Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) infection and latency-associated nuclear antigen (LANA-1) upregulate the multifunctional protein angiogenin (ANG). Our studies demonstrate that silencing ANG or inhibiting its nuclear translocation downregulates KSHV LANA-1 expression and ANG is necessary for KSHV latency, anti-apoptosis and angiogenesis (Sadagopan et al., J. Virol. 83:3342–3364, 2009; Sadagopan et al., J Virol. 85:2666–2685, 2011). Here we show that LANA-1 interacts with ANG and colocalizes in latently infected endothelial telomerase-immortalized human umbilical vein endothelial (TIVE-LTC) cells. Mass spectrometric analyses of TIVE-LTC proteins immunoprecipitated by anti-LANA-1 and ANG antibodies identified 28 common cellular proteins such as ribosomal proteins, structural proteins, tRNA synthetases, metabolic pathway enzymes, chaperons, transcription factors, antioxidants, and ubiquitin proteosome proteins. LANA-1 and ANG interaction with one of the proteins, annexin A2, was validated. Annexin A2 has been shown to play roles in cell proliferation, apoptosis, plasmin generation, exocytosis, endocytosis, and cytoskeleton reorganization. It is also known to associate with glycolytic enzyme 3-phosphoglyceratekinase in the primer recognition protein (PRP) complex that interacts with DNA polymerase α in the lagging strand of DNA during replication. A higher level of annexin A2 is expressed in KSHV+ but not in Epstein-Barr virus (EBV)+ B-lymphoma cell lines. Annexin A2 colocalized with several LANA-1 punctate spots in KSHV+ body cavity B-cell lymphoma (BCBL-1) cells. In triple-staining analyses, we observed annexin A2-ANG-LANA-1, annexin A2-ANG, and ANG-LANA-1 colocalizations. Annexin A2 appeared as punctate nuclear dots in LANA-1-positive TIVE-LTC cells. In LANA-1-negative TIVE-LTC cells, annexin A2 was detected predominately in the cytoplasm, with some nuclear spots, and colocalization with ANG was observed mostly in the cytoplasm. Annexin A2 coimmunoprecipitated with LANA-1 and ANG in TIVE-LTC and BCBL-1 cells and with ANG in 293T cells independent of LANA-1. This suggested that annexin A2 forms a complex with LANA-1 and ANG as well as a separate complex with ANG. Silencing annexin A2 in BCBL-1 cells resulted in significant cell death, downregulation of cell cycle-associated Cdk6 and of cyclin D, E, and A proteins, and downregulation of LANA-1 and ANG expression. No effect was seen in KSHV− lymphoma (BJAB and Ramos) and 293T cells. These studies suggest that LANA-1 association with annexin A2/ANG could be more important than ANG association with annexin A2, and KSHV probably uses annexin A2 to maintain the viability and cell cycle regulation of latently infected cells. Since the identified LANA-1- and ANG-interacting common cellular proteins are hitherto unknown to KSHV and ANG biology, this offers a starting point for further analysis of their roles in KSHV biology, which may lead to identification of potential therapeutic targets to control KSHV latency and associated malignancies.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV) (human herpesvirus 8 [HHV-8]) is an oncogenic DNA virus involved in the pathogenesis of Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and body cavity B-cell lymphoma (BCBL) and multicentric Castleman's disease (MCD) (16, 19). During latency, only a few genes, such as ORF73 (LANA-1), ORF72 (vCyclin), ORF71 (vFLIP), K12 (kaposins), and viral-encoded microRNAs (miRNAs), are expressed (14, 33, 37, 92). How KSHV, with the help of only a few expressed genes, is able to outsmart the complex mammalian cell network and persist for life in infected individuals is an area of active investigation. As an obligate intracellular parasite coevolved with the human host, KSHV has probably mastered the art of piracy and mimicry of host molecules to facilitate its intracellular parasitism and to survive in the complex eukaryotic environment.
LANA-1 is detected in all cells latently infected with KSHV and is often used as a marker of latency. It is a promiscuous protein that modulates the functions of diverse host proteins. For example, LANA-1 binds to and disrupts the tumor-suppressive functions of p53 and Rb proteins (34, 89). It recruits the EC5S ubiquitin complex for degradation of VHL which stabilizes hypoxia-inducible factor 1α (HIF1α) and promotes angiogenesis (13). By binding to and sequestering the β-catenin negative regulator glycogen synthase kinase 3β, LANA-1 stabilizes β-catenin and upregulates the transcription of c-myc, c-jun, and cyclin D genes (36). LANA-1 interactions with RING3/Brd2 have been hypothesized to promote the G1-S transition (37, 83, 85).
Our earlier studies showed that KSHV infection and LANA-1 expression induce angiogenin (ANG), a 14-kDa multifunctional angiogenic protein, first isolated from HT-29 human colon adenocarcinoma cell-conditioned medium based on its angiogenic activity and belonging to the RNase family (96). ANG has been shown to play a role in tumor angiogenesis. It is detected in human plasma at concentrations of 250 to 360 ng/ml (102). However, its expression is often upregulated in various cancers, including pancreatic, breast, prostate, cervical, ovarian, colon, colorectal, gastric, urothelial, and endometrial cancers, and is associated with cancer progression and poor outcomes (24, 25, 102, 113). Anti-angiogenin monoclonal antibodies used as antagonists inhibited the establishment, progression, and metastasis of human cancer cells inoculated in athymic mice (84).
The half-life of the angiogenin protein is estimated to be about 12 to 24 h. Angiogenin is a secreted protein and exerts multiple effects at the endothelial cell surface, nucleus, and nucleoli. Once secreted, ANG binds to actin on the cell surface, and this complex triggers a plasminogen activator-generating plasmin that helps in the basement membrane and extracellular matrix degradation that are required for the migration of endothelial cells (47, 48). ANG binding to the cell membrane also initiates signal transduction via phospholipase C (PLC), phospholipase A2 (PLA2), protein kinase B (PKR/AKT), and extracellular signal-regulated kinase 1/2 (ERK1/2) (10, 59, 72).
Nuclear translocation of angiogenin is dependent on cell type and cell density. It remains cytoplasmic in fibroblast cells but enters the nucleus and nucleolus of semiconfluent (<60%) and subconfluent (30%) endothelial cells, respectively (78). This specificity is lost in transformed cells, and ANG is detected in the nucleus even at a confluent state (107). In the nucleolus, ANG mediates rRNA transcription by binding to CT repeats abundant in the promoter region of the rRNA gene (113). Angiogenin also exerts its ribonucleolytic activity by catalyzing the generation of 18S and 28S rRNA (100). Nuclear translocation of angiogenin in endothelial cells has been shown to be necessary for the angiogenic potentials not only of angiogenin but also of vascular endothelial growth factor (VEGF) and basic fibroblast growth factor (bFGF) (61). Inhibition of nuclear translocation of angiogenin by the aminoglycoside antibiotic neomycin (46) or mutagenesis of ANG's nuclear localization sequence (78) both abolished the angiogenic activity of angiogenin. Activation of PLC-γ is required for nuclear translocation for angiogenin (46), and neomycin inhibits this nuclear translocation by inhibiting PLC-γ activation without affecting the viability of the cells or ERK1/2 phosphorylation. In contrast, paromomycin, an analogue aminoglycoside, does not inhibit nuclear translocation of angiogenin (46).
De novo KSHV infection of human microvascular dermal endothelial cells (HMVEC-d) resulted in increased secretion of several growth factors, cytokines, chemokines, and angiogenic factors, and angiogenin is one of them (94). KS tissue sections were positive for angiogenin, which suggested that angiogenin could play a role in KS pathogenesis. KSHV induced a time- and dose-dependent increase in angiogenin gene expression and protein secretion beginning as early as 8 h postinfection of HMVEC-d cells which lasted until the fifth day of the observation period. Telomerase-immortalized human umbilical vein endothelial (TIVE) cells latently infected with KSHV (TIVE-LTC) secreted high levels of angiogenin. Significant induction of angiogenin was observed in cells expressing ORF 73 (LANA-1, latent) and ORF74 (vGPCR, lytic) genes alone. KSHV infection induced ANG entered the nuclei and nucleoli of infected cells, stimulated 45S-rRNA gene transcription, proliferation, and tube formation, and had an anti-apoptotic effect in the infected cells (93).
Since we observed the presence of ANG in KSHV+ BCBL-1 (PEL) cells carrying KSHV in a latent state, we used quantitative real-time PCR assays and ANG enzyme-linked immunosorbent assays (ANG-ELISAs) to extend this observation to KSHV+ (BCBL-1 and BC-3) cells, KSHV+ and Epstein-Barr virus-positive (KSHV+ EBV+) (JSC-1) PEL cells, BJAB-KSHV cells, EBV+ (KSHV−) lymphoma cells (Akata-EBV and Raji), and EBV− KSHV− lymphoma cells (Akata, Loukes, Ramos, and BJAB). No significant ANG gene expression was observed in KSHV−, EBV−, and EBV+ cells; in contrast, we observed about 5.2-, 5.3-, 7.2-, and 6.7-fold increases in ANG expression in BJAB-KSHV, JSC-1, BCBL-1, and BC-3 cells, respectively, compared to the level in EBV− Akata cells (95). These results were confirmed by the detection of significant amounts of angiogenin (250 to 400 pg/ml) in BCBL-1, BC-3, BJAB-KSHV, and JSC-1 cell supernatants and less than 30 pg/ml from EBV− and EBV+ Akata cell supernatants (95). By immunofluorescence assay (IFA), significant ANG expression was detected in more than 80% of BCBL-1, BC-3, and JSC-1 cells, while ANG was barely detectable in <1 to 2% of EBV− and EBV+ Akata cells (95). These results clearly demonstrated that KSHV but not EBV infection is associated with increased angiogenin expression in B-lymphoma cells. This suggested a specific association of ANG in KSHV biology and the evolution of KSHV to exploit ANG for its advantage in maintaining its latency.
This notion was also supported by the observation that inhibition of nuclear translocation of ANG resulted in reduced BCBL-1 and TIVE-LTC cell survival and proliferation, while EBV− and EBV+ Akata cells were unaffected. ANG induced ORF73 (LANA-1) gene expression and PLC-γ and AKT phosphorylation (95), whereas silencing ANG and neomycin treatment inhibited them. Treatment with the conventional PLC-γ inhibitor U73122 also showed similar results (95). Blocking nuclear transport of ANG inhibited ORF73 (LANA-1) gene expression and increased lytic switch ORF50 (RTA) gene expression, both during de novo infection and in latently infected cells. Silencing ANG also had similar consequences. These extensive studies suggested that KSHV has evolved to exploit ANG for its advantage via a so-far-unexplored PLC-γ pathway for maintaining its latency.
Since LANA-1 induced ANG and we observed ANG-LANA-1 interaction, to further evaluate the relationship between ANG and KSHV biology, we carried out mass spectrometric analyses of proteins immunoprecipitated by anti-LANA-1 and ANG antibodies from TIVE-LTC cells. Both ANG and LANA-1 interacted with 28 common cellular proteins. We validated the interaction of one of the proteins, annexin A2. Studies presented here indicate a specific association of annexin A2 in KSHV biology and suggest that annexin A2 may be one of the crucial proteins utilized by KSHV to sustain the viability of latently infected PEL cells.
MATERIALS AND METHODS
Cells.
Telomerase-immortalized endothelial cells (TIVE) and long-term KSHV-infected TIVE cells (TIVE-LTC) were grown in endothelial basal medium (EBM) 2 with growth factors (Clonetics). HEK 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco BRL, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, and antibiotics. KSHV-carrying human PEL cells (BCBL-1, BC-3), EBV-positive B-lymphoma cells (Akata EBV+, Raji, LCL), EBV-negative B-lymphoma cells (Loukes, Ramos, Akata negative), and KSHV- and EBV-negative B-lymphoma cells (BJAB) were cultured in RPMI 1640 (Gibco BRL) medium with 10% heat-inactivated FBS (HyClone, Logan, UT), 2 mM l-glutamine, and antibiotics.
Antibodies, reagents, and growth factors.
Rabbit anti-LANA-1 antibody was raised in our laboratory (1). Mouse annexin A2 antibody was from BD Biosciences, San Diego, CA. Goat and rabbit polyclonal antibodies against human angiogenin, rabbit polyclonal antibodies against human cyclin E, and mouse monoclonal antibodies against green fluorescent protein (GFP), cyclin D, cyclin A, and cdk6 were from Santa Cruz Biotechnology Inc., Santa Cruz, CA. Phalloidin-linked Alexa 594-, Alexa 488-, or Alexa 594-linked anti-goat, anti-rabbit, and anti-mouse antibodies and anti-fade DAPI (4′,6′-diamidino-2-phenylindole) were from Molecular Probes, Eugene, OR. Antibodies against lamin B, tubulin, actin, bovine serum albumin (BSA), tetradecanoyl phorbol acetate (TPA), Triton X-100, and paraformaldehyde were from Sigma, St. Louis, MO.
Quantitative real-time RT-PCR.
Gene expression was measured by real-time reverse transcription (RT)-PCR using SYBR green techniques. Two micrograms of total RNA was treated with DNase for 1 h and then reverse transcribed into cDNA by using random hexamers with the SuperScript first-strand synthesis system for RT-PCR (Invitrogen, Carlsbad, CA). cDNA was used as a template with primers specific for 18S rRNA as an internal control. PCR was performed using the Applied Biosystems 7500 real-time PCR system with SYBR green PCR master mix (Applied Biosystems, Foster City, CA). The standard amplification program included 40 cycles of two steps each comprised of heating to 95°C and 60°C. Fluorescent product was detected at the last step of each cycle. The final mRNA levels of the genes studied were normalized using the comparative threshold cycle (CT) method. The primer sequences used were as follows (where Fw is forward and Rev is reverse): for annexin, Fw-CTCTACACCCCCAAGTGCAT and Rev-TCAGTGCTGATGCAAGTTCC; for angiogenin, Fw-CCGTTTCTGCGGACTTGTTC and Rev-GCCCATCACCATCTCTTCCA; and for 18S rRNA, Fw-CGGCTACCACATCCAAGGAA and Rev-GCTGGAATTACCGCGGCT.
Immunofluorescence assay (IFA).
TIVE-LTC cells in eight-well chamber slides (Nalge Nunc International, Naperville, IL) were fixed with 4% paraformaldehyde for 10 min at room temperature, permeabilized with 0.4% Triton X-100 for 10 min at room temperature, and stained with primary antibody for 1 h at 37°C. Target cells were washed and incubated with the appropriate dilution of secondary antibodies for 1 h at room temperature. Nuclei were visualized by using DAPI (Molecular Probes, Eugene, OR). Stained cells were washed and viewed with appropriate filters under a fluorescence microscope with the Nikon Metamorph digital imaging system. BCBL-1 cells were acetone fixed to glass slides, washed with phosphate-buffered saline (PBS) and treated with the indicated primary antibodies, and washed again with PBS, followed by Alexa 488 or Alexa 594 secondary antibody treatment. After the wash with PBS, cells were mounted with anti-fade reagent containing DAPI and observed under a fluorescence microscope equipped with the Nikon Metamorph digital imaging system.
Nuclear and cytoplasmic extract preparation.
Nuclear and cytoplasmic extracts from BCBL-1, Akata EBV-negative, TIVE, and TIVE-LTC cells were prepared using a nuclear extract kit (Active Motif Corp, Carlsbad, CA) as per the manufacturer's instructions. After measuring protein concentrations by BCA protein assay, extracts were stored at −80°C. The purity of the nuclear extracts was assessed by immunoblotting using anti-lamin B antibodies, and cytoskeletal contamination was checked by using anti-β-actin or anti-β-tubulin antibodies.
Immunoprecipitation (IP).
Equal protein concentrations of the whole-cell lysates or nuclear extracts from BCBL-1, BJAB, TIVE, and TIVE-LTC cells were immunoprecipitated with equal antibody concentrations of the respective antibody using 20 μl of protein G-Sepharose beads (GE Healthcare, Bio-Sciences). IP was carried out in radioimmunoprecipitation assay (RIPA) buffer for 2 h at 4°C in a rotating chamber. The beads were washed with RIPA buffer three times and suspended in 2× sample solubilizing buffer, heated to 95°C for 5 min, and resolved by SDS-PAGE.
Western blotting.
IP lysates, protein lysates, or nuclear extracts were resolved on SDS-PAGE, transferred to nitrocellulose membranes, blocked with 5% skim milk, and immunoblotted with antibodies. To confirm equal protein loading, blots were also probed with antibodies against human tubulin or actin. Species-specific horseradish peroxidase- or alkaline phosphatase-conjugated secondary antibodies were used for detection. Immunoreactive bands were developed by enhanced chemiluminescence reaction (NEN Life Sciences Products, Boston, MA).
Proteomics analysis.
TIVE-LTC cells were grown in T75 flasks. Approximately 2 × 106 cells were harvested in RIPA buffer (1% NP-40, 1.5% sodium deoxycholate, 0.1% SDS, 125 mM NaCl, 0.01 M sodium phosphate, 1 mM EDTA, 50 mM NaF, 1% protease inhibitor cocktail). The lysates were homogenized multiple times using a 22-gauge needle and syringe and centrifuged at 5,000 rpm (Eppendorf tabletop centrifuge) for 5 min, the supernatant was collected, and protein concentrations were measured. A 500-μg sample of cell lysate in RIPA buffer was mixed with 1 μg of either normal rabbit IgG, rabbit anti-angiogenin IgG, or rabbit anti-LANA-1 IgG antibodies and 20 μl of protein G-Sepharose for 2 h at 4°C in a rotating chamber. The beads were washed three times with RIPA buffer, suspended in 2× sample solubilizing buffer, and boiled at 95°C for 5 min. The eluted proteins were resolved by 4 to 20% gradient SDS-PAGE, and gels were stained with Coomassie G 250 dye for 2 h followed by destaining with destaining solution (43% acetic acid, 7% methanol) for 1 h. Visible protein bands unique to LANA-1 and angiogenin lanes were excised for proteomics analysis. Mass spectrometry was carried out using liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) methods at the Midwest Proteome Center, Rosalind Franklin University of Medical Sciences (RFUMS).
STRING analysis.
The pulldown proteins were annotated using GEDI (Gene Discovery/Genetic Discovery; http://gedi.ci.uchicago.edu), and the networks were generated using the STRING (Search Tool for the Retrieval of Interacting Genes) analysis tool. Each interaction in the database is annotated with a benchmarked numerical confidence score, which can be used to filter the interaction network at any desired stringency.
HEK 293T transfection studies.
Cells were transfected with 5 μg of full-length C-terminal GFP tagged angiogenin construct and pcDNA-GFP empty vector construct using the calcium phosphate precipitation method. The cells were incubated for 48 h under a 3% CO2 atmosphere, harvested in RIPA buffer, and subjected to immunoprecipitation or Western blotting.
Lentivirus production and infection of BCBL-1 cells.
Lentiviral infection was done as described before (109). Vesicular stomatitis virus G envelope-pseudotyped lentivirus was produced with a four-plasmid transfection system, as previously described (29, 77). Briefly, 293T cells were transfected with lentiviral constructs expressing short hairpin RNA (shRNA) to annexin A2, small interfering RNA (si)-annexin A2, clones 1, 2, 3, and 4 (si-Annex A2-1 to 4, respectively) (Open Biosystems, Huntsville, AL), si-GFP, and packaging plasmids by the calcium phosphate precipitation method. The sequences of 4 si-annexin A2 plasmids are as follows: Clone 1, CCGGCCTGCTTTCAACTGAATTGTTCTCGAGAACAATTCAGTTGAAAGCAGGTTTTTG; Clone 2, CCGGCTGTACTATTATATCCAGCAACTCGAGTTGCTGGATATAATAGTACAGTTTTTG; Clone 3, CCGGGCAGGAAATTAACAGAGTCTACTCGAGTAGACTCTGTTAATTTCCTGCTTTTTG; Clone 4, CCGGCGGGATGCTTTGAACATTGAACTCGAGTTCAATGTTCAAAGCATCCCGTTTTTG. The sequence of si-GFP used is TACAACAGCCACAACGTCTAT. The cells were incubated for 48 h at 3% CO2, culture supernatant containing the lentiviral virions was harvested, and cell debris was cleared by passing through a 0.45-μm-pore-size filter and either aliquoted directly or concentrated and stored at −80°C. 293T cells were infected with the desired amount of virus preparation to confirm knockdown. BCBL-1, BJAB, and Ramos cells were infected in culture for 8 h, and the medium was replaced with complete growth medium and incubated for 72 h without any selection. Total RNA and protein were extracted from lentivirus-transduced B cells and used for cDNA preparation and Western blotting.
Cell survival analysis by fluorescence-activated cell sorting (FACS).
BCBL-1, BJAB, and Ramos cells transduced with si-GFP or si-annexin A2 were collected after 3 days, washed, and stained with YO-PRO-1/propidium iodide (PI) (catalog no. V13243; Invitrogen) for 30 min. These were analyzed with an LSRII flow cytometer (Becton Dickinson) and ModFit Lt V3 software (Verity Software House) at the RFUMS flow cytometry core facility.
Cell cycle analysis by FACS.
BCBL-1, BJAB, and Ramos cells transduced with si-GFP or si-annexin A2 were collected after 3 days, washed, and fixed in 70% ethanol overnight. DNA was stained with propidium iodide at a final concentration of 50 μg/ml with RNase A (100 U/ml) prior to analysis by FACS and ModFit Lt V3 software. Ten thousand cells were analyzed.
RESULTS
KSHV LANA-1 interacts with angiogenin.
An et al. (2) infected telomerase-immortalized human umbilical vein endothelial (HUVEC-TIVE) cells with KSHV and isolated TIVE-LTC cells that were stably expressing LANA-1 and other KSHV latent genes. When quantitative real-time PCR was done using mRNA extracted from TIVE and TIVE-LTC cells, there was an approximately 5-fold increase in ANG mRNA expression levels in KSHV-positive TIVE-LTC cells compared to TIVE cells (93). ANG-ELISA performed using supernatants from TIVE and TIVE-LTC cells confirmed the increase in ANG levels in latently infected cells. We observed approximately six fold-higher ANG levels in TIVE-LTC cells (960 pg/ml) than in the control TIVE cells (160 pg/ml) (93). This further confirmed our results that KSHV infection induced the secretion of ANG and that latent viral gene expression could be involved in this induction.
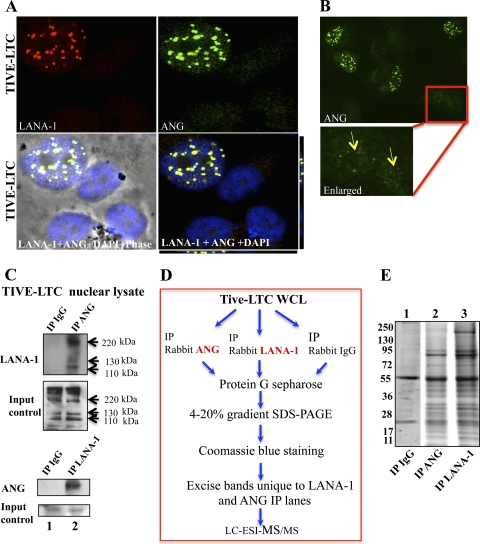
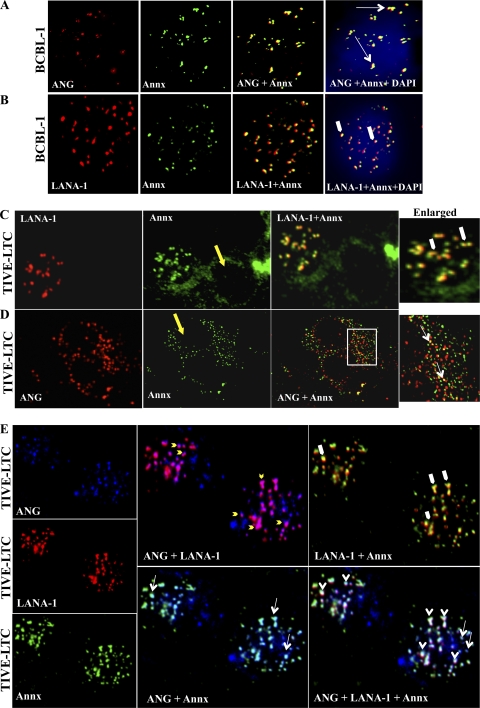
Since LANA-1 induced ANG, to evaluate the relationship between ANG and KSHV biology, we analyzed the TIVE-LTC cells. With its characteristic punctate nuclear fluorescence, LANA-1 was detected by confocal microscopy (Fig. 1A, red). ANG also appeared as punctate nuclear staining in LANA-1-positive cells (Fig. 1A and B, green), and >90% colocalization of ANG and LANA-1 was seen in these cells (Fig. 1A; see Fig. S1 in the supplemental material). In addition, noncolocalized LANA-1 and ANG were also detected in the nucleus. A more diffused cytoplasmic and nuclear staining of ANG was observed in LANA-1-negative cells (Fig. 1B, yellow arrows). Since colocalization is an indication of interaction, we carried out immunoprecipitation (IP) reactions to validate the interaction between ANG and LANA-1. Equal protein concentrations of TIVE-LTC lysates were immunoprecipitated with equal antibody concentrations of anti-ANG IgG or control IgG and probed with anti-LANA-1 antibody in Western blot reactions. LANA-1 was detected in ANG immunoprecipitates and vice versa (Fig. 1C, lanes 1 and 2). These results demonstrated that LANA-1 interacts either directly or indirectly with ANG in latently KSHV-infected TIVE-LTC cells.
Fig 1.
KSHV LANA-1 interacts with angiogenin (ANG). (A) TIVE-LTC cells were stained for angiogenin (green) and LANA-1 (red) after permeabilization. DAPI (blue) was used as a nuclear stain and merged with LANA-1 and ANG. Punctate ANG and LANA-1 colocalization can be readily seen. (B) TIVE-LTC cells were stained for ANG (green). The yellow arrows in the enlarged insert indicate diffused cytoplasmic and nuclear staining of ANG in uninfected, LANA-1-negative cells. (C) TIVE-LTC cells were grown to confluence, and nuclear extract was prepared. Equal protein quantities of each lysate were immunoprecipitated with equal concentrations of either rabbit anti-angiogenin IgG, rabbit LANA-1 IgG antibody, or control rabbit IgG antibody and Western blotted for LANA-1 and ANG. Input control for each IP is shown to demonstrate that an equal amount of LANA-1 and ANG were present in both the samples. (D) Schematics of the experimental design for proteomic analysis. Confluent TIVE-LTC cells were harvested with RIPA buffer, and 500 μg of cell lysates were immunoprecipitated using protein G-Sepharose beads and rabbit anti-angiogenin IgG, rabbit anti-LANA-1 IgG, or rabbit anti-IgG antibodies. The slurry was incubated at 4°C in a rotating chamber for 2 h. The beads were then washed three times with RIPA buffer, resuspended in 2× sample solubilizing buffer, boiled at 95°C for 5 min, and run on a 4% to 20% gradient SDS-PAGE gel. Protein bands unique to LANA-1- and angiogenin-immunoprecipitated lanes were excised and subjected to mass spectrometric analysis. (E) Representative samples of Coomassie stained gel used for mass spectrometric analysis. TIVE-LTC lysates were immunoprecipitated with rabbit IgG (lane 1), rabbit anti-ANG IgG (lane 2), and rabbit anti-LANA-1 IgG (lane 3) and analyzed as describe above. Molecular weight markers are shown on the left.
Proteomic analyses identify common proteins interacting with LANA-1 and angiogenin.
To determine whether the LANA-1-ANG complex contained other cellular proteins, equal protein concentrations of TIVE-LTC lysates were immunoprecipitated separately with 1 μg of anti-ANG IgG, anti-LANA-1, or control IgG and analyzed by SDS-PAGE (Fig. 1D and E). Protein bands visible by Coomassie staining that were unique to LANA-1 and ANG lanes (Fig. 1E, lanes 2 and 3) were excised for proteomic analysis using LC-ESI-MS methods at the Midwest Proteome Center, RFUMS (116). Repetition of MS analysis four times demonstrated that only about 30% of the proteins were consistently immunoprecipitated, which corroborated earlier reports (5). We considered only the proteins with a peak score percentage of 90 or higher (Table 1; see Tables S1, S2, and S3 in the supplemental material). The frequency of detection of the proteins listed in Table 1 is shown in Table S3 in the supplemental material. These experiments identified many proteins that were uniquely detected only in ANG-IP or in LANA-1-IP reactions (Tables S1 and S2, respectively). In addition, 28 proteins were immunoprecipitated by both angiogenin and LANA-1 antibodies (Table 1).
Table 1.
Common proteins immunoprecipitated by anti-ANG and LANA-1 IgG antibodies
| Functional category | Protein name |
|---|---|
| Structural protein/cytoskeletal | Myosin, heavy polypeptide 14, nonmuscle (MYH14) |
| ACTB, actin (beta, alpha) | |
| Alpha actinin 4 (ACTN4) | |
| Vimentin variant 3 (VIM) | |
| Ezrin (EZR) | |
| Ribosomal proteins (RP) | Ribosomal protein, large, P0 (RPLP0) |
| Ribosomal protein L4 (RPL4) | |
| Ribosomal protein S5 (RPS5) | |
| Ribosomal protein L11 (RPL11) | |
| Ribosomal protein S19 (RPS19) | |
| Chaperons | HSP90 |
| HSP70 | |
| Cyclophilin A complexed with dipeptide Gly-Pro, peptidyl prolyl isomerase A (CyPA) | |
| Transcription factor | Signal transducer and activator of transcription 1 (STAT 1) |
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein (14-3-3) (YWHAB) | |
| SP100, high-mobility-group box 1 (HMGB1) | |
| Calcium binding | Annexin A2 isoform 2 (ANXA2) |
| Enzyme-oxidoreductase | Phosphoglycerate dehydrogenase (PHGDH) |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | |
| Hexokinase (HK1) | |
| Pyruvate kinase M2 (PKM2) | |
| Aldolase A | |
| Ubiquitin proteosome system | Ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) |
| tRNA synthetase | Arginyl-tRNA synthetase (RARS) |
| JTV1, aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 (AIMP1) | |
| Leucyl tRNA synthetase (LARS) | |
| Alanyl-tRNA synthetase (AARS) | |
| Antioxidant | Peroxiredoxin (PRDX1) |
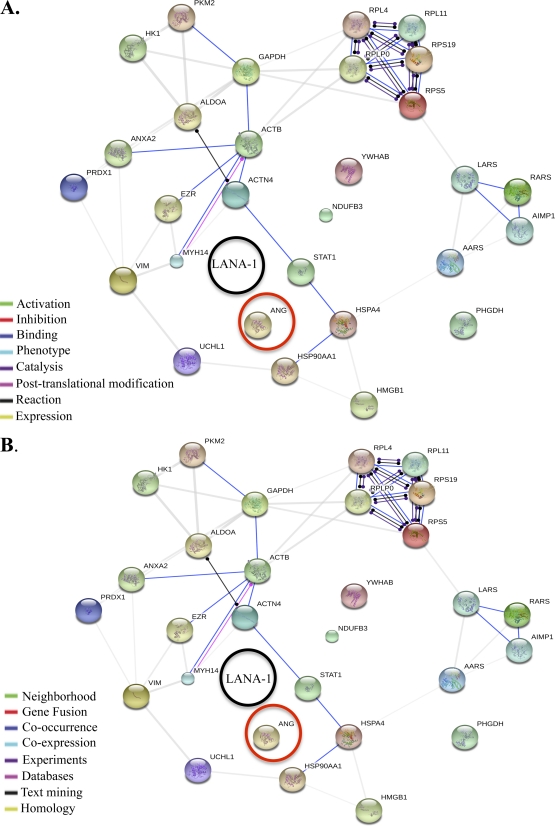
STRING analysis of common proteins immunoprecipitated by LANA-1 and ANG reveal a network of connections.
STRING (Search Tool for the Retrieval of Interacting Genes) is a predicted and known protein-protein interaction database program. It generates a network of interactions from a variety of sources, including different interaction databases, text mining, genetic interactions, and shared pathway interactions. The list of common proteins (from Table 1) pulled down by ANG and LANA-1 was used as the input to the STRING tool, and the resulting networks generated are shown in Fig. 2.
Fig 2.
STRING analysis of all the common proteins immunoprecipitated by angiogenin and LANA-1. The STRING program generates functional protein association networks. (A) Action view; uses different-colored lines to depict the types of interaction between proteins. (B) Evidence view; uses different-colored lines to depict the type of evidence that supports each interaction.
STRING provides four different viewing options to interpret the networks: confidence view, evidence view, action view, and interactive view. The action view is shown in Fig. 2A, and the evidence view is shown in Fig. 2B. The color-coded legend for the action view shows the nature of the interactions between proteins, for example, activation, inhibition, etc. The evidence view shows the type of evidence that supports the interaction, such as experimental, text mining, etc. This analysis (Fig. 2; see Fig. S2 in the supplemental material) shows that there has been no evidence or reports so far on the interaction of ANG with proteins detected in our MS analysis, thus making our study the first one to elucidate ANG binding partners. It is evident that there is a complex web of functional connections among these proteins. Communication is mostly achieved via binding (blue line), and some proteins also regulate each other's expression levels (yellow line).
Some of the proteins recognized in the LANA-1-IP (see Table S2 in the supplemental material) were also detected in an earlier report using a GST-LANA-1 pulldown approach from KSHV+ BC-3 PEL cells (54, 112) and in a recent study of LANA-1 IP in BC-3 cells (21) (see Fig. S3 in the supplemental material). The similarity of proteins recognized by LANA-1 using two different systems of analysis (GST-LANA-1 pulldown versus our direct IP reactions) and two different latent cell types (BC-3 versus TIVE-LTC) not only indirectly validated our results but also suggested the potential biological relevance of the identified proteins.
Biological implications of common proteins recognized by LANA-1 and ANG.
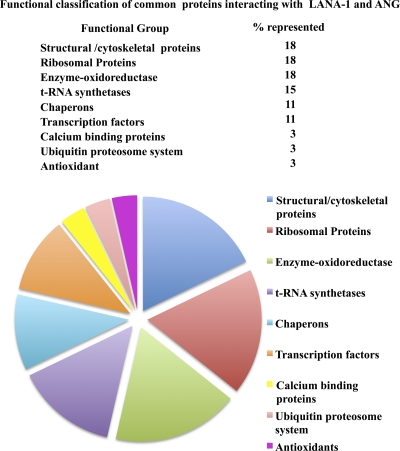
To explore the relevance of the 28 proteins immunoprecipitated by both angiogenin and LANA-1 (Table 1), an interactive exercise involving PubMed and a literature review was conducted. These proteins were classified according to their primary biological processes or molecular functions, although many nontraditional roles have also been identified (Fig. 3). Of the total proteins identified, the largest group of common proteins interacting with ANG and LANA-1 were ribosomal proteins (18%), structural proteins (18%), enzymes of the metabolic pathways (18%), and tRNA synthetases (14%). Other functional groups of proteins represented included chaperons, transcription factors, antioxidants, calcium binding proteins, and ubiquitin proteosome system proteins (Fig. 3).
Fig 3.
Pie chart showing the percentages of functionally classified proteins found to commonly interact with both angiogenin and LANA-1.
Validation of mass spectrometric results: a higher level of annexin A2 is expressed in KSHV-positive B-cell lines.
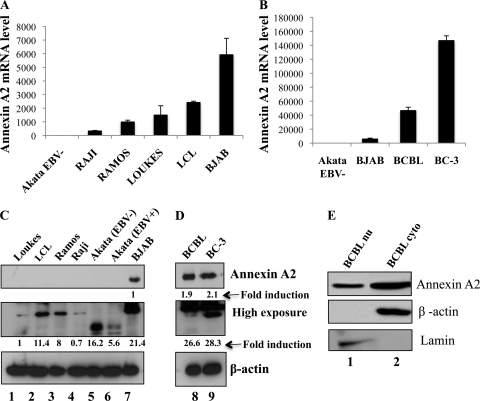
To validate the proteomic results, annexin A2 (calpactin I/lipocortin II/placental anticoagulant protein IV), a 36-kDa cytoplasmic/plasma membrane-associated, calcium-dependent, phospholipid-binding annexin family protein, was selected (39). When analyzed by real-time RT-PCR and Western blotting, low levels of annexin A2 mRNA (Fig. 4A) and protein (Fig. 4C) were detected in EBV−/KSHV− lymphoma cells (Loukes, Ramos, BJAB, Akata) and in EBV+ lymphoma (Raji and Akata) and lymphoblastoid (LCL) cells. In contrast, higher levels of annexin A2 mRNA and protein were detected in KSHV+/EBV− BCBL-1 and BC-3 cells (Fig. 4B and D). A lower-molecular-weight species of annexin A2 protein was detected in EBV− and EBV+ Akata cells during longer Western blot exposures. The EBV+ Akata cells seem to have lower levels of annexin A2 protein compared to EBV− Akata cells (Fig. 4C, lanes 5 and 6). When nuclear and cytoplasmic extracts of BCBL-1 cells were Western blotted, a substantial level of annexin A2 was also detected (Fig. 4E).
Fig 4.
Detection of annexin A2 in various B-cell lines. (A and B) RNA from the indicated B cells was extracted, and cDNA was prepared and used in real-time RT-PCR to determine annexin A2 gene expression using the SYBR green detection protocol. Results were normalized to 18S rRNA expression levels, and the annexin A2 level in the cell line with the lowest value was assigned a value of 1 for comparisons. (C and D) Equal concentrations of protein lysates from the indicated B-cell lines were separated by SDS-PAGE and Western blotted for annexin A2. β-Actin was blotted to show equal protein loading. (E) Nuclear and cytoplasmic extracts from BCBL-1 cells were prepared and Western blotted for annexin A2. Lamin and β-actin were blotted to check the purity of the extracts.
Triple staining detects annexin A2-ANG-LANA-1, annexin A2-ANG, and LANA-1-ANG colocalizations.
Annexin A2 appeared as granular spots in both the nucleus and cytoplasm of BCBL-1 cells, and some of these spots colocalized with ANG both in the cytoplasm and in the nucleus (Fig. 5A, white long arrows). Noncolocalized annexin A2 and ANG were also detected in the cytoplasm and nucleus. LANA-1 was detected predominately in the nucleus, and annexin A2 colocalized with several LANA-1 punctate spots in BCBL-1 cells (Fig. 5B, thick white arrows). Only about 10 to 20% of TIVE-LTC cells are infected with KSHV and exhibit LANA-1 staining (2). In TIVE-LTC cells, annexin A2 appeared as punctate nuclear dots only in LANA-1-positive cells, the results of which are similar to ANG profiles shown in Fig. 1A and B. Annexin A2 colocalized with LANA-1 in the TIVE-LTC cell nucleus as in BCBL-1 cells (Fig. 5C, enlarged, thick white arrows). In contrast, in LANA-1-negative cells (Fig. 5C and D, yellow arrows), annexin A2 was detected mostly in the cytoplasm, with some spots in the nucleus. ANG and annexin A2 colocalization dots were detected in the cytoplasm and a few spots in the nucleus (Fig. 5D, enlarged, white long arrows). This suggested that KSHV infection (LANA-1) is not a prerequisite for ANG-annexin A2 interaction.
Fig 5.
Colocalization of annexin A2-ANG and LANA-1 in KSHV-infected and noninfected cells. (A and B) Immunofluorescence colocalization of ANG and LANA-1 with annexin A2 in BCBL-1 cells. BCBL-1 cells were stained for ANG (red) and annexin A2 (green) in panel A and for LANA-1 (red) and annexin A2 (green) in panel B after permeabilization. DAPI (blue) was used as a nuclear stain and merged with ANG and annexin A2 or LANA-1 and annexin A2. Long white arrows indicate ANG-annexin A2 colocalization. LANA-1-annexin A2 colocalization is indicated by thick white arrows. (C and D) TIVE-LTC cells were stained for LANA-1 (red) and annexin A2 (green) in panel C and for ANG (red) and annexin A2 (green) in panel D after permeabilization. ANG-annexin A2 (long white arrows) and LANA-1-annexin A2 (thick white arrows) colocalizations are indicated. Long yellow arrows indicate the LANA-1-negative TIVE-LTC cells. Boxed area is enlarged. (E) TIVE-LTC cells were stained for ANG (blue), LANA-1 (red), and annexin A2 (green) and merged in different combinations. White arrowheads indicate ANG-LANA-1-annexin A2 triple colocalizations. Yellow short arrows indicate ANG-LANA-1 colocalizations. ANG-annexin A2 (long white arrows) and LANA-1-annexin A2 (thick white arrows) colocalizations are also indicated.
When TIVE-LTC cells were triple stained for ANG, LANA-1, and annexin A2, spots representing colocalization of all three proteins together were detected (Fig. 5E, white arrowheads; white color). This indicated that these proteins were in the same complex. In addition, complexes consisting of ANG-annexin A2 (Fig. 5E, white long arrows; cyan color) and ANG-LANA-1 (Fig. 5E, yellow short arrows; magenta color) were also separately detected in the nucleus of LANA-1-positive cells. Some noncolocalized ANG, LANA-1, and annexin A2 spots were also readily visible. Since most LANA-1 spots colocalized with ANG in infected TIVE-LTC cells, the annexin A2-LANA-1 colocalization spots probably all contained ANG as shown in the triple-staining panel. These observations of annexin A2-ANG-LANA-1, annexin A2-ANG, and ANG-LANA-1 colocalization suggested that annexin A2 forms a complex with LANA-1 and ANG as well as a separate complex with ANG.
Annexin A2 coimmunoprecipitates with ANG and LANA-1.
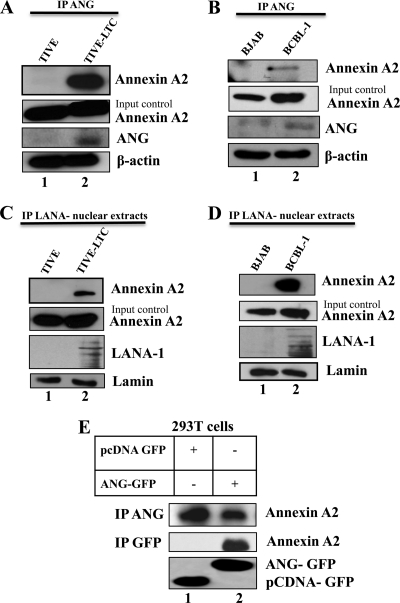
The above results suggested that annexin A2 interacts with LANA-1 and ANG in a complex. To further validate the annexin A2, ANG, and LANA-1 interactions, equal quantities of protein lysates of TIVE and TIVE-LTC cells and of BJAB and BCBL-1 cells were immunoprecipitated with anti-ANG antibody and probed for annexin A2. We observed a low level of annexin A2 coimmunoprecipitation with ANG in KSHV-negative TIVE and BJAB cells (Fig. 6A and B, lane 1). In contrast, a substantial level of annexin A2 was detected by ANG-IP from KSHV-infected cells (Fig. 6A and B, lane 2). When equal quantities of nuclear extracts of TIVE and TIVE-LTC and of BJAB and BCBL-1 cells were immunoprecipitated with anti-LANA-1 antibody and Western blotted for annexin A2, we observed that annexin A2 immunoprecipitated with LANA-1 as well (Fig. 6C and D, lane 2). We also performed IP using LANA-1 antibody in the cytoplasmic fractions of these cells but did not detect annexin A2 by Western blotting (data not shown). We have shown that LANA-1 induces ANG gene expression and secretion (93). As previously reported (93, 95), low levels of ANG were detected in KSHV-negative TIVE and BJAB cells (Fig. 6A and B, Input control, lanes 1), whereas higher levels of ANG were detected in KSHV-positive TIVE-LTC and BCBL-1 cells (Fig. 6A and B, Input control, lanes 2).
Fig 6.
Coimmunoprecipitation of annexin A2 with ANG and LANA-1. (A) TIVE and TIVE-LTC cells were grown to confluence, harvested in RIPA buffer, and immunoprecipitated using rabbit anti-angiogenin antibody and Western blotted for annexin A2. Annexin A2 and ANG input control and β-actin loading control are shown for each IP. (B) Same conditions as for panel A but with BJAB and BCBL-1 cells. (C) Nuclear extracts of TIVE and TIVE-LTC cells were prepared and immunoprecipitated using rabbit anti-LANA-1 antibody and Western blotted for annexin A2. Annexin A2 and LANA-1 input control and β-actin loading control are shown for each IP. (D) Same conditions as for panel C but with BJAB and BCBL cells. (E) 293T cells were transfected with an empty vector plasmid or full-length GFP-tagged angiogenin plasmid and harvested with RIPA buffer after 48 h. The lysates were immunoprecipitated with rabbit anti-angiogenin antibody and Western blotted for annexin A2. GFP input controls for pCDNA-GFP (lower band) and ANG-GFP (upper band) are shown.
ANG and annexin A2 were seen to colocalize in LANA-1-negative TIVE cells (Fig. 5D). To further validate their interaction in uninfected cells, we transfected 293T cells with pcDNA-GFP or pcDNA ANG-GFP. The cell lysates were then immunoprecipitated with anti-ANG antibody and Western blotted for annexin A2. Since 293T cells have high endogenous levels of ANG (approximately 1,600 pg/ml in the supernatant of 293T cells), annexin A2 and ANG were coimmunoprecipitated not only in ANG-GFP transfected cells but also in cells transfected with an empty vector (Fig. 6E). Upon immunoprecipitation with anti-GFP antibody, annexin A2 was detected only in the lysate transfected with ANG-GFP. This clearly shows that KSHV infection is not required for ANG-annexin A2 interaction. Taken together, these results demonstrated the interaction of annexin A2 with angiogenin and LANA-1 and validated our proteomic data.
Annexin A2 plays a role in the survival of KSHV-infected BCBL-1 cells: silencing annexin A2 results in significant cell death and a decrease in LANA-1 and ANG expression.
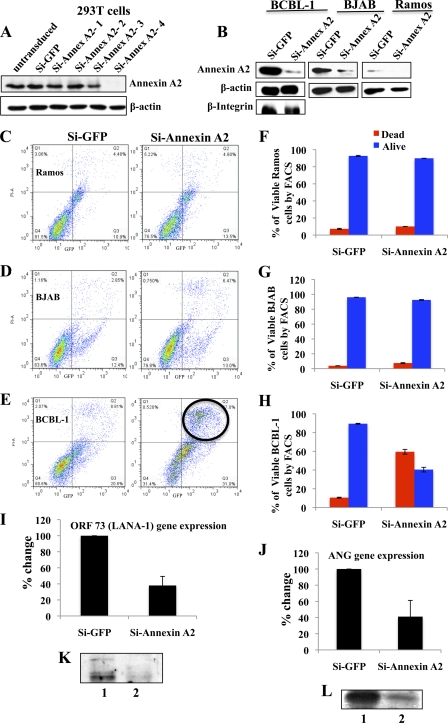
Annexin A2 is an abundant cellular protein that plays roles in cell proliferation, apoptosis (22), plasmin generation, exocytosis (96), endocytosis (31), lipid raft formation, and cytoskeleton reorganization (4, 81). Though KSHV infection is not a prerequisite for ANG-annexin A2 interaction (Fig. 5D and 6E), increased detection of annexin A2 in the nucleus along with ANG and LANA-1 (Fig. 5B, C, and E) suggested that LANA-1 association with annexin A2 could be more important than ANG association with annexin A2. We reasoned that KSHV must have evolved to utilize this multifunctional protein by targeting it via interaction with LANA-1 and ANG. To determine how annexin A2 may provide advantages for KSHV life cycle and latency in BCBL-1 cells, we used four lentivirus-based shRNAs targeting annexin A2 and one shRNA targeting GFP as a control and tested knockdown efficiency by Western blotting in 293T cells. A significant reduction in annexin A2 protein level was observed with si-annexin A2-4 (Fig. 7A) and was used in subsequent studies. BCBL-1 cells were transduced with si-annexin A2-4 and si-GFP lentivirus particles. In addition, we also transduced KSHV-negative B-lymphoma cell lines BJAB and Ramos with si-GFP and si-annexin A2-4 lentivirus particles. The cells were infected with GFP-tagged si-Renilla lentivirus particles as an indication of transduction efficiency. More than 90% of cells expressed GFP after 48 h of lentiviral infection. After 72 h, we observed about 89% reduction in annexin A2 protein levels in BCBL-1 and Ramos cells and about 80% reduction in BJAB cells (Fig. 7B). As shown in Fig. 4, a larger amount of annexin A2 was detected in BCBL-1 cells, followed by BJAB cells, while the Ramos cell line contained significantly less annexin A2 protein (Fig. 7B). The specificity of this knockdown was demonstrated by the absence of si-annexin A2 effect on the off-target proteins β-integrin and β-actin in BCBL-1 cells (Fig. 7B).
Fig 7.
Silencing annexin A2 affects viability and LANA-1 and ANG expression in BCBL-1 cells without affecting Ramos and BJAB cells. (A) 293T cells were infected with lentiviruses carrying four short hairpin RNAs targeting annexin A2 and one targeting GFP as a control. Annexin A2 knockdown was checked by Western blotting at 72 h post-lentivirus infection. (B) BCBL-1, BJAB, and Ramos cells were infected with si-GFP control or si-annexin A2-4 lentivirus particles and blotted for annexin A2 and β-actin at 72 h post-lentivirus infection. BCBL-1 cells were also blotted for β-integrin. (C, D, and E) Ramos (C), BJAB (D), and BCBL-1 (E) cells infected as described above were stained with YO-PRO-1/PI and analyzed by FACS at 72 h post-lentivirus infection. Cells were gated for propidium iodide (PI) and GFP. (F, G, and H) Quantitative analysis of YO-PRO staining. One of the representative samples from three independent experiments is shown here. The percentages of live and dead cells are represented for each cell type with si-GFP and si-annexin A2. The values were normalized to account for the differences in transduction efficiency. (I and J) RNA was extracted from BCBL-1 cells infected with si-GFP control or si-annexin A2 lentivirus particles as described above, and mRNA levels of LANA-1 and ANG were measured using real-time RT-PCR at 72 h post-lentivirus infection. (K and L) Protein levels of LANA-1 and ANG were measured by Western blotting BCBL-1 cells infected with si-GFP control or si-annexin A2 lentivirus particles as described above.
We used FACS to analyze the viability of BCBL-1 cells at 3 days after infection with si-annexin A2 lentivirus. The cells were stained with YO-PRO-1 and propidium iodide nucleic acid stains. YO-PRO-1 stain selectively passes through the plasma membranes of apoptotic cells and labels them with moderate green fluorescence. Necrotic cells exhibit red fluorescence with propidium iodide. We gated for GFP-positive (x axis) and propidium iodide (PI)-positive (y axis) cell populations. In Ramos and BJAB cell lines, silencing annexin A2 did not significantly affect the viability of the cells compared to the si-GFP control (Fig. 7C, D, F, and G). However, when annexin A2 was silenced in BCBL-1 cells, there was a remarkable decrease in viability, by about 55% (Fig. 7E and H). An average of three experiments at 3 days post si-annexin A2 transduction is shown in Fig. 7F to H, and the values were normalized to account for the differences in transduction efficiency. This suggested that annexin A2 is an important molecule that is required to sustain the viability of BCBL-1 cells. Although BJAB cells contained annexin A2 in larger amounts than Ramos cells, annexin A2 knockdown did not affect the viability of BJAB as well as Ramos cells. Silencing annexin A2 downregulated the level of ORF 73 (LANA-1) gene expression by about 60% after 3 days (Fig. 7I), and angiogenin was also downregulated to the same extent (Fig. 7J). The protein levels of LANA-1 and ANG were also significantly reduced (Fig. 7K and L). These results suggested that annexin A2 provides a specific survival advantage to cells latently infected with KSHV.
Silencing annexin A2 results in the downregulation of cell cycle-associated proteins in BCBL-1 cells but not in BJAB and Ramos cells.
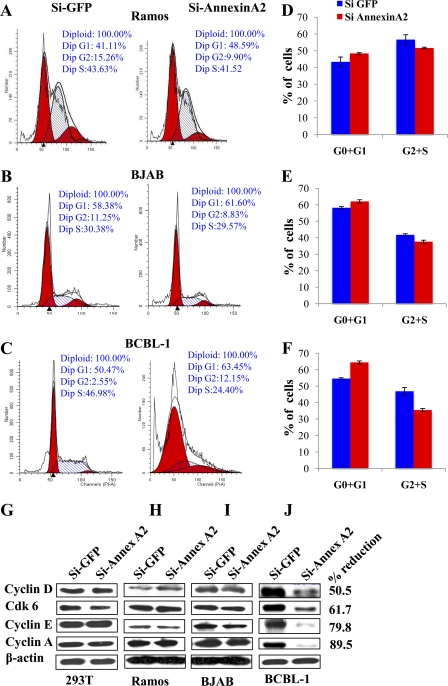
In mammalian cells the complex of cyclin and cyclin-dependent kinsaes (CDKs) regulate the transitions during the cell cycle. For example, cyclin D-CDK4/6 is involved in G1 progression, cyclin E-CDK2 is involved in G1-S transition, cyclin A-CDK2 is involved in S-phase progression, and cyclin A/B-Cdc2 regulates entry into M phase. During latency in PEL cells, KSHV genes such as ORF73 (LANA-1), ORFK10.5 (LANA-2;vIRF3), ORF72 (v-cyclin), ORF71 (v-FLIP), K12 (kaposins) and v-microRNAs function to override these cellular checkpoints and promote cell cycle leading into cell proliferation. To determine whether knocking down annexin A2 had any effects on the cell cycle, we used FACS to carry out cell cycle analyses of Ramos, BJAB, and BCBL-1 cells. Experiments were repeated three times; representative examples are shown in Fig. 8A to C, and the respective quantitative G0+G1- and G2+S-phase results are shown in Fig. 8D to F. When annexin A2 was silenced in Ramos and BJAB cells, no major difference in the cell cycle profile was seen (Fig. 8A, B, D, and E). The differences in G0+G1- and G2+S-phase results between the si-GFP and si-annexin A2 cell populations were equal to or less than 5%. As expected, more cell debris representing the dead cell population was apparent in the si-annexin A2-transduced BCBL-1 cell population than in si-GFP BCBL-1 cells (Fig. 8C). Knockdown of annexin A2 increased the G0+G1 population of BCBL-1 cells to 64.5% from 54.5% in the control cells. A consequent decrease in the G2+S or proliferative population was seen for si-annexin A2 BCBL-1 cells (36% [22% + 14%]) compared to the control cells (47%[43% + 3%]) (Fig. 8F).
Fig 8.
Silencing annexin A2 affects cell cycle progression in BCBL-1 cells but not in Ramos and BJAB cells. (A, B, and C) Ramos, BJAB, and BCBL-1 cells infected with si-GFP and si-annexin A2 lentivirus particles were stained with propidium iodide (PI) and analyzed by FACS at 72 h post-lentivirus infection for cell cycle distribution. One representative figure is shown. (D, E, and F) Quantitative analysis of cell cycle distribution of Ramos (D), BJAB (E), and BCBL-1 (F) cells treated with si-GFP or si-annexin A2 lentivirus from three independent experiments. The percentages of cells in G0+G1 and G2+S phases are graphed. (G, H, I, and J) 293T, Ramos, BJAB, and BCBL-1 cells treated as described above were harvested with RIPA buffer after 72 h. The lysates were Western blotted for cyclin D, Cdk6, cyclin E, cyclin A, and β-actin. The percent reduction of proteins in BCBL-1 cells is indicated.
We compared the levels of several cyclin proteins in si-GFP- and si-annexin A2-transduced 293T, Ramos, BJAB, and BCBL-1 cells. The protein levels of cyclin D, CDK 6, cyclin E, and cyclin A were comparable between si-GFP and si-annexin A2 samples from 293T, Ramos, and BJAB cells (Fig. 8G, H, and I). In contrast, the above-mentioned proteins were downregulated by about 51%, 62%, 80%, and 90%, respectively, in si-annexin A2 BCBL-1 cells (Fig. 8J) whereas the expression of β-actin was not affected. This also served as an indication of equal protein concentrations in these samples. These results demonstrated that si-annexin A2 profoundly affected the BCBL-1 cell cyclin proteins.
In summary, these studies clearly demonstrated that annexin A2 is a crucial protein important for cell survival and proliferation that is invariably targeted for regulation and manipulation by KSHV to establish and maintain its latency.
DISCUSSION
Our studies exposed the identity of several proteins not previously implicated in KSHV or angiogenin biology and offer a meaningful framework and starting point for further detailed analysis of the roles of these proteins in KSHV. The known functions of some of these proteins and why KSHV has evolved to interact with these proteins are discussed below.
Implications of LANA-1 interactions with annexin A2.
Increased levels of annexin A2 mRNA and protein were detected in B-lymphoma cells carrying KSHV (Fig. 4A). Annexin A2 is upregulated in many cancer types, such as colorectal, pancreatic, breast, gastric, kidney, vascular tumors, etc., where it has been hypothesized to be a promoter of tumor growth, angiogenesis, and metastasis (30, 32, 99, 104, 121). Interestingly, loss of annexin A2 has been observed in a few cancers such as prostatic adenocarcinoma and pulmonary metastases of osteosarcoma, where annexin A2 may have a role in suppressing tumor metastasis (40, 53, 71).
Annexin A2 exists in a hetero-tetrameric complex with S100A10/p11 proteins on the plasma membrane and acts as a receptor for tissue plasminogen activator (t-PA), resulting in the activation of plasminogen (44). In humans, overexpression of annexin A2 in acute promyelocytic leukemia leads to a hyperfibrinolytic bleeding diathesis reflective of excessive cell surface annexin A2-dependent generation of plasmin (76). The increased levels of annexin A2 could be contributing to the aggressive nature of PEL cells. Interestingly angiogenin, like annexin A2, is known to activate plasminogen by binding to the actins on the cell surface (48). Since B cells have very little cytoplasm, ANG and annexin A2 were seen to colocalize mostly in the nucleus; however, ANG and annexin A2 colocalization spots were also detected in the cytoplasm (Fig. 5A), which was more obvious in LANA-1-negative TIVE-LTC cells (Fig. 5D). Our study, demonstrating for the first time the interaction of ANG with annexin A2, is exciting, as this interaction may also be on the cell surfaces, where it could potentiate the action of t-PA. Since the interaction does not require LANA-1, this could have important consequences in the cancer cells where annexin A2 and angiogenin levels are usually upregulated.
We detected a substantial amount of annexin A2 in the nucleus and cytoplasm of BCBL-1 cells (Fig. 4E and Fig. 5). In TIVE-LTC cells, we detected distinctive differences in annexin A2 staining between LANA-1-positive and -negative cells, which was reminiscent of the result with angiogenin staining (Fig. 1B and 5D). Annexin A2 appeared as punctate nuclear dots and colocalized with LANA-1 in the nuclei of infected cells; in contrast, in LANA-1-negative cells, it was mostly cytoplasmic with some nuclear spots. In triple staining, LANA-1, ANG, and annexin A2 were all present in the same complex in the nucleus (Fig. 5E, white spots). However, not all annexin A2, ANG, and LANA-1 dots colocalized and few dots consisting of ANG and annexin A2 (cyan) and LANA-1-ANG (magenta) were detected. These results, together with increased annexin A2 levels in KSHV-infected cells, suggest that LANA-1 interaction with annexin A2 must have evolved for a specific advantage in KSHV biology.
Annexin A2 with S100A10/p11 in a tetrameric high-molecular-weight complex is localized to the cytoskeleton, while as a monomer it is cytosolic (62). In the nucleus, it is associated with the glycolytic enzyme 3-phosphoglyceratekinase in the primer recognition protein (PRP) complex that interacts with DNA polymerase α in the lagging strand of DNA during replication (52). This property is of great significance to DNA viruses such as KSHV. Since we observed the interaction between LANA-1, annexin A2, and ANG in the nucleus, we hypothesize that perhaps the interaction plays a role in enhancing viral/host DNA replication. Additionally, annexin A2 possesses a NES signal and is thought to be exported out of the nucleus rapidly (70). Since nuclear annexin A2 was distinctively seen in LANA-1-positive cells, LANA-1 and ANG could both be interacting with annexin A2 in the nucleus to keep it from being exported out and exploit its functions in DNA replication.
In EBV-infected cells, EBV latent membrane protein 1 (LMP 1) has been shown to mediate the phosphorylation and nuclear translocation of annexin A2 by activating a PKC pathway where it mediates DNA synthesis and cell proliferation (74, 115). We have shown that KSHV-induced ANG activates PLC-γ and AKT phosphorylation. Induction of the PLC-γ pathway by ANG could be also mediating the nuclear delivery of annexin A2 in BCBL-1 cells. Treatment with neomycin, PLC-γ inhibitor U73122, or silencing ANG resulted in cell death, inhibition of latent ORF73 (LANA-1) gene expression, and an increase in lytic switch ORF50 (RTA) gene expression (95). Nuclear translocation of annexin A2 was also shown to be inhibited by the PLC-specific inhibitor U73122 (74). Therefore, the decrease in KSHV-infected cell survival, LANA-1 expression, and KSHV latency upon neomycin treatment could be due to cumulative inhibition of both ANG and annexin A2 nuclear delivery.
When we silenced annexin A2 in BCBL-1 cells, we observed inhibition in cell growth and proliferation and a significant decrease in the viability of BCBL-1 cells despite the presence of latent KSHV, which offers survival advantages to the infected cells via various mechanisms. LANA-1 and ANG transcription levels as well as protein levels were significantly inhibited. This is not surprising, since we have shown that LANA-1 induces ANG (93). The levels of cyclin proteins that participate in cell cycle regulation were also downregulated. Given its role in DNA replication, knocking down annexin A2 probably initiated the G1 arrest and apoptosis. However, the knockdown of annexin A2 in 293T, Ramos, and BJAB cells did not have a significant effect on the viability or the cell cycle. Compared to 293T, BJAB, and BCBL-1 cells, Ramos cells have less annexin A2, and hence the lack of consequence seen could be due to its reduced reliance on annexin A2. BJAB and 293T cells, on the other hand, have a substantial amount of annexin A2, but the knockdown did not affect them as adversely as the BCBL-1 cells. Though KSHV infection is not a prerequisite for ANG-annexin A2 interaction (Fig. 5D and 6E), the fact that annexin A2 knockdown resulted in the downregulation of LANA-1 and ANG gene expression and protein levels (Fig. 7I to L), coupled with more nuclear ANG and annexin A2 along with LANA-1 in cells latently infected with KSHV (Fig. 1A and Fig. 5B, C and E) suggested that LANA-1 association with annexin A2 could be more important than ANG association with annexin A2, and there might be a specific association of annexin A2 with KSHV biology. Our studies demonstrating that annexin A2 provides a survival advantage for KSHV-infected cells suggest a potential reason why KSHV targets it via ANG and LANA-1.
Annexin A2 expression is induced in v-src-, v-H-ras-, v-mos-, or SV40-transformed cells (84), and by insulin, fibroblast, and epidermal growth factors (56). Annexin A2 expression itself is regulated during the cell cycle with maximal expression occurring as the cells enter S phase so as to facilitate DNA synthesis. A gradual reduction in steady-state levels of annexin A2 mRNA and protein occurs as cells progress through S phase (23). Our results show that annexin A2 is perhaps utilized by KSHV to regulate cell cycle progression in BCBL-1 cells. The effects seen with si-annexin A2 are most probably also due to combined consequences of downregulated LANA-1 and ANG. Although the mechanism by which interaction of LANA-1 and ANG with annexin A2 mediates BCBL-1 cell survival and proliferation is not yet clear, the observations that (i) LANA-1 induces ANG, (ii) LANA-1-positive cells have higher nuclear annexin A2, (iii) LANA-1 and ANG interact with each other and annexin A2, and (iv) knockdown of annexin A2 differentially decreases the viability of BCBL-1 cells and not that of BJAB, Ramos, or 293T cells strongly suggest that the effects seen could be due to the dependence of KSHV on annexin A2 functions. Because of its flexibility in location and function, annexin A2 probably serves as an excellent target for KSHV. Taken together, these results show that annexin A2 is an important and indispensable protein for KSHV biology and provide motive for LANA-1 and ANG to interact with it. However, further detailed studies need to be undertaken to fully understand the mechanisms behind the role of annexin A2 in KSHV biology, which are beyond the scope of current studies.
Our earlier findings indicated a highly interlinked loop between LANA-1 induction of ANG gene expression, PLC-γ/AKT phosphorylation, ANG nuclear translocation, ANG's ability to induce LANA-1 (as well as vCyclinD and vFLIP) expression, latency, and cell survival. Studies exploring the potential mechanisms by which ANG plays critical roles in KSVHV latency, anti-apoptosis, and angiogenesis are beyond the scope of this report and will be reported elsewhere. Nevertheless, our present study also exposes an interesting regulatory loop between KSHV induction of ANG expression, ANG's role in the establishment and maintenance of KSHV latency, and annexin A2's role in the survival of cells latently infected with KSHV. The observed effect of si-annexin A2 on cell survival in the context of KSHV-infected cells is probably not just due to annexin A2's role as the direct activator of these processes but is probably due to the combinatorial effects of reduction in KSHV latent gene expression and its downstream consequences, including reduction in angiogenin expression and cell survival. The observed regulatory loop of annexin A2 in KSHV biology opens up a new avenue that could potentially be exploited for effective control of KSHV and PEL. Further extensive studies are essential to fully comprehend these links and the roles of the various factors in KSHV genome epigenetics, latency, and cell survival.
Implications of LANA-1 and ANG interaction with ribosomal proteins.
About 80 ribosomal proteins (RPs) that form the 60S (large) and 40S (small) subunits of a ribosome are known to bind RNA. DNA-binding motifs have been found in ribosomal proteins, and many extraribosomal functions of these proteins have been highlighted in recent years (67, 111). Ribosomal proteins L5, L11, L23, and S7 have been shown to play a role in signaling pathways that communicate perturbations of ribosomal biogenesis to cell cycle control (20, 26, 51, 120). They bind to and inhibit the E3 ligase function of HDM2 and consequently stabilize and activate p53 (69, 73). Depletion of the ribonucleotide pool induced a p53-dependent cell cycle arrest without any DNA damage (69).
ANG translocates into the nucleoli of subconfluent endothelial cells and transcribes rRNA by binding to the abundant CT repeats present in the nontranscribed region of the rRNA gene (113). Association of ANG with the RPs could be a reflection of this function. Interestingly, previous studies using a similar proteomic approach also identified LANA-1 interaction with a number of ribosomal proteins (54). LANA-1 and ribosomal protein S9 have been found to interact with B23/NPM, which is implicated in ribosomal biosynthesis and chromatin remodeling (68, 111). Hence, LANA-1-ANG interactions with the ribosomal proteins could be a means to regulate ribosomal biosynthesis and therefore the rate of protein translation. Additionally, both ANG and LANA-1 are known to promote survival of cells latently infected with KSHV (93). It can be hypothesized that LANA-1 and ANG interact with RPs, such as L11, to inhibit its binding to MDM2 so that ubiquitination of p53 by MDM2 is not interrupted and hence p53 is not activated upon ribosomal stress.
Implications of LANA-1 and ANG association with tRNA synthetases.
Apart from their role in protein synthesis, many noncanonical functions have been attributed to aminoacyl-tRNA synthetases (aaRSs). Through cell signaling, aaRSs in higher eukaryotes have evolved to control complex cellular behaviors such as regulation of apoptosis, angiogenesis, and immune responses (12, 63, 65, 110). They also have the ability to physically interact with each other, as well as with additional cofactors forming macromolecular complexes (65). The multisynthetase complex (MSC) is composed of nine aaRSs and three aaRS-interacting multifunctional proteins (AIMP1, AIMP2, and AIMP3) (88), and their involvement in vital processes such as DNA replication, transcription and translation, signal transduction, and protein degradation has already been shown (3, 6, 25, 41, 55, 90). ANG and LANA-1 could be interacting with MSC as a whole or its components separately to enhance translation or to exploit their many auxiliary functions.
ANG is a member of the RNase A superfamily and has been shown to cleave tRNAs into tRNA halves to induce translational repression during stressful conditions (35, 114). LANA-1 could also be interacting with ANG to block its ribonucleolytic site to prevent tRNA cleavage so that translation goes undisturbed. AIMP2, a component of the MSC that was found to interact with LANA-1 and ANG, functions as a proapoptotic factor via p53 in response to DNA damage and also suppresses cell proliferation via downregulation of c-Myc (45, 60). It also has the ability to target TNF-receptor-associated factor 2 for degradation by ubiquitination and augment TNF-α-induced cell death (24). Since the anti-proliferative and proapoptotic functions of AIMP2 are not favorable for the maintenance of latency, LANA-1 and ANG could be binding to AIMP2 to change either its conformation to hinder its binding or its transactivation region to inhibit its functions.
Implications of LANA-1 and ANG association with metabolic pathway enzymes.
One of the common observations in cancer cells is an increase in glycolytic activity even in the presence of enough oxygen, known as the Warburg effect. KSHV infection was also shown to induce the Warburg effect in endothelial cells by upregulating the glucose transporter GLUT3 and the glycolytic enzyme hexokinase, increasing lactic acid production, and decreasing oxidative phosphorylation with sensitivity toward glycolytic inhibitors (28). Interaction of LANA-1 and ANG with glycolytic pathways enzymes could be an indication of a role for LANA-1 and ANG in the observed Warburg effect.
ANG, through PLC-γ signaling, helps in the establishment of latency. It also activates AKT pathways in the infected cells (95), which could assist in the induction of the Warburg effect and activation of hexokinases. LANA-1 and ANG could associate with these enzymes to create an energetically more favorable environment for the establishment of latency.
Implications of LANA-1 and ANG association with antioxidants.
Reactive oxygen species (ROS) are mainly generated during oxidative phosphorylation in mitochondria and are toxic at high levels to mammalian cells. Therefore, their levels need to be strictly monitored, which is accomplished with the help of different antioxidative systems in various cellular compartments. Peroxiredoxins (Prxs) are a ubiquitous and highly expressed family of antioxidant enzymes that contain an active-site cysteine that is oxidized to a sulfenic acid by the peroxide substrate (17). Prxs have strong peroxidase activity that can reduce and detoxify hydrogen peroxide (H2O2), peroxynitrite, and a wide range of organic hydroperoxides (ROOH) (50, 86). It also associates with PTEN, one of the powerful tumor suppressors, protecting its lipid phosphatase from H2O2-induced inactivation and thus preventing AKT-driven transformation (15). Prx I contains a consensus site (Thr90-Pro-Lys-Lys) for phosphorylation by cyclin-dependent kinases (CDKs) such as Cdc2 that reduce its peroxidase activity. The phosphorylation and inactivation of Prx I by Cdc2 occur only during mitosis, suggesting that the resulting intracellular accumulation of H2O2 might be important for progression of the cell cycle (18). Based on the functional background of this protein, LANA-1 and ANG probably associate with Prx-1 to inhibit its anti-proliferative functions. Oxidative stress induced by upregulated ROS has been shown to lead to KSHV reactivation or cell death in PEL cells (66). It is also possible that Prx-1 helps to maintain latency by protecting LANA-1 or ANG from ROS-induced modifications. Further studies need to be undertaken to understand the role of Prx1 in KSHV biology.
Implications of LANA-1 and ANG association with chaperons.
Heat shock proteins (HSPs), such as HSP60, HSC70, and HSP90, are constitutively expressed, while others, such as HSP27 and HSP70, are strongly induced by various stresses such as heat, oxidative stress, or anticancer drugs (38). HSPs directly interact with components of the apoptotic pathway at multiple points to ensure that stress-induced damage does not inappropriately trigger cell death (98). HSP90, HSP70, and cyclophilin A (CyPA) all have strong anti-apoptotic functions in the cell. CyPA also has proinflammatory effects on endothelial cells (57) and may play an important role in the pathogenesis of inflammatory diseases, such as Kaposi's sarcoma. Both LANA-1 and ANG were found to interact with chaperons like HSPs and cyclophilin A. These interactions could help to modulate antiviral stress responses or inflammatory responses or influence the apoptotic pathways to facilitate the establishment and maintenance of latency.
Implications of LANA-1 and ANG association with transcription factors (TF).
TFs can be activated or repressed by other regulatory proteins. STAT1 is a principal target of both type I and type II interferon (IFN) activation (27, 97) and mediates the growth inhibitory and proapoptotic effects of IFN (11, 79). SP100, a regulatory protein found in nuclear bodies (NBs), is highly induced by class I and class II interferons and has transcription regulatory functions (42). SP100-HMGB1 is one of the natural splice variants of SP 100 that contains the high-mobility-group box 1 (HMG1) protein sequence as a domain (43). Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, also known as 14-3-3, is a highly conserved protein known to interact with about 200 different, mostly phosphorylated proteins (75, 87), resulting in the activation, inhibition, or change in their conformation to control diverse biological processes such as cell signaling, cell cycle progression, signal transduction, intracellular trafficking/targeting, cytoskeletal structure, transcription, and apoptosis (108). LANA-1 is already known to modulate the transcriptional activity of a wide range of proteins, such as ATF, AP-1, ATF4/CREB2, CAAT, or Sp1, linked to a TATA box (37, 91). LANA-1 and ANG probably interact with the above mentioned TFs to modulate their functions in ways that favor latency.
LANA-1 interactions with STAT-1 are most likely to suppress its proapoptotic functions and interferon responses against the viral episome. Nuclear bodies have crucial antiviral activities, due to which many RNA and DNA viruses, including herpesviruses, adenoviruses, papovaviruses, papillomaviruses, and arenaviruses, express proteins that can localize and modify the composition and morphology of NBs (101), affecting cell cycle control, differentiation, apoptosis, DNA damage responses, cellular senescence, and angiogenesis (7). LANA-1 was not found to localize in NBs (105); however, existence of the SP100-HMGB1 variant suggests the possibility of a direct association of an NB protein with chromatin by DNA binding through the HMG box motifs. It can be hypothesized that LANA-1 interacts with SP100-HMG1 to tether the viral genome to the chromosome, which is one of the pivotal functions of LANA-1. KSHV-induced ANG could be recruited to the NB through its association with SP100-HMG to facilitate the processes.
Studies have found 14-3-3β to be highly expressed in KS tumor tissues compared to normal tissues from KS patients, implicating 14-3-3β in KS tumorigenesis (119). Latent protein LANA-2, detected exclusively in KSHV-infected B cells, has been shown to interact with 14-3-3 proteins and inhibit FOXO3α transcription factor (80). In another study, overexpression of xCT, an HHV-8 fusion-entry receptor, was shown to induce upregulation of 14-3-3β in KS (118). LANA-1 could interact with 14-3-3 in endothelial cells to function either in ways similar to those of LANA-2 or via a different mechanism to prevent cell cycle arrest. There have been no reports so far on ANG's ability to modulate other transcription factors. ANG could interact directly with these TFs or associate indirectly via LANA-1.
Implications of LANA-1 and ANG association with components of the ubiquitin-proteosome pathway.
Ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) belongs to a family of deubiquitinating enzymes (DUBs), capable of removing ubiquitin (Ub) from protein substrates to generate free monomeric ubiquitin (64, 82). UCHL1 possesses ubiquitin hydrolase as well as ubiquitin ligase activity and functions as a monoubiquitin stabilizer (82). Previous studies have shown that UCHL1 interacts with and promotes the degradation of cell cycle inhibitor p27kip1 (8). It also deubiquitinates and stabilizes Ikb, resulting in inactivation of NF-κB (102, 106) and p53 (117). UCHL1 is an upstream regulator of AKT and is known to enhance tumor cell invasion and migration via an AKT-mediated pathway (58). UCHL1 has been found to affect cell migration by changing cell morphology via the regulation of cell adhesion. UCHL1 and β-catenin/TCF/Lef can positively regulate each other and thus could have oncogenic potential (9).
The functional versatility of UCH L1 can be attributed to its capacity to function as both a ubiquitin hydrolase and a ubiquitin ligase, making it an attractive target for viral manipulation. Many viruses counter host defenses by either targeting key proteins for degradation or modulating their activity. Some viruses, such as the phycodnavirus infecting Feldmannia irregularis, mimivirus, and iridoviruses, encode a range of deubiquitinating enzymes (DUBs), implying that manipulation of the ubiquitination status of a protein by virally coded or host Ub and DUBs is a strategy extensively used by viruses in their interactions with their hosts (49). LANA-1 has previously been shown to be associated with components of the ubiquitin-proteosome pathway to induce enhancement of p53 and VHL ubiquitination and degradation (13, 103). Although no viral DUBs have been identified in KSHV so far, the interaction of LANA-1 and ANG with the cellular DUB UCHL1 indicates that perhaps KSHV utilizes cellular DUBs to target specific host cells to regulate their functions.
Most of the proteins detected in this study are ancient, highly conserved, and ubiquitous in nature. Perhaps due to these properties, they have been conscripted into performing diverse functions over the course of evolution. Their resourcefulness also makes them vulnerable to KSHV hijacking and manipulation, as it has had to evolve in their presence for a long time. Although many of the interactions found in the mass spectrometric analysis have not been validated yet, most of these proteins detected in our study have been shown to play important roles in signal transduction, cell cycle control, and apoptosis. LANA-1 is already known to interact with and manipulate other proteins with similar properties, making it more likely that such interactions do exist in KSHV-infected cells. Protein-protein interaction and complex formation are important biological events that govern various processes. Intermolecular associations between proteins could lead to conformation changes and new structures that could then be able to regulate more diverse intracellular events. The present study is the first of its kind to examine ANG-interacting proteins. A fraction of ANG's interactions could be mediated by LANA-1, while others could occur even in the absence of LANA-1. These results offer a glimpse into the complex multiprotein interactions taking place in the KSHV-infected cell environment to regulate vital functions. Further studies need to be undertaken to fully decipher the implications of these interactions. Our identification of binding partners of LANA-1 and ANG also provides important clues regarding potential targets for developing therapeutic remedies against KSHV infection and the associated malignancies.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by Public Health Service grant R56 AI091767 and the RFUMS-H. M. Bligh Cancer Research Fund to B.C. and by NIH NCRR S10 RR19325 grant to Marc J. Glucksman, RFUMS Midwest Proteome Center.
We thank Keith Philibert for critically reading the manuscript and Robert Dickinson for help in FACS analyses at the RFUMS FACS/Cell sorter core facility. We also thank Rolf Rene for his generous gifts of TIVE and TIVE-LTC cells.
Footnotes
Published ahead of print 30 November 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Akula SM, Pramod NP, Wang FZ, Chandran B. 2002. Integrin alpha3beta1 (CD 49c/29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108:407–419 [DOI] [PubMed] [Google Scholar]
- 2. An FQ, et al. 2006. Long-term-infected telomerase-immortalized endothelial cells: a model for Kaposi's sarcoma-associated herpesvirus latency in vitro and in vivo. J. Virol. 80:4833–4846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Asano K, Merrick WC, Hershey JW. 1997. The translation initiation factor eIF3-p48 subunit is encoded by int-6, a site of frequent integration by the mouse mammary tumor virus genome. J. Biol. Chem. 272:23477–23480 [DOI] [PubMed] [Google Scholar]
- 4. Babiychuk EB, Monastyrskaya K, Burkhard FC, Wray S, Draeger A. 2002. Modulating signaling events in smooth muscle: cleavage of annexin 2 abolishes its binding to lipid rafts. FASEB J. 16:1177–1184 [DOI] [PubMed] [Google Scholar]
- 5. Bader GD, Hogue CW. 2002. Analyzing yeast protein-protein interaction data obtained from different sources. Nat. Biotechnol. 20:991–997 [DOI] [PubMed] [Google Scholar]
- 6. Benkovic SJ, Valentine AM, Salinas F. 2001. Replisome-mediated DNA replication. Annu. Rev. Biochem. 70:181–208 [DOI] [PubMed] [Google Scholar]
- 7. Bernardi R, Pandolfi PP. 2007. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 8:1006–1016 [DOI] [PubMed] [Google Scholar]
- 8. Bheda A, Shackelford J, Pagano JS. 2009. Expression and functional studies of ubiquitin C-terminal hydrolase L1 regulated genes. PLoS One 4:e6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bheda A, et al. 2009. Positive reciprocal regulation of ubiquitin C-terminal hydrolase L1 and beta-catenin/TCF signaling. PLoS One 4:e5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bicknell R, Vallee BL. 1988. Angiogenin activates endothelial cell phospholipase C. Proc. Natl. Acad. Sci. U. S. A. 85:5961–5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bromberg JF, Horvath CM, Wen Z, Schreiber RD, Darnell JE., Jr 1996. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc. Natl. Acad. Sci. U. S. A. 93:7673–7678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buddha MR, Keery KM, Crane BR. 2004. An unusual tryptophanyl tRNA synthetase interacts with nitric oxide synthase in Deinococcus radiodurans. Proc. Natl. Acad. Sci. U. S. A. 101:15881–15886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cai QL, Knight JS, Verma SC, Zald P, Robertson ES. 2006. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS Pathog. 2:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cai X, et al. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. U. S. A. 102:5570–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cao J, et al. 2009. Prdx1 inhibits tumorigenesis via regulating PTEN/AKT activity. EMBO J. 28:1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186–1191 [DOI] [PubMed] [Google Scholar]
- 17. Chae HZ, et al. 1994. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc. Natl. Acad. Sci. U. S. A. 91:7017–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang TS, et al. 2002. Regulation of peroxiredoxin I activity by Cdc2-mediated phosphorylation. J. Biol. Chem. 277:25370–25376 [DOI] [PubMed] [Google Scholar]
- 19. Chang Y, et al. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869 [DOI] [PubMed] [Google Scholar]
- 20. Chen D, et al. 2007. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene 26:5029–5037 [DOI] [PubMed] [Google Scholar]
- 21. Chen W, Dittmer DP. 2011. Ribosomal protein S6 interacts with the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 85:9495–9505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chiang Y, Rizzino A, Sibenaller ZA, Wold MS, Vishwanatha JK. 1999. Specific down-regulation of annexin II expression in human cells interferes with cell proliferation. Mol. Cell. Biochem. 199:139–147 [DOI] [PubMed] [Google Scholar]
- 23. Chiang Y, Schneiderman MH, Vishwanatha JK. 1993. Annexin II expression is regulated during mammalian cell cycle. Cancer Res. 53:6017–6021 [PubMed] [Google Scholar]
- 24. Choi JW, et al. 2009. AIMP2 promotes TNFalpha-dependent apoptosis via ubiquitin-mediated degradation of TRAF2. J. Cell Sci. 122:2710–2715 [DOI] [PubMed] [Google Scholar]
- 25. Conaway JW, Bradsher JN, Tan S, Conaway RC. 1993. Transcription factor SIII: a novel component of the RNA polymerase II elongation complex. Cell Mol. Biol. Res. 39:323–329 [PubMed] [Google Scholar]
- 26. Dai MS, Lu H. 2004. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 279:44475–44482 [DOI] [PubMed] [Google Scholar]
- 27. Darnell JE, Jr, Kerr IM, Stark GR. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415–1421 [DOI] [PubMed] [Google Scholar]
- 28. Delgado T, et al. 2010. Induction of the Warburg effect by Kaposi's sarcoma herpesvirus is required for the maintenance of latently infected endothelial cells. Proc. Natl. Acad. Sci. U. S. A. 107:10696–10701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dull T, et al. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Duncan R, Carpenter B, Main LC, Telfer C, Murray GI. 2008. Characterisation and protein expression profiling of annexins in colorectal cancer. Br. J. Cancer. 98:426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emans N, et al. 1993. Annexin II is a major component of fusogenic endosomal vesicles. J. Cell Biol. 120:1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Esposito I, et al. 2006. Tenascin C and annexin II expression in the process of pancreatic carcinogenesis. J. Pathol. 208:673–685 [DOI] [PubMed] [Google Scholar]
- 33. Fakhari FD, Dittmer DP. 2002. Charting latency transcripts in Kaposi's sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J. Virol. 76:6213–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Friborg J, Jr, Kong W, Hottiger MO, Nabel GJ. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889–894 [DOI] [PubMed] [Google Scholar]
- 35. Fu H, et al. 2009. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 583:437–442 [DOI] [PubMed] [Google Scholar]
- 36. Fujimuro M, et al. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300–306 [DOI] [PubMed] [Google Scholar]
- 37. Ganem D. 2007. Kaposi's sarcoma-associated herpesvirus, 5th edition. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 38. Garrido C, Gurbuxani S, Ravagnan L, Kroemer G. 2001. Heat shock proteins: endogenous modulators of apoptotic cell death. Biochem. Biophys. Res. Commun. 286:433–442 [DOI] [PubMed] [Google Scholar]
- 39. Gerke V, Moss SE. 2002. Annexins: from structure to function. Physiol. Rev. 82:331–371 [DOI] [PubMed] [Google Scholar]
- 40. Gillette JM, Chan DC, Nielsen-Preiss SM. 2004. Annexin 2 expression is reduced in human osteosarcoma metastases. J. Cell. Biochem. 92:820–832 [DOI] [PubMed] [Google Scholar]
- 41. Glickman MH, et al. 1998. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94:615–623 [DOI] [PubMed] [Google Scholar]
- 42. Guldner HH, Szostecki C, Grotzinger T, Will H. 1992. IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J. Immunol. 149:4067–4073 [PubMed] [Google Scholar]
- 43. Guldner HH, et al. 1999. Splice variants of the nuclear dot-associated Sp100 protein contain homologies to HMG-1 and a human nuclear phosphoprotein-box motif. J. Cell Sci. 112(Pt 5):733–747 [DOI] [PubMed] [Google Scholar]
- 44. Hajjar KA, Krishnan S. 1999. Annexin II: a mediator of the plasmin/plasminogen activator system. Trends Cardiovasc. Med. 9:128–138 [DOI] [PubMed] [Google Scholar]
- 45. Han JM, et al. 2008. AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53. Proc. Natl. Acad. Sci. U. S. A. 105:11206–11211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu GF. 1998. Neomycin inhibits angiogenin-induced angiogenesis. Proc. Natl. Acad. Sci. U. S. A. 95:9791–9795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hu GF, Riordan JF. 1993. Angiogenin enhances actin acceleration of plasminogen activation. Biochem. Biophys. Res. Commun. 197:682–687 [DOI] [PubMed] [Google Scholar]
- 48. Hu GF, Strydom DJ, Fett JW, Riordan JF, Vallee BL. 1993. Actin is a binding protein for angiogenin. Proc. Natl. Acad. Sci. U. S. A. 90:1217–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Iyer LM, Balaji S, Koonin EV, Aravind L. 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117:156–184 [DOI] [PubMed] [Google Scholar]
- 50. Jacobson FS, Morgan RW, Christman MF, Ames BN. 1989. An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage. Purification and properties. J. Biol. Chem. 264:1488–1496 [PubMed] [Google Scholar]
- 51. Jin A, Itahana K, O'Keefe K, Zhang Y. 2004. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 24:7669–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jindal HK, Chaney WG, Anderson CW, Davis RG, Vishwanatha JK. 1991. The protein-tyrosine kinase substrate, calpactin I heavy chain (p36), is part of the primer recognition protein complex that interacts with DNA polymerase alpha. J. Biol. Chem. 266:5169–5176 [PubMed] [Google Scholar]
- 53. Kang JS, et al. 2002. Dysregulation of annexin I protein expression in high-grade prostatic intraepithelial neoplasia and prostate cancer. Clin. Cancer Res. 8:117–123 [PubMed] [Google Scholar]
- 54. Kaul R, Verma SC, Robertson ES. 2007. Protein complexes associated with the Kaposi's sarcoma-associated herpesvirus-encoded LANA. Virology 364:317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kerjan P, Cerini C, Semeriva M, Mirande M. 1994. The multienzyme complex containing nine aminoacyl-tRNA synthetases is ubiquitous from Drosophila to mammals. Biochim. Biophys. Acta 1199:293–297 [DOI] [PubMed] [Google Scholar]
- 56. Keutzer JC, Hirschhorn RR. 1990. The growth-regulated gene 1B6 is identified as the heavy chain of calpactin I. Exp. Cell Res. 188:153–159 [DOI] [PubMed] [Google Scholar]
- 57. Kim H, et al. 2005. Cyclophilin A may contribute to the inflammatory processes in rheumatoid arthritis through induction of matrix degrading enzymes and inflammatory cytokines from macrophages. Clin. Immunol. 116:217–224 [DOI] [PubMed] [Google Scholar]
- 58. Kim HJ, et al. 2009. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene 28:117–127 [DOI] [PubMed] [Google Scholar]
- 59. Kim HM, Kang DK, Kim HY, Kang SS, Chang SI. 2007. Angiogenin-induced protein kinase B/Akt activation is necessary for angiogenesis but is independent of nuclear translocation of angiogenin in HUVE cells. Biochem. Biophys. Res. Commun. 352:509–513 [DOI] [PubMed] [Google Scholar]
- 60. Kim MJ, et al. 2003. Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat. Genet. 34:330–336 [DOI] [PubMed] [Google Scholar]
- 61. Kishimoto K, Liu S, Tsuji T, Olson KA, Hu GF. 2005. Endogenous angiogenin in endothelial cells is a general requirement for cell proliferation and angiogenesis. Oncogene 24:445–456 [DOI] [PubMed] [Google Scholar]
- 62. Klee CB. 1988. Ca2+-dependent phospholipid- (and membrane-) binding proteins. Biochemistry 27:6645–6653 [DOI] [PubMed] [Google Scholar]
- 63. Ko YG, et al. 2001. Glutamine-dependent antiapoptotic interaction of human glutaminyl-tRNA synthetase with apoptosis signal-regulating kinase 1. J. Biol. Chem. 276:6030–6036 [DOI] [PubMed] [Google Scholar]
- 64. Larsen CN, Price JS, Wilkinson KD. 1996. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: identification of two active site residues. Biochemistry 35:6735–6744 [DOI] [PubMed] [Google Scholar]
- 65. Lee SW, Cho BH, Park SG, Kim S. 2004. Aminoacyl-tRNA synthetase complexes: beyond translation. J. Cell Sci. 117:3725–3734 [DOI] [PubMed] [Google Scholar]
- 66. Li X, Feng J, Sun R. 2011. Oxidative stress induces reactivation of Kaposi's sarcoma-associated herpesvirus and death of primary effusion lymphoma cells. J. Virol. 85:715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lindstrom M. S. 2011. NPM1/B23: a multifunctional chaperone in ribosome biogenesis and chromatin remodeling. Biochem. Res. Int. 2011:195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lindstrom MS, Zhang Y. 2008. Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J. Biol. Chem. 283:15568–15576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Linke SP, Clarkin KC, Di Leonardo A, Tsou A, Wahl GM. 1996. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 10:934–947 [DOI] [PubMed] [Google Scholar]
- 70. Liu J, Vishwanatha JK. 2007. Regulation of nucleo-cytoplasmic shuttling of human annexin A2: a proposed mechanism. Mol. Cell. Biochem. 303:211–220 [DOI] [PubMed] [Google Scholar]
- 71. Liu JW, et al. 2003. Annexin II expression is reduced or lost in prostate cancer cells and its re-expression inhibits prostate cancer cell migration. Oncogene 22:1475–1485 [DOI] [PubMed] [Google Scholar]
- 72. Liu S, Yu D, Xu ZP, Riordan JF, Hu GF. 2001. Angiogenin activates Erk1/2 in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 287:305–310 [DOI] [PubMed] [Google Scholar]
- 73. Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. 2003. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3:577–587 [DOI] [PubMed] [Google Scholar]
- 74. Luo W, et al. 2008. Epstein-Barr virus latent membrane protein 1 mediates serine 25 phosphorylation and nuclear entry of annexin A2 via PI-PLC-PKCalpha/PKCbeta pathway. Mol. Carcinog. 47:934–946 [DOI] [PubMed] [Google Scholar]
- 75. Meek SE, Lane WS, Piwnica-Worms H. 2004. Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J. Biol. Chem. 279:32046–32054 [DOI] [PubMed] [Google Scholar]
- 76. Menell JS, et al. 1999. Annexin II and bleeding in acute promyelocytic leukemia. N. Engl. J. Med. 340:994–1004 [DOI] [PubMed] [Google Scholar]
- 77. Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. 1998. Development of a self-inactivating lentivirus vector. J. Virol. 72:8150–8157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Moroianu J, Riordan JF. 1994. Nuclear translocation of angiogenin in proliferating endothelial cells is essential to its angiogenic activity. Proc. Natl. Acad. Sci. U. S. A. 91:1677–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Muller M, et al. 1993. Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-alpha and -gamma signal transduction pathways. EMBO J. 12:4221–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Munoz-Fontela C, et al. 2007. Latent protein LANA2 from Kaposi's sarcoma-associated herpesvirus interacts with 14-3-3 proteins and inhibits FOXO3a transcription factor. J. Virol. 81:1511–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Oliferenko S, et al. 1999. Analysis of CD44-containing lipid rafts: Recruitment of annexin II and stabilization by the actin cytoskeleton. J. Cell Biol. 146:843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Osaka H, et al. 2003. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum. Mol. Genet. 12:1945–1958 [DOI] [PubMed] [Google Scholar]
- 83. Ottinger M, et al. 2006. Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J. Virol. 80:10772–10786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ozaki T, Sakiyama S. 1993. Molecular cloning of rat calpactin I heavy-chain cDNA whose expression is induced in v-src-transformed rat culture cell lines. Oncogene 8:1707–1710 [PubMed] [Google Scholar]
- 85. Platt GM, Simpson GR, Mittnacht S, Schulz TF. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789–9795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Poole LB, Ellis HR. 1996. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 1. Purification and enzymatic activities of overexpressed AhpF and AhpC proteins. Biochemistry 35:56–64 [DOI] [PubMed] [Google Scholar]
- 87. Pozuelo Rubio M, et al. 2004. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem. J. 379:395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Quevillon S, Robinson JC, Berthonneau E, Siatecka M, Mirande M. 1999. Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein-protein interactions and characterization of a core protein. J. Mol. Biol. 285:183–195 [DOI] [PubMed] [Google Scholar]
- 89. Radkov SA, Kellam P, Boshoff C. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121–1127 [DOI] [PubMed] [Google Scholar]
- 90. Ramakrishnan V, White SW. 1998. Ribosomal protein structures: insights into the architecture, machinery and evolution of the ribosome. Trends Biochem. Sci. 23:208–212 [DOI] [PubMed] [Google Scholar]
- 91. Renne R, et al. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:458–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Renne R, Lagunoff M, Zhong W, Ganem D. 1996. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J. Virol. 70:8151–8154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sadagopan S, et al. 2009. Kaposi's sarcoma-associated herpesvirus upregulates angiogenin during infection of human dermal microvascular endothelial cells, which induces 45S rRNA synthesis, antiapoptosis, cell proliferation, migration, and angiogenesis. J. Virol. 83:3342–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sadagopan S, et al. 2007. Kaposi's sarcoma-associated herpesvirus induces sustained NF-kappaB activation during de novo infection of primary human dermal microvascular endothelial cells that is essential for viral gene expression. J. Virol. 81:3949–3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sadagopan S, Valiya Veettil M, Paudel N, Bottero V, Chandran B. 2011. Kaposi's sarcoma-associated herpesvirus-induced angiogenin plays roles in latency via the phospholipase C gamma pathway: blocking angiogenin inhibits latent gene expression and induces the lytic cycle. J. Virol. 85:2666–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sarafian T, Pradel LA, Henry JP, Aunis D, Bader MF. 1991. The participation of annexin II (calpactin I) in calcium-evoked exocytosis requires protein kinase C. J. Cell Biol. 114:1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE., Jr 1992. Proteins of transcription factor ISGF-3: one gene encodes the 91- and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc. Natl. Acad. Sci. U. S. A. 89:7836–7839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Schmitt E, Gehrmann M, Brunet M, Multhoff G, Garrido C. 2007. Intracellular and extracellular functions of heat shock proteins: repercussions in cancer therapy. J. Leukoc. Biol. 81:15–27 [DOI] [PubMed] [Google Scholar]
- 99. Sharma MR, Koltowski L, Ownbey RT, Tuszynski GP, Sharma MC. 2006. Angiogenesis-associated protein annexin II in breast cancer: selective expression in invasive breast cancer and contribution to tumor invasion and progression. Exp. Mol. Pathol. 81:146–156 [DOI] [PubMed] [Google Scholar]
- 100. St Clair DK, Rybak SM, Riordan JF, Vallee BL. 1988. Angiogenin abolishes cell-free protein synthesis by specific ribonucleolytic inactivation of 40S ribosomes. Biochemistry 27:7263–7268 [DOI] [PubMed] [Google Scholar]
- 101. Sternsdorf T, Grotzinger T, Jensen K, Will H. 1997. Nuclear dots: actors on many stages. Immunobiology. 198:307–331 [DOI] [PubMed] [Google Scholar]
- 102. Strayhorn WD, Wadzinski BE. 2002. A novel in vitro assay for deubiquitination of I kappa B alpha. Arch. Biochem. Biophys. 400:76–84 [DOI] [PubMed] [Google Scholar]
- 103. Suzuki T, Isobe T, Kitagawa M, Ueda K. 2010. Kaposi's sarcoma-associated herpesvirus-encoded LANA positively affects on ubiquitylation of p53. Biochem. Biophys. Res. Commun. 403:194–197 [DOI] [PubMed] [Google Scholar]
- 104. Syed SP, Martin AM, Haupt HM, Arenas-Elliot CP, Brooks JJ. 2007. Angiostatin receptor annexin II in vascular tumors including angiosarcoma. Hum. Pathol. 38:508–513 [DOI] [PubMed] [Google Scholar]
- 105. Szekely L, et al. 1999. Human herpesvirus-8-encoded LNA-1 accumulates in heterochromatin-associated nuclear bodies. J. Gen. Virol. 80(Pt 11):2889–2900 [DOI] [PubMed] [Google Scholar]
- 106. Takami Y, et al. 2007. Ubiquitin carboxyl-terminal hydrolase L1, a novel deubiquitinating enzyme in the vasculature, attenuates NF-kappaB activation. Arterioscler. Thromb. Vasc. Biol. 27:2184–2190 [DOI] [PubMed] [Google Scholar]
- 107. Tsuji T, et al. 2005. Angiogenin is translocated to the nucleus of HeLa cells and is involved in ribosomal RNA transcription and cell proliferation. Cancer Res. 65:1352–1360 [DOI] [PubMed] [Google Scholar]
- 108. van Hemert MJ, Steensma HY, van Heusden GP. 2001. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays 23:936–946 [DOI] [PubMed] [Google Scholar]
- 109. Vart RJ, et al. 2007. Kaposi's sarcoma-associated herpesvirus-encoded interleukin-6 and G-protein-coupled receptor regulate angiopoietin-2 expression in lymphatic endothelial cells. Cancer Res. 67:4042–4051 [DOI] [PubMed] [Google Scholar]
- 110. Wakasugi K, et al. 2002. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl. Acad. Sci. U. S. A. 99:173–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wool IG. 1996. Extraribosomal functions of ribosomal proteins. Trends Biochem. Sci. 21:164–165 [PubMed] [Google Scholar]
- 112. Xiao B, et al. 2010. Bub1 and CENP-F can contribute to Kaposi's sarcoma-associated herpesvirus genome persistence by targeting LANA to kinetochores. J. Virol. 84:9718–9732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Xu ZP, Tsuji T, Riordan JF, Hu GF. 2003. Identification and characterization of an angiogenin-binding DNA sequence that stimulates luciferase reporter gene expression. Biochemistry 42:121–128 [DOI] [PubMed] [Google Scholar]
- 114. Yamasaki S, Ivanov P, Hu GF, Anderson P. 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 185:35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Yan G, et al. 2007. Epstein-Barr virus latent membrane protein 1 mediates phosphorylation and nuclear translocation of annexin A2 by activating PKC pathway. Cell Signal. 19:341–348 [DOI] [PubMed] [Google Scholar]
- 116. Yang X, et al. 2009. Proteomic analysis for process development and control of therapeutic protein separation from human plasma. Electrophoresis 30:1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Yu J, et al. 2008. Epigenetic identification of ubiquitin carboxyl-terminal hydrolase L1 as a functional tumor suppressor and biomarker for hepatocellular carcinoma and other digestive tumors. Hepatology 48:508–518 [DOI] [PubMed] [Google Scholar]
- 118. Zeng Y, et al. 2010. Overexpression of xCT induces up-regulation of 14-3-3beta in Kaposi's sarcoma. Biosci. Rep. 30:277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Zeng Y, et al. 2007. Detection of 14-3-3beta, PI and AI in Kaposi's sarcoma. Chin. J. Dermatol. Venereol. 21:385–387 [Google Scholar]
- 120. Zhang Y, et al. 2003. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 23:8902–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zimmermann U, et al. 2004. Expression of annexin II in conventional renal cell carcinoma is correlated with Fuhrman grade and clinical outcome. Virchows Arch. 445:368–374 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.