1 | 6.2 |
| MC23 | 1.6 | 0.260 |
| DL11 | 0.004 | 0.312 |
| 1D3 | 0.390 | 6.2 |
| MC4 | >25 | >25 |
| MC10 | >25 | >25 |
| MC14 | >25 | >25 |
| DL6 | >25 | >25 |
| BD66 | >25 | >25 |
| BD78 | >25 | >25 |
Data are expressed as amounts of IgG needed to neutralize 50% of the visible plaques formed by HSV on Vero cells at 48 h postinfection. The starting number of plaques was 100.
As we hypothesize that gD has a post-receptor-binding, fusion-promoting function (19, 30), we set out to identify gD-specific antibodies that would neutralize virus but still allow binding to both receptors. Since we lacked gD2-specific MAbs in our collection, we utilized gD2(s322) as the immunogen to generate 23 new MAbs designated MC1 through MC23. We characterized each antibody and constructed an antibody “tree” (Fig. 1A), ultimately relating them to each other and our previously characterized antibodies. The majority of the MC MAbs were type common, but five reacted either preferentially or exclusively with a truncated form of gD2, gD2(306t) (Fig. 1A and 2). MAbs that reacted with denatured gD were further mapped with overlapping synthetic peptides (data not shown) as previously reported (6, 8). MAbs in type-common groups IIa and IIc recognized amino acids 262 to 272 (Fig. 1A), and none had virus-neutralizing activity. Among the MAbs in group IIa, MC4, MC10, and MC14 were chosen as negative controls in later experiments.
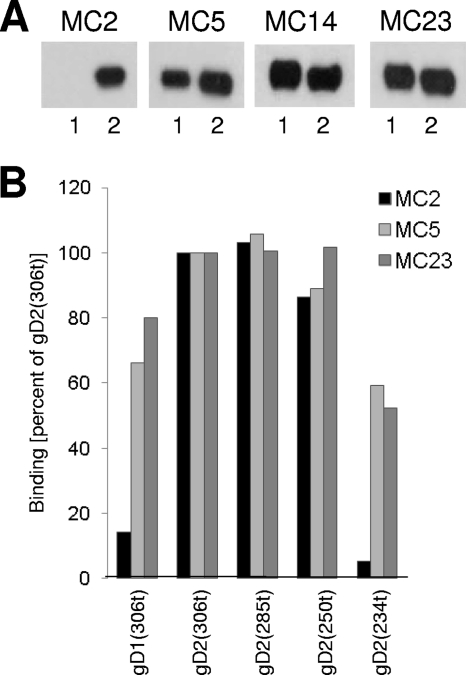
Fig 2.
Mapping of antibody epitopes on gD for the MC MAbs. (A) Western blot analysis of MC2, MC5, MC14, and MC23. Purified truncated gD1(306), indicated as 1, or gD2(306), indicated as 2, were subjected to SDS-PAGE under nondenaturing conditions. Western blots were probed with the indicated MAbs. (B) ELISA. Truncated gD1(306t), as well as gD2 truncated at amino acid 306, 285, 250, or 234, was reacted with the same IgG concentration of MC2, MC5, or MC23 for 1 h at RT. Bound antibody was detected by an anti-mouse secondary antibody and ABTS [2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid)] substrate. Absorbance values were normalized by setting the values obtained for each MAb binding to gD2(306t) as 100%.
Most importantly, only MC2, MC5, and MC23 exhibited virus-neutralizing activity (Fig. 1A and Table 1) and all three recognized only nondenatured gD. Since MC5 and MC23 were type common but MC2 was type 2 specific (Fig. 2A), studies for this report used HSV-2, gD2, and gD2 peptides. We then pursued three goals with these three MAbs: (i) to define the epitopes recognized by each; (ii) to test whether any of them could still bind gD in the presence of receptor; and (iii) to test whether any functioned by blocking gD binding to one or both receptors.
Mapping studies of conformation-dependent MAbs.
We first tested MC2, MC5, and MC23 against truncated forms of gD2 and linker insertion mutants (11) and then further grouped them by competition studies.
We found that MC5 and MC23 were able to bind to all truncations tested, including gD2(234t), whereas the shortest truncation seen by MC2 was gD2(250t) (Fig. 2B). This identified the epitopes for MC5 and MC23 as being upstream of residue 234 in the linear gD sequence. For MC2, the epitope is (at least in part) between amino acids 234 and 250. Interestingly, the only amino acid difference between gD1 and gD2 in this stretch of amino acids is at residue 246 (proline in gD2 and alanine in gD1). Thus, this amino acid might be in the MC2 epitope. We also examined binding of each of the MAbs to linker insertion mutants (11) and found that MC5 failed to recognize a gD2 mutant with an insertion at residue 77 (Fig. 1A, data not shown). Thus, part of the MC5 epitope may include this residue. Importantly, MC2 and MC23 were able to bind to this mutant, suggesting that their epitopes were distinct from that seen by MC5.
As a further means of mapping epitopes, we attempted to isolate mutants resistant to neutralization by MC2, MC5, and MC23 (21, 37). We succeeded in isolating such a monoclonal antibody-resistant (mar) mutant for MC23; it has a single mutation (threonine to methionine) at residue 213 (data not shown). Since other group Ia mar mutants map at residue 216 (35, 38), close to 213 in the gD crystal structure (Fig. 1B) (10), we tentatively assigned MC23 to this group.
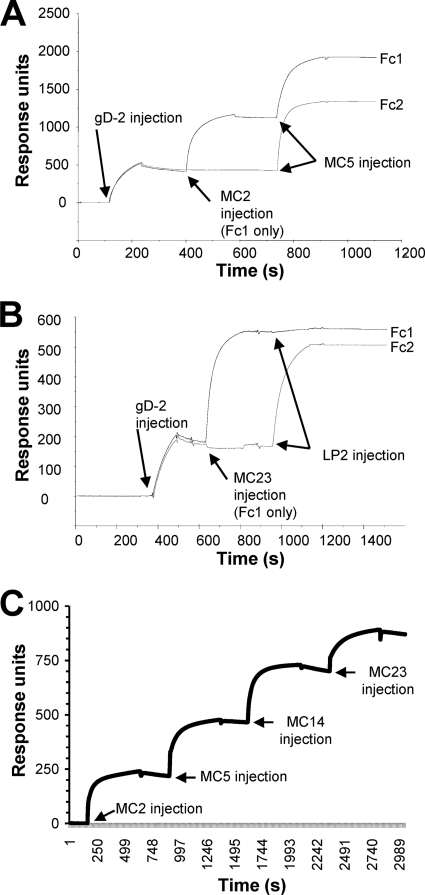
Biosensor analysis of MC2, MC5, and MC23.
Because we obtained only limited information about the epitopes for these MAbs, we used surface plasmon resonance to determine the relationships of MC2, MC5, and MC23 to each other and to the control MAb MC14 (Fig. 3). Three sets of data illustrate how we used the biosensor for MAb competition. This assay worked well, since the MAbs had low off rates. Since MC2 is type 2 specific, gD2(306t) was bound to the sensor chip, and we examined binding of each MAb. Figure 3A shows that both MC5 and MC2 bound well to gD2(306t) when tested individually (the flow cells are abbreviated Fc1 and Fc2). We then injected MC5 over the chip containing MC2 bound to gD (Fc1), and MC5 still bound well to gD. This indicated that the epitope for MC5 on gD was not obscured by the presence of MC2. Likewise, MC2 still bound well when injected over a chip containing MC5 bound to gD (data not shown). These data suggest that MC2 and MC5 recognize different sites on gD. In similar studies, neither MC2 nor MC5 competed with any of the other neutralizing MAbs (e.g., 1D3, DL11, or LP2) for binding to gD (Fig. 1, data not shown).
Fig 3.
MAb blocking of gD binding to receptors measured by surface plasmon resonance. Anti-His MAb was coupled to a CM5 biosensor chip, and gD2(306t) was captured via its C-terminal His tag on flow cell 1 (Fc1) and Fc2. The first MAb was injected across Fc1 for 2 min at 5 μl/min (control flow cell). The next MAb was then injected across both Fc1 and Fc2 for 2 min. (A) Sensorgram obtained using MAbs MC2 (1st MAb) and MC5 (2nd MAb). Since both bind to gD, there was no competition. (B) Sensorgram obtained using MC23 (1st MAb) and LP2 (2nd MAb). In this case, when MC23 was injected first, LP2 failed to bind, indicating they competed for the same epitope on gD. (C) Sensorgram of MC2, MC5, MC14, and MC23, each added sequentially to gD in Fc1. Binding of gD2(306t) to the anti-His MAb is not shown. In this case, each MAb bound to gD independently of whether the other MAbs were present, indicating that all four recognize distinct epitopes on gD.
Second, we examined whether MC23 would compete with LP2, a prototypical group Ia MAb (35). In this case, when MC23 was bound to gD, binding of LP2 was blocked by more than 90% (Fig. 3B). This blocking was reciprocal, i.e., when LP2 was bound first, MC23 no longer bound to gD2(306t) (data not shown). Since the two MAbs competed with each other for binding to gD, the data indicated that their epitopes overlap and that MC23 is a group Ia MAb. Moreover, these data predicted that MC23 functions by interfering with nectin-1 binding to gD. As a third example, we first injected MC2 across gD2. Once binding reached a plateau, we injected MC5, then MC14, and finally MC23 (Fig. 3C). In each case, the newly injected MAb bound to gD2(306t), showing that these four MAbs recognize distinct epitopes on gD. Since MC23 (like other group Ia MAbs) likely functions by blocking nectin-1 binding, we turned our attention to MC2 and MC5 to determine whether they functioned independently of receptor binding.
Does MC2 or MC5 block gD binding to receptors?
We took several approaches to address this question.
(i) Coimmunoprecipitation of gD and nectin-1 in the absence or presence of gD MAbs.
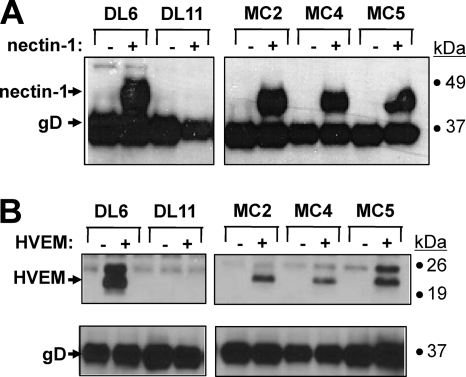
In previous studies, we showed that the nonneutralizing MAb DL6 could coprecipitate gD bound to HVEM (40). In contrast, neutralizing MAbs 1D3 (group VII) and DL11 (group Ib) did not coprecipitate the complex of gD and HVEM (40). Here, using a similar approach, we preincubated gD2(306) with nectin-1 (Fig. 4A) or HVEM (Fig. 4B), followed by addition of MAbs to these mixtures. For both receptors, we used DL6 as a positive control because it coprecipitates gD with HVEM and nectin-1; DL11 was used as a negative control because it fails to coprecipitate either receptor (28, 40). When gD was preincubated with nectin-1 or HVEM, MAbs MC2, MC4, and MC5 each coprecipitated both proteins, showing that the epitopes for these MAbs were still accessible when gD was bound to either receptor. Since both MC2 and MC5 neutralize virus (Table 1), these results suggest that both MAbs block a critical gD function other than receptor binding.
Fig 4.
Coimmunoprecipitation of HVEM(200t) and nectin-1(346t) by anti-gD MAbs. Reaction mixtures of gD2(285t) in the presence (+) or absence (−) of purified truncated nectin-1(346t) (A) or HVEM(200t) (B) were immunoprecipitated by the indicated MAbs. DL6 and DL11 were used as positive and negative controls, respectively. For nectin-1, the Western blots were probed with anti-gD and anti-nectin-1 polyclonal Abs. For HVEM, two gels, one for HVEM and the other for gD, were processed after the coimmunoprecipitation, and each blot was probed. Secondary Abs were then added, and enhanced chemiluminescence was used to visualize the bands.
(ii) ELISA.
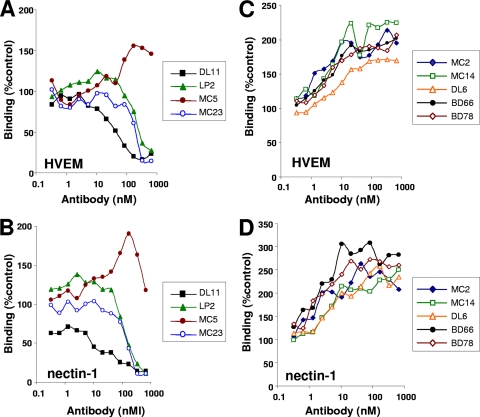
We next determined the extent to which these antibodies could prevent receptors from binding to gD. To do this, we incubated a set amount of gD2(306t) with increasing amounts of each antibody, allowed them to bind, and then added the mixtures to ELISA plates coated with HVEM (Fig. 5A and C) or nectin-1 (Fig. 5B and D). In this case, MAbs DL11, LP2, and MC23 effectively blocked the binding of gD to HVEM or nectin-1 in a dose-dependent manner. However, in contrast, MC5 did not block gD binding to either receptor. In fact, at higher concentrations, MC5 appeared to enhance binding of gD to both receptors. MC2 significantly enhanced binding of either HVEM or nectin-1 to gD (Fig. 5C and D). To our surprise, several nonneutralizing MAbs, including MC14, DL6, BD66, and BD78, had this property. The data suggest that binding of MC2 as well as these nonneutralizing antibodies alters gD structure in some way to allow better access of receptors to associate with their binding sites.
Fig 5.
Effect of MAbs on binding of gD2(306t) to HVEM and nectin-1. Serial 2-fold dilutions of each MAb IgG were incubated for 1 h with a constant amount of gD2(306t). The gD-IgG mixture was then added to an ELISA plate on which either HVEM (A and C) or nectin-1 (B and D) had previously been immobilized. Binding of gD was then detected with polyclonal antibody R8 followed by secondary antibody and substrate. The results are presented in separate panels for clarity (note the differences in the y-axis scale values). Results were normalized to the amount of gD bound following incubation with a control IgG (anti-myc). Results of a representative experiment are shown; each antibody was tested a minimum of three times.
To what extent do MC2, MC5, and MC14 enhance gD binding to receptor?
Previously, we found that the affinity of gD(306t) for both HVEM and nectin-1 is in the micromolar range whereas the affinity of gD(285t) for these receptors is enhanced significantly (approximately 50-fold) (44, 60). This is explained by crystallographic data that showed that the C-terminal residues of the gD ectodomain lie across the core of gD and interact with the N terminus, thereby impeding receptor binding (16, 30, 60). Thus, removal of the residues from 286 to 306 likely relieves the impediment to receptor binding. Additionally, we showed that a mutant form of gD with a single amino acid change at residue 294 (from a tryptophan to alanine) also has very high affinity for receptors, suggesting that it acts as a linchpin that helps to stabilize the C-term in a “closed” position across the gD core (16, 30). In the crystal structure, W294 fits into a groove near the gD N terminus and forms hydrogen bonds with several key residues in that region. Thus, receptor binding requires that the C-term be in an “open” conformation.
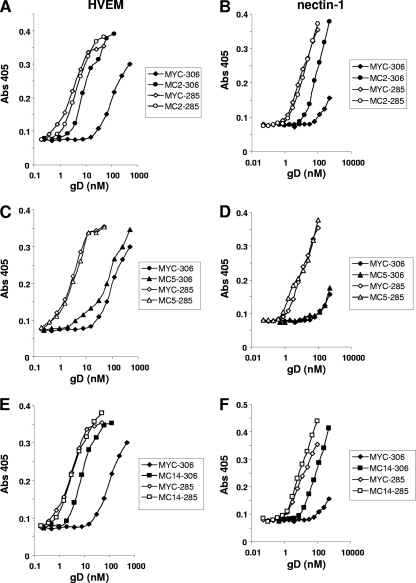
To quantify the extent of the enhancement of receptor binding, a single (excess) concentration of MAb MC2, MC5, MC14, or MYC IgG was incubated with increasing amounts of gD2(306t) or with gD2(285t). Each mixture was then added to an ELISA plate coated with HVEM (Fig. 6A, C, and E) or nectin-1 (Fig. 6B, D, and F). The anti-MYC antibody control reflects the normal binding capacity of each form of gD to receptors. As expected (44, 60), gD2(285t) bound to HVEM and nectin-1 better than gD2(306t). Interestingly, MC2 enhanced the binding of gD2(306t) to HVEM almost as well as the C-terminal truncation [gD2(285t)] (Fig. 6A). This antibody also enhanced gD2(306t) binding to nectin, but the effect was not as dramatic as with HVEM (Fig. 6B). These results support our concept that binding of MC2 affects the position of the C-term of gD. In contrast, MC5 had a much more modest enhancing effect on binding of gD2(306t) to HVEM (Fig. 6C) and had no effect at all on gD2(306t) binding to nectin-1 (Fig. 6D). As predicted, neither MC2 nor MC5 enhanced the binding of gD(285t) to HVEM or nectin-1. Finally, MC14 enhanced binding of gD2(306t) to both HVEM and nectin-1 (Fig. 6E and F) and the extent of this enhancement mirrored that seen with MC2. Biosensor analysis indicated that the increased affinity of gD2(306t) for HVEM and nectin-1 caused by MC2 or MC14 was due to an increase in the rate of association (kon), with little change in dissociation (koff) (data not shown). This pattern was identical to that seen for the kinetics of gD2(285t) binding to HVEM and nectin-1 (44, 60). These experiments support the concept that binding of MC2 or MC14 to gD induces a more “open” conformation of the C-term, thereby enhancing the ability of gD to bind the receptor.
Fig 6.
Effect of MAbs MC2 and MC5 on receptor binding to gD2(306t) or gD2(285t). Serial 2-fold dilutions of the two forms of gD2 were mixed with antibody MC2 (A and B), MC5 (C and D), or MC14 (E and F) at a concentration of 0.5 μM for 1 h. The gD-IgG mixture was then incubated with purified receptor that had been immobilized to an ELISA plate. Binding of gD was detected with polyclonal anti-gD antibody R8 and anti-rabbit secondary antibody. Results of a representative experiment are shown; each antibody was tested a minimum of three times.
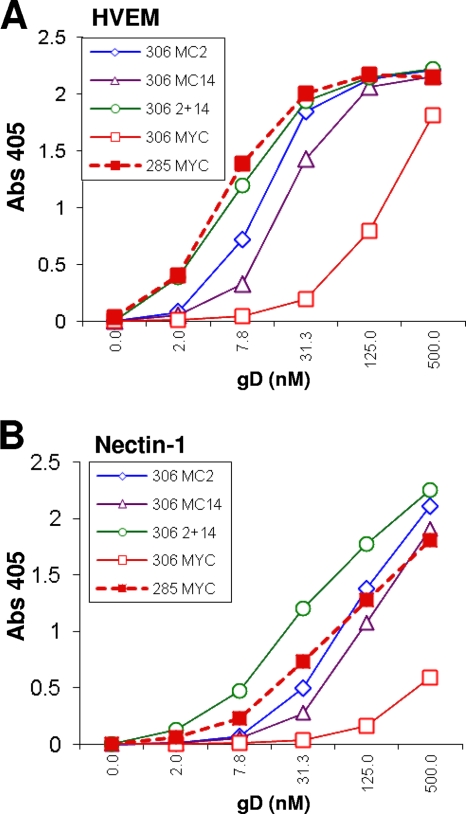
Since MC2 and MC14 recognize different epitopes, we wondered whether binding of gD2(306t) to receptors would be even more enhanced if we mixed the two MAbs. If so, this would suggest that there may be two different mechanisms to alter the C-term. Therefore, we tested the effect of a combination of MC2 with MC14 on receptor binding (Fig. 7). As shown before (Fig. 6), the presence of MC2 or MC14 alone enhanced the binding of gD2(306t) to either HVEM (Fig. 7A) or nectin-1 (Fig. 7B). The combination of the two MAbs enhanced gD2(306t) binding to HVEM and nectin-1 even more. Overall, our data suggest that the combination of MC2 with MC14 has a greater enhancing effect on receptor binding to gD than does either MAb alone. This supports the idea that MC2 and MC14 may affect gD conformation in different ways. A similar effect was seen for MC4 or MC10 in combination with MC2 (data not shown). More importantly, our observations about enhancement of receptor binding led us to wonder whether the nonneutralizing MAbs MC4, MC10, and MC14 would enhance either the extent or kinetics of virus neutralization by MC2 (46).
Fig 7.
Effect of mixtures of MAbs MC2 and MC14 on receptor binding of gD2(306t) or gD2(285t). (A) Serial 2-fold dilutions of the two forms of gD2 were mixed with MYC (control antibody) or MC2 or MC14 or a mixture of MC2 and MC14 and then incubated with purified HVEM that had been immobilized on an ELISA plate. (B) The procedure was the same as described for panel A, except that nectin-1 was bound to the ELISA plate. Binding of gD was detected with polyclonal anti-gD antibody R8 and anti-rabbit secondary antibody. Results of a representative experiment are shown; each antibody was tested a minimum of three times.
Effect of MC4, MC10, and MC14 on HSV-2 neutralization by MC2.
Sanna et al. (46) showed that the combination of H170, a group VII MAb, and H128, a group 1b MAb (Fig. 1), was more effective at neutralizing HSV-2 infection than was either MAb alone both in terms of efficiency (50% endpoint) and kinetics (the rate of neutralization as a function of time of exposure of virus to MAb). However, in that study, Sanna and colleagues combined two virus-neutralizing MAbs that function by interfering with receptor binding. Having shown that MC2 does not interfere with receptor binding but still neutralizes HSV2, we used a similar approach to investigate the extent and kinetics of neutralization by MC2 in the presence or absence of MAb MC4, MC10, or MC14. Additionally, we tested BD66 and DL6, two other nonneutralizing MAbs with epitopes in the C-term that enhanced receptor binding (Fig. 5D).
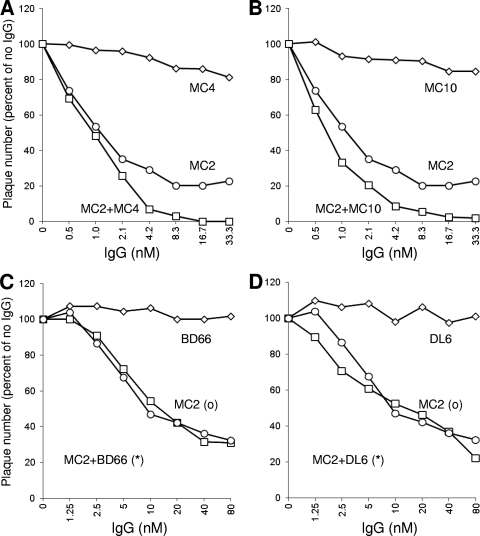
First, we examined the effect of MAbs that mapped to the C-term on neutralization by MC2 (Fig. 8). MAbs such as MC4, MC10 (Fig. 8A and B), and MC14 (data not shown) enhanced neutralization by MC2. These MAb combinations increased the amount of virus neutralized (to nearly 100%), whereas, in multiple experiments, MC2 alone never neutralized more than 80% of the input virus. Interestingly, neither DL6 (22) nor BD66 (22) had any enhancing effect on neutralization by MC2 (Fig. 8C and D). Thus, the enhancing effect may be due to how each MAb to a linear epitope along the C-term is oriented in relation to the orientation of MC2 (Fig. 1). No MAb enhanced neutralization by MC5, again emphasizing that it neutralizes virus differently than MC2 (data not shown). Thus, there is specificity among the MAbs that enhance receptor binding. In addition to MC4, MC10, and MC14, several other MAbs that map to this epitope in particular worked in concert with MC2 to enhance neutralization.
Fig 8.
Effect of MAbs to the C-term of gD on virus neutralization by MC2. In each experiment, serial 2-fold dilutions of MC2 were mixed either with antibody IgGs or with a control lacking Ig (“no IgG”) and then incubated with 100 PFU of virus for 1 h at 37°C. The virus-antibody mixtures were added to Vero cells, and plaques were allowed to develop. (A) Neutralization by MC4 or MC2 or a combination of MC4 (33.3 nM) plus MC2. (B) Neutralization by MC10 or MC2 or a combination of MC10 (33.3 nM) plus MC2. (C) Neutralization by BD66 or MC2 or a combination of BD66 (80 nM) plus MC2. (D) Neutralization by DL6 or MC2 or a combination of DL6 (80 nM) plus MC2. The results are expressed as percentages compared to the no-Ig control. Results of a representative experiment are shown; each antibody was tested a minimum of three times.
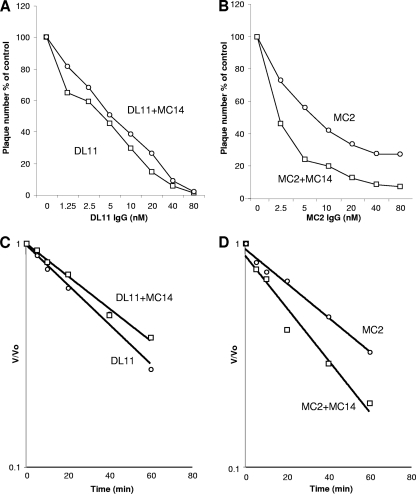
Having determined that MC4, MC10, and MC14 enhance the extent of neutralization by MC2, we tested how rapidly they neutralized virus compared with antibodies such as DL11 that block receptor binding. First, we repeated the neutralization experiment shown in Fig. 8, testing whether MC14 would enhance neutralization by DL11 (Fig. 9A and B). Again, we adjusted the IgG concentration and measured plaque reduction at 1 h after the addition of DL11 alone or in combination with MC14 (Fig. 9A and B). In contrast to what was seen with MC2 (Fig. 9B), MC14 had no effect on virus neutralization by DL11. We next compared the kinetics of neutralization by DL11 and MC2 in the presence or absence of MC14 (Fig. 9C and D). For this experiment, the amount of antibody (IgG) was held constant at 10 nM for both MC2 and DL11 and plaques were counted over time once the antibody virus mixture was added to the cells (46). We used 80 nM MC14 to be certain that any enhancement of kinetics would not be limited by the MC14 concentration. We then adjusted the time of incubation of the MAbs with virus (from 5 min to 1 h) before adding the mixture to cells. We plotted the data as the number of plaques at each time point divided by the number of plaques at time zero (no MAb) on the y axis (V/Vo) against time on the x axis (46). Thus, we could determine the rate from the slope of each line; the steeper the slope, the higher the rate of neutralization. As expected, antibody neutralization followed first-order kinetics in each case (i.e., a straight line). For DL11, the slopes (rate) were nearly identical in the presence or absence of MC14 (Fig. 9C). Similar results were found for MC23 or MC5 plus or minus MC14 (data not shown). However, in the case of MC2, the rate of neutralization was twice as high when both MC14 and MC2 were present compared with MC2 alone (Fig. 9D). We found the same enhancement in the kinetics of neutralization when we used different concentrations of MC2 (data not shown). This suggests that the kinetics of neutralization by MC2 and the enhancing effect of MC14 are independent of MC2 concentration over a wide range. Together, these data suggest that MAb MC14 which is directed at residues 262 to 272 enhances both the extent and the rate of neutralization by MC2.
Fig 9.
Effect of MAb MC14 on the rate of virus neutralization by DL11 or MC2. (A and B) Neutralization with antibody concentrations. Comparison of DL11 plus or minus 80 nM MC14 (A) versus MC2 plus or minus 80 nM MC14 (B). In each experiment, serial 2-fold dilutions of DL11 or MC2 were mixed either with MC14 or with a control lacking Ig and then incubated with 100 PFU of virus for 1 h at 37°C. The virus-antibody mixtures were added to Vero cells, and plaques were allowed to develop. The results are expressed as percentages compared to the control lacking Ig. (C and D) Kinetics of neutralization. For this assay, the concentrations of MAbs were fixed and the time during which MAbs and virus were incubated was adjusted (0 to 60 min). At each time point, the virus-antibody mixture was rapidly diluted in growth medium and added to cells to measure the virus neutralization over time. The results are plotted as V/V0, where V is the number of plaques remaining at each time point and V0 is the number of plaques at time zero. This number was plotted against time. A straight-line relationship between V/V0 and time represents first-order kinetics, and the slope of the line represents the rate of virus neutralization. Results of a representative experiment are shown; each antibody was tested a minimum of three times.
DISCUSSION
In previous studies of MAbs to HSV gD, we identified a number that have virus-neutralizing activity. In all cases, the antibodies prevented binding of gD to the HSV receptors HVEM and nectin-1. We concluded that this activity accounted for neutralization of HSV by gD antibodies. However, by solving the structures of gD bound to HVEM (10, 30) and gD bound to nectin-1 (16), we found that approximately 50 of the C-terminal amino acids (the C-term) of the gD ectodomain occluded the binding site for nectin-1 and also prevented formation of the N-terminal hairpin of gD needed for HVEM binding (16, 30). We hypothesized that in both cases, receptor binding necessitated a common conformational change in gD whereby the C-term moved away from the core, allowing receptors to bind. We postulated that gD with the C-term in this “open” position is an activated form of gD that is responsible for triggering events carried out by gH/gL and/or gB. These events then result in fusion of the virion envelope with a cell membrane to allow exposure of the capsid. Indeed, a gD mutant, which locked much but not all of the C-term of the gD ectodomain against the gD core, was still able to bind both gD receptors and yet was unable to trigger cell-cell or virus-cell fusion (31). This hypothesis was ultimately supported by ultrastructural studies that revealed the normal position of the C-term and proved that binding of either HVEM or nectin-1 to gD requires the same movement of the C-term to reveal the receptor-binding sites for both HVEM and nectin-1 (10, 16, 30). Presumably, these “open” sites on gD were the target of virus-neutralizing antibodies that blocked binding of gD to receptor.
As another means of testing our hypothesis, we searched for anti-gD MAbs that would not interfere with receptor binding but still neutralized virus infection. In screening a newly made panel of anti-gD2 MAbs, we found three that had virus-neutralizing activity. Of these, MC23 blocked gD binding to receptors and competed with another group Ia MAb (LP2). However, two others MAbs in the new panel, MC2 and MC5, bound to gD in the presence of the HVEM and nectin-1 receptors and neither MAb prevented these receptors from binding to gD. Thus, MAbs MC2 and MC5 had unique properties that might interfere with a gD function other than receptor binding.
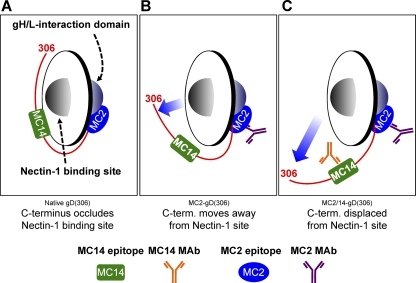
We have modeled antibody-induced changes to gD in Fig. 10, using MC2 and MC14 sites as examples of what happens when MAbs such as MC2 and MC14 bind. For the sake of simplicity, we did not include MC5 or MAbs to the C-term such as MC4 and MC10. We hypothesize that MC2 and MC5 neutralize virus by occluding residues needed to trigger later events. To test our hypothesis, we studied the properties of these MAbs in more detail. First, we found that binding of receptors to gD was modestly enhanced by binding of MC5 to gD but was dramatically enhanced by binding of MC2 to gD. This led us to propose that gD is normally in a “closed conformation,” with the C-term bound to the core of gD (Fig. 10A). Binding of MC2 induces the C-term of gD2(306) to assume an “open” conformation similar to that caused by receptor (Fig. 10B). MC2 interferes with post-receptor-binding events that are critical for triggering virus-cell fusion. Based on previous results (2), we think that MC2 binds to and masks the gH/gL interaction domain (Fig. 10). Antibodies to residues along the C-term such as MC14 also cause gD to assume an open conformation, thereby enhancing the binding of both HVEM and nectin-1. We believe that this is the first report of anti-gD antibodies that neutralize HSV without blocking the binding of one or both of its receptors. It is also the first report showing that MAbs to gD can enhance receptor binding.
Fig 10.
Model depicting the receptor-binding and fusion activation domains of gD relative to the MC2 and MC14 epitopes. The Ig-like core of gD (residues 56 to 184) is represented as a disc. Each side of the disc represents a distinct protein interaction site. One side of gD interacts with cellular receptors (i.e., nectin-1), while the opposite side interacts with downstream elements of the HSV membrane fusion machinery (i.e., gH/gL). The C terminus (residues 240 to 306) is depicted as a red line extending from one side of gD to the other around the base of the Ig-like gD core. The MC2 and MC14 epitopes are represented by a blue oval and a green rectangle, respectively. (A) Native gD with the C terminus positioned across the nectin-1 binding site. In this conformation, receptor access is subjected to competition by the gD C-term. (B) The binding of either MC2 (shown) or MC14 to gD alters the gD conformation such that receptor binding is enhanced. The most likely explanation is that the C-term is displaced from its native conformation (movement is shown by the blue arrow). (C) When MC2 and MC14 are both bound to gD, the effect on gD structure is more profound. One manifestation of this structural alteration is an even greater increase in nectin-1 binding. We hypothesize that the neutralizing activity of MC2 is a result of the proximity of its epitope to the fusion activation site. Perhaps MC14 augments MC2-mediated neutralization by imposing additional conformational constraints on the fusion activation domain. Alternatively, MC14 may simply enhance MC2 binding or Ab:Ag complex stability.
Second, we found that the epitopes for MC2 and MC5 are distinct from each other, as shown by three kinds of data: (i) MC2 is type 2 specific and MC5 is type common; (ii) MC2 can bind a form of gD truncated to amino acid 234, whereas MC2 cannot bind this form but does bind a form truncated at amino acid 250; and (iii) biosensor data show that MC2 and MC5 can bind to gD at the same time (no competition). We cannot precisely map where they bind, since both are conformation dependent, and we were unable to generate mar mutants for either MAb. Thus far, our data suggest that a portion of the MC5 epitope is found downstream of residue 250 but may involve amino acids surrounding amino acid 77, which is near residue 250 in the 3-dimensional structure. In the case of MC2, a portion of its epitope is between amino acids 234 and 250. Because MC2 is type 2 specific and residue 246 is the only residue that differs between 234 and 250 of gD1 and gD2, we speculate that it is critical for the MC2 epitope. However, further experiments need to be done to define other amino acids involved in binding of both MC2 and MC5.
Enhancement of receptor binding by MC14 and other MAbs.
To our surprise, we found that binding of gD to a receptor is significantly enhanced not only by MC2 but also by several nonneutralizing MAbs, including MC14 as well as its sister MAbs MC4 and MC10, all of which recognize and bind to residues 262 to 272 in the C-term (Fig. 10B and C). In addition, receptor binding is enhanced by other MAbs, such as DL6, whose epitopes are along the C-term of gD, but these MAbs do not enhance the effect of MC2. It should be noted that none of these MAbs would be predicted to interfere with receptor binding, and the fact that they do not neutralize virus infection or neutralize only weakly means that they do not block a downstream function. However, we believe that our general hypothesis explains why particular MAbs enhance receptor binding, i.e., they all cause the C-term to move away from gD and “open up” the receptor-binding sites (Fig. 10C).
Which function in the pathway to fusion is blocked by MC2?
Previously, we proposed that HSV-induced cell fusion occurs in a series of steps that ultimately activate gB into a fusogenic state (2). Movement of the gD C-term also reveals a domain that activates downstream steps, the first of which we proposed to be an interaction with gH/gL. This interaction with gD activates gH/gL into a form that enables it to help convert gB into a fusogenic state. Briefly, cells engineered to express nectin-1 and transfected to express gB could be triggered to fuse by addition of soluble forms (ectodomains) of gD and gH/gL. This experiment showed that neither gD nor gH/gL needs to be membrane bound to function. We further showed that brief exposure of nectin-1-bearing cells transfected to express gH/gL to soluble gD enabled those cells to fuse with cells transfected to express gB. Together, these results suggested that the pathway to fusion consists of three major steps: (i) binding of gD to receptor; (ii) activation of gH/gL by receptor-activated gD; and (iii) activation of gB into a fusogen by activated gH/gL. Although MC2 could interfere with either step i or ii, we currently favor the idea that it interferes with the interaction between gD and gH/gL. Thus far, using the technique of bimolecular complementation (24, 25), we have found that MC2 blocks the interaction between half-enhanced-yellow-fluorescent-protein (EYFP)-tagged forms of gB and gH/gL as well as fusion (data not shown and reference 3). However, when we used half-EYFP-tagged forms of gD and gH along with untagged gB and gL, we detected an interaction between gD and gH/gL, as evidenced by EYFP fluorescence, but fusion was blocked (3). This might imply that there is an unstable interaction between gD and gH/gL and that forming the irreversible EYFP-complemented complex might block the ability of gH/gL to activate gB. However, it could also mean that the complex formed in this setting did not represent the appropriate one to trigger gB to cause fusion. In any event, we were unable to use this technique to assess the ability of MC2 to block an interaction between gH/gL and gD that would lead to fusion. Alternative approaches are being explored to determine whether MC2 blocks a functional interaction between gD and gH/gL or another downstream event that normally leads to fusion.
It is of some interest that those MAbs that neutralize by blocking receptor binding appear to cluster on one face of the three-dimensional structure of gD, the same face where receptors bind, whereas MC2 and MC5 bind to epitopes on the opposing face (Fig. 1B and 10). We suggest that the uncovered residues of the gD core directly contact a site on gH/gL that upregulates its activity (2).
The combination of MC2 with MC14 enhances neutralization by MC2.
It was previously noted that antibodies to two distinct epitopes on gD can behave synergistically, but in that experiment, both antibodies blocked receptor binding, i.e., H170 (group VII) blocked binding of HVEM and H128 (group 1b) blocked both. We know from earlier studies that group VII and 1b MAbs do not compete with each other (11, 13, 17, 36). Here, we found an enhancing effect on both the extent and rate of neutralization by combining the neutralizing MAb MC2 with a nonneutralizing MAb such as MC4, MC10, or MC14. This indicates that nonneutralizing MAbs can contribute, albeit indirectly, to the overall neutralization seen in a polyclonal response to gD. The data suggest that binding of these nonneutralizing antibodies alters gD structure such that access of receptors is enhanced (Fig. 10). Since the MC2 epitope appears to be located away from receptor-binding sites, we suggest that MC14 allows better exposure of the MC2 epitope on gD, thereby accounting for the enhanced effect on the extent and rate of neutralization.
Virus neutralization by antibodies that block receptor binding.
Interference with receptor binding is a common mechanism by which antibodies block virus infection (49, 61). For instance, a previous study showed that a combination of neutralizing human MAbs against severe acute respiratory syndrome (SARS) coronavirus acted synergistically by targeting different epitopes of the spike (S) protein (61). However, the authors of that study did not determine whether any MAbs allowed receptor binding and did not examine whether the MAbs interfered with a preattachment step or one that occurred after attachment. In a recent study, Sabo et al. studied a large panel of MAbs to the attachment protein of hepatitis C virus (45) and found some that blocked preattachment and others that blocked at a postattachment step. In that study, the receptor was not known, so one does not know which of the MAbs interfered with receptor binding itself. In our studies, we have been able to determine which MAbs to gD interfere with receptor binding and which do not. We did not test pre- versus postattachment effects of these MAbs, because attachment to the cell surface, as opposed to receptor binding, is largely due to interaction of the virus with heparan sulfate (5, 23, 53). However, since the neutralization assays were done on Vero cells, the effect of these MAbs did not depend on a step that occurs only in endosomes, since virus entry into Vero cells occurs at the plasma membrane (34).
Neutralization of other viruses by other antibodies that block at a post-receptor-binding step.
For other enveloped viruses, there are a number of reports of MAbs that block entry at a post-receptor-binding step, e.g., ones directed at SU, the murine leukemia virus (MuLV) receptor-binding protein (7). In this case, the antibodies are thought to block interactions between the receptor-binding protein and the fusion protein that are important for triggering fusion. Indeed, it has been recognized for a number of years that virus neutralization can occur both at the receptor-binding step and at any other step that precedes “the first major biosynthetic event in the viral replicative cycle” (26). Since many viruses are taken into endocytic compartments, bound antibodies are also taken into the cell, allowing them to act not just at the cell surface but “inside” the cell. For example, Vogt et al. (56, 57) showed that certain MAbs to West Nile virus (WNV) neutralize at a postattachment step. Moreover, as Klasse and Sattentau stress (26), the mechanisms of neutralization should block an event in the replicative cycle and the kinetics of neutralization may vary due to differences in which event is blocked.
Recently, an MAb (E53) to WNV envelope protein E was discovered to block the acid-catalyzed fusion step required for release of the nucleocapsid from the endocytic compartment it is taken into (54). It does so by binding to the fusion loop of the E protein, thereby preventing WNV from binding to the membrane bilayer. In another study with this MAb, it was discovered that the kinetics of neutralization between the virus and MAb increased over the time of incubation (43). Those authors suggested that the MAb actually engages an epitope (the fusion loop) that is not well exposed on the virion surface. In their study, they examined the kinetics of neutralization by incubating the virus and MAb for many hours. According to our hypothesis, MAbs that enhance receptor binding to gD cause the C-term of gD to move, thereby making receptor-binding sites more accessible. Perhaps we may find that the kinetics of MC2 or of the combination of MC2 with MC14 is no longer linear if we extend the time of incubation from 1 h to a longer period.
Some gD mutants can bind alternative receptors. How can we reconcile them with our hypothesis regarding open and closed forms of the C-term?
Other studies have shown that large portions of gD can be deleted and replaced by domains that bind new receptors (9). This has led to some skepticism about our model for receptor-induced activation of gD. A recently published study of gD bound to nectin-1 (16) pointed out that, even when a large portion of the gD core was replaced with a single-chain antibody, the N- and C-terminal extensions to the Ig core were maintained. Thus, the regions of gD that are needed for its activation and binding to other viral glycoproteins may still be maintained in this hybrid construct. Therefore, we predict that even in these cases, receptor binding would necessitate displacement of the C-term. This could be tested by measuring the affinity of these mutated forms of gD for their receptors when the C-term is present and when it is absent. The resulting virus was highly attenuated, and it was found that “HSV evolved to unmask the gD C terminus only upon receptor binding to ensure efficient tropism in the host.”
Such studies are beyond the scope of the present report but are ones that should be considered, particularly by those who are interested in using gD with altered receptor tropism for gene therapy. By knowing whether displacement of the C-term is always needed for gD to activate downstream events, one could predict which type of receptor-binding residues would be likely to produce a form of gD that was functional.
Finally, we believe that our hypothesis of how MC2 functions is well supported by ultrastructural studies that imply that the C-term of gD must move out of the way in order for nectin-1 to bind and for gD to form the loop that binds HVEM (10, 16, 30). One caveat is that MC2 might prevent binding of the C-term to gH/gL or gB, not necessarily linked to the displacement of the C-term, but as a consequence of a local perturbation. Likewise, MC14 might alter the capacity of receptor-bound gD to bind gH/gL and/or gB. The latter possibility, though not yet tested, would be important to explore. Ultimately, however, it would be of great value to know how the structure of gD bound to neutralizing MAbs that block receptor binding, e.g., DL11, compares with the structure of gD2 bound to MC2 and how the latter structure compares with those of gD bound to nectin-1 and gD bound to HVEM. Such structures are clearly ones we would be greatly interested in knowing and would help us understand much more about how MC2 works.
ACKNOWLEDGMENTS
We thank all the members of the Cohen-Eisenberg laboratory for helpful discussions of the work reported here. We particularly thank Huan Lou for providing purified soluble glycoproteins. Special thanks to Leslie B. King of the School of Veterinary Medicine at the University of Pennsylvania for careful editing of the manuscript.
This work was supported by NIH grants R01-AI-056045 and R01-AI-076231 to R.J.E. and by NIH grant R37-AI-18289 to G.H.C. E.L. was supported by NIH T32-07234. C.K. was supported by NIH grant R21-AI-073384 and by the Joseph and Josephine Rabinowitz award for Research Excellence at the UPENN School of Dental Medicine.
Experiments were carried out by E.L., J.C.W., S.H.W., M.P.-D.-L., T.M.C., and Y.Z. Intellectual input and manuscript preparation were done by E.L., R.J.E., G.H.C., C.K., T.M.C., and J.C.W.
Footnotes
Published ahead of print 30 November 2011
REFERENCES
- 1. Aldaz-Carroll L, et al. 2005. Epitope-mapping studies define two major neutralization sites on the vaccinia virus extracellular enveloped virus glycoprotein B5R. J. Virol. 79:6260–6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. 2010. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J. Virol. 84:12292–12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atanasiu D, et al. 2007. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc. Natl. Acad. Sci. U. S. A. 104:18718–18723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Backovic M, Longnecker R, Jardetzky TS. 2009. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 106:2880–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banfield BW, Leduc Y, Esford L, Schubert K, Tufaro F. 1995. Sequential isolation of proteoglycan synthesis mutants by using herpes simplex virus as a selective agent: evidence for a proteoglycan-independent virus entry pathway. J. Virol. 69:3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bender FC, et al. 2007. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J. Virol. 81:3827–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burkhart MD, Kayman SC, He Y, Pinter A. 2003. Distinct mechanisms of neutralization by monoclonal antibodies specific for sites in the N-terminal or C-terminal domain of murine leukemia virus SU. J. Virol. 77:3993–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cairns TM, et al. 2006. Epitope mapping of herpes simplex virus type 2 gH/gL defines distinct antigenic sites, including some associated with biological function. J. Virol. 80:2596–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campadelli-Fiume G, et al. 2011. Rethinking herpes simplex virus: the way to oncolytic agents. Rev. Med. Virol. 21:213–226 [DOI] [PubMed] [Google Scholar]
- 10. Carfí A, et al. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169–179 [DOI] [PubMed] [Google Scholar]
- 11. Chiang HY, Cohen GH, Eisenberg RJ. 1994. Identification of functional regions of herpes simplex virus glycoprotein gD by using linker-insertion mutagenesis. J. Virol. 68:2529–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chowdary TK, et al. 2010. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat. Struct. Mol. Biol. 17:882–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cohen GH, Isola VJ, Kuhns J, Berman PW, Eisenberg RJ. 1986. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with Western blotting. J. Virol. 60:157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cohen J. 2010. Immunology. Painful failure of promising genital herpes vaccine. Science 330:304. [DOI] [PubMed] [Google Scholar]
- 15. Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. 2011. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 9:369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Di Giovine P, et al. 2011. Structure of herpes simplex virus glycoprotein D bound to the human receptor nectin-1. PLoS Pathog. 7:e1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eisenberg RJ, et al. 1985. Localization of epitopes of herpes simplex virus type 1 glycoprotein D. J. Virol. 53:634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harrison SC. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heldwein EE, Krummenacher C. 2008. Entry of herpesviruses into mammalian cells. Cell. Mol. Life Sci. 65:1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heldwein EE, et al. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217–220 [DOI] [PubMed] [Google Scholar]
- 21. Highlander SL, et al. 1987. Neutralizing monoclonal antibodies specific for herpes simplex virus glycoprotein D inhibit virus penetration. J. Virol. 61:3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Isola VJ, et al. 1989. Fine mapping of antigenic site II of herpes simplex virus glycoprotein D. J. Virol. 63:2325–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karger A, Saalmuller A, Tufaro F, Banfield BW, Mettenleiter TC. 1995. Cell surface proteoglycans are not essential for infection by pseudorabies virus. J. Virol. 69:3482–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kerppola TK. 2008. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37:465–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kerppola TK. 2008. Bimolecular fluorescence complementation: visualization of molecular interactions in living cells. Methods Cell Biol. 85:431–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klasse PJ, Sattentau QJ. 2002. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J. Gen. Virol. 83:2091–2108 [DOI] [PubMed] [Google Scholar]
- 27. Krummenacher C, et al. 2000. Localization of a binding site for herpes simplex virus glycoprotein D on herpesvirus entry mediator C by using antireceptor monoclonal antibodies. J. Virol. 74:10863–10872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krummenacher C, et al. 1998. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J. Virol. 72:7064–7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krummenacher C, et al. 1999. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC. J. Virol. 73:8127–8137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krummenacher C, et al. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 24:4144–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lazear E, et al. 2008. Engineered disulfide bonds in herpes simplex virus type 1 gD separate receptor binding from fusion initiation and viral entry. J. Virol. 82:700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luftig MA, et al. 2006. Structural basis for HIV-1 neutralization by a gp41 fusion intermediate-directed antibody. Nat. Struct. Mol. Biol. 13:740–747 [DOI] [PubMed] [Google Scholar]
- 33. Matsuura H, Kirschner AN, Longnecker R, Jardetzky TS. 2010. Crystal structure of the Epstein-Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. Proc. Natl. Acad. Sci. U. S. A. 107:22641–22646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Milne RS, Nicola AV, Whitbeck JC, Eisenberg RJ, Cohen GH. 2005. Glycoprotein D receptor-dependent, low-pH-independent endocytic entry of herpes simplex virus type 1. J. Virol. 79:6655–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Minson AC, et al. 1986. An analysis of the biological properties of monoclonal antibodies against glycoprotein D of herpes simplex virus and identification of amino acid substitutions that confer resistance to neutralization. J. Gen. Virol. 67:1001–1013 [DOI] [PubMed] [Google Scholar]
- 36. Muggeridge MI, et al. 1988. Antigenic analysis of a major neutralization site of herpes simplex virus glycoprotein D, using deletion mutants and monoclonal antibody-resistant mutants. J. Virol. 62:3274–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muggeridge MI, Roberts SR, Isola VJ, Cohen GH, Eisenberg RJ. 1990. Herpes simplex virus, p 459–481 In VanRegenmortel MHV, Neurath AR. (ed), Immunochemistry of viruses, vol II. The basis for serodiagnosis and vaccines. Elsevier Biochemical Press, Amsterdam, The Netherlands [Google Scholar]
- 38. Muggeridge MI, Wilcox WC, Cohen GH, Eisenberg RJ. 1990. Identification of a site on herpes simplex virus type 1 glycoprotein D that is essential for infectivity. J. Virol. 64:3617–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Muggeridge MI, et al. 1990. Antigenic and functional analysis of a neutralization site of HSV-1 glycoprotein D. Virology 174:375–387 [DOI] [PubMed] [Google Scholar]
- 40. Nicola AV, et al. 1998. Monoclonal antibodies to distinct sites on herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J. Virol. 72:3595–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nicola AV, Willis SH, Naidoo NN, Eisenberg RJ, Cohen GH. 1996. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J. Virol. 70:3815–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pereira L, Klassen T, Baringer JR. 1980. Type-common and type-specific monoclonal antibody to herpes simplex virus type 1. Infect. Immun. 29:724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pierson TC, Doms RW. 2003. HIV-1 entry and its inhibition. Curr. Top. Microbiol. Immunol. 281:1–27 [DOI] [PubMed] [Google Scholar]
- 44. Rux AH, et al. 1998. Functional region IV of glycoprotein D from herpes simplex virus modulates glycoprotein binding to the herpesvirus entry mediator. J. Virol. 72:7091–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sabo MC, et al. 2011. Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step. J. Virol. 85:7005–7019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sanna PP, Ramiro-Ibanez F, De Logu A. 2000. Synergistic interactions of antibodies in rate of virus neutralization. Virology 270:386–396 [DOI] [PubMed] [Google Scholar]
- 47. Serafini-Cessi F, Dall'Olio F, Malagolini N, Pereira L, Campadelli-Fiume G. 1988. Comparative study on O-linked oligosaccharides of glycoprotein D of herpes simplex virus types 1 and 2. J. Gen. Virol. 69:869–877 [DOI] [PubMed] [Google Scholar]
- 48. Sisk WP, et al. 1994. High-level expression and purification of secreted forms of herpes simplex virus type 1 glycoprotein gD synthesized by baculovirus-infected insect cells. J. Virol. 68:766–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531–569 [DOI] [PubMed] [Google Scholar]
- 50. Spear PG, Eisenberg RJ, Cohen GH. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1–8 [DOI] [PubMed] [Google Scholar]
- 51. Spear PG, et al. 2006. Different receptors binding to distinct interfaces on herpes simplex virus gD can trigger events leading to cell fusion and viral entry. Virology 344:17–24 [DOI] [PubMed] [Google Scholar]
- 52. Stanberry LR, et al. 2002. Glycoprotein-D-adjuvant vaccine to prevent genital herpes. N. Engl. J. Med. 347:1652–1661 [DOI] [PubMed] [Google Scholar]
- 53. Tal-Singer R, et al. 1995. Interaction of herpes simplex virus glycoprotein gC with mammalian cell surface molecules. J. Virol. 69:4471–4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thompson BS, et al. 2009. A therapeutic antibody against West Nile virus neutralizes infection by blocking fusion within endosomes. PLoS Pathog. 5:e1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vanarsdall AL, Ryckman BJ, Chase MC, Johnson DC. 2008. Human cytomegalovirus glycoproteins gB and gH/gL mediate epithelial cell-cell fusion when expressed either in cis or in trans. J. Virol. 82:11837–11850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vogt MR, et al. 2011. Poorly neutralizing cross-reactive antibodies against the fusion loop of West Nile virus envelope protein protect in vivo via Fc-{gamma} receptor and complement-dependent effector mechanisms. J. Virol. 85:11567–11580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vogt MR, et al. 2009. Human monoclonal antibodies against West Nile virus induced by natural infection neutralize at a postattachment step. J. Virol. 83:6494–6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Whitbeck JC, et al. 1999. The major neutralizing antigenic site on herpes simplex virus glycoprotein D overlaps a receptor-binding domain. J. Virol. 73:9879–9890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Whitbeck JC, et al. 1997. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J. Virol. 71:6083–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Willis SH, et al. 1998. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, using surface plasmon resonance. J. Virol. 72:5937–5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zeng F, et al. 2006. Quantitative comparison of the efficiency of antibodies against S1 and S2 subunit of SARS coronavirus spike protein in virus neutralization and blocking of receptor binding: implications for the functional roles of S2 subunit. FEBS Lett. 580:5612–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]