Abstract
Sensory neurons latently infected with bovine herpesvirus 1 (BHV-1) abundantly express latency-related (LR) RNA (LR-RNA). Genetic evidence indicates that LR protein expression plays a role in the latency-reactivation cycle, because an LR mutant virus that contains three stop codons downstream of the first open reading frame (ORF2) does not reactivate from latency. The LR mutant virus induces higher levels of apoptotic neurons in trigeminal ganglia, and ORF2 interferes with apoptosis. Although ORF2 is important for the latency-reactivation cycle, other factors encoded by the LR gene are believed to play a supportive role. For example, two microRNAs (miRNAs) encoded within the LR gene are expressed in trigeminal ganglia of latently infected calves. These miRNAs interfere with bICP0 protein expression and productive infection in transient-transfection assays. In this report, we provide evidence that the two LR miRNAs cooperate with poly(I·C), interferon (IFN) regulatory factor 3 (IRF3), or IRF7 to stimulate beta interferon (IFN-β) promoter activity. Both miRNAs also stimulated IFN-β promoter activity and nuclear factor-kappa B (NF-κB)-dependent transcription when cotransfected with a plasmid expressing retinoic acid-inducible gene I (RIG-I). In the presence of RIG-I, the LR miRNAs enhanced survival of mouse neuroblastoma cells, which correlated with activation of the antiapoptosis cellular transcription factor, NF-κB. Immunoprecipitation assays demonstrated that both miRNAs stably interact with RIG-I, suggesting that this interaction directly stimulates the RIG-I signaling pathway. In summary, the results of these studies suggest that interactions between LR miRNAs and RIG-I promote the establishment and maintenance of latency by enhancing survival of infected neurons.
INTRODUCTION
Bovine herpesvirus 1 (BHV-1) is an Alphaherpesvirinae subfamily member that causes significant economical losses to the cattle industry. Infection of cattle with BHV-1 can lead to conjunctivitis, pneumonia, genital disorders, abortions, and bovine respiratory disease complex, a life-threatening upper respiratory tract infection (39, 41). The ability of BHV-1 to induce immune suppression in cattle is important for its pathogenic potential (reviewed in reference 39).
Following acute replication in mucosal epithelium, BHV-1 establishes lifelong latency in ganglionic neurons within the peripheral nervous system (84). The latency-related (LR) gene encodes a transcript that is abundantly expressed in trigeminal ganglia (TG) of latently infected calves (37, 40, 41, 49). LR-RNA is antisense with respect to the bICP0 gene (38, 40), which encodes the major BHV-1 transcriptional trans-activator. The LR gene encodes two or more proteins, and these proteins can be detected in a subset of latently infected neurons (28, 35). LR protein expression is necessary for the latency reactivation cycle (31). One of the LR proteins (open reading frame 2 [ORF2]) inhibits apoptosis (12, 54, 72) and regulates certain viral promoters, in part, by interacting with cellular transcription factors (56, 85).
A recent study demonstrated that the LR gene encodes two families of small noncoding RNAs (sncRNAs) (34). These sncRNAs are believed to be precursors for two mature microRNAs (miRNAs) that are expressed in TG of latently infected calves but not in TG of latently infected calves treated with dexamethasone (DEX) to initiate reactivation from latency (36). The LR gene-encoded sncRNAs or the mature miRNAs inhibit bICP0 gene expression and productive infection (36). It is not currently known whether all latently infected neurons express LR-encoded proteins and miRNAs or whether a subset of latently infected neurons express just LR-encoded protein or only the miRNAs. We predict that these noncoding RNAs have additional functions and play a supportive role in the lifelong latency-reactivation cycle of BHV-1.
Although viral genes play a role in regulating the latency-reactivation cycle, host factors also play a key role in these complex virus-host interactions (17, 38). For example, a cell-mediated immune response persists in TG of cattle infected with BHV-1 (58, 84) or mice infected with herpes simplex virus type 1 (HSV-1) (2, 7, 24, 74). Furthermore, type I alpha/beta interferon (IFN-α/β) can be detected in TG of mice latently infected with HSV-1 (9), suggesting that innate immune factors regulate the latency-reactivation cycle. Support for this prediction comes from the finding that type I interferon limits HSV-1 replication in cultured neurons (19) and small-animal models of infection (26, 57, 65).
The retinoic acid-inducible gene (RIG-I) encodes a cytosolic protein that detects viral double-stranded RNA (dsRNA) inside the cell and initiates signaling pathways that produce type I IFNs (IFN-α/β) and inflammatory cytokines (88). RIG-I contains an N-terminal caspase recruitment domain (CARD) and a C-terminal DExD/H-box RNA helicase domain (88). The helicase domain recognizes viral dsRNA, and the CARD activates downstream signaling through the adaptor mitochondrial antiviral signaling protein (MAVS) (87). RIG-I is believed to exist in an inactive state: the C-terminal regulatory domain of RIG-I interacts with the N-terminal CARD, preventing its association with MAVS. RNA binding to the C-terminal helicase induces conformational changes and exposes the CARD of RIG-I, leading to an interaction with MAVS. The interaction between RIG-I and MAVS activates two transcription factors, nuclear factor kappa B (NF-κB) and IFN regulatory factor 3 (IRF3). NF-κB and IRF3 induce transcription of type I IFN and other innate immune modulatory genes (88). In vitro, RIG-I recognizes RNAs containing a 5′ triphosphate moiety and partially double-stranded regions (27, 43, 62, 70, 78). In the context of a viral infection, RIG-I preferentially associates with shorter viral RNAs that contain 5′ triphosphates and/or dsRNA regions (3). RIG-I stimulates innate immune responses independently of the presence of toll-like receptor 3 (TLR3), which recognizes viral dsRNA at the endosomal compartment and induces a type I IFN response (1).
In this study, we tested whether LR-encoded miRNAs stimulated type I IFN signaling pathways. Both miRNAs stimulated IFN-β promoter activity in the presence of low levels of the synthetic dsRNA poly(I·C), IRF3, or IRF7. Additional studies provided evidence that when the miRNAs were cotransfected with RIG-I, IFN-β promoter activity and RNA levels were increased. The LR miRNAs also stimulated NF-κB-dependent transcription and cell survival when cotransfected with RIG-I. NF-κB is stimulated by RIG-I-dependent signaling pathways and can interfere with apoptosis by stimulating expression of genes that inhibit the intrinsic and extrinsic pathways of apoptosis (reviewed in reference 55). When cotransfected with RIG-I, both miRNAs enhanced cell survival after apoptosis was induced by cold shock treatment. Immune precipitation studies provided evidence that both LR-miRNAs were associated with RIG-I, suggesting that these interactions directly stimulated RIG-I-dependent signaling pathways. Collectively, these results suggest that the ability of the LR-encoded miRNAs to stimulate RIG-I signaling promotes the establishment and maintenance of latency by promoting survival of infected neurons.
MATERIALS AND METHODS
Cells.
Low-passage-number bovine testicle (BTest) cells, mouse neuroblastoma cells (Neuro-2A), and human embryonic kidney cells (293) were cultured in Earle's modified Eagle's medium (EMEM) supplemented with 10% fetal calf serum (FCS), penicillin (10 U/ml), and streptomycin (100 μg/ml) in a humidified 5% CO2 atmosphere at 37°C. BTest cells were prepared from healthy calves by the use of standard procedures.
Viruses.
The Cooper strain of BHV-1 (wild-type [wt] virus) was obtained from the National Veterinary Services Laboratory, Animal and Plant Health Inspection Services, Ames, IA. The BHV-1 LR mutant virus contains three stop codons at the beginning of ORF2 and lacks 25 bp of the wild-type sequence at the beginning of ORF2 (32). BHV-1 stocks were prepared in bovine cells (CRIB).
Plasmids.
The bovine IFN-β promoters were described previously (16). The human IRF3 and IRF7 expression constructs were obtained from Luwen Zhang (University of Nebraska, Lincoln, NE). LR-encoded sncRNA or miRNA or the LR control construct was cloned into pSilencer 2.1-U6 neo vector (Ambion) (34). The FLAG-tagged RIG-I constructs, including full-length RIG-I (pFE-BOS RIG-I) and the constitutively active RIG-I C-terminal deletion mutant (pEF-BOS N-RIG-I) (77), were obtained from M. Gale (University of Washington). The p5X-NF-κB-luciferase reporter construct (1) was obtained from R. Flavell (Yale University School of Medicine), and the Renilla luciferase (pRL-TK) reporter construct was purchased from Promega. For convenience, the full-length RIG-I (pFE-BOS RIG-I) and the constitutively active RIG-I (pEF-BOS N-RIG-I) are referred to here as RIG-I and N-RIG-I, respectively.
Reporter assays.
Chloramphenicol acetyltransferase (CAT) IFN-β reporter assays were performed using BTest or 293 cells cultured in 60-mm-diameter dishes as described previously (16). The CAT assays were repeated at least three times to confirm the results.
The NF-κB-luciferase reporter assays were performed using human 293 cells (1 × 106 cells) that were seeded in 60-mm-diameter dishes containing EMEM plus 10% FCS at approximately 24 h before transfection. Two hours before transfection, EMEM with 10% FCS was replaced with fresh EMEM containing 0.5% FCS, which kept basal levels of NF-κB promoter activity consistently low. Cells were then transfected with a plasmid containing the firefly luciferase gene that is regulated by a simple promoter with 5 consensus NF-κB binding sites (p5X-NF-κB-luciferase) (1 μg), a plasmid encoding Renilla luciferase under the control of the herpesvirus TK promoter (pRL-TK) (0.033 μg), the indicated LR expression plasmids (2 μg), and the designated RIG-I expression plasmid (1 μg) by the use of TransIT (Mirus; catalog no. MIR2000) transfection reagent. Forty hours after transfection, cells were harvested and subjected to the dual-luciferase assay by using a commercially available kit (Promega; catalog no. E1910) according to the manufacturer's instructions.
Cell survival studies.
Cold shock-induced apoptosis was performed as previously described (8, 72, 73). In brief, approximately 3 × 105 Neuro-2A cells (mouse neuroblastoma cells) were plated in six-well culture plates containing 10% FCS–EMEM 24 h prior to transfection. Cells were cotransfected with a plasmid carrying the β-galactosidase (β-Gal) gene (0.025 μg), the RIG-I expression plasmid (1 μg), and 2 μg of pSilencer carrying an LR miRNA1 or miRNA2. As a negative control, the LR control plasmid or the empty pSilencer plasmid was used. To maintain equal amounts of DNA for transfection, the empty pSilencer plasmid was used. After transfection for 24 h, cells were seeded in 24-well plates containing EMEM with 10% FCS and incubated for 12 h, and then EMEM with 2% FCS was added to the cultures. After 12 h in 2% FCS–EMEM, Neuro-2A cells were placed on ice for 1 h, and the culture plates were sealed with Parafilm. After 1 h on ice, the Parafilm was removed and plates were incubated at 37°C for 3.5 h. Cells were then fixed and stained for β-Gal expression assays. To calculate the relative level of cell survival after cold shock-induced cell death, the number of β-Gal-positive cells was counted as previously described (12, 13, 25, 60, 61). The number of blue cells in cultures transfected with the empty vector plus β-Gal was set as 1. The number of blue cells in cultures transfected with RIG-I along with empty vector or the LR miRNA constructs was divided by the number of blue cells in cultures transfected with the empty vector plus β-Gal. The results represent averages determined for three independent experiments.
RNA extraction and RT-PCR.
Total RNA was prepared from BTest cells using TRIzol reagent (Invitrogen; catalog no. 15596-018), according to the manufacturer's instructions. RNA was treated with amplification grade DNase I (Invitrogen; catalog no. 18068-015). Reverse transcription (RT) was carried out using SuperScript III reverse transcriptase (Invitrogen; catalog no. 18080-044) and oligo(dT) primers (Invitrogen; catalog no. 18418-020). Five percent of the resulting cDNA was subjected to PCR with primers specific to the three bovine IFN-β genes, as previously described (59). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control for equivalent sample loadings (forward primer, 5′-CCATGGAGAAGGCTGGGG-3′; reverse primer, 5′-CAAAGTTGTCATGGATGACC-3′). PCR products were subjected to electrophoresis on 1.5% agarose gels and stained with ethidium bromide.
Immunoprecipitation and amplification of LR miRNAs associated with RIG-I.
Human 293 cells (4 × 106) were cotransfected with 5 μg of full-length RIG-I plasmid, the C-terminal deletion mutant N-RIG-I, or an empty FLAG expression vector plus 5 μg of plasmids expressing miRNA1, miRNA2, or the LR control plasmid. Forty-eight hours after transfection, cells were harvested in hypotonic buffer (10 mM Tris [pH 7.5], 10 mM KCl, 0.5 mM EGTA, 1.5 mM MgCl2, plus protease inhibitor) and subjected to immunoprecipitation (IP) as previously described (11) with minor modifications. Briefly, cells were lysed by Dounce homogenization and centrifuged at 13,000 rpm for 15 min at 4°C. The supernatant was then incubated with normal rabbit serum for 1 h on ice and cleared with protein G agarose beads (Santa Cruz; catalog no. 2002). Immunoprecipitation of RIG-I complexes was carried out with 4 μg of anti-FLAG antibody (Sigma; catalog no. F1804) for 2 h at 4°C, and the antibody-antigen complex was collected using protein G agarose beads. A fraction of the beads was reserved for testing the efficiency of the immunoprecipitation by Western blot analysis. The remaining beads were suspended in 1× TBS buffer, and RNA bound to the RIG-I antibody complex was extracted from the beads by the use of acid-phenol:chloroform (Ambion catalog no. AM9720). The total extracted RNA (100 ng) was used with a Global MicroRNA amplification kit (SBI System Biosciences; catalog no. RA400A-1) to amplify total sncRNAs according to the manufacturer's instructions. One percent of the amplified cDNA served as a template for amplification with the reverse primers specific for miRNA1 (5′-ACACAAAGCCCCGTCTAGACA-3′), miRNA2 (5′-GGGGTTTGGTCTGGCCTATAC-3′), or the LR control (5′-TCGCGCATGCGCAATCAATAA-3′) and the forward primer specific for the 5′ adaptor provided in the kit.
RESULTS
LR mutant virus induces higher levels of IFN-β than wt BHV-1 in low-passage-number bovine testicle cells.
In contrast to humans or mice, cattle contain three IFN-β genes (IFN-β1, IFN-β2, and IFN-β3) that are regulated by distinct promoters (16, 80, 82). BHV-1 infection increases IFN-β1 and IFN-β3 RNA levels in established bovine cell lines (16, 59) but not in low-passage-number bovine cells (16). When de novo protein synthesis is inhibited by cycloheximide (CHX) treatment, infection of low-passage-number bovine cells with BHV-1 leads to increased IFN-β1, but not IFN-β3, RNA levels (16). Relative to wt BHV-1, the LR mutant virus induced higher levels of IFN-β3 and other IFN-stimulated genes in established bovine cells (59), suggesting that expression of an LR-encoded protein interfered with IFN-β signaling. However, we were unable to provide evidence that a protein encoded by the LR gene has an effect on IFN-β signaling. The LR mutant virus prematurely expresses high levels of LR-RNA relative to wt BHV-1, which correlates with higher levels of IFN-β mRNA induced by the LR mutant virus (59). Although these results implied that LR-RNA stimulated the IFN-β response, we felt it was important to confirm these findings in low-passage-number bovine cells before attempting to identify LR gene products that stimulate IFN-β signaling pathways.
To test whether the LR mutant virus induced higher IFN-β RNA levels in low-passage-number bovine cells, bovine testicle (BTest) cells were infected with the LR mutant virus or wild-type BHV-1 (multiplicity of infection [MOI] of 5) in the absence or presence of CHX (100 μg/ml). Infection with the LR mutant virus (Fig. 1A), but not with wt BHV-1 (Fig. 1B), consistently increased IFN-β1 mRNA levels 8-fold at 8 h after infection. IFN-β2 RNA levels were increased 3.5-fold at 8 h after infection, whereas a modest 2-fold increase was detected at 4 h after infection with wt BHV-1. Treatment with CHX led to higher levels of IFN-β1 and IFN-β2 mRNA by 4 h after infection with the LR mutant virus or wt BHV-1, which was consistent with a previous study (16). These studies demonstrated that IFN-β1 RNA levels were readily detected in low-passage-number bovine cells following infection with the LR mutant virus but not with wt BHV-1.
Fig 1.
The LR mutant virus induces higher levels of IFN-β than wild-type (wt) BHV-1 in low-passage-number bovine testicle cells (BTest). BTest cells (3 × 106) were mock infected (lane M) or infected with the LR mutant virus (A) or wt BHV-1 (B) at an MOI of 5. Where indicated, cultures were treated with 100 μg/ml of CHX. Cells were harvested, and total RNA was extracted at the indicated times (in hours) after infection. cDNA synthesis was subsequently performed by RT-PCR. The resultant cDNA was used as a template for PCR with primers specific for each of the three bovine IFN-β genes, as previously described (59). Genomic DNA extracted from bovine kidney cells (CRIB) served as template for the positive-control PCR (+). GAPDH cDNA amplification served as a loading control. The relative RNA levels were quantified using a biomolecular imager (Bio-Rad), and density values were normalized to a GAPDH control. The mock-infection value was set as 1. The values represent averages of the results of three independent experiments.
LR-encoded miRNAs enhance IFN-β promoter activity in the presence of other IFN-β activators.
The LR gene (Fig. 2A and B) encodes two families of sncRNAs (LR sncRNA1 and sncRNA2), which are believed to be precursors for two miRNAs (34) (Fig. 2C). Although the sncRNAs and miRNAs interfere with bICP0 protein expression (34), they do not overlap bICP0 coding sequences (Fig. 2B). Since the LR mutant virus induced higher levels of IFN-β RNA during productive infection (59) (Fig. 1), we predicted that LR-RNA may stimulate the IFN-β response following infection of cultured cells or ganglionic neurons. Since other viruses encode sncRNAs or miRNAs that regulate immune responses (4, 33, 53, 67, 76), we initially tested whether the LR sncRNAs or miRNAs regulate IFN-β signaling pathways.
Fig 2.
Schematic of LR gene and locations of sncRNAs and miRNAs. (A) Locations of the LR-RNA and bICP0 genes in the context of the BHV-1 genome. The bICP0 gene, but not the intact LR gene, is present in both repeats. The unique short (US) and unique long (UL) regions of the genome are denoted. (B) A partial restriction map of the LR gene is shown. The numbering system of the LR gene was derived from a previous study (49). The locations of the LR-RNA in cultured cells and TG are denoted relative to the bICP0 stop codon and 3′ terminus of bICP0 mRNA. The positions where the LR miRNA1 and miRNA2 are proposed to base pair with bICP0 mRNA are denoted by the black circles and were previously determined (34). The numbers in parentheses represent the BHV-1 genomic locations (GenBank accession number AJ004801). (C) Two families of LR sncRNAs were identified (34): LR sncRNA1 and LR sncRNA2. All of the sncRNA1 clones begin at position 525 of the LR gene sequence. The 5′ terminus of the sncRNA2 family begins at position 591 of the LR gene sequence. The 3′ terminus of the respective sncRNAs is variable. The positions and nucleotide sequences of the respective miRNAs within the sncRNAs are also denoted.
To test whether the LR miRNAs regulate IFN-β signaling pathways, BTest cells or human 293 cells were cotransfected with the designated bovine IFN-β promoter constructs plus plasmids that express the respective LR miRNAs and promoter activity was measured. Plasmids expressing sncRNA1 and sncRNA2 containing miRNA1 or miRNA2, respectively, were used for comparison to the respective miRNAs. The bovine IFN-β1 and IFN-β3 promoters resemble the human IFN-β promoter and are activated by synthetic dsRNA [poly(I·C)], IRF7, or IRF3 (16). Although IFN-β3 RNA was not readily detected during productive infection in BTest cells (Fig. 1), IFN-β3 RNA levels are increased in established bovine cell lines following infection with BHV-1 (16, 59) and the IFN-β3 promoter is strongly activated by virus-induced transcription factors (IRF3 or IRF7) or poly(I·C) (16). A previous study demonstrated that IRF7 strongly activates the IFN-β1 promoter whereas IRF3 activates only the IFN-β3 promoter (16). As expected, IRF7 or IRF3 trans-activated the IFN-β1 or IFN-β3 promoter, respectively, at least 8-fold in BTest cells (Fig. 3A). The plasmid encoding sncRNA1 stimulated the IFN-β1 and IFN-β3 promoters 3- to 4-fold in BTest cells (Fig. 3A). Plasmids expressing sncRNA2 or the two miRNAs also induced IFN-β1 or IFN-β3 promoter activity approximately 2-fold in BTest cells (Fig. 3A). In human 293 cells, the sncRNAs or miRNAs had essentially no effect on IFN-β1 or IFN-β3 promoter activity (Fig. 3B). As expected, poly(I·C) stimulated IFN-β1 or IFN-β3 promoter activity more than 15-fold in 293 cells.
Fig 3.
LR gene-encoded sncRNAs and miRNAs enhance IFN-β promoter activity stimulated by poly(I·C), IRF7, or IRF3. (A and B) BTest cells (A) or 293 cells (B) (3 × 106) were cotransfected with 1 μg of CAT reporter plasmids regulated by the IFN-β1 or IFN-β3 promoter, 2 μg of pSilencer expressing the LR sncRNAs (sncRNA1 or -2), or miRNAs (miRNA1 or -2). IRF3 (1 μg), IRF7 (1 μg), or synthetic dsRNA poly(I·C) (0.5 μg) served as a positive control for the indicated cell lines and IFN-β promoters. Plasmid DNA was maintained at the same concentration by including an empty expression vector. Two days after transfection, cells were harvested and analyzed for CAT expression. The results presented represent the fold increase relative to cells transfected with the IFN-β CAT reporter plasmids plus empty pSilencer (Empty), whose value was arbitrarily set as 1. (C and D) BTest cells (C) or 293 cells (D) (3 × 106) were cotransfected with 1 μg of IFN-β1 or IFN-β3 CAT plasmids, factors known to stimulate the IFN-β promoter [IRF3, IRF7 or poly(I·C)], 1 or 2 μg of LR-encoded sncRNA (sncRNA1 or -2), and 1 or 2 μg of miRNA (miRNA1 or -2). Two days after transfection, cells were harvested, and CAT activity was measured. The results shown represent the fold increase relative to the value obtained using IRF3-, IRF7-, or poly(I·C)-induced IFN-β promoter activity, whose value was arbitrarily set as 1.
LR miRNAs and sncRNAs have the potential to form partially dsRNA if they interact with a cellular mRNA (34). Innate immune sensors of dsRNA include TLR3 (1), RIG-I, and MDA5 (27, 43, 70, 71), and they are IFN inducible (46, 75, 87). Therefore, we tested whether the LR sncRNAs or miRNAs regulated IFN-β signaling in the presence of an IFN response. To this end, BTest cells were cotransfected with plasmids expressing the LR sncRNA or miRNA, the bovine IFN-β1 or IFN-β3 promoter constructs, and plasmids expressing IRF3 or IRF7 (Fig. 3C). For these studies, IFN-β1 or IFN-β 3 promoter activity in the presence of IRF3 or IRF7, respectively, was normalized to 1 and the effect of the respective LR-encoded sncRNAs was compared to these values. We have consistently observed in BTest cell studies that miRNA1 or miRNA2 enhanced IFN-β1 or IFN-β3 promoter activity approximately 2-fold in the presence of IRF7 or IRF3, respectively (Fig. 3C). Plasmids encoding the sncRNA1 or sncRNA2 had slightly higher effects on IFN-β1 or IFN-β3 promoter activity.
When poly(I·C) stimulated the IFN-β signaling pathway in 293 cells, miRNA1 increased IFN-β1 or IFN-β3 promoter activity 3- or 4-fold, respectively (Fig. 3D). A 4- to 7-fold enhancement of the respective IFN-β1 or IFN-β3 promoter activity relative to the effects observed with just poly(I·C) was detected in 293 cells expressing miRNA2 (Fig. 3D). Plasmids expressing sncRNA1 or either sncRNA2 were not as efficient as miRNA1 or miRNA2 with respect to activating IFN-β promoter activity. In summary, these studies indicated that miRNA1 or miRNA2 enhanced the ability of IRF3, IRF7, or poly(I·C) to stimulate IFN-β promoter activity.
RIG-I induces IFN-β promoter activity in the presence of LR sncRNAs and miRNAs.
We hypothesized that the LR sncRNAs and/or miRNAs, due to their potential to interact with cellular mRNAs or small noncoding RNAs, were recognized by RIG-I as short dsRNA and that IFN-β signaling was consequently activated. The rationale for this hypothesis is that TLR3 is not normally expressed in 293 cells (1, 42), which suggested that TLR3 was not required for activation of IFN-β by the LR sncRNAs and miRNAs. Furthermore, MDA5 primarily recognizes long dsRNA molecules (>2,000 nucleotides) (43), suggesting that it does not play an important role in sensing LR sncRNAs and/or miRNAs. Consequently, the effect of LR sncRNAs and miRNAs on RIG-I signaling was examined in BTest and 293 cells.
Plasmids expressing miRNA1, miRNA2, or sncRNA2 induced IFN-β1 promoter activity more than 5-fold when BTest cells were cotransfected with a plasmid expressing wt RIG-I (Fig. 4A). A plasmid expressing sncRNA1 induced IFN-β1 promoter activity more than 10-fold when cotransfected with wt RIG-I. As expected, IRF7 stimulated IFN-β1 promoter activity approximately 15-fold. IFN-β3 promoter activity was stimulated 83- or 68-fold when BTest cells were cotransfected with the plasmid expressing RIG-I and miRNA1 or miRNA2, respectively (Fig. 4B), which was higher than the effect elicited by IRF3. LR sncRNAs also stimulated RIG-I-mediated IFN-β3 promoter activity to levels similar to those induced by the miRNAs. RIG-I contains an inactive conformation in the absence of an agonist RNA (3, 14, 44, 62, 78, 88) and, as expected, did not stimulate IFN-β1 (Fig. 4A) or IFN-β3 (Fig. 3B) promoters. Although N-RIG-I was expected to be constitutively active, we did not observe higher levels of IFN-β1 or IFN-β3 promoter activity in BTest cells when they were cotransfected with the plasmid expressing N-RIG-I. The LR control plasmid encodes LR-RNA downstream of the region encoding the sncRNAs (34) (Fig. 2 C) and served as a negative control. The LR control RNA was unable to stimulate IFN-β1 (Fig. 4A) or IFN-β3 (Fig. 4B) promoter activity when cotransfected with RIG-I, indicating that not all sncRNAs activate RIG-I.
Fig 4.
The LR sncRNAs or miRNAs induced IFN-β promoter activity in BTest cells when cotransfected with RIG-I. BTest cells (3 × 106) were transfected with CAT plasmids containing IFN-β1 (A) (1 μg) or IFN-β3 promoter (B) (1 μg), RIG-I (1 μg), and 2 μg of a plasmid expressing the sncRNAs (sncRNA1 or -2) or miRNAs (miRNA1 or -2). Poly(I·C) (0.5 μg) and N-RIG-I (1 μg) served as positive controls. A pSilencer control construct containing an LR gene sequence adjacent to the regions encoding the LR sncRNAs was used as a negative control (LR control; see Fig. 2C). Equivalent amounts of DNA were used for transfection by addition of an empty expression vector. Two days after transfection, cells were harvested and CAT activity was measured. The results are shown as fold increases relative to cells transfected with the IFN-β CAT reporter plasmid plus the empty pSilencer plasmid (Empty), whose value was arbitrarily set as 1. The data shown represent averages of the results of three independent experiments.
In 293 cells, plasmids expressing miRNA1 or miRNA2 stimulated IFN-β1 promoter activity between 10- and 15-fold when cotransfected with the RIG-I expression plasmid (Fig. 5A). Conversely, RIG-I had little or no effect on IFN-β1 promoter activity when cotransfected with sncRNA1 and sncRNA2. Plasmids encoding miRNA1 or miRNA2 stimulated IFN-β3 promoter activity more than 50-fold when cotransfected with RIG-I (Fig. 5B). As with the IFN-β1 promoter in 293 cells, the sncRNAs had little or no effect on IFN-β3 promoter activity when cotransfected with RIG-I. Poly(I·C) and the constitutively active RIG-I (N-RIG-I) served as positive controls, and both activated IFN-β1 or IFN-β3 promoter activity in 293 cells (Fig. 5A and B, respectively). As expected, the plasmid expressing full-length RIG-I was unable to stimulate IFN-β1 or IFN-β3 promoter activity. The plasmid expressing the LR control RNA did not activate IFN-β1 or IFN-β3 promoter activity when cotransfected with RIG-I, which was consistent with the results obtained in BTest cells. In summary, these results indicated that the LR miRNAs and RIG-I synergistically activated IFN-β promoter activity in BTest or 293 cells. The sncRNAs stimulated IFN-β promoter activity only when cotransfected with RIG-I in BTest cells.
Fig 5.
The LR sncRNAs or miRNAs induced IFN-β promoter activity in 293 cells when cotransfected with RIG-I. Human 293 cells (3 × 106) were transfected with CAT plasmids containing IFN-β1 (A) (1 μg) or IFN-β3 promoter (B) (1 μg), RIG-I (1 μg), and 2 μg of a plasmid expressing the sncRNAs (sncRNA1 or -2) or miRNAs (miRNA1 or -2). Poly(I·C) (0.5 μg) and N-RIG-I (1 μg) served as positive controls. A pSilencer control construct containing an LR gene sequence adjacent to the regions encoding the small and microRNAs was used as a negative control (LR control). Equivalent amounts of DNA were used for transfection by addition of an empty expression vector. Two days after transfection, cells were harvested and CAT activity was measured. The results shown represent fold increases relative to cells transfected with the IFN-β CAT reporter plasmid plus the empty pSilencer plasmid (Empty), whose value was arbitrarily set as 1. The data shown represent averages of the results of three independent experiments.
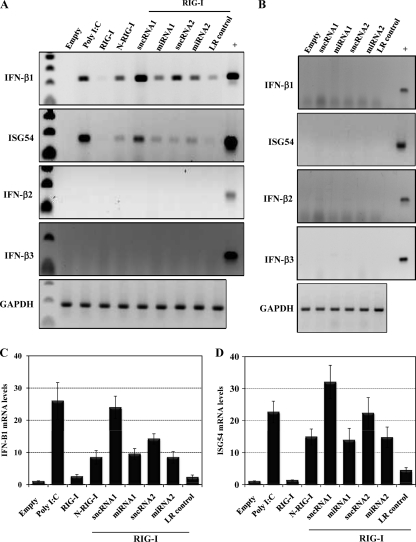
RIG-I induces endogenous IFN-β and ISG54 mRNAs in the presence of LR sncRNAs and miRNAs.
Semiquantitative RT-PCR was performed to confirm the results obtained with the reporter assays. Endogenous levels of the three bovine IFN-β mRNAs and interferon-stimulated gene 54 (ISG54) mRNA were analyzed after transfection of low-passage-number BTest cells with RIG-I and the viral sncRNAs or miRNAs. IFN-β1 and ISG54 mRNA levels were readily detected when BTest cells were cotransfected with RIG-I and the sncRNA1 (Fig. 6A, C, and D). In addition, sncRNA2 and the two miRNAs also induced IFN-β1 and ISG54 RNA at levels similar to those induced by constitutively active RIG-I (N-RIG-I) (Fig. 6A, C, and D). However, in several experiments the level of induction was not as high as observed with sncRNA1. In some experiments, the LR control slightly increased IFN-β1 RNA levels, but the induction was not as high as observed with the miRNAs or sncRNAs (Fig. 6A and C). The LR sncRNAs or miRNAs did not dramatically increase IFN-β1 mRNA levels when RIG-I was not included in the transfection (Fig. 6B). None of the constructs had an effect on the IFN-β2 or IFN-β3 mRNA levels (Fig. 6A). In summary, these results demonstrated that IFN-β1 mRNA levels were increased when low-passage-number BTest cells were transfected with RIG-I plus the LR sncRNAs or miRNAs, which subsequently induced ISG54 RNA levels.
Fig 6.
LR-encoded sncRNAs and miRNAs increase bovine IFN-β1 and ISG54 mRNA levels when cotransfected with RIG-I. (A and B) BTest cells were transfected with the LR sncRNAs and miRNAs (2 μg) along with 1 μg of a RIG-I expression vector (A) or an empty vector (B). Poly(I·C) (0.5 μg) and N-RIG-I (1 μg) served as positive controls, and the LR control plasmid (LR control) was used as a negative control. Forty hours after transfection, cells were harvested and total RNA was extracted. cDNA synthesis was subsequently performed by RT-PCR. The resultant cDNA was used as a template for PCR with primers specific for the three bovine IFN-β genes or ISG54, as previously described (59). Genomic DNA extracted from bovine CRIB cells served as a template for the positive-control PCR (+). GAPDH amplification was used as a loading control. These results are representative of at least three independent studies. (C and D) The IFN-β1 (C) and ISG54 (D) RNA levels in agarose gels were examined using a Bio-Rad Molecular Imager FX (Molecular Dynamics, Sunnyvale, CA). The values shown are expressed as fold increases relative to the band from mock-infected cells, whose value was arbitrarily set as 1.
LR miRNAs increase NF-κB-dependent transcription.
Since the NF-κB cellular transcription factor is also activated by RIG-I and NF-κB is important for stimulating IFN-β promoter activity (reviewed in reference 87), we subsequently tested whether LR miRNAs stimulated NF-κB-dependent transcription in 293 cells. A luciferase reporter gene containing a simple promoter and 5 consensus NF-κB binding sites (p5X-NF-κB) was used to measure NF-κB-dependent transcription. The LR-encoded miRNA1 and miRNA2 stimulated NF-κB-dependent transcription at least 20-fold when cotransfected with RIG-I (Fig. 7). When the respective miRNAs were cotransfected with an empty vector, NF-κB-dependent transcription was not stimulated. The controls N-RIG-I and poly(I·C) activated NF-κB-dependent transcription approximately 7-fold, which was consistent with a previous study (88). The LR control plasmid was unable to stimulate NF-κB-dependent transcription alone or when cotransfected with RIG-I (Fig. 7). These studies add further evidence that LR-encoded miRNAs stimulated the RIG-I signaling pathway.
Fig 7.
LR miRNAs stimulate NF-κB-dependent transcription when cotransfected with RIG-I. Approximately 2 × 106 293 cells were transfected with the 5× NF-κB luciferase reporter construct (1 μg), pRL-TK (0.033 μg), RIG-I (1 μg), 2 μg of an empty vector, the indicated LR miRNAs, or the LR control. N-RIG-I (1 μg) and poly(I·C) (0.5 μg) served as positive controls. Cells were harvested at 40 h after transfection, and the dual-luciferase assay was performed. The data represent the relative firefly luciferase activity normalized to Renilla luciferase activity. The values are expressed as fold differences relative to the pSilencer empty vector (Empty), whose value was arbitrarily set as 1.
LR miRNAs stimulate cell survival following cold shock-induced apoptosis.
Transient-transfection assays were performed to test whether LR miRNAs enhance cell survival, because NF-κB can promote cell survival (21, 23, 55). Plasmids expressing the respective LR miRNAs, RIG-I, and a CMV β-Gal plasmid were transfected into Neuro-2A cells, and cold shock-induced apoptosis was performed as previously described (8, 72, 73). The β-Gal cotransfection assay has been successfully used to measure the effects of various genes on apoptosis (12, 25, 30, 36, 61). When a known apoptosis stimulator is used, the number of surviving cells can be accurately measured. Neuro-2A cells, but not 293 cells, are sensitive to cold shock-induced apoptosis. BTest cells are not appropriate for these studies, because transfection efficiency is relatively low in these cells. At 36 h after transfection, cells were starved in 2% fetal calf serum for 12 h; cultures were then incubated on ice for 1 h and finally returned to 37°C for 3.5 h. Neuro-2A cells or Neuro-2A cells placed on ice for 1 h contain little to no detectable DNA laddering (72, 73). However, extensive DNA laddering, indicative of apoptosis, occurs when Neuro-2A cells are returned to 37°C for 3 or 6 h (72, 73). After 3.5 h at 37°C, it was clear that cold shock-induced apoptosis effectively reduced cell survival, as judged by the number of β-Gal-positive Neuro-2A cells (Fig. 8A). When LR miRNA1 or miRNA2 was cotransfected with RIG-I, the number of surviving cells increased dramatically relative to cells transfected with the miRNAs alone or RIG-I alone (Fig. 8B and C). It was also clear that the LR control had little or no effect on cell survival regardless of whether RIG-I was included in the transfection mix. In summary, these studies indicated that the LR miRNA1 or miRNA2 cooperated with RIG-I to enhance survival of Neuro-2A cells after cold shock-induced apoptosis.
Fig 8.
LR miRNAs interfere with cold shock-induced apoptosis. (A) Neuro-2A cells were transfected with a plasmid encoding the β-galactosidase (β-Gal) gene (0.025 μg) and 3 μg of the empty pSilencer. Images of cells before and after cold shock-induced apoptosis are shown. Cold shock-induced apoptosis was performed as described in Materials and Methods. (B) Top panels: Neuro-2A cells were cotransfected with a plasmid encoding the β-galactosidase (β-Gal) gene (0.025 μg), 1 μg of empty pSilencer, and 2 μg of pSilencer encoding the LR miRNAs (miRNA1 or -2) or the RIG-I expression plasmid, as indicated. Bottom panels: Neuro-2A cells were cotransfected with a plasmid encoding the β-galactosidase (β-Gal) gene (0.025 μg), RIG-I (1 μg), and 2 μg of pSilencer encoding the LR miRNAs (miRNA1 or -2) or LR control, as indicated. At 48 h after transfection, Neuro-2A cells were submitted to cold shock-induced apoptosis and subsequently recovered for 3.5 h at 37°C. Cold shock-induced apoptosis was performed, and pictures of the cultures were taken. (C) The number of β-Gal-positive cells was determined, and cell survival was compared to that seen with the empty vector, whose value was arbitrarily set at 1. The values represent averages of the results of three independent studies.
The two LR miRNAs are stably associated with RIG-I.
Although the results presented above suggested that LR-encoded miRNAs stimulate RIG-I-dependent signaling, they do not allow us to conclude whether the effect was direct or indirect. When RIG-I interacts with short dsRNA, the RIG-I signaling pathway is activated (43, 70, 71, 88). If the LR miRNAs directly activate the RIG-I signaling pathway, LR miRNAs should be stably associated with the RIG-I protein. If LR miRNAs indirectly activate the RIG-I signaling pathway, the respective miRNAs would not be expected to stably associate with RIG-I.
To test whether LR-encoded miRNAs were associated with RIG-I, 293 cells were cotransfected with RIG-I, N-RIG-I, or an empty vector plus plasmids expressing miRNA1, miRNA2, or the LR control RNA (see Fig. 9A for a schematic of the study). N-RIG-I lacks the RNA binding domain of RIG-I, and thus the LR-encoded miRNAs would not be expected to associate with N-RIG-I. Forty-eight hours after transfection, 293 cells were lysed and RIG-I/RNA or N-RIG-I/RNA complexes were immunoprecipitated with a FLAG antibody. Immuno precipitation of RIG-I or N-RIG-I was efficient, because approximately half of the total FLAG-tagged RIG-I proteins were precipitated from cell lysate (Fig. 9B). Following immunoprecipitation (IP), the RNA associated with RIG-I or N-RIG-I was recovered by acid phenol extraction and converted into cDNA by performing RT-PCR using adapter-specific primers. The resulting cDNA was subsequently used as a template for PCR with primers specific for the miRNA1, miRNA2, or LR control as described in Materials and Methods. A band of approximately 50 bp was amplified from cDNA prepared from the RIG-I/RNA complex of cells cotransfected with RIG-I and miRNA1 or miRNA2 but not the LR control (Fig. 9C). In contrast, the amplified band was not detected in cells transfected with N-RIG-I. In summary, these results suggested that LR-encoded miRNA1 and miRNA2 interacted with RIG-I or a complex containing RIG-I and that the RNA binding domain of RIG-I was necessary for interactions with LR-encoded miRNAs.
Fig 9.
The LR-encoded miRNAs interact with full-length RIG-I. (A) Schematic of experimental steps used to immunoprecipitate (IP) and amplify RIG-I-associated RNAs. (B) Monolayers containing 4 × 106 293 cells were transfected with 5 μg of an empty FLAG vector (Empty) or FLAG vectors expressing RIG-I or N-RIG-I along with 5 μg of plasmids expressing the LR miRNA1 or miRNA2 or LR control plasmid. Forty-eight hours after transfection, cells were harvested in hypotonic buffer and the RIG-I IP was performed using the FLAG monoclonal antibody. The efficiency of the IP was monitored by Western blot (WB) analysis. (C) RNA associated to the RIG-I complex was extracted by the acid-phenol:chloroform method and subsequently converted into cDNA by RT-PCR. The resultant cDNA served as a template for amplification by the reverse primers specific for the LR miRNA1 or miRNA2 or the LR control and the forward primer specific for the 5′ adaptor. These results are representative of 2 independent experiments. The first lane (M) corresponds to molecular weight markers.
DISCUSSION
Although it is well established that RNA viruses stimulate RIG-I due, in large part, to accumulation of dsRNA during the replication cycle (44, 77, 87, 88), this study suggested that two BHV-1 miRNAs expressed during latency also stimulated the RIG-I signaling pathway. Although we do not understand the mechanism by which RIG-I recognizes the LR miRNAs, we suggest that the ability of the LR-encoded miRNAs to base pair with a cellular mRNA leads to production of a partially double-stranded template that is recognized by RIG-I. Not all of the substrates that are recognized by RIG-I have been identified. For example, RIG-I recognizes RNA of various lengths and appears to prefer dsRNA with or without 5′ triphosphates (3, 27, 62, 69–71, 87). RIG-I has structural similarity to Dicer, an RNase III-type nuclease that mediates RNA interference (89). Dicer requires dsRNA binding protein partners, PACT for example, for maximal activity (47, 52). Interestingly, RIG-I also directly binds to PACT, and this interaction facilitates innate antiviral responses (48). Due to the complexity of the process by which RIG-I recognizes its substrates, it is not clear how the LR-encoded miRNAs were recognized by RIG-I. Regardless of the mechanism by which RIG-I recognizes the LR-encoded miRNAs, we have provided evidence that both LR-encoded miRNAs interact with RIG-I or a stable RIG-I complex, which correlated with increased IFN-β promoter activity, IFN-β1 RNA levels, and ISG54 RNA levels. Epstein-Barr virus (EBV) abundantly expresses two nonpolyadenylated sncRNAs (EBER1 and EBER2) that are recognized by RIG-I and activate type I IFN signaling (67), suggesting that RIG-I plays an important role during the latency-reactivation cycle of more than one herpesvirus.
In low-passage-number BTest cells, the sncRNAs stimulated RIG-I-dependent activation of IFN-β promoter activity with efficiency similar to that of constructs expressing the respective miRNAs. Conversely, in human 293 cells, only the miRNAs stimulated RIG-I-induced IFN-β promoter activity. We suggest that the sncRNAs may be more efficiently processed in BTest cells or that novel proteins in BTest cells facilitate the association between RIG-I and the sncRNAs. It is also conceivable that novel RNA sensors expressed in low-passage-number bovine cells, but not 293 cells, recognized the LR sncRNAs. LR-RNA initiates at two major start sites, one that is predominantly utilized during productive infection of cultured cells and another that is in TG and is 200 to 300 bp upstream from that utilized during productive infection (20, 29) (Fig. 2B). Considering that the major start site for transcription during productive infection was mapped by primer extension, minor start sites of LR-RNA transcription, especially 200 to 300 nucleotides upstream of the major start site, may not have been readily detected. If this is true, low levels of the sncRNAs and/or the miRNAs (34) would be expressed during productive infection. Since the major start site of LR-RNA expressed in TG includes sequences that encompass the LR-encoded sncRNAs and miRNAs, the sncRNAs and miRNAs would be expressed at higher levels in infected sensory neurons. Thus, LR-encoded sncRNA and miRNAs would appear to have a bigger impact on amplifying RIG-I signaling in infected TG neurons than in productively infected nonneuronal cells.
The finding that overexpression of RIG-I was necessary for the miRNAs to inhibit cold shock-induced apoptosis in Neuro-2A cells correlated with the ability of the miRNAs to stimulate NF-κB. It is well established that NF-κB directly stimulates expression of several genes that encode antiapoptosis proteins (6, 51, 55, 81). For example, NF-κB activates expression of c-FLIP (6), Bcl-2 family members (Bcl-X and Bfl-1/A1) (51), and the inhibitor of apoptosis (IAP) family members (66). Interferon induction has also been linked to apoptosis induction (10, 50), in part by inducing p53 expression (79). It is generally assumed that cell type-specific factors play a role in determining whether IFN signaling pathways result in apoptosis or antiapoptosis signaling. Following transfection of Neuro-2A cells with an LR miRNA and RIG-I, we detected many cells that survived cold shock-induced apoptosis that were not β-Gal positive. Although we believe that this result was due to RIG-I and the miRNA stimulating production of IFN-β, which was secreted to surrounding cells, we cannot rule out the possibility that the LR miRNAs were released from transfected cells and taken up by surrounding cells. Interestingly, EBER RNAs have been reported to be released from EBV-infected cells and can activate IFN signaling pathways in surrounding cells (33). Many viruses, including EBV and human herpesvirus 8, usurp NF-κB for their lytic and latent infections (reviewed in reference 18). However, a role for NF-κB in the latency-reactivation cycle of BHV-1 has not been described.
Infection of calves with the LR mutant virus results in reduced viral genomes in TG, in part because of increased levels of apoptosis during the establishment of latency (31, 54). Since the LR mutant virus does not express ORF2 (35) and ORF2 inhibits apoptosis (72), we believe that ORF2 is an important viral factor that promotes survival of infected neurons. We suspect that not all latently infected neurons constitutively express ORF2 and that certain latently infected neurons, including sncRNAs and miRNAs, may express only LR-RNA. In neurons that do not express ORF2, we predict that LR-encoded sncRNAs and/or miRNAs could, in fact, promote survival of infected neurons by more than one mechanism. For example, LR sncRNAs and/or miRNAs may interfere with productive infection in sensory neurons by reducing bICP0 protein levels (34) and enhancing IFN-β responses by interacting with RIG-I. Second, the ability of both miRNAs to cooperate with RIG-I and activate NF-κB would reduce the incidence of apoptosis in infected cells, as discussed above. Finally, NF-κB signaling promotes neurite formation in rat PC-12 cells stimulated with nerve growth factor (21) suggesting that LR miRNAs maintain the mature neuronal phenotype after infection. Interestingly, dexamethasone, which stimulates BHV-1 reactivation from latency, impairs NF-κB signaling (22, 86). Heat stress stimulates HSV-1 in vivo reactivation from latency in mice (68) and, like dexamethasone, impairs NF-κB signaling (5, 63). Thus, there appears to be a link between disrupting NF-κB signaling, inducing apoptosis, and increasing the incidence of reactivation from latency.
In the context of the latency-reactivation cycle, we predict that the ability of the LR-encoded miRNAs to interact with RIG-I is particularly important during the establishment of latency, because BHV-1 stimulates IFN responses (16, 59), RIG-I is an IFN-inducible gene (15, 45), and amplification of IFN signaling would dampen viral gene expression and enhance neuronal survival. During the maintenance of latency, low levels of viral gene expression may periodically occur in a subset of latently infected neurons, which we operationally define as “spontaneous reactivation.” During spontaneous reactivation, IFN-β signaling and increased RIG-I protein levels would likely occur as a result of lytic cycle viral gene expression. Since a potent reactivation stimulus is lacking during spontaneous reactivation, LR miRNA expression may not be repressed and interactions between RIG-I and LR miRNAs would contribute to reestablishing latency. During dexamethasone-induced reactivation from latency, the reactivation stimulus is much stronger because extensive lytic cycle viral gene expression occurs (64, 83) and expression of LR gene products is repressed, including LR miRNA expression (34, 64). Although RIG-I is IFN inducible, RIG-I protein expression can be detected in many unstimulated cell types (45), suggesting that LR-encoded sncRNAs or miRNAs interact with RIG-I in the absence of viral gene activity and consequently enhance neuronal survival. Support for this prediction comes from the finding that the LR sncRNA1 stimulated IFN-β promoter activity in BTest cells even when RIG-I was not overexpressed. It is unlikely that LR-encoded sncRNAs or miRNAs directly influence reactivation from latency, because the LR miRNAs are not detectable in TG of latently infected calves at 6 h after DEX treatment (34). In summary, our studies suggest that interactions between RIG-I and LR-encoded miRNAs or sncRNAs promote latency by “sensing” viral activity during acute infection or spontaneous reactivation. To directly examine the role that LR miRNAs play in the latency-reactivation cycle, it would be necessary to determine whether expression of the LR-encoded miRNAs affects the latency-reactivation cycle of BHV-1 in cattle.
ACKNOWLEDGMENTS
This research was supported by grants from the USDA, Agriculture and Food Research Initiative Competitive Grants Program (09-01653). A grant to the Nebraska Center for Virology (1P20RR15635) supported certain aspects of these studies.
Footnotes
Published ahead of print 30 November 2011
REFERENCES
- 1. Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413: 732–738 [DOI] [PubMed] [Google Scholar]
- 2. Allen SJ, et al. 2011. The role of LAT in increased CD8+ T cell exhaustion in trigeminal ganglia of mice latently infected with herpes simplex virus 1. J. Virol. 85: 4184–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baum A, Sachidanandam R, Garcia-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A. 107: 16303–16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bauman Y, et al. 2011. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe 9: 93–102 [DOI] [PubMed] [Google Scholar]
- 5. Belardo G, Santoro MG. 2010. Heat stress triggers apoptosis by impairing NF-kappaB sirvival signaling in malignant B cells. Leukemia 24: 187–196 [DOI] [PubMed] [Google Scholar]
- 6. Benayoun B, et al. 2008. NF-kB-dependent expression of the antiapoptotic factor c-FLIP is regulated by calpain 3, the protein involved in limb-girdle muscular dystrophy type 2A. FASEB J. 22: 1521–1529 [DOI] [PubMed] [Google Scholar]
- 7. Cantin EM, Hinton DR, Chen J, Openshaw H. 1995. Gamma interferon expression during acute and latent nervous system infection by herpes simplex virus type 1. J. Virol. 69: 4898–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carpenter D, et al. 2007. Stable cell lines expressing high levels of the herpes simplex virus type 1 LAT are refractory to caspase 3 activation and DNA laddering following cold shock induced apoptosis. Virology 369: 12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carr DJ, Veress LA, Noisakran S, Campbell IL. 1998. Astrocyte-targeted expression of IFN-alpha1 protects mice from acute ocular herpes simplex virus type 1 infection. J. Immunol. 161: 4859–4865 [PubMed] [Google Scholar]
- 10. Chawla-Sarkar M, et al. 2003. Apoptosis and interferons: role of intrferon-stimulated genes as mediators of apoptosis. Apoptosis 8: 237–249 [DOI] [PubMed] [Google Scholar]
- 11. Chiu YH, Macmillan JB, Chen ZJ. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138: 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ciacci-Zanella J, Stone M, Henderson G, Jones C. 1999. The latency-related gene of bovine herpesvirus 1 inhibits programmed cell death. J. Virol. 73: 9734–9740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ciacci-Zanella JR, Jones C. 1999. Fumonisin B1, a mycotoxin contaminant of cereal grains, and inducer of apoptosis via the tumour necrosis factor pathway and caspase activation. Food Chem. Toxicol. 37: 703–712 [DOI] [PubMed] [Google Scholar]
- 14. Cui S, et al. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29: 169–179 [DOI] [PubMed] [Google Scholar]
- 15. Cui X-F, Imaizumi T, Yoshida H, Borden EC, Satoh K. 2004. Retinoic acid-inducible gene-I is induced by interferon-gamma and regulates the expression of interferon-gamma stimulated gene 15 in MCF-7 cells. Biochem. Cell Biol. 82: 401–405 [DOI] [PubMed] [Google Scholar]
- 16. da Silva LF, Jones C. 2011. Infection of cultured bovine cells with bovine herpesvirus 1 (BHV-1) or Sendai virus induces different beta interferon subtypes. Virus Res. 157: 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Decman V, Freeman ML, Kinchington PR, Hendricks RL. 2005. Immune control of HSV-1 latency. Viral Immunol. 18: 466–473 [DOI] [PubMed] [Google Scholar]
- 18. DE Oliveira DE, Ballon G, Caserman E. 2010. NF-kB signaling modulation by EBV and KSHV. Trends Microbiol. 18: 248–267 [DOI] [PubMed] [Google Scholar]
- 19. De Regge N, Van Opdenbosch N, Nauwynck HJ, Efstathiou S, Favoreel HW. 2010. Interferon alpha induces establishment of alphaherpesvirus latency in sensory neurons in vitro. PLoS One 5: e13076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Devireddy LR, Jones C. 1998. Alternative splicing of the latency-related transcript of bovine herpesvirus 1 yields RNAs containing unique open reading frames. J. Virol. 72: 7294–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Foehr ED, et al. 2000. NF-kB signaling promotes both cell survival and neurite process formation in nerve growth factor-stimulated PC12 cells. J. Neurosci. 20: 7556–7563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garside H, et al. 2004. Glucocorticoid ligands specify different interactions with NF-kB by allosteric effects on the glucocorticoid receptor DNA binding domain. J. Biol. Chem. 279: 50050–50059 [DOI] [PubMed] [Google Scholar]
- 23. Goodkin ML, Ting AT, Blaho JA. 2003. NF-kB is required for apoptosis prevention during herpes simplex virus type 1 infection. J. Virol. 77: 7261–7280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Halford WP, Gebhardt BM, Carr DJ. 1996. Persistent cytokine expression in trigeminal ganglion latently infected with herpes simplex virus type 1. J. Immunol. 157: 3542–3549 [PubMed] [Google Scholar]
- 25. Henderson G, et al. 2002. Regulation of caspase 8- and caspase 9-induced apoptosis by the herpes simplex virus latency-associated transcript. J. Neurovirol. 8: 103–111 [DOI] [PubMed] [Google Scholar]
- 26. Hendricks RL, et al. 1991. Endogenously produced interferon alpha protects mice from herpes simplex virus type 1 corneal disease. J. Gen. Virol. 72(Pt 7): 1601–1610 [DOI] [PubMed] [Google Scholar]
- 27. Hornung V, et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314: 994–997 [DOI] [PubMed] [Google Scholar]
- 28. Hossain A, Schang LM, Jones C. 1995. Identification of gene products encoded by the latency-related gene of bovine herpesvirus 1. J. Virol. 69: 5345–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hossain A, Holt T, Ciacci-Zanella J, Jones C. 1997. Analysis of cyclin-dependent kinase activity after herpes simplex virus type 2 infection. J. Gen. Virol. 78: 3341–3348 [DOI] [PubMed] [Google Scholar]
- 30. Inman M, et al. 2001. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. J. Virol. 75: 3636–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Inman M, Lovato L, Doster A, Jones C. 2002. A mutation in the latency-related gene of bovine herpesvirus 1 disrupts the latency reactivation cycle in calves. J. Virol. 76: 6771–6779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Inman M, Lovato L, Doster A, Jones C. 2001. A mutation in the latency-related gene of bovine herpesvirus 1 leads to impaired ocular shedding in acutely infected calves. J. Virol. 75: 8507–8515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iwakiri D, et al. 2009. Epstein-Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. J. Exp. Med. 206: 2091–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jaber T, Workman A, Jones C. 2010. Small noncoding RNAs encoded within the bovine herpesvirus 1 latency-related gene can reduce steady-state levels of infected cell protein 0 (bICP0). J. Virol. 84: 6297–6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang Y, Inman M, Zhang Y, Posadas NA, Jones C. 2004. A mutation in the latency related gene of bovine herpesvirus 1 (BHV-1) inhibits protein expression of a protein from open reading frame 2 (ORF-2) and an adjacent reading frame during productive infection. J. Virol. 78: 3184–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jin L, et al. 2003. Identification of herpes simplex virus type 1 (HSV-1) latency associated transcript (LAT) sequences that both inhibit apoptosis and enhance the spontaneous reactivation phenotype. J. Virol. 77: 6556–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jones C. 1998. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv. Virus Res. 51: 81–133 [DOI] [PubMed] [Google Scholar]
- 38. Jones C. 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin. Microbiol. Rev. 16: 79–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jones C. 2009. Regulation of innate immune responses by bovine herpesvirus 1 and infected cell protein 0. Viruses 1: 255–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jones C, et al. 2006. Functional analysis of bovine herpesvirus 1 (BHV-1) genes expressed during latency. Vet. Microbiol. 113: 199–210 [DOI] [PubMed] [Google Scholar]
- 41. Jones C, Chowdhury S. 2007. A review of the biology of bovine herpesvirus type 1 (BHV-1), its role as a cofactor in the bovine respiratory disease complex and development of improved vaccines. Anim. Health Res. Rev. 8: 187–205 [DOI] [PubMed] [Google Scholar]
- 42. Karikó K, Ni H, Capodici J, Lamphier M, Weissman D. 2004. mRNA is an endogenous ligand for Toll-like receptor 3. J. Biol. Chem. 279: 12542–12550 [DOI] [PubMed] [Google Scholar]
- 43. Kato H, et al. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205: 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kato H, et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441: 101–105 [DOI] [PubMed] [Google Scholar]
- 45. Kawaguchi S, et al. 2009. Retinoic acid-inducible gene-I is constitutively expressed and involved in IFN-gamma-stimulated CXCL9-11 production in intestinal epithelial cells. Immunol. Lett. 123: 9–13 [DOI] [PubMed] [Google Scholar]
- 46. Khoo JJ, Forster S, Mansell A. 2011. Toll-like receptors as interferon-regulated genes and their role in disease. J. Interferon Cytokine Res. 31: 13–25 [DOI] [PubMed] [Google Scholar]
- 47. Kok KH, Ng MHJ, Ching YP, Jin DY. 2007. Human TRBP and PACT interact with each other and associate with Dicer to facilitate the production of siRNA. J. Biol. Chem. 282: 17649–17657 [DOI] [PubMed] [Google Scholar]
- 48. Kok KH, et al. 2011. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate immune antiviral response. Cell Host Microbe 9: 299–309 [DOI] [PubMed] [Google Scholar]
- 49. Kutish G, Mainprize T, Rock D. 1990. Characterization of the latency-related transcriptionally active region of the bovine herpesvirus 1 genome. J. Virol. 64: 5730–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leaman DW, et al. 2002. Identification of X-linked inhibitor of apoptosis-associated factor-1 as an interferon-stimulated gene that augments TRAIL Apo2L-induced apoptosis. J. Biol. Chem. 277: 28504–28511 [DOI] [PubMed] [Google Scholar]
- 51. Lee HH, Dadgostar H, Cheng Q, Shu J, Cheng G. 1999. NF-kB-mediated up-regulation of Bcl-X and Bfl-1/A1 is required for CD40 survival signaling in B lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 96: 9136–9141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee Y, et al. 2006. The role of PACT in the RNA silencing pathway. EMBO J. 25: 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liang D, et al. 2011. A human herpesvirus miRNA attenuates interferon signaling and contributes to maintenance of viral latency by targeting IKKepsilon. Cell Res. 21: 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lovato L, Inman M, Henderson G, Doster A, Jones C. 2003. Infection of cattle with a bovine herpesvirus 1 (BHV-1) strain that contains a mutation in the latency related gene leads to increased apoptosis in trigeminal ganglia during the transition from acute infection to latency. J. Virol. 77: 4848–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mattson MP, Meffert MK. 2006. Roles for NF-kB in nerve cell survival, plasticity, and disease. Cell Death Differ. 13: 852–860 [DOI] [PubMed] [Google Scholar]
- 56. Meyer F, et al. 2007. A protein encoded by the bovine herpes virus 1 (BHV-1) latency related gene interacts with specific cellular regulatory proteins, including the CCAAT enhancer binding protein alpha (C/EBP-a). J. Virol. 81: 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mikloska Z, Cunningham AL. 2001. Alpha and gamma interferons inhibit herpes simplex virus type 1 infection and spread in epidermal cells after axonal transmission. J. Virol. 75: 11821–11826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Perez S, Lovato L, Zhou J, Doster A, Jones C. 2006. Comparison of inflammatory infiltrates in trigeminal ganglia of cattle infected with wild-type bovine herpesvirus 1 versus a virus strain containing a mutation in the LR (latency-related) gene. J. Neurovirol. 12: 392–397 [DOI] [PubMed] [Google Scholar]
- 59. Perez S, Meyer F, Saira K, Doster A, Jones C. 2008. Premature expression of the latency-related RNA encoded by bovine herpesvirus type 1 correlates with higher levels of beta interferon RNA expression in productively infected cells. J. Gen. Virol. 89: 1338–1345 [DOI] [PubMed] [Google Scholar]
- 60. Perng G-C, et al. 2002. A gene capable of blocking apoptosis can substitute for the herpes simplex virus type 1 latency-associated transcript gene and restore wild-type reactivation levels. J. Virol. 76: 1224–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Perng G-C, et al. 2000. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript (LAT). Science 287: 1500–1503 [DOI] [PubMed] [Google Scholar]
- 62. Pichlmair A, et al. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314: 997–1001 [DOI] [PubMed] [Google Scholar]
- 63. Piva R, Belardo G, Santoro MG. 2000. 6. NF-kappaB: a stress-regulated switch for cell survival. Antioxid. Redox Signal. 8: 478–486 [DOI] [PubMed] [Google Scholar]
- 64. Rock D, Lokensgard J, Lewis T, Kutish G. 1992. Characterization of dexamethasone-induced reactivation of latent bovine herpesvirus 1. J. Virol. 66: 2484–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sainz B, Jr, Halford WP. 2002. Alpha/beta interferon and gamma interferon synergize to inhibit the replication of herpes simplex virus type 1. J. Virol. 76: 11541–11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Salvesen GS, Duckett CS. 2002. IAP proteins: blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 3: 401–410 [DOI] [PubMed] [Google Scholar]
- 67. Samanta M, Iwakiri D, Kanda T, Imaizumi T, Takada K. 2006. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 25: 4207–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sawtell NM, Thompson RL. 1992. Rapid in vivo reactivation of herpes simplex virus in latently infected murine ganglionic neurons after transient hyperthermia. J. Virol. 66: 2150–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schlee M, et al. 2009. Approaching the RNA ligand for RIG-I? Immunol. Rev. 227: 66–74 [DOI] [PubMed] [Google Scholar]
- 70. Schlee M, et al. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schmidt A, et al. 2009. 5′-Triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. U. S. A. 106: 12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shen W, Jones C. 2008. Open reading frame 2, encoded by the latency-related gene of bovine herpesvirus 1, has antiapoptotic activity in transiently transfected neuroblastoma cells. J. Virol. 82: 10940–10945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shen W, et al. 2009. Two small RNAs encoded within the first 1.5 kilobases of the herpes simplex virus type 1 latency-associated transcript (LAT) can inhibit productive infection and cooperate to inhibit apoptosis. J. Virol. 83: 9131–9139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shimeld C, et al. 1995. Immune cell infiltration and persistence in the mouse trigeminal ganglion after infection of the cornea with herpes simplex virus type 1. J. Neuroimmunol. 61: 7–16 [DOI] [PubMed] [Google Scholar]
- 75. Sirén J, Pirhonen J, Julkunen I, Matikainen S. 2005. IFN-alpha regulates TLR-dependent gene expression of IFN-alpha, IFN-beta, IL-28, and IL-29. J. Immunol. 174: 1932–1937 [DOI] [PubMed] [Google Scholar]
- 76. Stern-Ginossar N, et al. 2007. Host immune system gene targeting by a viral miRNA. Science 317: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sumpter R, Jr, et al. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79: 2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Takahasi K, et al. 2008. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell 29: 428–440 [DOI] [PubMed] [Google Scholar]
- 79. Takaoka A, et al. 2003. Integration of interferon-a/b signalling to p53 responses in tumour suppression and antiviral defence. Nature 424: 516–523 [DOI] [PubMed] [Google Scholar]
- 80. Valarcher J-F, et al. 2003. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J. Virol. 77: 8426–8439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. 1996. Suppression of TNF-alpha-induced apoptosis by NF-kB. Science 274: 787–789 [DOI] [PubMed] [Google Scholar]
- 82. Wilson V, Jeffreys AJ, Barrie PA. 1983. A comparison of vertebrate interferon gene families detected by hybridization with human interferon DNA. J. Mol. Biol. 166: 457–475 [DOI] [PubMed] [Google Scholar]
- 83. Winkler MT, Doster A, Sur JH, Jones C. 2002. Analysis of bovine trigeminal ganglia following infection with bovine herpesvirus 1. Vet. Microbiol. 86: 139–155 [DOI] [PubMed] [Google Scholar]
- 84. Winkler MTC, Doster A, Jones C. 1999. Bovine herpesvirus 1 can infect CD4(+) T lymphocytes and induce programmed cell death during acute infection of cattle. J. Virol. 73: 8657–8668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Workman A, Sinani D, Pittayakhajonwut D, Jones C. 2011. A protein (ORF2) encoded by the latency related gene of bovine herpesvirus 1 interacts with Notch1 and Notch3. J. Virol. 85: 2536–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yamamoto Y, Gaynor RB. 2001. Therapeutic potential of inhibition of the NF-kB pathway in the treatment of inflammation and cancer. J. Clin. Invest. 107: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yoneyama M, Fujita T. 2009. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 227: 54–65 [DOI] [PubMed] [Google Scholar]
- 88. Yoneyama M, et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5: 730–737 [DOI] [PubMed] [Google Scholar]
- 89. Zou J, Chang M, Nie P, Secombes CJ. 2009. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol. Biol. 9: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]