Abstract
Dengue viruses (DENV) are characterized by extensive genetic diversity and can be organized in multiple, genetically distinct lineages that arise and die out on a regular basis in regions where dengue is endemic. A fundamental question for understanding DENV evolution is the relative extent to which stochastic processes (genetic drift) and natural selection acting on fitness differences among lineages contribute to lineage diversity and turnover. Here, we used a set of recently collected and archived low-passage DENV-1 isolates from Thailand to examine the role of mosquito vector-virus interactions in DENV evolution. By comparing the ability of 23 viruses isolated on different dates between 1985 and 2009 to be transmitted by a present-day Aedes aegypti population from Thailand, we found that a major clade replacement event in the mid-1990s was associated with virus isolates exhibiting increased titers in the vector's hemocoel, which is predicted to result in a higher probability of transmission. This finding is consistent with the hypothesis that selection for enhanced transmission by mosquitoes is a possible mechanism underlying major DENV clade replacement events. There was significant variation in transmission potential among isolates within each clade, indicating that in addition to vector-driven selection, other evolutionary forces act to maintain viral genetic diversity. We conclude that occasional adaptive processes involving the mosquito vector can drive major DENV lineage replacement events.
INTRODUCTION
Worldwide, dengue viruses (DENV) are the most important mosquito-borne viral pathogens of humans. The four antigenically distinct DENV serotypes (DENV-1 to -4) cause a broad spectrum of clinical manifestations. An estimated 50 million people experience dengue illness each year, approximately 500,000 of which are associated with severe, life-threatening disease (18). In addition, a significant portion of infections can be inapparent and thus go undetected by surveillance programs (15). Despite the large disease burden imposed by dengue on the human population, there is currently no commercially available DENV vaccine or antiviral therapy (46). In regions where dengue is endemic and multiple serotypes cocirculate, DENV epidemiological dynamics are characterized by complex oscillations in incidence and serotype prevalence (6, 32, 43). A variety of ecological (10, 24) and immunological factors (1, 36) are thought to govern these complex spatiotemporal dynamics. There is also compelling evidence for the influence of virological factors in disease incidence and severity (reviewed in reference 37). DENV are single-stranded, positive-sense RNA viruses of the genus Flavivirus (family Flaviviridae) with extensive genetic diversity (21). Each serotype can be divided into large, genetically diverse phylogenetic clusters, which, in turn, consist of multiple, distinct lineages (22). Here, we use the terms clade and lineage interchangeably. In the last 2 decades, in-depth phylogenetic analyses have significantly improved understanding of DENV epidemiological and evolutionary dynamics (reviewed in references 35, 45).
One of the most striking features of DENV evolutionary dynamics is that viral lineages within serotypes arise and die out on a regular basis (22). This lineage turnover is detected in phylogenetic analyses in two different, nonmutually exclusive forms: (i) continuous, ladderlike temporal structure within clades and (ii) more dramatic, major clade replacement events. Within a clade, the majority of sublineages present at a specific time point are not detected at later time points, resulting in a ladderlike tree topology (8, 13, 26). Occasionally, an entire clade that persisted for a number of years at a given location goes extinct as an entirely new clade takes over. Such major clade replacement events have been recurrently documented at regional scales, for example, DENV-3 in Sri Lanka in the late 1980s (30), DENV-4 in Puerto Rico during the 1980s and 1990s (8), DENV-1 in Thailand in the mid-1990s (48), DENV-3 in Thailand in the early 1990s (47), DENV-1 in Myanmar in the late 1990s (31), DENV-2 in Vietnam in the early 2000s (43), and DENV-1 in Cambodia in the early 2000s (14). Major clade replacement events have also been reported at larger scales. For example, DENV-2 lineages from Southeast Asia displaced a native DENV-2 lineage in the Americas during the early 1990s (38). Although successive clades may be temporally nonoverlapping (9), in most cases there is a transition period of cocirculation (14, 30, 31, 43, 47, 48).
Understanding the causes of DENV lineage birth and death has important implications for dengue epidemiology and control, because it is often associated with changes in disease incidence and severity (16, 17, 30, 31, 38, 40). Elucidating the mechanisms underlying lineage turnover helps to retrospectively understand spatiotemporal patterns of dengue incidence and to make predictions about the risk of future epidemics (43). It also has implications for vaccine design because DENV lineages may differ in their antigenic properties (39, 44).
Despite their epidemiological significance, the evolutionary forces underlying major clade replacement events are especially unclear. In particular, the extent to which clade replacement events result from the random sampling of viral variants (genetic drift) due to the stochastic nature of DENV transmission and/or from differences in fitness among variants (i.e., adaptive evolution) is an unresolved question. Analyses of DENV gene sequence data have shown that clade replacement events are sometimes associated with signals of positive natural selection, such as an increase in the rate of nonsynonymous substitutions (7), but not always (26, 47). Some phylogenetic studies have concluded that major clade replacements were primarily due to stochastic events (31, 47), whereas others found potentially causal differences in viral fitness between lineages, such as a higher viremia level in the human host (43) or enhanced infectivity to mosquito vectors (3, 5, 19).
Here, we addressed the question whether DENV evolution could be driven by adaptation to mosquito vectors using a set of 23 low-passage DENV-1 isolates from Thailand collected during the period of 1985 to 2009. The 23 isolates were recovered from human serum as part of the routine dengue diagnostic and surveillance services performed at the Armed Forces Research Institute of Medical Sciences (AFRIMS) in Bangkok. We compared the ability of the 23 isolates to be transmitted by Aedes aegypti, the principal DENV vector species, in experimental in vivo vector competence assays. We hypothesized that if viruses are positively selected for enhanced mosquito transmission, a relative increase in transmission potential should be observed for viruses collected over advancing years. All experiments were carried out from 2009 to 2010 using the first laboratory-reared generation of mosquitoes derived from a wild A. aegypti population collected in Kamphaeng Phet, Thailand. Although we cannot be sure that these mosquitoes exactly reproduce the vector-virus interactions that occurred at different dates in the past, they are the most relevant vector population of reference available to test our hypothesis. We estimated virus transmissibility with two distinct, complementary vector competence indices: the proportion of A. aegypti females that developed a DENV infection that disseminated from their midgut into their hemocoel, a requirement for virus transmission by a mosquito, and the infectious titer of disseminated virus (27).
MATERIALS AND METHODS
Mosquitoes.
Wild A. aegypti immatures (larvae and pupae) were collected from a variety of artificial containers in several households in the Nhong Pling, Kon Tee, Nakorn Choom, Na Bo Kham, and Thep Na Korn subdistricts, Muang district, Kamphaeng Phet Province, Thailand, during July 2009 (experiment 1) and March 2010 (experiment 2). In a preliminary experiment, immatures were collected from containers in villages of the Ladkrabang district, Bangkok, and the Muang district, Kamphaeng Phet Province, in January 2009. F0 adults were allowed to emerge in the laboratory, mate randomly, and feed on defibrinated sheep blood (National Laboratory Animal Center, Mahidol University, Bangkok, Thailand) through a membrane feeding system. F1 eggs, which were collected and stored on dry pieces of paper towel and maintained under high humidity, were hatched synchronously by placing them under low pressure for 30 min. Larvae were reared in 24- by 34- by 9-cm plastic trays filled with 2.0 liters of dechlorinated tap water at a density of approximately 200 first instars per tray and fed a standard diet of approximately 1.0 g of fish food pellets (C.P. Hi Pro; Perfect Companion Group Co. Ltd., Bangkok, Thailand) per tray. After emergence, F1 adults were housed in plastic 30- by 30- by 30-cm cages (Megaview Science Education Service Co. Ltd., Taichung, Taiwan) with permanent access to 10% sucrose. They were maintained under standard insectary conditions at 28 ± 1°C and 80% humidity and with a 12:12 h light-dark cycle.
Virus isolates.
Viruses were originally isolated and archived as seed stocks from serum samples collected during routine surveillance for diagnostic public health testing at AFRIMS from clinically ill dengue patients attending Kamphaeng Phet Provincial Hospital in Kamphaeng Phet (experiment 1) and Queen Sirikit National Institute of Child Health in Bangkok (experiment 2). Virus isolation and identification was performed as previously described (25). Each isolate underwent 3 to 4 passages in cell culture (Table 1), which, according to standard procedures at the AFRIMS laboratory, is the minimum required to obtain a viral titer sufficiently high to infect mosquitoes orally using an artificial blood meal.
Table 1.
DENV-1 isolates used in experimental mosquito infectionsa
| Expt | Isolation yr | Isolate ID | Source | Passage history | Clade |
|---|---|---|---|---|---|
| 1 | 1986 | KD86-035 | KPPH | TS-1, C6/36-3 | Early |
| 1990 | KD90-157 | KPPH | TS-1, C6/36-2 | Early | |
| 1992 | KD92-080 | KPPH | TS-1, C6/36-2 | Early | |
| 1995 | 30399/95 | KPPH | C6/36-4 | New | |
| 1997 | 30231/97 | KPPH | C6/36-4 | New | |
| 2000 | 30529/00 | KPPH | C6/36-4 | New | |
| 2003 | 30247/03 | KPPH | C6/36-4 | New | |
| 2005 | 30230/05 | KPPH | C6/36-4 | New | |
| 2006 | 30118/06 | KPPH | C6/36-4 | New | |
| 2009 | 30015/09 | KPPH | C6/36-4 | New | |
| 2009 | 30025/09 | KPPH | C6/36-4 | New | |
| 2 | 1985 | D85-372 | QSNICH | TS-1, C6/36-3 | Early |
| 1986 | D86-412 | QSNICH | TS-1, C6/36-3 | Early | |
| 1987 | D87-116 | QSNICH | TS-1, C6/36-3 | Early | |
| 1990 | D90-1197 | QSNICH | TS-1, C6/36-3 | Early | |
| 1992 | 03881/92 | QSNICH | TS-1, C6/36-3 | New | |
| 1995 | 00407/95 | QSNICH | TS-1, C6/36-3 | Early | |
| 1997 | 00616/97 | QSNICH | TS-1, C6/36-3 | New | |
| 2000 | 04805/00 | QSNICH | TS-1, C6/36-3 | New | |
| 2003 | 01417/03 | QSNICH | TS-1, C6/36-3 | New | |
| 2005 | 00442/05 | QSNICH | TS-1, C6/36-3 | New | |
| 2007 | 02128/07 | QSNICH | C6/36-4 | New | |
| 2009 | 00132/09 | QSNICH | C6/36-4 | New |
For each isolate, the year of virus isolation, location of origin, number of passages in cell culture, and phylogenetic group (clade) are indicated. KPPH, Kamphaeng Phet Provincial Hospital; QSNICH, Queen Sirikit National Institute for Child Health (Bangkok); TS, Toxorhynchites splendens mosquitoes; C6/36, Aedes albopictus cells.
Oral challenge.
Two sets of 2-day-old confluent cultures of Aedes albopictus cells (C6/36, ATCC CRL-1660) in 25-cm2 flasks (approximately 107 cells/flask) were inoculated with 1.0 ml of stock virus per flask and incubated at 35°C under 5% CO2. A similar titer can be reached by growing virus in C6/36 cells at 35°C for 4 to 6 days instead of the usual 7 to 10 days at 28°C. In experiment 1, supernatant was harvested at 5 and 6 days postinoculation to prepare the infectious blood meal of experimental blocks 1 and 2, respectively. In experiment 2, supernatant was harvested at 4 days postinoculation to prepare the infectious blood meal of both experimental blocks. The protocol difference between experiments 1 and 2 was due to logistical constraints. The artificial blood meal consisted of a 1:1 mix of defibrinated sheep blood (National Laboratory Animal Center, Mahidol University, Bangkok, Thailand) and virus suspension. Three- to 7-day-old A. aegypti F1 females deprived of sucrose and water for 24 h were offered an infectious blood meal for 30 min through pieces of desalted porcine intestine stretched over water-jacketed glass feeders maintained at 37°C. Samples of the blood meal were saved for subsequent titration by plaque assay. After blood feeding, mosquitoes were briefly sedated with CO2 from dry ice, and fully engorged females were transferred to clean paper cups. Unfed or partially fed females were discarded. Engorged females were maintained under standard insectary conditions and provided cotton soaked with 10% sucrose ad libitum.
Vector competence.
The ability of DENV isolates to be transmitted by A. aegypti was assessed at 7 and 14 days after blood meal (pbm) with two vector competence phenotypes: (i) the proportion of mosquitoes that developed a disseminated infection and (ii) the infectious titer of disseminated virus. These two complementary indices of vector competence represent two successive, nonoverlapping aspects of the infection process in mosquitoes that lead to their ability to transmit virus, namely, that virus disseminates beyond the midgut and the magnitude of infectious titer in the hemocoel. As such, they were analyzed separately. The two periods of extrinsic incubation (7 and 14 days pbm) were chosen to represent early and late phases of dissemination kinetics (28). The relationship between the infectious titer of disseminated virus and the potential for transmission in vitro was established in a preliminary experiment (see below). Upon harvest, the head of each female was cut off and placed individually in 1.0 ml of mosquito diluent (MD), consisting of RPMI 1640 medium with 10% heat-inactivated fetal bovine serum (FBS) with 100 units/ml penicillin and 100 μg/ml streptomycin. Bodies were kept separately in 1.0 ml of MD. Samples were stored at −70°C before processing. Body and head samples were quickly thawed in a water bath at 35 ± 2°C and homogenized in a mixer mill (Qiagen) at 24 cycles/s for 2 min. Infected bodies were screened by serotype-specific reverse transcription (RT)-PCR (experiment 1) or plaque assay (experiment 2). In experiment 1, total RNA was extracted from 140 μl of body homogenates using the QIAamp viral RNA minikit (Qiagen) according to the manufacturer's instructions. RT-PCR was performed with 5 μl of extracted RNA following a standard protocol (29) with the following modifications: (i) 1× PCR buffer II supplied with AmpliTaq DNA polymerase (Applied Biosystems) was used instead of the original buffer (50 mM KCl, 10 mM Tris [pH 8.5], and 0.01 mM gelatin) in both the first-round RT-PCR and the second-round PCR (nested PCR); (ii) the first-round RT-PCR mixture contained Avian myeloblastosis virus reverse transcriptase (Promega) instead of rav-2 recombinant reverse transcriptase; (iii) the 1:50 dilution of the first-round RT-PCR product was used as the template in the nested PCR; (iv) the nested PCR mixture contained 12.5 pmol of each primer instead of 50 pmol; and (v) the number of the nested PCR cycles was increased from 20 cycles to 25 cycles (29). Plaque assay was performed in rhesus monkey kidney cells (LLC-MK2, ATCC CCL-7) as described previously (42). Briefly, the homogenized samples were passed individually through a 0.22-μm syringe filter unit and 1:2, 1:10, and 1:100 dilutions were prepared in MD. The samples were placed in an ice bath, and 100 μl/well was inoculated into a monolayer of LLC-MK2 cells in 24-well plates. The virus was adsorbed for 1 h at room temperature (20 to 28°C) on a rocker platform. The inoculum was removed and 0.5 ml/well of a first overlay of medium was added. The cells were incubated for 5 days at 35 ± 1°C in a 5 ± 0.5% CO2 incubator. The cells were stained with a second overlay of medium containing 4% neutral red (Sigma). Plaques were counted and PFU/ml were calculated. Head samples of infected bodies were titrated by plaque assay in LLC-MK2 cells. Mosquitoes whose bodies were negative by RT-PCR (experiment 1) or plaque assay (experiment 2) were considered uninfected, and their heads were not processed further.
In vitro transmission.
A preliminary experiment was carried out to define the relationship between the titer of disseminated virus and the potential for in vitro virus transmission. This experiment involved two F1 populations of A. aegypti sampled during January and February 2009 in Bangkok and Kamphaeng Phet, respectively. Both populations were experimentally exposed to two 2009 DENV-1 isolates from Bangkok (ID numbers 00076/09 and 00088/09) and two 2009 DENV-1 isolates from Kamphaeng Phet (ID numbers 30015/09 and 30025/09). The relationship between the titer of disseminated virus and in vitro transmission success was evaluated both at 8 and 14 days pbm. Mosquitoes were anesthetized with triethylamine (Sigma-Aldrich), and their legs were removed individually and placed into 1.0 ml of MD. Saliva samples were collected using a forced salivation technique (2). Briefly, mosquito mouthparts were inserted for 15 min into a glass microcapillary tube filled with approximately 10 μl of FBS. After 15 min, the contents of the microcapillary tube were placed into 0.3 ml of MD and snap-frozen in dry ice. The remainders of the mosquito bodies were placed individually into 1.0 ml of MD, and all samples were stored at −70°C before processing. Body and leg samples were quickly thawed in a water bath at 35 ± 2°C and homogenized in a mixer mill. Infected bodies were screened by serotype-specific RT-PCR as described above. The titer of disseminated virus in the legs of mosquitoes whose bodies were positive by RT-PCR was determined by plaque assay in LLC-MK2 cells as described above. Mosquitoes whose bodies were negative by RT-PCR were considered uninfected, and their legs were not processed further. The presence of virus in the saliva samples from mosquitoes with a disseminated infection was tested by plaque assay in LLC-MK2 cells.
Sequencing and phylogenetic analysis.
Viral RNA extracted by the QIAamp viral RNA minikit was converted to cDNA using the Transcriptor high-fidelity kit (Roche) and random hexamer oligonucleotides according to the manufacturer's instructions. Sequencing of E genes and complete genomes was performed following previously described methods (48). The DNA fragments of the E gene and the overlapping DNA fragments covering the entire DENV-1 genome were amplified using AmpliTaq DNA polymerase (Applied Biosystems) and purified using the QIAquick PCR purification kit and QIAquick gel extraction kit (Qiagen). Purified DNA fragments were used as templates in cycle sequencing reactions using the DYEnamic ET dye terminator sequencing kit (GE Healthcare) according to the manufacturer's instructions. The sequencing primers were described previously (48). The sequencing products were purified by standard ethanol precipitation before sequencing in a MegaBACE 500 automated DNA sequencer (GE Healthcare). The overlapping sequences obtained from forward and reverse primers were combined for analysis and edited by using the Sequencher software (Gene Code Corporation). Phylogenetic trees were constructed with PAUP* version 4.0 (41) using a maximum likelihood (ML) method. ML trees included the new sequences generated in this study and 17 “background” DENV-1 sequences from GenBank. The best model of nucleotide substitution was chosen with jModelTest (33) based on the lowest Akaike information criterion (AIC) value (34). The TIM2+Γ and GTR+Γ models of nucleotide substitution were applied to construct the trees for the E gene and complete genome sequences, respectively. Reliability of particular phylogenetic groupings was calculated using bootstrap resampling analysis with 1,000 replicate neighbor-joining (NJ) trees under the ML substitution models.
Data analysis.
All statistical analyses of vector competence data included the confounding effects of the experiment and the experimental block. The study was run in two separate experiments that involved two different sets of DENV isolates and used a population of A. aegypti that was sampled at two different times. In both experiments, the same batch of mosquitoes was exposed twice to the same set of DENV isolates on two successive days (i.e., two experimental blocks). In the two blocks, viruses came from the same passage in cell culture but were harvested separately (see above). The effect of the block was thus nested within each experiment and accounted for the difference in titer between the two harvests, the effect of the 1-day difference in mosquito age, and the effect of any uncontrolled differences between the 2 days. The isolate effect was nested within each experiment and clade, because each isolate belonged to only one phylogenetic clade and each experiment involved a different set of isolates. Blood meal titers were log transformed (to ensure normal distribution of the residuals) and compared using a multifactorial analysis of variance (MANOVA) accounting for the effects of clade, experiment, block, and their interactions. The proportion of mosquitoes with a disseminated infection was analyzed with a nominal logistic regression that included the effects of blood meal titer, experiment, experimental block, clade, isolate, and their interactions. The titer of disseminated virus was log transformed (to ensure normal distribution of the residuals) and analyzed with an MANOVA that included the effects of blood meal titer, experiment, experimental block, clade, isolate and their interactions. Differences were considered statistically significant at P values of <0.05. Analyses were performed with the software JMP version 5.1.2.
Nucleotide sequence accession numbers.
All sequences have been submitted to GenBank and assigned accession numbers JN638322 to JN638344.
RESULTS
Estimation of transmission potential.
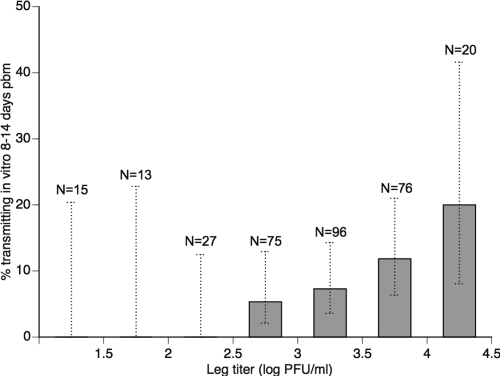
Transmission potential was defined as the presence of infectious virus in the mosquito's saliva. In a preliminary experiment, a total of 322 individual A. aegypti females that developed a disseminated infection following oral challenge were tested for virus titer in their legs and virus presence in their saliva, collected in vitro. The data included two F1 A. aegypti populations (sampled in Bangkok and Kamphaeng Phet, respectively), four DENV-1 isolates (two from Bangkok and two from Kamphaeng Phet), and two extrinsic incubation periods (8 and 14 days pbm). Leg titers ranged from 1 × 101 to 2.3 × 104 PFU/ml, and infectious virus was detected in 7.5% of saliva samples. The probability of detecting infectious virus in saliva was positively correlated with the infectious titer of disseminated virus (Fig. 1). Logistic regression showed that the leg titer was a highly significant predictor of virus detection in saliva (df = 1; likelihood ratio [L-R] χ2 = 15.48; P < 0.0001). In particular, no virus was detected in any of the saliva samples from mosquitoes with a leg titer <1 × 103 PFU/ml. For the rest of the study, the infectious titer of disseminated virus was used as an estimate of transmission potential. Because the number of mosquito legs may vary among individuals (they can lose them over time), the head titer was used instead of the leg titer. Previous data showed that the leg titer and the head titer are strongly correlated in individual A. aegypti (27).
Fig 1.
Mosquito transmission potential strongly correlates with infectious titer of disseminated DENV-1. The percentage of A. aegypti females tested positive for infectious virus in saliva samples collected in vitro is shown as a function of infectious virus titers in their legs at either 8 or 14 days pbm. Bars represent individuals with a leg titer greater than the lower value of the corresponding interval on the x axis. Data shown is pooled from vector competence assays involving two mosquito populations exposed to four different DENV-1 isolates (see Materials and Methods for details). Dotted, vertical lines indicate the 95% confidence intervals of percentages. Sample sizes are indicated above the bars.
Phylogenetic relationships between DENV isolates.
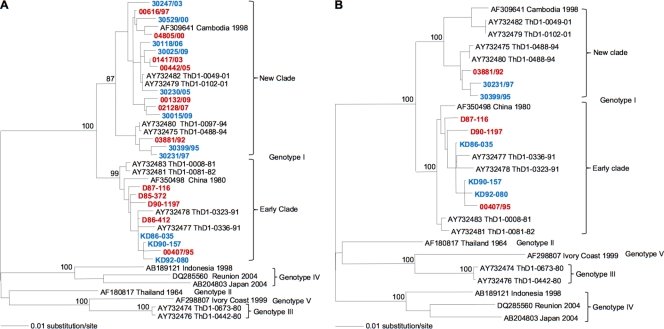
All but two of the 23 DENV-1 isolates were passaged four times in cell culture (Table 1) prior to their use in experimental mosquito infections; two were passaged three times. The study was run in two separate experiments involving two different sets of virus isolates (11 in the first experiment and 12 in the second) and A. aegypti mosquitoes sampled at two different times. Phylogenetic analysis based on the complete DENV E gene sequence indicated that the virus isolates could be divided into two major phylogenetic clades within the genotype I of DENV-1 (Fig. 2A). The first clade included 8 isolates sampled between 1985 and 1995; the second clade included 15 isolates sampled between 1992 and 2009. Here, the first and second clades are termed “early clade” and “new clade,” respectively, in reference to the chronological order of their detection. Complete genome sequences obtained for a selection of 9 isolates sampled between 1986 and 1997 (6 from the early clade and 3 from the new clade) confirmed the phylogenetic relationships inferred using the E gene sequence (Fig. 2B).
Fig 2.
Phylogenetic relationships among DENV-1 isolates. Maximum likelihood trees based on E gene (A) or complete genome (B) sequences. Sequences from GenBank are in black font, and sequences generated in this study are in color. Blue and red fonts indicate isolates used in experiment 1 and 2, respectively. All horizontal branch lengths are drawn to scale, with bootstrap support values shown next to relevant nodes.
Variation in transmission potential among DENV isolates.
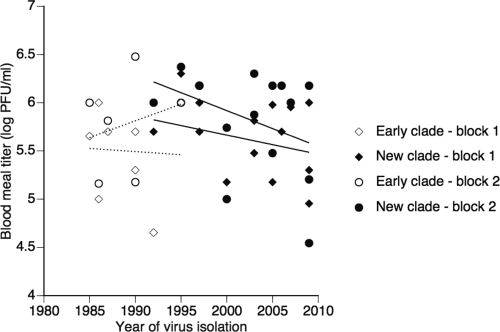
Vector competence assays included a total of 738 individual A. aegypti females. Transmission potential was characterized based on 7 to 44 (mean 32) individuals per isolate. Infectious titer of artificial blood meals used in vector competence assays varied among isolates (Fig. 3), ranging from 3.5 × 104 PFU/ml to 3.0 × 106 PFU/ml. Blood meal titers were significantly higher in the second experiment (sum of squares [SS] = 3.251; df = 1; F = 19.63; P < 0.0001), but they were not significantly different between blocks within experiments (SS = 0.574; df = 2; F = 1.734; P = 0.191). The higher blood meal titers in experiment 2 may have resulted from the different harvest date postinoculation in cell culture (day 4) compared to that of experiment 1 (day 5 to 6). Importantly, blood meal titers did not differ between clades (SS = 0.439; df = 1; F = 2.649; P = 0.112) (Fig. 2). In the early clade, blood meal titers ranged from 4.5 × 104 PFU/ml to 3.0 × 106 PFU/ml (mean, 7.0 × 105 PFU/ml). In the new clade, blood meal titers ranged from 3.5 × 104 PFU/ml and 2.4 × 106 PFU/ml (mean, 8.2 × 105 PFU/ml).
Fig 3.
Variation of artificial DENV-1 blood meal titers in vector competence assays. Infectious titers of the artificial blood meals measured by plaque assay are shown for each isolate as a function of their date of isolation and phylogenetic group. Each isolate was tested twice in two experimental blocks (for details, see Materials and Methods). Lines represents linear y = f(x) regressions to give a graphical indication of temporal trends. Lower lines correspond to experimental block 1; upper lines correspond to experimental block 2.
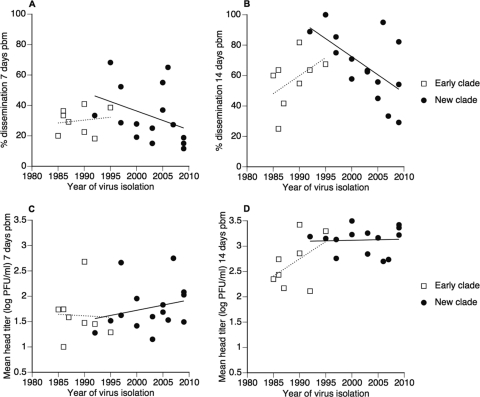
Overall, about two-thirds of mosquitoes were infected (65.6% at 7 days pbm and 69.6% at 14 days pbm). Nearly all infected females developed disseminated infections within the first 2 weeks pbm (52.9% at 7 days pbm and 94.6% at 14 days pbm). The percentage of females with a disseminated infection (including uninfected individuals) varied significantly among isolates at both 7 and 14 days pbm (Table 2). It ranged from 0 to 81% at 7 days pbm (mean, 32%) and from 20 to 100% (mean, 64%) at 14 days pbm. The percentage of females with a disseminated infection was not significantly influenced by the blood meal titer, but it was significantly higher in the first than in the second experiment at 14 days pbm (Table 2). The higher level of dissemination in the second experiment may have been due to either the different geographical origin of viruses (Bangkok versus Kamphaeng Phet) or any uncontrolled, environmental difference between the two experiments. There was no significant difference between the early and new clade in the mean percentage of females with a disseminated infection at either 7 or 14 days pbm (Table 2). Dissemination tended to increase as a function of the year of virus isolation in the early clade, whereas it tended to decrease as a function of the year of virus isolation in the new clade, both at 7 days pbm (Fig. 4A) and 14 days pbm (Fig. 4B). These trends may have been a consequence of variation in blood meal titers, which showed similar patterns (Fig. 3).
Table 2.
Logistic regression analysis of dissemination prevalence 7 and 14 days pbm among mosquitoes experimentally exposed to different DENV-1 isolatesa
| Source | df | Dissemination 7 days pbm |
Dissemination 14 days pbm |
||
|---|---|---|---|---|---|
| L-R χ2 | P value | L-R χ2 | P value | ||
| Blood meal titer | 1 | 1.451 | 0.228 | 0.749 | 0.387 |
| Expt | 1 | 1.058 | 0.304 | 11.45 | 0.001** |
| Clade | 1 | 0.877 | 0.349 | 3.619 | 0.057 |
| Expt*clade | 1 | 0.443 | 0.506 | 0.461 | 0.497 |
| Block (within expt) | 2 | 1.604 | 0.448 | 11.34 | 0.003** |
| Block*clade (within expt) | 2 | 0.080 | 0.961 | 4.236 | 0.120 |
| Isolate (within expt, clade) | 19 | 34.82 | 0.015* | 46.21 | 0.001** |
L-R, likelihood ratio; *, P < 0.05; **, P < 0.01.
Fig 4.
Variation in vector competence indices among DENV-1 isolates. In the upper panels, the percentage of females with a disseminated infection is indicated at 7 days pbm (A) and 14 days pbm (B) as a function of the year of virus isolation and phylogenetic group (clade). In the lower panels, mean infectious titer in heads of females with a disseminated infection is indicated for each isolate at 7 days pbm (C) and 14 days pbm (D) as a function of virus isolation year and phylogenetic group. Each isolate is represented by the mean of two experimental blocks (for details, see Materials and Methods). Lines represents linear y = f(x) regressions to give a graphical indication of temporal trends.
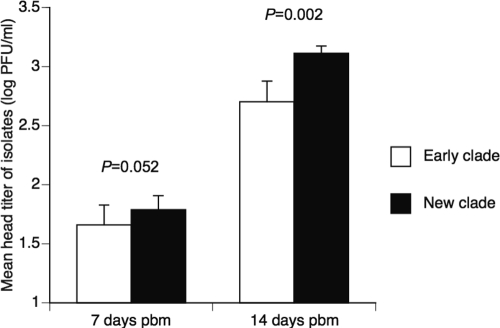
Among females with a disseminated infection, head titers ranged from 1.0 × 101 to 2.3 × 104 PFU/ml at 7 days pbm (n = 128; mean, 5.4 × 102 PFU/ml; median, 3.0 × 101 PFU/ml) and from 1.0 × 101 to 2.9 × 104 PFU/ml at 14 days pbm (n = 243; mean, 3.4 × 103 PFU/ml; median, 1.5 × 103 PFU/ml). The titer of disseminated virus was not significantly influenced by the blood meal titer; it differed significantly among isolates at 14 days pbm (Table 3). The mean head titer of the new clade was significantly higher than that of the early clade at 14 days pbm (Table 3; Fig. 5). At 7 days pbm, the new clade had a higher mean head titer than the early clade, but the difference was marginally insignificant (Table 3; Fig. 5). The statistical power at 7 days pbm, however, was reduced compared to that at 14 days pbm, because the number of mosquitoes with a disseminated infection was half as large. At 7 days pbm, the head titer tended to increase slightly as a function of the year of virus isolation in the new clade, whereas it did not vary temporally in the early clade (Fig. 4C). At 14 days pbm, the head titer tended to increase as a function of the year of virus isolation in the early clade, whereas it did not vary temporally in the new clade (Fig. 4D).
Table 3.
Analysis of variance of head titers at 7 and 14 days pbm among mosquitoes experimentally exposed to different DENV-1 isolatesa
| Source | Head titer 7 days pbm |
Head titer 14 days pbm |
||||||
|---|---|---|---|---|---|---|---|---|
| df | SS | F ratio | P value | df | SS | F ratio | P value | |
| Blood meal titer | 1 | 2.005 | 4.086 | 0.046* | 1 | 0.027 | 0.048 | 0.828 |
| Expt | 1 | 2.651 | 5.403 | 0.022* | 1 | 0.353 | 0.633 | 0.427 |
| Clade | 1 | 1.898 | 3.869 | 0.052 | 1 | 5.494 | 9.838 | 0.002** |
| Expt*clade | 1 | 0.235 | 0.479 | 0.491 | 1 | 0.007 | 0.013 | 0.908 |
| Block (within expt) | 2 | 0.787 | 0.802 | 0.451 | 2 | 0.483 | 0.433 | 0.649 |
| Block*clade (within expt) | 2 | 3.215 | 3.276 | 0.042* | 2 | 3.153 | 2.823 | 0.062 |
| Isolate (within expt, clade) | 19 | 15.60 | 1.673 | 0.054 | 19 | 26.76 | 2.522 | 0.001** |
| Error | 100 | 49.06 | 215 | 120.1 | ||||
Titers were log transformed to satisfy statistical assumptions. The analysis is performed on individual females with a disseminated infection (i.e., excluding those that did not develop a disseminated infection). SS, sum of squares; *, P < 0.05; **, P < 0.01.
Fig 5.
DENV-1 isolates in the new clade disseminate at higher titers in orally challenged A. aegypti mosquitoes. Bars show the mean head titer of isolates belonging to the early and new clades at 7 and 14 days pbm. Error bars indicate the standard errors of the means. Statistical significance of the difference between clades is given by P values derived from a complete multifactorial analysis of variance accounting for the effects of infectious dose (blood meal titer), experiment, block, clade, and isolates; the analysis includes only mosquitoes with a disseminated infection.
DISCUSSION
By comparing the ability of 23 DENV-1 isolates from Thailand spanning a 24-year period to infect and be transmitted by A. aegypti, we found that a major clade replacement event in the mid-1990s was associated with a higher transmission potential of the isolates belonging to the new clade. Higher transmissibility was due mainly to a higher infectious titer of virus in the vector's hemocoel, which is predicted to result in a higher probability of transmission. This finding supports the hypothesis that major clade replacement events can be driven by natural selection and emphasizes the potentially important role of vector-virus interactions in DENV evolution.
The major clade replacement event revealed by the phylogenetic analysis of the 23 isolates of our study was described previously (48). In their study, Zhang et al. (48) observed that the DENV-1 clade replacement was associated with a decline in DENV-1 prevalence and a concomitant rise of DENV-4 in Thailand. They speculated that clade replacement events might result from differential susceptibility to cross-reactive immune responses in the human host. Here, we provide a nonmutually exclusive, alternative explanation. Our data suggest that this clade replacement event may have been driven, at least in part, by adaptation to mosquito vectors. Even though it occurred in a context of declining DENV-1 incidence, the new clade may have outcompeted the early clade through enhanced mosquito transmission. Isolates in the new clade had a higher dissemination titer, which is expected to result in higher viral fitness, because the probability of transmission by a mosquito is positively correlated with the quantity of virus that circulates in the insect's hemocoel.
Our estimate of DENV transmission potential relied on an experiment demonstrating a positive correlation between the titer of disseminated virus and the probability of virus detection in the mosquito's saliva (Fig. 1). Virus presence in the saliva was determined using an established in vitro method (2) that likely underestimates transmission probability, because it may fail to detect small but transmissible amounts of virus. Indeed, infectious virus was detected only in the saliva of 7.5% of females with a disseminated infection. Our analysis showed, however, that the probability to detect infectious virus in saliva samples was positively correlated with the infectious titer of disseminated virus, across two different A. aegypti populations and four different DENV-1 isolates. In particular, there was a lower threshold titer below which none of the mosquitoes with a disseminated infection had a positive saliva sample. Although the actual threshold may be lower than we estimate due to the relatively low sensitivity of the method, our data established that the titer of disseminated virus is a strong predictor of transmission probability in this system.
It is worth noting that we did not consider other entomological parameters underlying DENV transmission, such as vector survival or biting rate, which could potentially vary among viruses. Future studies will determine whether variation of these traits contribute to fitness differences among viruses. Another limitation of the study was that adaptation to cell culture could have occurred during the 3 to 4 passages of virus amplification prior to their use in experimental infections of mosquitoes. The selection regime, however, was the same for all isolates; thus, comparisons among vector competence phenotypes remained meaningful. The mechanisms underlying the phenotypic differences we observed between the two clades remain unknown. Authors of a recent study suggested that variation in vector competence among DENV-2 lineages does not depend on mosquito midgut binding affinity but rather on replication ability (12).
Investigators who preceded us and studied DENV-2 and DENV-3 indicated that vector-driven selection could play an important role in DENV evolution. By comparing the ability of three DENV-3 isolates of a native lineage and three DENV-3 isolates of an invasive lineage to be transmitted by A. aegypti, Hanley et al. (19) showed that a clade replacement event in Sri Lanka during the 1980s was associated with enhanced mosquito transmission potential for the invasive lineage. Enhanced transmissibility was associated with a higher body titer and an increased proportion of A. aegypti females with a disseminated infection (19). At a larger geographic scale, Rico-Hesse and others suggested in a series of studies that displacement of a native DENV-2 lineage in the Americas by Southeast Asian DENV-2 lineages during the early 1990s was driven in part by adaptation to mosquitoes (3–5, 11). In both the DENV-3 and DENV-2 cases, invasive lineages were associated with more severe disease in humans, which highlights the critical need to more fully explore the connection between DENV evolutionary biology and dengue epidemiology.
Our study helps to understand the respective contributions of stochastic and adaptive processes in DENV evolution. Frequent introductions of new lineages (gene flow) are known to maintain substantial DENV genetic diversity even at a very local scale (23). In contrast, elimination of deleterious mutations (purifying selection) strongly limits in situ evolution of lineages (23). Purifying selection is likely the dominant evolutionary force in DENV evolution (20), and nucleotide fixation events are due primarily to genetic drift (13, 20, 26). Although stochastic processes are undoubtedly important in DENV evolution, the present study indicates that more dramatic major clade replacement events may have an adaptive basis (43). The fact that extinction of the early DENV-1 clade was concomitant with the rise of the new clade, with an overlapping period in the early 1990s (48), was suggestive of a fitness difference between the two clades. The relatively modest average fitness difference between the two clades (a 0.5-log difference in head titer) may explain why they cocirculated for at least 4 years before the early clade went extinct. It is worth noting that we also observed significant variation in vector competence indices within each clade (Fig. 4), indicating that other evolutionary forces than vector-driven selection promote DENV genetic diversity. In particular, the temporal pattern within each clade did not provide a clear indication that adaptation to mosquitoes occurs continuously through time (Fig. 4). We speculate that the temporal trends observed within clades resulted from uncontrolled variation in experimental blood meal titers (Fig. 3), but this merits further investigation.
Although other selective forces may be important drivers of DENV evolution, especially those occurring in the human host that we did not examine, we provide evidence for the possible role of adaptation to mosquito vectors in a major DENV-1 clade replacement event in Thailand in the early 1990s. Our results reinforce the idea that vector-virus interactions can play a critical role in DENV epidemiological and evolutionary dynamics and suggest that the role of vector-mediated selection be investigated in other instances of clade replacement.
ACKNOWLEDGMENTS
We acknowledge the invaluable contributions of the clinical, laboratory, and entomological personnel of AFRIMS and the Kamphaeng Phet AFRIMS Virology Research Unit (KAVRU).
This study was primarily supported by grant R01 GM-083224 from the National Institutes of Health. L.L. was supported by grant ANR-09-RPDOC-007-01 from the French Agence Nationale de la Recherche. T.W.S. received support from the Research and Policy for Infectious Disease Dynamics Program of the Science and Technology Directorate, Department of Homeland Security, and Fogarty International Center, National Institutes of Health.
We are grateful to Robert Gibbons, Timothy Endy, and Brian Evans for their continuous support. We thank Eddie Holmes, Robert Gibbons, and four anonymous reviewers for helpful comments on a previous version of the manuscript.
The opinions or assertions contained herein are the private views of the authors and are not to be construed as reflecting the official views of the United States Army, the Royal Thai Army, or the United States Department of Defense.
L.L. conceived and coordinated the study, designed the research, analyzed the data, and wrote the manuscript. T.F. and A.P. performed the field collection of mosquitoes and vector competence assays. B.T. supervised the isolation, amplification, and titration of viruses. C.K. supervised the molecular detection and sequencing of viruses and performed the phylogenetic analyses. J.H.R. and A.P. coordinated the field collection of mosquitoes and vector competence assays. R.G.J. coordinated the collection of virus isolates, virological assays, and phylogenetic analyses. T.W.S. conceived and coordinated the study and helped to design the research and write the manuscript. All authors read and approved the final manuscript.
Footnotes
Published ahead of print 30 November 2011
REFERENCES
- 1. Adams B, et al. 2006. Cross-protective immunity can account for the alternating epidemic pattern of dengue virus serotypes circulating in Bangkok. Proc. Natl. Acad. Sci. U. S. A. 103: 14234–14239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aitken THG. 1977. An in vitro feeding technique for artificially demonstrating virus transmission by mosquitoes. Mosq. News. 37: 130–133 [Google Scholar]
- 3. Anderson JR, Rico-Hesse R. 2006. Aedes aegypti vectorial capacity is determined by the infecting genotype of dengue virus. Am. J. Trop. Med. Hyg. 75: 886–892 [PMC free article] [PubMed] [Google Scholar]
- 4. Armstrong PM, Rico-Hesse R. 2001. Differential susceptibility of Aedes aegypti to infection by the American and Southeast Asian genotypes of dengue type 2 virus. Vector Borne Zoonotic Dis. 1: 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Armstrong PM, Rico-Hesse R. 2003. Efficiency of dengue serotype 2 virus strains to infect and disseminate in Aedes aegypti. Am. J. Trop. Med. Hyg. 68: 539–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bennett SN, et al. 2010. Epidemic dynamics revealed in dengue evolution. Mol. Biol. Evol. 27: 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bennett SN, et al. 2006. Molecular evolution of dengue 2 virus in Puerto Rico: positive selection in the viral envelope accompanies clade reintroduction. J. Gen. Virol. 87: 885–893 [DOI] [PubMed] [Google Scholar]
- 8. Bennett SN, et al. 2003. Selection-driven evolution of emergent dengue virus. Mol. Biol. Evol. 20: 1650–1658 [DOI] [PubMed] [Google Scholar]
- 9. Carrillo-Valenzo E, et al. 2010. Evolution of dengue virus in Mexico is characterized by frequent lineage replacement. Arch. Virol. 155: 1401–1412 [DOI] [PubMed] [Google Scholar]
- 10. Cazelles B, Chavez M, McMichael AJ, Hales S. 2005. Nonstationary influence of El Nino on the synchronous dengue epidemics in Thailand. PLoS Med. 2: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cologna R, Armstrong PM, Rico-Hesse R. 2005. Selection for virulent dengue viruses occurs in humans and mosquitoes. J. Virol. 79: 853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cox J, Brown HE, Rico-Hesse R. 2011. Variation in vector competence for dengue viruses does not depend on mosquito midgut binding affinity. PLoS Negl. Trop. Dis. 5: e1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Descloux E, Cao-Lormeau VM, Roche C, De Lamballerie X. 2009. Dengue 1 diversity and microevolution, French Polynesia 2001–2006: connection with epidemiology and clinics. PLoS Negl. Trop. Dis. 3: e493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Duong V, et al. 2 July 2011. Genetic diversity and lineage dynamic of dengue virus serotype 1 (DENV-1) in Cambodia. Infect. Genet. Evol. [Epub ahead of print.] doi: 10.1016/j.meegid.2011.06.019 [DOI] [PubMed] [Google Scholar]
- 15. Endy TP, et al. 2011. Determinants of inapparent and symptomatic dengue infection in a prospective study of primary school children in Kamphaeng Phet, Thailand. PLoS Negl. Trop. Dis. 5: e975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gubler DJ, Reed D, Rosen L, Hitchcock JR., Jr 1978. Epidemiologic, clinical, and virologic observations on dengue in the Kingdom of Tonga. Am. J. Trop. Med. Hyg. 27: 581–589 [DOI] [PubMed] [Google Scholar]
- 17. Gubler DJ, Suharyono W, Lubis I, Eram S, Gunarso S. 1981. Epidemic dengue 3 in central Java, associated with low viremia in man. Am. J. Trop. Med. Hyg. 30: 1094–1099 [DOI] [PubMed] [Google Scholar]
- 18. Guzman MG, et al. 2010. Dengue: a continuing global threat. Nat. Rev. Microbiol. 8: S7–S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanley KA, Nelson JT, Schirtzinger EE, Whitehead SS, Hanson CT. 2008. Superior infectivity for mosquito vectors contributes to competitive displacement among strains of dengue virus. BMC Ecol. 8: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Holmes EC. 2003. Patterns of intra and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J. Virol. 77: 11296–11298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holmes EC, Burch SS. 2000. The causes and consequences of genetic variation in dengue virus. Trends Microbiol. 8: 74–77 [DOI] [PubMed] [Google Scholar]
- 22. Holmes EC, Twiddy SS. 2003. The origin, emergence and evolutionary genetics of dengue virus. Infect. Genet. Evol. 3: 19–28 [DOI] [PubMed] [Google Scholar]
- 23. Jarman RG, et al. 2008. Microevolution of dengue viruses circulating among primary school children in Kamphaeng Phet, Thailand. J. Virol. 82: 5494–5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johansson MA, Dominici F, Glass GE. 2009. Local and global effects of climate on dengue transmission in Puerto Rico. PLoS Negl. Trop. Dis. 3: e382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klungthong C, et al. 2007. Dengue virus detection using whole blood for reverse transcriptase PCR and virus isolation. J. Clin. Microbiol. 45: 2480–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klungthong C, Zhang C, Mammen MP, Jr, Ubol S, Holmes EC. 2004. The molecular epidemiology of dengue virus serotype 4 in Bangkok, Thailand. Virology 329: 168–179 [DOI] [PubMed] [Google Scholar]
- 27. Lambrechts L, et al. 2009. Genetic specificity and potential for local adaptation between dengue viruses and mosquito vectors. BMC Evol. Biol. 9: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lambrechts L, et al. 2011. Impact of daily temperature fluctuations on dengue virus transmission by Aedes aegypti. Proc. Natl. Acad. Sci. U. S. A. 108: 7460–7465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. 1992. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J. Clin. Microbiol. 30: 545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Messer WB, Gubler DJ, Harris E, Sivananthan K, de Silva AM. 2003. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg. Infect. Dis. 9: 800–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Myat Thu H, et al. 2005. Lineage extinction and replacement in dengue type 1 virus populations are due to stochastic events rather than to natural selection. Virology 336: 163–172 [DOI] [PubMed] [Google Scholar]
- 32. Nisalak A, et al. 2003. Serotype-specific dengue virus circulation and dengue disease in Bangkok, Thailand from 1973 to 1999. Am. J. Trop. Med. Hyg. 68: 191–202 [PubMed] [Google Scholar]
- 33. Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25: 1253–1256 [DOI] [PubMed] [Google Scholar]
- 34. Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53: 793–808 [DOI] [PubMed] [Google Scholar]
- 35. Pybus OG, Rambaut A. 2009. Evolutionary analysis of the dynamics of viral infectious disease. Nat. Rev. Genet. 10: 540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Recker M, et al. 2009. Immunological serotype interactions and their effect on the epidemiological pattern of dengue. Proc. Biol. Sci. 276: 2541–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rico-Hesse R. 2003. Microevolution and virulence of dengue viruses. Adv. Virus Res. 59: 315–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rico-Hesse R, et al. 1997. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology 230: 244–251 [DOI] [PubMed] [Google Scholar]
- 39. Shrestha B, et al. 2010. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathog. 6: e1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Steel A, Gubler DJ, Bennett SN. 2010. Natural attenuation of dengue virus type-2 after a series of island outbreaks: a retrospective phylogenetic study of events in the South Pacific three decades ago. Virology 405: 505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Swofford DL. 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), version 4.0 Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 42. Thomas SJ, et al. 2009. Dengue plaque reduction neutralization test (PRNT) in primary and secondary dengue virus infections: how alterations in assay conditions impact performance. Am. J. Trop. Med. Hyg. 81: 825–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vu TT, et al. 2010. Emergence of the Asian 1 genotype of dengue virus serotype 2 in Vietnam: in vivo fitness advantage and lineage replacement in South-East Asia. PLoS Negl. Trop. Dis. 4: e757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wahala WM, et al. 2010. Natural strain variation and antibody neutralization of dengue serotype 3 viruses. PLoS Pathog. 6: e1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weaver SC, Vasilakis N. 2009. Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect. Genet. Evol. 9: 523–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wilder-Smith A, Ooi EE, Vasudevan SG, Gubler DJ. 2010. Update on dengue: epidemiology, virus evolution, antiviral drugs, and vaccine development. Curr. Infect. Dis. Rep. 12: 157–164 [DOI] [PubMed] [Google Scholar]
- 47. Wittke V, et al. 2002. Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology 301: 148–156 [DOI] [PubMed] [Google Scholar]
- 48. Zhang C, et al. 2005. Clade replacements in dengue virus serotypes 1 and 3 are associated with changing serotype prevalence. J. Virol. 79: 15123–15130 [DOI] [PMC free article] [PubMed] [Google Scholar]