Abstract
The 5′-untranslated regions (5′ UTRs) of picornavirus genomes contain an internal ribosomal entry site (IRES) that promotes the end-independent initiation of translation. Picornavirus IRESs are classified into four structurally distinct groups, each with different initiation factor requirements. Here, we identify a fifth IRES class in members of Kobuvirus, Salivirus, and Paraturdivirus genera of Picornaviridae: Aichi virus (AV), bovine kobuvirus (BKV), canine kobuvirus (CKoV), mouse kobuvirus (MKoV), sheep kobuvirus (SKV), salivirus A (SV-A), turdivirus 2 (TV2), and TV3. The 410-nucleotide (nt)-long AV IRES comprises four domains (I to L), including a hairpin (L) that overlaps a Yn-Xm-AUG (pyrimidine tract/spacer/initiation codon) motif. SV-A, CKoV, and MKoV also contain these four domains, whereas BKV, SKV, and TV2/TV3 5′ UTRs contain domains that are related to domain I and equivalent to domains J and K but lack an AV-like domain L. These IRESs are located at different relative positions between a conserved 5′-terminal origin of replication and divergent coding sequences. Elements in these IRESs also occur elsewhere: domain J's apical subdomain, which contains a GNRA tetraloop, matches an element in type 1 IRESs, and eIF4G-binding motifs in domain K and in type 2 IRESs are identical. Other elements are unique, and their presence leads to unique initiation factor requirements. In vitro reconstitution experiments showed that like AV, but in contrast to other currently characterized IRESs, SV-A requires the DExH-box protein DHX29 during initiation, which likely ensures that the initiation codon sequestered in domain L is properly accommodated in the ribosomal mRNA-binding cleft.
INTRODUCTION
The genomes of RNA viruses contain structural elements that have important roles in translation, replication, and encapsidation. Their activities depend on structural integrity and on specific sequence motifs, thus limiting sequence variation, so that functionally related elements in different viruses commonly contain conserved sequence motifs and have a common architecture in which covariant substitutions maintain base-pairing patterns. Internal ribosomal entry sites (IRESs) are large, structured RNA elements that mediate the end-independent initiation of translation (28). Each of the five classes of viral IRES that have been identified to date has a different characteristic structure and promotes initiation by a distinct mechanism.
The simplest mechanism is used by the ∼180-nucleotide (nt)-long intergenic region (IGR) IRESs of dicistroviruses, which consist of three pseudoknots (52). They bind directly to the ribosome and mediate initiation without the involvement of eukaryotic initiation factors (eIFs) (61, 88). The second IRES class is epitomized by the ∼330-nt-long IRESs of hepatitis C virus (HCV) and pestiviruses (designated HP) (47) and includes structurally related HP-like IRESs that occur in numerous picornaviruses, including porcine kobuvirus (PKV), a member of the Kobuvirus genus, members of the Teschovirus, Sapelovirus, Senecavirus, Tremovirus, and Avihepatovirus genera of Picornaviridae, seal picornavirus 1, and turkey hepatitis virus (8, 14, 21, 26, 35, 37, 73). The presence of related IRESs in unrelated viruses suggests that these elements have been exchanged by recombination (21). HP-like IRESs bind directly to eIF3 and to the ribosomal 40S subunit, promoting 48S initiation complex formation without the factors that are required for the cap-dependent recruitment of 43S preinitiation complexes to mRNA (eIF4A, eIF4B, eIF4E, or eIF4G) or for scanning (eIF1 and eIF1A) (10, 57, 58, 66, 69).
The three other structurally defined classes of IRESs all occur in picornavirus 5′ untranslated regions (UTRs), which have a modular organization in which the origin of replication (ori) is followed by the IRES. Type 1 IRESs occur only in the Enterovirus genus (5). They are ∼450 nt long and contain five domains (II to VI), and their 3′ borders are marked by a Yn-Xm-AUG motif in which Yn, a pyrimidine tract (n; 8 to 10 nt), is separated by a spacer (m; 18 to 20 nt) from an AUG triplet (29, 63). This motif is the presumed point of ribosomal entry into the 5′ UTR (64) and is separated from the initiation codon by a nonconserved spacer (<30 nt in rhinoviruses but >150 nt in poliovirus). Conserved, functionally important nucleotides occur at the base of domain II, in the cruciform domain IV (including a GNRA tetraloop and a C-rich loop in adjacent apical arms), and in the central loop and basal half of domain V (3, 5, 11, 19). This region of domain V interacts specifically with eIF4G and eIF4A, an associated RNA helicase, which together promote the recruitment of 43S complexes to the IRES (11).
Type 2 IRESs occur in the Aphthovirus, Cardiovirus, and Parechovirus genera (5); divergent members occur in the Erbovirus and human Cosavirus genera (23, 34). They are ∼450 nt long and also have a 3′-terminal Yn-Xm-AUG motif, but in contrast to type 1 IRESs, the AUG triplet may be the initiation codon. Their five principal domains (H, I, J, K, and L) are unrelated to domains in type 1 IRESs, with the exception of domain I, which, like domain IV of type 1 IRESs, contains a C-rich loop and a functionally important GNRA tetraloop (5). However, in contrast to domain IV of type 1 IRESs, domain I in type 2 IRESs has a patriarchal cross (i.e., double-barred) conformation, and the stem-loops in which these motifs are located are not adjacent. Initiation on the IRESs of encephalomyocarditis virus (EMCV), a cardiovirus, and foot-and-mouth disease (FMDV), an aphthovirus, requires the specific binding of eIF4G and eIF4A to the Y-shaped J-K domain (65, 68), an interaction that is dependent on a conserved sequence/structural motif at the apex of domain J (4, 9, 39, 94). The IRESs from some EMCV isolates require only eIF4G/eIF4A to recruit a 43S complex consisting of eIF3, eIF2-GTP/initiator tRNA, and a 40S subunit to the initiation codon to form a 48S complex (62, 65). Type 2 IRESs therefore can function without eIF4E and factors implicated in ribosomal scanning, such as eIF1 and eIF1A, although some additionally require one or more IRES trans-acting factors (ITAFs), which are cellular RNA-binding proteins, such as the pyrimidine tract-binding protein (PTB) (54). All type 1 IRESs that have been characterized to date are dependent on ITAFs which, apart from PTB, are distinct from those used by type 2 IRESs (54).
The type 3 IRES occurs only in hepatitis A virus (HAV); its ∼410-nt-long core comprises two major domains (IV and V) followed by a Yn-Xm-AUG motif, but its function is significantly enhanced by the upstream ∼175-nt-long domains II and III (7). There are major structural and mechanistic differences between it and type 1 and type 2 IRESs, such as the length and sequence of core structural elements and the exceptional requirement of HAV IRES for the cap-binding protein eIF4E (2).
The existence of these structurally and mechanistically distinct classes of IRESs raises the question of whether other unrelated IRESs remain to be identified. To address this possibility, we recently characterized the 5′ UTR of Aichi virus (AV), a member of the Kobuvirus genus that causes acute gastroenteritis in humans (13), and found that it contains an ∼410-nt-long IRES (Fig. 1) that is structurally distinct from the different classes of IRESs described above (95). The AV IRES also has distinct factor requirements: unlike all other IRESs that have been characterized to date, 48S complex formation on it is dependent on DHX29 (95), a ribosome-associated DExH-box helicase that has been implicated in the correct accommodation of structured mRNA in the mRNA-binding channel of the 40S subunit (1, 71). The Kobuvirus genus currently comprises six accepted or proposed species: bovine kobuvirus (BKV), canine kobuvirus (CKoV), mouse kobuvirus (MKoV), sheep kobuvirus (SKV), AV, and PKV (33, 41, 67, 74, 90, 93). It is distantly related to salivirus A (formerly known as klassevirus) of the proposed Salivirus genus (17, 25, 42, 81) and to turdivirus 2 (TV2) and turdivirus 3 (TV3) of the proposed Paraturdivirus genus (89).
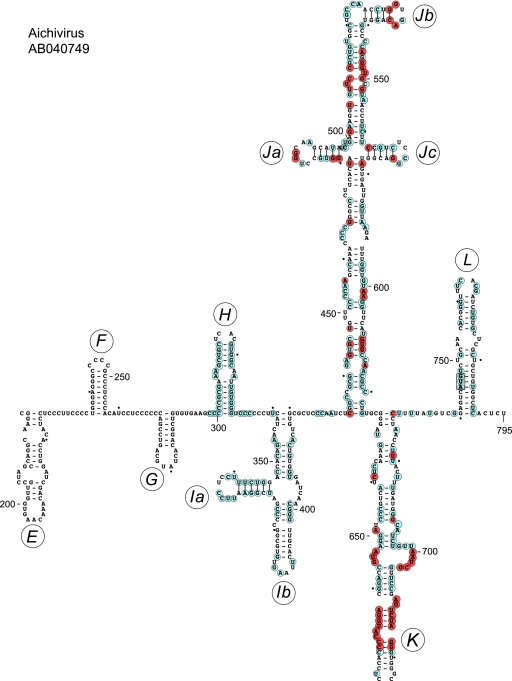
Fig 1.
Model of the structure of the 5′ UTR and adjacent coding region of Aichi virus downstream of domains A to C and the pseudoknot. This model was derived as described previously (95), and domains/subdomains are labeled sequentially from E to L. Nucleotides shown in cyan circles are conserved in all or all but one of the AV, CKoV, MkoV, and SV-A sequences listed in Materials and Methods; nucleotides shown in orange circles, which are deliberately limited to the highly conserved domains I and J, match these criteria and also are present in all currently available BKV, SKV, TV2, and TV3 sequences. The AV initiation codon AUG745 is boxed.
Previous analyses had identified a Yn-Xm-AUG motif in the AV 5′ UTR (n = 7; m = 11) and in two SV-A isolates (Y12-X8-AUG and Y12-X4-AUG) that included the initiation codon, which was taken as an indication that these viruses contain type 2 IRESs (17, 42, 91). However, the BKV and TV3 5′ UTRs contain divergent Y7-X37-AUG and Y6-X34-AUG motifs, respectively (89, 90), and several reports had noted that the secondary structures of these 5′ UTRs were unknown (81), could not be determined (90), or were not similar to those of cardioviruses and aphthoviruses (17, 74, 89). We therefore reanalyzed these elements using methods that had previously been used to identify HP-like IRESs in diverse picornaviruses and to model the structure of the AV IRES (21, 95), and we identified AV-like IRES structures in each of these genomes. The extensive similarities between these predicted structures and the experimentally verified AV IRES indicate that these RNA elements constitute a novel class of viral IRESs.
MATERIALS AND METHODS
Sequences.
Sequences were analyzed from the Aichi virus strains A846/88 (GenBank accession numbers AB040749 and AB010145), BAY/1/03/DEU (AY747174), Goiania/GO/03/01/Brazil (DQ028632), Chshc7 (FJ890523), D/VI2169/2004 (GQ927704), D/VI2359/2004 (GQ927705), D/VI2321/2004 (GQ927706), D/VI2582/2004 (GQ927707), D/VI2524/2004 (GQ927708), D/VI2528/2004 (GQ927709), D/VI2287/2004 (GQ927711), D/VI2244/2004 (GQ927712), and D/VI2535/2004 (GQ927710); bovine kobuvirus strain U-1 (AB084788); canine kobuvirus AN211D (JN387133); canine kobuvirus PC0082 (JN088541); mouse kobuvirus (JF755427); porcine kobuvirus strains swine/S-1-HUN/2007/Hungary (EU787450), swine/K-30-HUN/2008/Hungary (GQ249161), pig/JY-2010a/CHN/Y-1-CHI (GU292559), and pig/Ch-kobu/2008/CHN (GU298977); salivirus A isolates NG-J1 (GQ179640), 02394-01 (GQ184145), SH1 (China) (GU245894), and 1 to 9 (GU376738 to GU376746); sheep kobuvirus (GU245693.2); TV2 strains 10717 (GU182408) and 007167 (GU182409); and TV3 strains 10878 (GU182410) and 00742 (GU182411).
Sequence alignment.
Viruses containing AV IRES-like sequences in their 5′ UTRs were identified using BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/) of the GenBank database. For short (11-nt) sequences, the parameters used were the following: E (expect), 1,000; word size, 7; reward match/mismatch, 1/−3; gap extension, 5; and gap penalty, 2. Searches done with long sequences (>100 nt) used the following parameters: E, 10; word size, 11; reward match/mismatch, 1/−1; gap extension, 0; and gap penalty, 2. Searches done using more stringent parameters (e.g., match/mismatch, 2/−3; gap extension, 5; gap penalty, 2) identified shorter segments of the same sequences. Nucleotide sequences were aligned with CLUSTAL-W2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) using the default parameters (DNA weight matrix, CLUSTALW; gap open penalty, 10.0; gap extension penalty, 0.01).
Modeling of secondary and tertiary RNA structures.
Secondary structure elements were modeled using complementary probabilistic (Pfold; http://www.daimi.au.dk/∼compbio/pfold/) (36) and posterior decoding approaches (CentroidFold; http://www.ncrna.org/centroidfold) (80) and were verified and refined by free energy minimization using Mfold (http://mfold.rna.albany.edu/?q=mfold) (96) and RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) (18), in all instances using default parameters. Computational analysis was, in all cases, done using series of overlapping viral RNA sequences because of the limits on the number of nucleotides that can be processed via these web servers. Tertiary structures in picornavirus 5′ UTRs were modeled using pKnotsRG (http://bibiserv.techfak.uni-bielefeld.de/pknotsrg/submission.html) (72).
Assembly and analysis of initiation complexes.
A vector for the transcription of mRNA of SV-A nt 1 to 1498 was made (GenScript) by inserting DNA into pUC57, which corresponds to a 5′-terminal T7 promoter, two G residues, and a variant of SV-A nt 1 to 1498 (GenBank no. GQ184145) followed by two UAA stop codons. The SV-A sequence contained single substitutions that eliminated a BamHI restriction site at nt 1241 to 1246 and introduced AUG triplets at positions corresponding to codons 137, 179, 212, 221, 237, and 253 of the 259-amino-acid-long SV-A LΔVP0 coding sequence. Further mutations were introduced into this SV-A transcription vector by NorClone Biotech Laboratories (London, Ontario, Canada). SV-A mRNA was transcribed from this vector, and AV and EMCV IRES-containing mRNAs were transcribed from previously described vectors (15, 95) using T7 RNA polymerase. 40S subunits and native eIF2 and eIF3 were purified from rabbit reticulocyte lysate (RRL) (Green Hectares, Oregon, WI). Recombinant eIF1, eIF1A, eIF4A (wild-type [wt]), eIF4AR362Q, and eIF4G736-1115 (designated eIF4Gm) and mutant variants thereof, PTB, DHX29, and Escherichia coli methionyl tRNA synthetase were expressed and purified from E. coli as described previously (38, 43, 57, 62, 65, 83). tRNAMeti was transcribed in vitro using T7 RNA polymerase and a previously described vector (60) and was aminoacylated using recombinant E. coli methionyl tRNA synthetase (43). 48S complexes were assembled on SV-A, AV, and EMCV mRNA transcripts and analyzed by primer extension using avian myeloblastosis virus reverse transcriptase and 32P-labeled primer 5′-GAAAGAAGATGAGTGAGGAG-3′ (complementary to SV-A nt 788 to 807), 5′-GCCTATCATAGCAGTCAAGG-3′ (complementary to AV nt 790 to 809), or 5′-GTCAATAACTCCTCTGG-3′ (complementary to EMCV nt 957 to 974) as appropriate, essentially as described previously (70).
In vitro translation.
Monocistronic SV-A mRNAs (0.1 μg) were translated using the Flexi RRL system (Promega) (20-μl reaction volume) supplemented with 0.5 mCi/ml [35S]methionine (43.5 TBq/mmol) for 60 min at 37°C. Purified recombinant eIF4AR362Q (56) was added to translation reactions as described previously (95). Translation products were analyzed by electrophoresis using NuPAGE 4 to 12% Bis-Tris gels (Invitrogen), followed by autoradiography.
Directed hydroxyl radical cleavage.
Wild-type eIF4G736-1115 and a panel of single-cysteine mutant forms of eIF4G736-1115 (termed eIF4Gm) were derivatized with Fe(II)-1-(p-bromoacetamidobenzyl)-EDTA (BABE) as described previously (38, 39) by incubating 3,000 pmol of a protein with 1 mM Fe(II)-BABE in 100 μl buffer containing 80 mM HEPES (pH 7.5), 300 mM KCl, and 10% glycerol for 30 min at 37°C. Derivatized proteins were separated from unincorporated reagent by buffer exchange on Microcon YM-30 filter units and stored at −80°C.
To investigate hydroxyl radical cleavage, 5 pmol of wt or mutant SV-A mRNA (nt 1 to 1498) was incubated at 37°C for 10 min in 50 μl buffer A (20 mM HEPES [pH 7.6], 100 mM KCl, 2.5 mM MgCl2, and 5% glycerol) with 10 pmol [Fe(II)-BABE]-eIF4Gm in the presence of 10 pmol unmodified eIF4A. To generate hydroxyl radicals, reaction mixtures were supplemented with 0.05% H2O2 and 5 mM ascorbic acid and incubated on ice for 10 min. Reactions were quenched by adding 20 mM thiourea. Sites of hydroxyl radical cleavage were determined by primer extension using avian myeloblastosis virus reverse transcriptase and the 32P-labeled primer 5′-GAAAGAAGATGAGTGAGGAG-3′ (complementary to SV-A nt 788 to 807). cDNA products were resolved in a 6% sequencing gel.
RESULTS
Sequence similarities between the Aichi virus IRES and other picornavirus 5′ UTRs.
The AV IRES (nt 338 to 747) consists of three major domains, designated I, J, and K, the Yn-Xm-AUG motif, and domain L, which contains the initiation codon (Fig. 1) (95). In BLAST searches of viral mRNAs in the nonredundant GenBank nucleotide database (July 2011 version), the sequences that most closely matched this element occurred in the 5′ UTRs of other AV isolates (97 to 99% identity across the entire sequence), in SV-A 5′ UTRs (72% identity to AV nt 342 to 732), and in the BKV 5′ UTR (71% identity to AV nt 484 to 722). Subsequent analysis identified matches with mouse kobuvirus (67) (80% identity between MKoV nt 1 to 610 and AV nt 144 to 732), CKoV isolate AN211D (41) (73% identity between CKoV isolate AN211D nt 1 to 539 and AV nt 189 to 724), the closely related canine kobuvirus isolate PC0082 (33) (76% identity between CKoV nt 1 to 690 and AV nt 33 to 724), and sheep kobuvirus (SKV) (75) (69% identity between SKV nt 410 to 644 and AV nt 484 to 710).
The apex of AV domain K contains two highly conserved sequences, AGGUACCCC (nt 662 to 670) and GGGAUCUGA (nt 681 to 689), that form a functionally important base-paired motif (95). BLAST searches of picornavirus 5′ UTRs with these and flanking nucleotides identified 12 other AV isolates, SV-A, BKV, CKoV, MKoV, SKV, two TV3 isolates, >225 Aphthovirus isolates, and >175 Cardiovirus isolates. Slightly divergent variants of this motif, differing at one or two positions in each strand, were identified in 11 cosavirus isolates (data not shown). The same bipartite motif is a functionally important element in type 2 IRESs (4, 9), and its presence in the TV3 5′ UTR prompted us to identify picornavirus sequences that are related to this 5′ UTR.
The 5′ UTRs of TV3 strains 00742 and 1087 are 94% identical across their entire lengths and are ∼65% identical to nt 157 to 523 and nt 157 to 584 of the 5′ UTRs of TV2 strains 007617 and 10717, respectively. Shorter sequences related to the TV3 5′ UTR occur in the 5′ UTRs of Aichi viruses (nt 585 to 716; 62 to 63% identity), BKV (nt 572 to 670; 66% identity), SV-A (nt 655 to 712; 70% identity), and Saffold virus and other cardioviruses (49 to 74 nt; 70 to 75% identity). These matching sequences are located in the apex of domain J of the cardiovirus IRES (5), whereas the larger Aichi virus sequences mapped to the 3′-terminal half of domain J and the adjacent region of domain K. Homology between these regions of kobuvirus, SV-A, and turdivirus 5′ UTRs and type 2 IRESs will be discussed below.
Three distinct subgroups of AV-like IRESs.
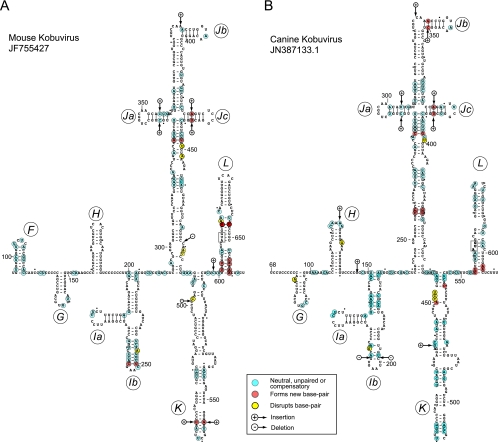
BLAST searches thus identified extensive related elements in the 5′ UTRs of members of three genera of Picornaviridae. Pairwise comparisons confirmed that the AV (744 nt long) and SV-A (718 nt long) 5′ UTRs are 68% identical in their latter two thirds (AV nt 293 to 744) as reported previously (17); moreover, sequence identity of more than 50% extends for ∼300 nt into the coding region. Similarly, MKoV and CKoV 5′ UTRs are highly homologous to the AV 5′ UTR across their entire length (as described above), and both are followed by coding sequences that are ∼70% identical to ∼300 nt of the equivalent AV coding sequence.
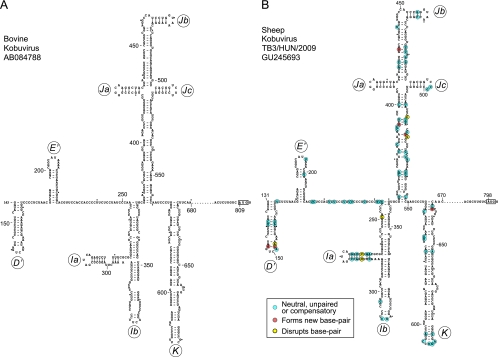
Nucleotides 315 to 675 of the 807-nt-long BKV 5′ UTR are 64% identical to AV nt 347 to 726, nt 410 to 644 of the 797-nt-long SKV 5′ UTR are 68% identical to AV nt 484 to 720, and nt 204 to 499 of the 643-nt-long TV3 5′ UTR are 54% identical to AV nt 431 to 729. BKV and SKV 5′ UTRs are 82% identical across their entire lengths, and similarly, TV2 and TV3 5′ UTRs align well with each other (60% identity across the entire 5′ UTR), with the exception of a 26-nt-long 5′-terminal sequence that is present only in the latter (which implies that the TV2 5′ UTR is incomplete) and several insertions, for example, of 16 and 15 nt in the TV2 5′ UTR at the equivalent of TV3 nt 139 and nt l45, respectively. The 5′ UTRs of BKV and SKV and of TV2 and TV3 therefore are more closely related to each other than to the AV 5′ UTR. BKV, SKV, and TV3 5′ UTRs contain insertions of ∼120, 120, and 150 nt upstream of the initiation codon, respectively, relative to AV and SV-A 5′ UTRs.
The mosaic nature of Kobuvirus, Salivirus, and Paraturdivirus 5′ UTRs.
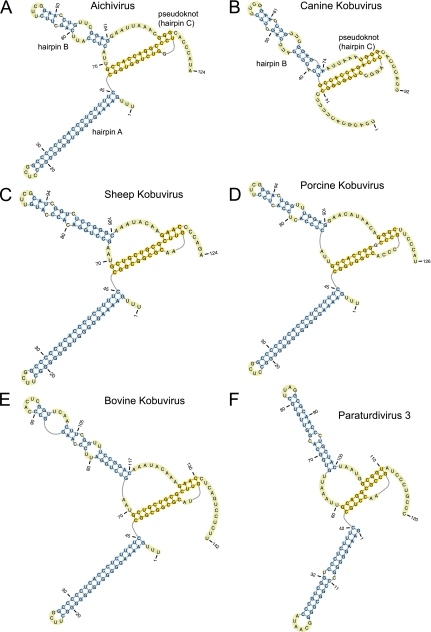
The 5′-terminal regions of AV and BKV 5′ UTRs form a domain that in AV has been found to be required for replication and encapsidation (51, 78, 79, 80). It consists of a series of 5′-terminal hairpins (A and C) and a pseudoknot encompassing hairpin B, corresponding to AV nt 1 to 116 (Fig. 2A) and BKV nt 1 to 124 (Fig. 2E) (17, 51, 78, 90). The equivalent SKV structure is similar to that of BKV, except that the apex of hairpin B is truncated (Fig. 2C). The 5′-terminal regions of SV-A 5′ UTRs may be incomplete, but despite limited sequence similarity with these Kobuvirus sequences, they also can form structures equivalent to the hairpin B pseudoknot and to hairpin C (25, 42). The CKoV isolate PC0082 sequence likely also is incomplete but is similarly capable of forming equivalent structures (Fig. 2B). The equivalent 5′-terminal regions of porcine kobuvirus (PKV) and of TV3 genomes also have the potential to form three similar terminal hairpins, the first of which is closely related to hairpin A of the AV 5′ UTR and the second of which forms part of pseudoknots that are comparable to those of AV and SV-A (Fig. 2D and F). As noted previously, the PKV 5′ UTR downstream of this element does not resemble 5′ UTRs from members of the Kobuvirus genus but instead aligns with segments of picornavirus 5′ UTRs that have the potential to adopt HP IRES-like structures (73, 93).
Fig 2.
Secondary and tertiary 5′-terminal structures of Aichi virus (GenBank accession number AB040749) (A), canine kobuvirus (GenBank accession number JN088541) (B), sheep kobuvirus (GenBank accession number GU245693) (C), porcine kobuvirus (GenBank accession number EU787450) (D), bovine kobuvirus (GenBank accession number AB084788) (E), and paraturdivirus 3 strain 10878 (GenBank accession number GU182410) (F), all predicted using pKnotsRG (72). This model of a segment of the AV 5′ UTR and analogous data concerning the BKV 5′ UTR have been described elsewhere (17, 90). Domains A, B, and C are labeled in panels A and B, and nucleotides are numbered throughout.
The region adjacent to the 5′ UTR in each of these genomes encodes a leader (L) protein, which ranges in size from 53 amino acid residues in TV2 to 195 amino acid residues in PKV and which, apart from those of MKoV and CKoV, shares low sequence identity with the AV L protein. However, the L proteins of BKV, SKV, and PKV are similar in size (188 to 195 amino acids in length), share significant (38 to 56%) pairwise sequence identity, and in the case of BKV and PKV contain a C-X2-C-X21-C-X2-H zinc finger motif that closely resembles the C-X2-C-X21-C-X2-C zinc finger motif in the AV, MKoV, and CKoV L proteins. SV-A, TV2, and TV3 L proteins are much shorter and do not contain zinc finger motifs. BKV/SKV and PKV therefore contain similar ori noncoding and L protein coding sequences flanking two completely unrelated types of IRESs, the presence of which is indicative of recombination leading to lateral gene transfer, as suggested previously to account for the presence of related HP-like IRESs in unrelated viruses (21).
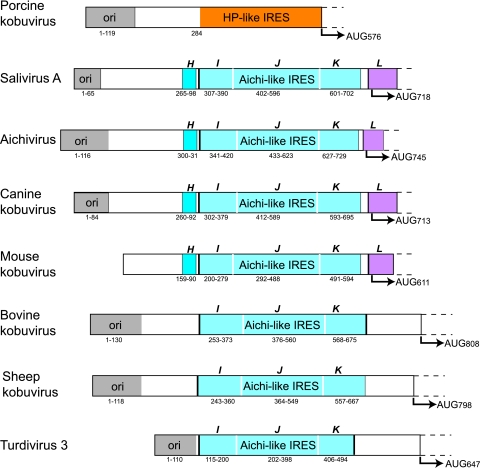
These picornavirus 5′ UTRs therefore all contain similar 5′-terminal elements that likely are required for replication and encapsidation, followed by an IRES that may either be HP-like (in PKV) or that has a sequence related to that of the AV IRES (BKV, CKoV, MKoV, SV-A, SKV, TV2, and TV3), which may in turn be followed by unrelated spacer elements (BKV/SKV and TV3) (Fig. 3) and divergent L coding sequences.
Fig 3.
Schematic representations of the 5′ UTRs and adjacent coding regions of porcine kobuvirus, SV-A (isolate 02394-01), Aichi virus, canine kobuvirus isolate PC0082, mouse kobuvirus, bovine kobuvirus, sheep kobuvirus, and turdivirus 3 (strain 10878). 5′ UTRs are divided into domains and are shaded to show those that are structurally related to domains A, B, and C and the pseudoknot, which together constitute the 5′-terminal ori region of Aichi virus (51, 78) (gray); domain H (dark blue); domains I, J, and K (light blue); and domain L (purple). The ori region and domains H, I, J, K, and L are labeled, and the initiation codon for each polyprotein and the nucleotides that make up each domain of the 5′ UTR are indicated. The 5′ UTRs of SV-A (GenBank accession number GQ184145) (17), canine kobuvirus (GenBank accession no. JN088541), and mouse kobuvirus (GenBank accession no. JF755427) are suspected to be incomplete.
Structural model of the salivirus A IRES.
The data described above indicate that BKV, SV-A, TV2, and TV3 5′ UTRs contain sequences that are related to the AV IRES. Structural models of these 5′ UTRs were derived using the same complementary methods that had previously been used to obtain models of picornavirus HP-like IRESs (21) and of the AV IRES (95). The models of these IRESs that were derived in this way are supported by the results of chemical and enzymatic probing, validating this approach (14, 87, 95).
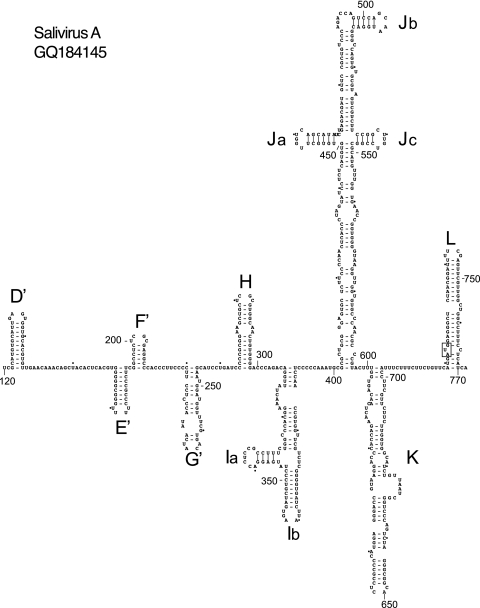
To derive a model of the SV-A 5′ UTR, analysis initially was limited to the complete or near-complete 5′ UTR sequences of SV-A isolate 02394-01 and the closely related SH1 and NG-J1 isolates, and then it was extended to include nine partial SV-A 5′ UTR sequences (17, 42, 81). Sequences were aligned and then analyzed using Pfold (36), which applies an explicit evolutionary model to aligned sequences to generate a probabilistic structural model of RNAs. These results then were validated using CentroidFold, a posterior decoding approach (80), and by free energy minimization, e.g., Mfold (18, 96). Individual structural elements identified using these approaches are listed in Table 1. Pfold identified all major structural elements except for domain C, domain G, the base of domain J (which were modeled using Mfold and, in the case of domain B, also with pKnotsRG), and subdomain Ja (which was identified using CentroidFold). Domain A is absent, confirming that the SV-A sequence likely is incomplete at its 5′ terminus (17). The SV-A 5′ UTR downstream of the putative ori (domains B, C, and the pseudoknot) comprises five small hairpins (domains D′ to G′ and H), an ∼90-nt-long Y-shaped domain (I), the ∼200-nt-long cruciform domain J, and an ∼100-nt-long interrupted hairpin (domain K) which is separated by a pyrimidine tract from the ∼70-nt-long hairpin domain L, which contains the initiation codon (Fig. 4). To maintain a standardized nomenclature, elements that are related to segments of the AV 5′ UTR are named in the same way, whereas unrelated elements are also named sequentially from the putative ori but are designated divergent domains by a prime symbol, starting with domain D′. Domains D′ to G′ (nt 122 to 254) are structurally distinct from equivalent domains in the AV IRES, but consistently with the high level of sequence homology downstream of nt 293 with the AV 5′ UTR (see above), the structure of this region of the SV-A 5′ UTR closely resembles that of the AV 5′ UTR, with only minor differences, such as the presence of an additional 5-bp helix at the base of domain I (Fig. 1 and 4). Sequence alignments indicate that the only significant insertions (of 4 to 5 nt) or deletions (of 3 nt) in this domain relative to AV domain I occur at its borders (data not shown). Sequence identity is concentrated in domains H, J, K, and L (Fig. 1), but the structures of other regions, such as domain I and the domain J apex, are maintained by extensive compensatory mutations (Fig. 1 and 4).
Table 1.
Structural elements identified using Pfold, CentroidFold, and Mfold in salivirus A, bovine kobuvirus, and turdivirus 3 5′ UTRsa
| Structural elementb | Result for indicated virus and method |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Salivirus A |
Bovine kobuvirus |
Turdivirus 3 |
|||||||
| Pfold | Centroid | Mfold | Pfold | Centroid | Mfold | Pfold | Centroid | Mfold | |
| A | NA | NA | NA | + | + | + | + | + | |
| B | + | + | + | + | + | + | + | + | |
| C | + | + | + | + | + | + | + | ||
| D (D′) | + | + | + | + | + | + | |||
| E (E′) | + | + | + | + | + | ||||
| F (F′) | + | + | + | ||||||
| G (G′) | + | ||||||||
| H | + | + | + | ||||||
| Ia | + | + | + | + | + | + | |||
| Ib | + | + | + | + | + | + | + | ||
| Ja | + | + | + | + | + | ||||
| Jb | + | + | + | + | + | + | + | + | + |
| Jc | + | + | + | + | + | + | + | + | |
| K | + | + | + | + | + | + | + | + | + |
| L | + | + | + | ||||||
Fig 4.
Model of the structure of the 5′ UTR and adjacent coding region of salivirus A downstream of domains B and C and the pseudoknot, derived as described in the text. Domains are labeled D′ to G′ and H to L, and the initiation codon for the viral protein (AUG722) is boxed. Nucleotides are numbered at 50-nucleotide intervals, and every 20th nucleotide starting from nt 160 is marked by a small solid circle.
Structural model of the bovine kobuvirus IRES.
The observation that the BKV and TV2/TV3 5′ UTRs contain sequences that resemble elements of the AV IRES prompted us to use the complementary approaches described above to derive structural models of these 5′ UTRs downstream of the putative ori. This approach identified five additional major domains in the BKV 5′ UTR (Table 1). As for SV-A, domains in the BKV 5′ UTR that are related to elements in the AV 5′ UTR are named in the same way, whereas two unrelated elements which form hairpin-like structures interrupted by small internal loops (Fig. 5A) are (in this case) designated domains D′ and E′. Although their sizes are similar to those of the domains D to H that occur between the AV ori and IRES elements, their sequences are distinct from those of these AV domains. Domain I (120 nt long) is significantly larger than the equivalent domain in the AV IRES (∼80 nt long), but it similarly adopts a Y-shaped structure, whereas domains J (184 nt) and K (117 nt) are comparable in size and structure to equivalent elements in the AV IRES, and there is significant sequence homology between them (see below). The ∼180-nt-long pyrimidine-rich region downstream of BKV domain K includes the initiation codon (AUG809), but neither its sequence nor its potential structure resemble domain L of AV and SV-A.
Fig 5.
Models of the structure of the 5′ UTR and adjacent coding region of bovine kobuvirus (A) and sheep kobuvirus (B) downstream of domains B and C and the pseudoknot, derived as described in the text. Domains are labeled D′, E′, and Ia-K, and the initiation codons for the viral proteins (BKV AUG809 and SKV AUG798) are boxed. Nucleotides are numbered at 50-nucleotide intervals, and every 20th nucleotide starting from nt 160 is marked by a small solid circle. To maintain the continuity of nomenclature for this group of IRESs, we have designated the first two hairpins domains D′ and E′, whereas elements that are related to segments of the AV 5′ UTR are designated subdomains/domains Ia, Ib, Ja, Jb, Jc, and K. The predicted structural elements between domain K and the initiation codon are not shown, and the omitted nucleotides (BKV nt 681 to 798 and SKV nt 671 to 788) are represented by ellipses. Nucleotide differences between BKV and SKV are classified as indicated on the inset key and are shown on the SKV model.
Structural model of the turdivirus IRES.
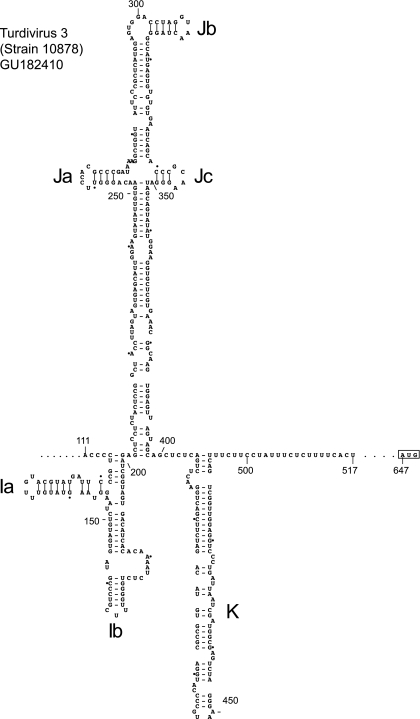
The 5′ UTRs of TV2 and TV3 are closely related except for minor relative insertions and deletions, and their sequences therefore were aligned and used to derive a structural model for only one of them. TV3 was chosen because its 5′ UTR apparently is complete, so that a model would complement the intact 5′-terminal domains A to C and the pseudoknot (Fig. 2F). These putative ori sequences are followed immediately by a domain that is similar in size (87 nt) to domain I of the AV IRES, has 44% sequence identity to it, but diverges somewhat from its regular Y-shaped conformation (Fig. 6). The size and structure of the downstream domains J (196 nt) and K (89 nt) are comparable to equivalent elements in the AV IRES, and there is a high degree of sequence homology between them (see below). TV3 domain K is followed immediately by a 19-nt-long pyrimidine (Yn) tract, but there is no sequence or structural similarity between the part of the TV 5′ UTR between the Yn tract and the initiation codon (AUG647) and either the equivalent noncoding region of the BKV 5′ UTR or the proximal coding region of the AV genome.
Fig 6.
Model of the structure of the 5′ UTR and adjacent coding region of turdivirus 3 (strain 10878) downstream of domains B and C and the pseudoknot, derived as described in the text. Domains are labeled Ia to K, and the initiation codon for the viral polyprotein (AUG647) is boxed. Nucleotides are numbered at 50-nucleotide intervals, and every 20th nucleotide starting from nt 120 is marked by a small solid circle. Nucleotides 518 to 646 between the pyrimidine (Yn) tract and the initiation codon have been omitted and are represented by ellipses.
In addition to similar 5′-terminal ori structures, BKV and TV2/TV3 5′ UTRs therefore also contain large segments that closely resemble the essential J and K domains of the AV IRES, and they all are flanked by related elements, namely, the upstream Y-shaped domain I and the downstream Yn tract. However, these BKV and TV 5′ UTRs differ from the closely related AV and SV-A 5′ UTRs, in that the ori and domain I are more than 100 nt closer to each other in the BKV 5′ UTR than in the AV 5′ UTR, and there is almost no separation between them in the TV3 5′ UTR; moreover, both BKV and TV3 lack an equivalent of AV domain L and instead contain large insertions between the domain K/Yn tract tandem and the initiation codon. The structural and sequence homology between the core AV IRES (domains I, J, and K) and these other picornavirus elements suggests that they are functional IRESs but are separated from the initiation codon by a spacer that can vary significantly in length and sequence. Interestingly, these spacers do not contain AUG triplets, thus they resemble the spacers found at the same relative location in type 1 IRESs, which also vary in length and sequence and lack AUG triplets (5).
Conservation of structure in picornavirus AV-like IRESs.
Several picornaviruses have recently been reported with 5′ UTRs that are closely related to either Aichi virus or to bovine kobuvirus IRESs, including MKoV (67), SKV (75), and the closely related CKoV isolate AN211D (41) and canine Aichi virus isolate JN088541 (33). Since these sequences were not used to build the structural models of AV and KBV IRESs, their compatibility with them was used as a test of their validity.
The few insertions or deletions in MKoV and CKoV IRESs relative to the sequence of the AV IRES, and in the SKV IRES relative to the BKV IRES, mapped either to unpaired regions or occurred in tandem in both strands of helical elements, thus leading to increases in the length of helices in domains Ja and Jc and the apex of domain K in the MKoV IRES, and in domains Ja, Jb, and Jc of the CKoV IRES (Fig. 7A and B). Further pairwise comparisons of MKoV and CKoV IRESs to the AV IRES (Fig. 1 and 7A and B) and of the SKV IRES to the BKV IRES (Fig. 5A and B) indicated that in each case, ∼83% of nucleotide differences were covariant (e.g., A-U⇔G-C double substitutions), neutral (e.g., A-U⇔G-U and G-U⇔G-C transitions), or occurred in unpaired regions (Table 2). These differences thus would not disrupt base pairing in predicted helices. A few nucleotide differences between each pair of IRESs would disrupt base pairing (disruptive differences, e.g., A-U⇔A-C), but interestingly, many of these nucleotide differences were located in regions of weak base pairing and in the immediate proximity of substitutions that would lead to the appearance of new base pairs. Accordingly, the nucleotide differences observed in these IRESs both tend to maintain the base pairing of helical elements and preserve the potential for the intrinsic flexibility of weakly base-paired regions. The potential for additional base pairing also was noted in MKoV and CKoV IRESs at the base of domain L, which thus may be longer than that in the AV IRES. The frequencies of compensatory mutations in these IRESs were higher, and those of disruptive mutations were lower, than those in a control set of viral IRES domains with a similar GC content generated by random evolution in silico (21). The observed pattern of sequence variation in AV-like and BKV/SKV IRESs provides strong support for the proposed pattern of the folding of the entire IRES and for the existence of the proposed individual structural elements.
Fig 7.
Models of the structures of the 5′ UTR and adjacent coding region of mouse kobuvirus (MKoV) downstream of domain E (A) and that of canine kobuvirus (CKoV) isolate AN211D downstream of domain F (B). The nomenclature of domains is the same as that used for Fig. 1, and the initiation codons for the viral polyproteins (MKoV AUG611 and CKoV AUG562) are boxed. Nucleotides are numbered at 50-nucleotide intervals, and every 20th nucleotide starting from MKoV nt 120 and CKoV nt 80 is marked by a small solid circle. Nucleotide differences between MKoV and AV (A) and CKoV and AV (B) are classified as indicated in the inset key.
Table 2.
Relative frequencies of different categories of nucleotide differences in AV, BKV, CKoV, MkoV, and SKV 5′ UTRs
| Picornavirus 5′ UTR fragments used for comparisons | No. (%) of nucleotide differences in indicated category |
||||||
|---|---|---|---|---|---|---|---|
| Total | Covariant | Neutral | Unpaired | Additional | Disruptive | ||
| BKV (nt 143-675) | SKV (nt 131-667) | 83 | 32 (39) | 8 (10) | 30 (36) | 6 (7) | 7 (8) |
| AV (nt 223-790) | CKoV-AN211d (nt 68-606) | 139 | 60 (43) | 18 (13) | 37 (27) | 17 (12) | 7 (5) |
| AV (nt 26-790) | MkoV (nt 68-606) | 116 | 53 (46) | 13 (11) | 30 (26) | 15 (13) | 3 (3) |
| CKoV-AN211d (nt 70-610) | CKoV (US-PC0082) (nt 231-761) | 18 | 0 | 2 (11) | 10 (56) | 5 (28) | 1 (5) |
Sequence and structural similarities between AV-like IRESs and their relationships with other IRESs.
Detailed sequence and structural comparisons between the 5′ UTR elements described above were undertaken to identify their distinguishing characteristics and any similarities with other IRESs. This analysis (see below) showed that these elements constitute a distinct, coherent group.
Domain H occurs in the AV, MKoV, CKoV, and SV-A 5′ UTRs, and it is highly conserved (Fig. 1). Interestingly, the sequence of its apical stem-loop (e.g., AV nt 307 to 324) is identical to that of domain III of some type 1 IRESs, including those of isolates of coxsackievirus B3 (CV-B3), CV-B4, echovirus 21 (EV-21), and EV-30 (data not shown). Despite its strong conservation, domain H enhances but is not required for AV IRES function, at least in cell extracts (95). This finding parallels observations that domain III is lacking in some type 1 IRESs, and that it is not required by the type 1 poliovirus IRES (12). The strong sequence conservation in domain H suggests that it has an accessory role in IRES function that is apparent only in cells or during viral infection or that is involved in some other aspect of viral multiplication.
Aichi virus domain I does not have obvious sequence or structural homologues in other classes of IRESs, but it is a well-conserved element of the 5′ UTRs of SV-A, MKoV, CKoV, and of other AV isolates (Fig. 1). Moreover, it has significant sequence homology with domain I of BKV (54% identity), SKV (60% identity), and TV3 (47% identity), which all adopt folds that are related to that of domain I in AV, MKoV, CKoV, and SV-A IRESs. This domain is thus an element that distinguishes AV-like IRESs from other viral IRESs.
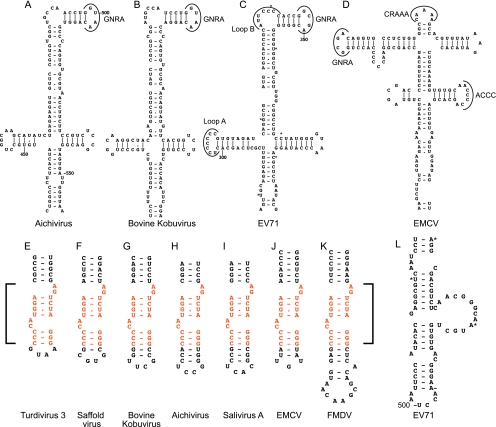
Systematic pairwise comparisons of AV, BKV, CKoV, MKoV, SV-A, TV2, and TV3 sequences showed that sequence identity in the J-K domain region ranged from 94% for the closely related SV-A to 52% for BKV and TV2 (Table 3). This level of sequence conservation is comparable to that in type 1 and type 2 IRESs, which also can diverge from each other in pairwise comparisons by as much as 50% but nevertheless contain related structures (5). There is ∼22% absolute conservation of nucleotides within the strongly conserved J and K domains in the eight AV-like IRESs characterized here (which in total comprised more than 30 sequences), and these conserved residues are concentrated in only a few locations. They include the four-way helical junction and a subapical helical region in domain J that is required for AV IRES function (95), the major internal loop of domain K, and the apex of this domain (Fig. 1). As noted above, the conserved apical region of domain K of AV-like IRESs shares significant homology with the apex of domain J of type 2 IRESs: both contain a conserved bipartite motif (Fig. 8E to K) that is located at a similar distance from the base of the domain and the adjacent polypyrimidine tract in the different IRESs. This motif is a functionally important element in type 2 IRESs (4, 9) and is critical for the activity of the AV IRES (95): it is a key determinant of eIF4G's interaction with both classes of IRESs. Although eIF4G also binds specifically to domain V of type 1 IRESs (11), there is no obvious similarity between the sequence or structure of this region and the apical regions of domain J of type 2 IRESs and domain K of AV-like IRESs (compare Fig. 8L to E to K).
Table 3.
Comparison of nucleotide sequence identities between J and K domains of the 5′ UTR of Aichi virus and related picornaviruses
| Virus domain | Pairwise nucleotide identity (%) to: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Aichivirus | Salivirus A 02394-01 | Salivirus A NG-J1 | Bovine kobuvirus | Sheep kobuvirus | Mouse kobuvirus | Canine kobuvirus | Turdivirus 2 | Turdivirus 3 | |
| Turdivirus 3 (strain 10878; nt 202-494) | 56 | 56 | 55 | 57 | 57 | 55 | 55 | 63 | |
| Turdivirus 2 (strain 10717; nt 215-507) | 56 | 53 | 53 | 52 | 56 | 56 | 53 | ||
| Canine kobuvirus (nt 241-544) | 76 | 66 | 66 | 67 | 64 | 77 | |||
| Mouse kobuvirus (nt 292-594) | 65 | 50 | 65 | 63 | 65 | ||||
| Sheep kobuvirus (nt 364-668) | 64 | 63 | 63 | 68 | |||||
| Bovine kobuvirus (nt 376-675) | 67 | 67 | 64 | ||||||
| Salivirus A (strain NG-J1; nt 307-605) | 73 | 94 | |||||||
| Salivirus A (strain 02394-01; nt 433-623) | 75 | ||||||||
| Aichivirus (nt 433-729) | |||||||||
Fig 8.
Conservation of sequence and structure in apical regions of picornavirus 5′ UTR domains. Models of apical regions of Aichi virus domain J (A), bovine kobuvirus domain J (B), enterovirus 71 (EV71) domain IV (C), EMCV domain I (D), turdivirus 3 domain K (E), Saffold virus domain J (F), bovine kobuvirus domain K (G), Aichi virus domain K (H), salivirus A domain K (I), EMCV domain J (J), foot-and-mouth disease virus (FMDV) domain J (K), and EV71 domain V (L). Panels are annotated to show the GNRA tetraloop in domain J of the AV and BKV 5′ UTRs (A and B); loop A, loop B, and GNRA loops in domain IV of the EV71 IRES (C); and the GNRA, CRAAAA, and ACCC loops in domain I of the EMCV IRES (D). The nucleotides that make up the conserved discontinuous sequence motif present in domain J of type 2 IRESs and domain K of AV-like IRESs are in orange and are indicated by square brackets.
Whereas these observations point to a specific similarity between type 2 and AV-like IRESs, the structure of the apical cruciform region of domain J in the latter (Fig. 8A and B) is quite distinct from the structure of the apical region of domain I of type 2 IRESs (Fig. 8D), instead it resembles the structure of the corresponding apical region of domain IV in type 1 IRESs, such as those of enterovirus 71 (Fig. 8C) and CV-B3 (3). Similarities include the overall cruciform pattern of folding, the sizes of the Jb subdomain and of its equivalent in type 1 IRESs, and the presence of both a GRNA tetraloop at the apex of this subdomain in a similar structural context, viz., adjacent to a 5-bp hairpin, and a 5- to 6-nt-long internal bulge. However, neither this internal bulge nor the apical loop of subdomain Ja on AV-like IRESs are C rich, in contrast to the corresponding loop B and loop A equivalents in type 1 IRESs (Fig. 8C), which are binding sites for the poly(rC) binding protein 2, an ITAF (16). Moreover, subdomains Ja and Jc are significantly smaller than the equivalent hairpins in type 1 IRESs (Fig. 8A to C). The implication that there are functional differences between these apical cruciform regions of type 1 and AV-like IRESs is supported by the observation that whereas the substitution of conserved nucleotides in the apical tetraloop severely impairs type 1 IRES function (32), equivalent substitutions did not reduce the efficiency of initiation on the AV IRES (95).
Initiation on the salivirus A IRES is dependent on eIF4A and requires specific binding of eIF4G.
Initiation on each class of IRES occurs by a distinct mechanism and thus has distinct initiation factor requirements. Salivirus A is representative of the Aichi-like class of IRESs identified here, and we therefore characterized aspects of the mechanism of initiation on it and compared them to initiation on other IRESs. Initiation on mRNAs that require eIF4F (or its eIF4A and eIF4G subunits) for the recruitment of 43S complexes (such as the type 1 PV IRES and the type 2 EMCV IRES) is inhibited by a dominant-negative eIF4AR362Q mutant (56), whereas initiation by mechanisms that are eIF4A independent (such as on the classical swine fever IRES) is not (59, 66). The translation of SV-A mRNA in rabbit reticulocyte lysate was completely abrogated by the inclusion of eIF4AR362Q in translation reactions (Fig. 9B, lanes 2 and 5), indicating that this IRES is strongly dependent on eIF4A/eIF4G.
Fig 9.
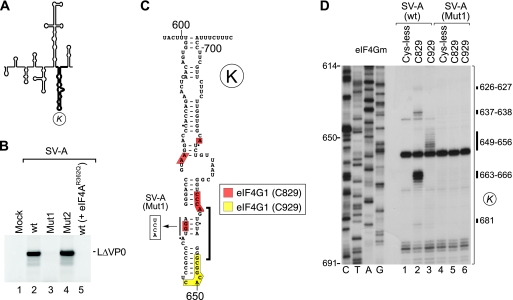
Interaction of the salivirus A IRES with eIF4G. (A) Model of the SV-A IRES, with domain K in boldface. (B) The activity of SV-A mRNA in the presence of eIF4A(R362Q) (lane 5) or containing destabilizing mutations in domain K (Mut1) (lane 3) or domain L (Mut2) (lane 4) compared to wt SV-A mRNA (lane 2) and assayed by translation in RRL. (C) Secondary structure of SV-A IRES domain K showing the conserved motif (bracket) that is required for interaction with eIF4G in the EMCV IRES. The destabilizing AGGU→UCCA mutation in this motif in SV-A (Mut1) mRNA is indicated. Domain K has been annotated to show sites of hydroxyl radical cleavage from unique surface-exposed cysteine residues on eIF4Gm (D), color coded as described in the inset key. (D) Primer extension analysis of directed hydroxyl radical cleavage of wt and Mut2 SV-A IRESs from Fe(II)-tethered eIF4Gm in the presence of eIF4A.
The requirement for eIF4G/eIF4A in initiation on type 1 IRESs, type 2 IRESs, and the AV IRES in each case involves the specific binding of eIF4G to the IRES in a manner that is enhanced by eIF4A (21, 44, 62, 65, 95). The ability of eIF4Gm to bind specifically to the SV-A IRES was characterized using the directed hydroxyl radical probing technique, in which Fe(II) tethered to unique surface-exposed cysteine residues on eIF4Gm via the linker 1-(p-bromoacetamidobenzyl)-EDTA (BABE) is used to generate hydroxyl radicals that cleave the IRES in its immediate vicinity. Cleavage sites then were mapped by primer extension inhibition. eIF4Gm consists of five pairs of stacked α-helices (48), and we used a cysteineless (Cys-less) variant and two previously validated eIF4Gm mutants with single surface-exposed cysteines (11, 39, 94, 95): a substitution mutant with a novel cysteine at residue 829 (located between helices 2b and 3a) and a D928→D928C insertion mutant, designated C929, with a novel cysteine residue located after helix 4b (numbering as in the revised sequence NM_182917). Hydroxyl radicals from Cys829 cleaved the SV-A IRES in ternary eIF4Gm/eIF4A/IRES complexes at nt 626 and 627, 637 and 638, 645, and 681 and strongly at nt 663 to 666, and hydroxyl radicals from C929 cleaved the IRES at nt 649 to 656 (Fig. 9D). These sites of cleavage mapped to opposite sides of domain K (Fig. 9C), overlapping the conserved motif identified near the apex of this domain that is identical to a determinant of eIF4G's interaction with type 2 IRESs (Fig. 1 and 8E to K).
The disruption of this element in SV-A (Mut1) mRNA abrogated both its translation (Fig. 9B, lanes 2 and 3, and C) and its cleavage by Fe(II)-eIF4Gm (Fig. 9D, compare lanes 2 and 3 to 5 and 6). These results are directly comparable to the results of the mutation of this conserved element in the AV IRES (95) and similarly suggest that the loss of function of the mutant SV-A IRES in promoting translation initiation is due to the loss of its ability to bind stably to eIF4G.
Initiation on salivirus A IRES is dependent on DHX29.
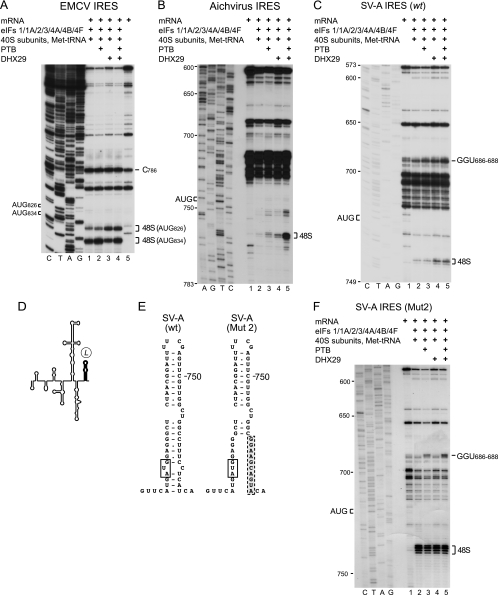
A distinguishing characteristic of initiation on the AV IRES is its strong dependence on the DExH-box protein DHX29 (Fig. 10B, lanes 2 to 5) (95). DHX29 almost abrogates initiation on IGR and HP-like IRESs, whereas on the type 2 EMCV IRES it promotes 48S complex formation at AUG826, which is upstream of the initiation codon for the viral polyprotein (AUG834) (71) (Fig. 10A, lanes 1 and 3). The function of DHX29 in initiation is to ensure that structured mRNA is properly unwound and accommodated in the mRNA-binding cleft of the 40S subunit (1, 71), and in the case of the AV IRES, its dependence on DHX29 is due to the sequestration of the initiation codon in domain L, a stable hairpin. Our analysis of the SV-A IRES indicated that it has a similar structure (Fig. 4), suggesting that it, too, is dependent on DHX29.
Fig 10.
Initiation on the salivirus A IRES is dependent on DHX29. Toeprinting analysis of 48S complex formation on EMCV (A), Aichi virus (B), (C) wt SV-A, and (F) Mut2 SV-A mRNAs in reaction mixtures containing 40S subunits, eIF1, eIF1A, eIF2, eIF3, eIF4A, eIF4B, eIF4F, initiator tRNA, PTB, and DHX29 as indicated. AUG codons are indicated on the left, and toeprints that correspond to 48S complexes are indicated on the right. Lanes C, T, A, and G depict corresponding DNA sequences. (D) Model of the SV-A IRES, with domain L in boldface. (E) Structures of the L domain of the wt SV-A IRES and an SV-A IRES mutant containing destabilizing substitutions in domain L.
This was tested by the in vitro reconstitution of 48S complex formation on the SV-A IRES from individual purified components of the translation apparatus, using primer extension inhibition to map the position of this complex on the mRNA. Aminoacylated initiator tRNA, ribosomal 40S subunits, and eIFs 1, 1A, 2, 3, 4A, 4B, and 4F, with or without PTB, led to a very low level of 48S complex formation at AUG722 on the SV-A IRES (Fig. 10C, lanes 1 to 3). 48S complex formation was strongly enhanced by the inclusion of DHX29 (Fig. 10C, lanes 1, 4, and 5). The presence of PTB in initiation reactions enhanced toeprints at nt 686 to 688, which map to a helical element in domain K (Fig. 10C, lanes 3 and 5, and F, lanes 3 and 5). The binding of PTB to the AV IRES led to the appearance of toeprints at an identical location (95).
To determine whether the sequestration of the SV-A initiation codon in domain L, a stable hairpin (ΔG = −23.3 kcal/mol), accounts for the requirement for DXH29, we characterized the effect of a disruptive mutation that reduced the stability of the residual domain to ΔG = −14.3 kcal/mol (SV-A Mut2) (Fig. 10D and E). These substitutions enhanced SV-A IRES-mediated translation in RRL (Fig. 9B, lanes 2 and 4), and the in vitro reconstitution of initiation on SV-A Mut2 mRNA yielded a greater level of 48S complex formation than that on the equivalent wild-type mRNA. Importantly, 48S complex formation was independent of DHX29 (Fig. 10F, compare lanes 2 to 5).
Taken together, these observations indicate that the factor requirements for initiation on the SV-A IRES correspond to the requirements for initiation on the AV IRES, but they differ from the requirements for initiation on type 2, IGR, and HP-like IRESs.
DISCUSSION
Structural and functional analyses indicate that the 5′ UTR of Aichi virus, the type member of the Kobuvirus genus of picornaviruses, contains an IRES that has a structure and which uses a mechanism that are in many respects distinct from those of established classes of IRESs (95). Sequence analysis and modeling of secondary structures reported here have now identified elements in the 5′ UTRs of BKV, CKoV, MKoV, and SKoV (also members of the Kobuvirus genus), SV-A (of the proposed Salivirus genus), and TV types 2 and 3 (members of the proposed Paraturdivirus genus) that are closely related to the AV IRES and that together constitute a new class of IRES. Further members of this group will likely be identified, such as tortoise testivirus 1 (which, on the basis of the 115 nt preceding the initiation codon, has a 5′ UTR that is related to that of AV) (22).
Sequence and structural characteristics of AV-like IRESs.
AV-like IRESs contain unique structural elements (domain I, the basal region of domain K, and domain L), elements that are either highly homologous or structurally related to elements in type 1 IRESs (i.e., the apical hairpin of AV domain H and the apical cruciform region of domain J), an element (the apex of AV domain K) that is highly homologous to an element in type 2 IRESs, and elements that have direct sequence/structural counterparts in type 1 and type 2 IRESs (such as the GNRA tetraloop) or in type 1, type 2, and type 3 IRESs (the Yn tract). The essential core of the AV IRES consists of domains I, J, K, and the adjacent Yn tract (95), and these domains constitute the most highly conserved elements of the different AV-like IRESs identified here. Domains H and L are highly conserved in all AV, CKoV, MKoV, and SV-A isolates (Fig. 1), but the former is divergent and the latter does not occur at all in BKV and TV2/TV3 5′ UTRs. It thus may be appropriate to assign these IRESs to two subclasses. Sequence identity within the core of AV-like IRESs is high and is concentrated in discrete locations that include the apical cruciform region of domain J and the apical region of domain K (Fig. 1).
Specific functions have been assigned to several elements in AV-like IRESs, including some that distinguish them from other classes of IRES (95). The well-conserved domain I (Fig. 1) contains binding sites for PTB, which enhances initiation on the AV IRES. Domain K of AV-like IRESs has a structure that in its entirety is also distinct from other domains in members of established IRES groups, although it does contain a conserved apical element that is identical to a motif in type 2 IRESs (Fig. 8). Data reported here for SV-A (Fig. 9) and elsewhere for the AV IRES (95) revealed that this motif is a key determinant of the interaction of these IRESs with eIF4G and is essential for their function. The fact that this motif has the same key role of binding eIF4G during initiation on type 2 IRESs constitutes a point of significant similarity between these two types of IRESs (4, 9, 39, 94, 95). Footprinting and directed hydroxyl radical probing experiments have shown that the basal region of domain K, which is not related to any part of the type 2 IRES, constitutes a second major site of interaction with PTB, and that PTB interacts with this and other parts of the AV IRES (95) in a way that is distinct from its modes of binding to type 1 and type 2 IRESs (30, 31). Domain L in AV, CKoV, MKoV, and SV-A IRESs contains the initiation codon for the viral polyprotein, and data reported here (Fig. 10) and elsewhere (95) indicate that this domain's stability accounts for the AV and SV-A IRESs' unusual requirement for DHX29, a DExH-box protein that has been implicated in the correct accommodation of structured mRNA in the mRNA-binding channel of the 40S subunit for scanning through highly structured 5′ UTRs (1, 71). This requirement for DHX29 is in contrast to all other IRESs that have been characterized to date (71). A further point of difference concerns domain J of AV-like IRESs, which has a global fold that resembles domain IV of type 1 IRESs and, to a lesser extent, domain I of type 2 IRESs. The specific role of these large central domains in internal initiation is not known, but mutational analyses have established that the requirement for conserved sequence elements within them differs. AV IRES function depends on the integrity of subdomain Jb, which contains a highly conserved central element (Fig. 1), but not on the sequence of its GNRA tetraloop (95), whereas the activities of type 1 and type 2 IRESs depend strongly on the nature of the sequence of this tetraloop motif (32, 45, 76).
Taken together, there are therefore global structural and mechanistic similarities between type 1, type 2, and AV-like IRESs that distinguish them from IGR IRESs and HP-like IRESs, but there also are clear differences between AV-like IRESs and the other two major classes of picornavirus IRESs that justify the classification of them into separate groups.
Recombination as a source of diversity in viral IRESs.
Recombination enables genetic diversity to be generated rapidly over a broad sequence space, and it therefore contributes significantly to the divergence and evolution of many viruses. In picornaviruses, recombination occurs primarily, but not exclusively, in regions of the genome encoding nonstructural proteins and usually is limited to members of the same or closely related species (82). One factor that likely accounts for the majority of viable picornavirus recombinants deriving from parental strains with high levels of sequence identity is the requirement for the maintenance of sequence-specific interactions between viral proteins or between viral proteins and RNA. Recombinants that disrupt interactions within a protein or RNA structure or assembly are likely to be rapidly lost from viral populations by purifying selection. On the other hand, the exchange of modular elements between genomes leading to the appearance of viable recombinants with novel combinations of characteristics (6) may be more feasible if such elements do not interact extensively with sequences elsewhere in the genome. In the case of RNA, such elements would be self contained, in the sense of adopting the same optimal structure whether they exist in isolation or as part of a longer sequence (85). IRESs fit this criterion, and they function in the absence of interaction with other elements of the genome; indeed, a key test for their activity is for them to mediate initiation in the context of synthetic dicistronic mRNAs in which they are flanked by heterologous sequences (5).
We have previously suggested that the presence of related HP-like IRES elements in distinct, deeply branched genera of Picornaviridae and in distinct genera of Flaviviridae, juxtaposed against unrelated coding sequences, suggest that IRES modules have been exchanged between these viruses by recombination (21), which might represent an example of lateral gene transfer (24). The observation that the genomes of BKV and SKV on the one hand and PKV on the other contain structurally and mechanistically distinct IRES elements located between related noncoding ori and L protein-coding sequences (Fig. 3) (73, 75) provide further strong support for the hypothesis that IRESs can be considered independent functional modules that can be transferred between related or unrelated genomes. The example of BKV/SKV and PKV may be an extreme case in which unrelated elements in picornaviruses have been exchanged by recombination, but the possibility that related IRESs can be exchanged between kobuviruses is supported by the recent determination that the Aichi virus isolate Chshc7 has a mosaic structure in which nt 261 to 852 (i.e., the IRES and part of the L coding region) may derive from a different isolate than the remainder of the genome (20). Moreover, the observation that the core AV-like IRES elements identified here are located at positions relative to the ori or the initiation codon that are unique in each case further supports the possibility that recombination occurs in the 5′ UTR to exchange functional noncoding RNA elements between viruses from different genera. Thus, the spacer between ori and domain I of the IRES ranges in length from 5 (TV3) to ∼240 nt (SV-A) in the examples considered here, and even in the closely related AV and SV-A it differs in terms of length and sequence, whereas at the 3′ border of the IRES the initiation codon may be part of a conventional Yn-Xm-AUG motif or may, as in BKV and TV3, be separated by a long spacer. Significantly, a growing number of interspecies and intraspecies recombination events involving picornavirus 5′ UTRs has been reported (see, for example, references 27, 46, 49, 55, 84, and 92). The suggestion that IRES elements can be exchanged in toto between picornavirus genomes by recombination does not preclude the possibility that this mechanism also leads to the transfer of individual IRES domains between genomes. In this respect, the identification of homology between the apical regions of domain J in AV-like IRESs and domain J in cardio- and aphthovirus IRESs, which are otherwise distinct, is particularly intriguing: this region includes the highly conserved eIF4G-binding motif and in some instances extends over as much as 74 nt.
The analysis of the genomes of plant and other RNA viruses provides additional support for the hypothesis that terminal noncoding RNA regions of viral genomes contain functional, independently folded structural elements that can be transferred between species, genera, and even families by recombination. There is direct evidence for the recombinational transfer of the ∼300-nt-long 3′ UTR of grapevine chrome mosaic virus RNA-1 to tomato black ring virus RNA-2 (40), and a similar phenomenon could account for the presence of structurally related and functionally exchangeable translational enhancers known as Barley yellow dwarf virus (BYDV)-like translation elements in the 3′ UTRs of members of Luteovirus, Dianthovirus, Necrovirus, and Umbravirus genera (50, 86), of the sequence- and structurally related 3′-cap-independent translational enhancer (3′CITE) in the 3′ UTRs of members of Tombusvirus, Aureusvirus, and Carmovirus genera (53), and of the stem-loop 2-like motif (s2m) in the 3′ end of the genome of some species in Astroviridae, Caliciviridae, Coronaviridae, and Picornaviridae (37, 77).
ACKNOWLEDGMENT
This work was supported by National Institutes of Health grant AI-51340.
Footnotes
Published ahead of print 23 November 2011
REFERENCES
- 1. Abaeva IS, Marintchev A, Pisareva VP, Hellen CU, Pestova TV. 2011. Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning. EMBO J. 30:115–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ali IK, McKendrick L, Morley SJ, Jackson RJ. 2001. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 75:7854–7863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bailey JM, Tapprich WE. 2007. Structure of the 5′ nontranslated region of the coxsackievirus B3 genome: chemical modification and comparative sequence analysis. J. Virol. 81:650–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bassili G, et al. 2004. Sequence and secondary structure requirements in a highly conserved element for foot-and-mouth disease virus internal ribosome entry site activity and eIF4G binding. J. Gen. Virol. 85:2555–2565 [DOI] [PubMed] [Google Scholar]
- 5. Belsham GJ, Jackson RJ. 2000. Translation initiation on picornavirus RNA, p. 869–900 In Sonenberg N, Hershey JWB, Mathews MB. (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 6. Botstein D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484–490 [DOI] [PubMed] [Google Scholar]
- 7. Brown EA, Zajac AJ, Lemon SM. 1994. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J. Virol. 68:1066–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chard LS, Bordeleau ME, Pelletier J, Tanaka J, Belsham GJ. 2006. Hepatitis C virus-related internal ribosome entry sites are found in multiple genera of the family Picornaviridae. J. Gen. Virol. 87:927–936 [DOI] [PubMed] [Google Scholar]
- 9. Clark AT, Robertson ME, Conn GL, Belsham GJ. 2003. Conserved nucleotides within the J domain of the encephalomyocarditis virus internal ribosome entry site are required for activity and for interaction with eIF4G. J. Virol. 77:12441–12449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Breyne S, Yu Y, Pestova TV, Hellen CU. 2008. Factor requirements for translation initiation on the Simian picornavirus internal ribosomal entry site. RNA 14:367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CUT. 2009. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl. Acad. Sci. U. S. A. 106:9197–9202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dildine SL, Semler BL. 1989. The deletion of 41 proximal nucleotides reverts a poliovirus mutant containing a temperature-sensitive lesion in the 5′ noncoding region of genomic RNA. J. Virol. 63:847–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drexler JF, et al. 2011. Aichi virus shedding in high concentrations in patients with acute diarrhea. Emerg. Infect. Dis. 17:1544–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Easton LE, Locker N, Lukavsky PJ. 2009. Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus-like internal ribosome entry sites. Nucleic Acids Res. 37:5537–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Evstafieva AG, Ugarova TY, Chernov BK, Shatsky IN. 1991. A complex RNA sequence determines the internal initiation of encephalomyocarditis virus RNA translation. Nucleic Acids Res. 19:665–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gamarnik AV, Andino R. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Greninger AL, et al. 2009. The complete genome of klassevirus–a novel picornavirus in pediatric stool. Virol. J. 6:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 36:W70–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haller AA, Semler BL. 1992. Linker scanning mutagenesis of the internal ribosome entry site of poliovirus RNA. J. Virol. 66:5075–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Han X, Zhang W, Xue Y, Shao S. 2011. Sequence analysis reveals mosaic genome of Aichi virus. Virol. J. 8:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hellen CU, de Breyne S. 2007. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 81:5850–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heuser W, Kaleta E, Giesow K, Keil GM, Knowles NJ. 2010. Genome sequence of virus X, a picornavirus isolated from a spur-thighed tortoise (Testudo graeca). Abstr. 16th Meet. Eur. Study Group Mol. Biol. Picornaviruses, abstr. H15. [Google Scholar]
- 23. Hollister JR, Vagnozzi A, Knowles NJ, Rieder E. 2008. Molecular and phylogenetic analyses of bovine rhinovirus type 2 shows it is closely related to foot-and-mouth disease virus. Virology 373:411–425 [DOI] [PubMed] [Google Scholar]
- 24. Holmes EC. 2009. The evolution and emergence of RNA viruses. Oxford series in ecology and evolution (OSEE). Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 25. Holtz LR, et al. 2009. Klassevirus 1, a previously undescribed member of the family Picornaviridae, is globally widespread. Virol. J. 6:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Honkavuori KS, et al. 2011. Novel picornavirus in Turkey poults with hepatitis, California, U. S. A. Emerg. Infect. Dis. 17:480–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang T, et al. 2009. Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS One 4:e6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jackson RJ, Hellen CUT, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11:113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jang SK, Pestova TV, Hellen CUT, Witherell GW, Wimmer E. 1990. Cap-independent translation of picornavirus RNAs: structure and function of the internal ribosomal entry site. Enzyme 44:292–309 [DOI] [PubMed] [Google Scholar]
- 30. Kafasla P, et al. 2009. Polypyrimidine tract binding protein stabilizes the encephalomyocarditis virus IRES structure via binding multiple sites in a unique orientation. Mol. Cell 34:556–568 [DOI] [PubMed] [Google Scholar]
- 31. Kafasla P, Morgner N, Robinson CV, Jackson RJ. 2010. Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J. 29:3710–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaminski A, Hunt SL, Gibbs CL, Jackson RJ. 1994. Internal initiation of mRNA translation in eukaryote, p. 115–155 In Setlow JK. (ed.), Genetic engineering, vol. 16 Plenum Press, New York, NY: [PubMed] [Google Scholar]
- 33. Kapoor A, et al. 2011. Characterization of a canine homolog of human Aichivirus. J. Virol. 85:11520–11525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kapoor A, et al. 2008. A highly prevalent and genetically diversified Picornaviridae genus in south Asian children. Proc. Natl. Acad. Sci. U. S. A. 105:20482–20487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kapoor A, et al. 2008. A highly divergent picornavirus in a marine mammal. J. Virol. 82:311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Knudsen B, Hein J. 2003. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 31:3423–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kofstad T, Jonassen CM. 2011. Screening of feral and wood pigeons for viruses harbouring a conserved mobile viral element: characterization of novel astroviruses and picornaviruses. PLoS One 6:e25964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kolupaeva VG, de Breyne S, Pestova TV, Hellen CU. 2007. In vitro reconstitution and biochemical characterization of translation initiation by internal ribosomal entry. Methods Enzymol. 430:409–439 [DOI] [PubMed] [Google Scholar]
- 39. Kolupaeva VG, Lomakin IB, Pestova TV, Hellen CU. 2003. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol. Cell. Biol. 23:687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Le Gall O, Candresse T, Dunez J. 1995. Transfer of the 3′ non-translated region of grapevine chrome mosaic virus RNA-1 by recombination to tomato black ring virus RNA-2 in pseudorecombinant isolates. J. Gen. Virol. 76:1285–1289 [DOI] [PubMed] [Google Scholar]
- 41. Li L, et al. 2011. Viruses in diarrhetic dogs include novel kobuviruses and sapoviruses. J. Gen. Virol. 92:2534–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li L, et al. 2009. A novel picornavirus associated with gastroenteritis. J. Virol. 83:12002–12006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV. 2006. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 25:196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lomakin IB, Hellen CUT, Pestova TV. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20:6019–6029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. López de Quinto S, Martínez-Salas E. 1997. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J. Virol. 71:4171–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lukashev AN, et al. 2005. Recombination in circulating human enterovirus B: independent evolution of structural and non-structural genome regions. J. Gen. Virol. 86:3281–3290 [DOI] [PubMed] [Google Scholar]
- 47. Lukavsky PJ. 2009. Structure and function of HCV IRES domains. Virus Res. 139:166–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marcotrigiano J, et al. 2001. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell 7:193–203 [DOI] [PubMed] [Google Scholar]
- 49. McIntyre CL, McWilliam Leitch EC, Savolainen-Kopra C, Hovi T, Simmonds P. 2010. Analysis of genetic diversity and sites of recombination in human rhinovirus species C. J. Virol. 84:10297–10310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Meulewaeter F, et al. 2004. Conservation of RNA structures enables TNV and BYDV 5′ and 3′ elements to cooperate synergistically in cap-independent translation. Nucleic Acids Res. 32:1721–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nagashima S, Sasaki J, Taniguchi K. 2005. The 5′-terminal region of the Aichi virus genome encodes cis-acting replication elements required for positive- and negative-strand RNA synthesis. J. Virol. 79:6918–6931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nakashima N, Uchiumi T. 2009. Functional analysis of structural motifs in dicistroviruses. Virus Res. 139:137–147 [DOI] [PubMed] [Google Scholar]
- 53. Nicholson BL, Wu B, Chevtchenko I, White KA. 2010. Tombusvirus recruitment of host translational machinery via the 3′ UTR. RNA 16:1402–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Niepmann M. 2009. Internal translation initiation of picornaviruses and hepatitis C virus. Biochim. Biophys. Acta 1789:529–541 [DOI] [PubMed] [Google Scholar]
- 55. Oberste MS, Peñaranda S, Pallansch MA. 2004. RNA recombination plays a major role in genomic change during circulation of coxsackie B viruses. J. Virol. 78:2948–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pause A, Methot N, Svitkin Y, Merrick WC, Sonenberg N. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13:1205–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pestova TV, Borukhov SI, Hellen CU. 1998. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394:854–859 [DOI] [PubMed] [Google Scholar]
- 58. Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CUT. 2008. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 27:1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pestova TV, Hellen CUT. 1999. Internal initiation of translation of bovine viral diarrhea virus RNA. Virology 258:249–256 [DOI] [PubMed] [Google Scholar]
- 60. Pestova TV, Hellen CUT. 2001. Preparation and activity of synthetic unmodified mammalian tRNAi(Met) in initiation of translation in vitro. RNA 7:1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pestova TV, Hellen CUT. 2003. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 17:181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pestova TV, Hellen CUT, Shatsky IN. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pestova TV, Hellen CU, Wimmer E. 1991. Translation of poliovirus RNA: role of an essential cis-acting oligopyrimidine element within the 5′ nontranslated region and involvement of a cellular 57-kilodalton protein. J. Virol. 65:6194–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pestova TV, Hellen CU, Wimmer E. 1994. A conserved AUG triplet in the 5′ nontranslated region of poliovirus can function as an initiation codon in vitro and in vivo. Virology 204:729–737 [DOI] [PubMed] [Google Scholar]
- 65. Pestova TV, Shatsky IN, Hellen CUT. 1996. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16:6870–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CUT. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Phan TG, et al. 2011. The fecal viral flora of wild rodents. PLoS Pathog. 7:e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pilipenko EV, et al. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14:2028–2045 [PMC free article] [PubMed] [Google Scholar]
- 69. Pisarev AV, et al. 2004. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 78:4487–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pisarev AV, Unbehaun A, Hellen CU, Pestova TV. 2007. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 430:147–177 [DOI] [PubMed] [Google Scholar]
- 71. Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. 2008. Translation initiation on mammalian mRNAs with structured 5′ UTRs requires DExH-box protein DHX29. Cell 135:1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Reeder J, Steffen P, Giegerich R. 2007. pknotsRG: RNA pseudoknot folding including near-optimal structures and sliding windows. Nucleic Acids Res. 35:W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Reuter G, Boldizsár A, Pankovics P. 2009. Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch. Virol. 154:101–108 [DOI] [PubMed] [Google Scholar]
- 74. Reuter G, Boros A, Pankovics P. 2011. Kobuviruses–a comprehensive review. Rev. Med. Virol. 21:32–41 [DOI] [PubMed] [Google Scholar]
- 75. Reuter G, Boros A, Pankovics P, Egyed L. 2010. Kobuvirus in domestic sheep, Hungary. Emerg. Infect. Dis. 16:869–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Robertson ME, Seamons RA, Belsham GJ. 1999. A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA 5:1167–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Robertson MP, Igel H, Baertsch R, Haussler D, Ares M, Jr., Scott WG. 2005. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol. 3:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sasaki J, et al. 2001. Construction of an infectious cDNA clone of Aichi virus (a new member of the family Picornaviridae) and mutational analysis of a stem-loop structure at the 5′ end of the genome. J. Virol. 75:8021–8030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sasaki J, Taniguchi K. 2003. The 5′-end sequence of the genome of Aichi virus, a picornavirus, contains an element critical for viral RNA encapsidation. J. Virol. 77:3542–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sato K, Hamada M, Asai K, Mituyama T. 2009. CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res. 37:W277–W280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shan T, et al. 2010. Picornavirus salivirus/klassevirus in children with diarrhea, China. Emerg. Infect. Dis. 16:1303–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Simmonds P. 2006. Recombination and selection in the evolution of picornaviruses and other mammalian positive-stranded RNA viruses. J. Virol. 80:11124–11140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Skabkin MA, et al. 2010. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 24:1787–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Smura T, et al. 2007. Enterovirus surveillance reveals proposed new serotypes and provides new insight into enterovirus 5′-untranslated region evolution. J. Gen. Virol. 88:2520–2526 [DOI] [PubMed] [Google Scholar]
- 85. Wagner A, Stadler PF. 1999. Viral RNA and evolved mutational robustness. J. Exp. Zool. 285:119–127 [PubMed] [Google Scholar]
- 86. Wang Z, Kraft JJ, Hui AY, Miller WA. 2010. Structural plasticity of Barley yellow dwarf virus-like cap-independent translation elements in four genera of plant viral RNAs. Virology 402:177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Willcocks MM, et al. 2011. Structural features of the Seneca Valley virus internal ribosome entry site element; a picornavirus with a pestivirus-like IRES. J. Virol. 85:4452–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wilson JE, Pestova TV, Hellen CUT, Sarnow P. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102:511–520 [DOI] [PubMed] [Google Scholar]
- 89. Woo PC, et al. 2010. Comparative analysis of six genome sequences of three novel picornaviruses, turdiviruses 1, 2 and 3, in dead wild birds, and proposal of two novel genera, Orthoturdivirus and Paraturdivirus, in the family Picornaviridae. J. Gen. Virol. 91:2433–2448 [DOI] [PubMed] [Google Scholar]
- 90. Yamashita T, et al. 2003. Isolation and characterization of a new species of kobuvirus associated with cattle. J. Gen. Virol. 84:3069–3077 [DOI] [PubMed] [Google Scholar]
- 91. Yamashita T, et al. 1998. Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans. J. Virol. 72:8408–8412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yozwiak NL, et al. 2010. Human enterovirus 109: a novel interspecies recombinant enterovirus isolated from a case of acute pediatric respiratory illness in Nicaragua. J. Virol. 84:9047–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yu JM, et al. 2011. Analysis and characterization of the complete genome of a member of a new species of kobuvirus associated with swine. Arch. Virol. 156:747–751 [DOI] [PubMed] [Google Scholar]
- 94. Yu Y, Abaeva IS, Marintchev A, Pestova TV, Hellen CUT. 2011. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 39:4851–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yu Y, et al. 2011. The mechanism of translation initiation on Aichi virus RNA mediated by a novel type of picornavirus IRES. EMBO J. 30:4423–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]