Abstract
The essential Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) minor capsid protein VP80 has been recently shown to interact with the virus-triggered, nuclear F-actin cytoskeleton. A role for VP80 in virus morphogenesis has been proposed in the maturation of progeny nucleocapsids and in their egress from the virogenic stroma toward the nuclear periphery by a mechanism, which also includes F-actin filaments. We performed functional mapping of VP80 demonstrating that its highly conserved C-terminal region plays a crucial role in virion morphogenesis. Protein database mining identified a putative basic helix-loop-helix (bHLH) domain, a DNA-binding module typical for eukaryotic transcription factors, in the essential C-terminal region of VP80. Using a molecular modeling approach, we predicted the three-dimensional structure of this domain, revealing some unique properties. Biochemical assays proved that VP80 can form homodimers, a critical prerequisite of DNA-binding bHLH proteins. The ability of VP80 to bind DNA was subsequently confirmed by an electrophoretic mobility shift assay. We further show that AcMNPV DNA replication occurs in the absence of VP80. Immunolabeling of VP80 in baculovirus-infected cells rather points toward its involvement in nucleocapsid maturation. The competence of VP80 to interact with both F-actin and DNA provides novel insight into baculovirus morphogenesis.
INTRODUCTION
Virus morphogenesis is a complex process in which viral structural proteins, genetic information carried by either viral DNA or RNA, and occasionally, depending on the type of virus, also lipid bilayers are assembled into viral infectious units, the so-called virions. The formation of virions occurs for the most part inside infected cells and is frequently supported by various host cell factors, for instance, cytoskeletal elements (see references 5, 38, and 41) for reviews).
Baculoviruses, a group of enveloped viruses with a circular double-stranded DNA genome, usually over 100 kbp in size (36), loaded in a rod-shaped nucleocapsid, require the actin cytoskeleton for successful productive infection of their arthropod hosts (see reference 38 for a review). Baculoviruses replicate their DNA genomes in the nuclei of infected cells, where progeny nucleocapsids are also assembled. A typical baculovirus infection includes the formation of two virion types: (i) extracellular budded virus (BV) formed from nucleocapsids leaving the cell nucleus and budding through the plasma membrane and (ii) occlusion-derived virus (ODV) built up from nucleocapsids accumulated in the nuclear periphery, where envelopment occurs prior to embedding into viral occlusion bodies (OBs) (see reference 32 for a review). BVs are responsible for the spread of infection within the body of insect larvae, while ODVs encapsulated in OBs mediate horizontal virus transmission between insects via oral infection.
Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV), the prototype of the genus Alphabaculovirus of the family Baculoviridae, encodes numerous proteins that interact with and manipulate the host actin cytoskeleton toward viral needs. BVs release their nucleocapsid into the cytoplasm after clathrin-dependent endocytosis (16). In the cytoplasm, the nucleocapsids are immediately transported toward the cell nucleus by actin-based movement, which is orchestrated by the host actin-polymerizing Arp2/3 complex and the viral, nucleocapsid-associated phosphoprotein P78/83 (open reading frame [ORF] 1629), which shows homology to the Wiskott-Aldrich syndrome protein (WASP) (29, 39). Nuclear pores have been recently shown to be involved in translocation of incoming nucleocapsids into the nucleus, where their uncoating is followed by viral early gene transcription (29).
During early viral transcription, the actin rearrangement factor 1 (Arif-1) is synthesized and targeted to the plasma membrane in order to disrupt the continuous filamentous actin (F-actin) cortex (6), a typical feature of uninfected insect cells. Six other early expressed viral proteins, namely, IE1, PE38, HE65, Ac004, Ac102, and Ac152, are involved in accumulation of a globular-actin (G-actin) pool in the nucleus (27). In the late phase of infection, four viral actin-interacting proteins are newly synthesized. First, the major capsid protein VP39 binds both G- and F-actins but shows higher affinity for G-actin (10). Two additional structural proteins, the above-mentioned P78/83 (8) and BV/ODV-C42 (15), are involved in polymerization of G-actin monomers into F-actin filaments, thus shifting the G- and F-actin levels from abundant G to abundant F.
Recently, a fourth AcMNPV structural protein, VP80, has been shown to colocalize and associate with the virus-triggered, nuclear F-actin scaffold (22). AcMNPV VP80 is an end-linked nucleocapsid protein (22) that is essential for the formation of both virion types, BVs and ODVs (23). The necessity of the vp80 gene was also confirmed in the related Bombyx mori NPV (BmNPV) (34). Although deletion of the AcMNPV vp80 gene did not prevent assembly of nucleocapsids, these appeared to be aberrantly filled with DNA and especially were not able to migrate from the viral replication factory, the so-called virogenic stroma, to the nuclear periphery to form viral progeny (23, 34). In virus-infected cells, VP80 is entirely localized in the nucleus, adjacent to the virus-triggered F-actin scaffold that forms a highly organized three-dimensional (3D) network permeating and connecting the virogenic stroma physically with the nuclear envelope (22). Moreover, AcMNPV and BmNPV VP80 proteins possess sequence motifs related to invertebrate paramyosin proteins (22). Based on these facts, we logically concluded that cooperation of VP80 with the host actin-myosin (acto-myosin) complex provide traffic connections between the virogenic stroma and the nuclear periphery to facilitate egress of progeny nucleocapsids (22). In addition, confocal imaging also provided evidence for apparent colocalization of VP80 with DNA in distinct areas of the virogenic stroma, signifying a potential interaction between VP80 and DNA (22).
In the present study, we provide evidence for the crucial functional role of the evolutionary conserved C-terminal region of the VP80 protein. Bioinformatic searches identified a putative DNA-binding basic helix-loop-helix (bHLH) domain in the essential C-terminal region of VP80. We performed several biochemical assays to show the competence of VP80 to form homodimers that bind DNA in vitro. The finding that VP80 interacts with both F-actin and DNA provides key insight into baculovirus morphogenesis.
MATERIALS AND METHODS
Bioinformatics and phylogeny reconstruction.
The AcMNPV VP80 sequence was searched against the nonredundant protein database at the National Center for Biotechnology Information (3) with the iterated PSI-BLAST algorithm (1) to identify VP80 homologues. The presence of a bHLH domain was predicted by a SMART (14) search. Multiple sequence alignments were constructed using ClustalX (11) with default parameters and manually edited in GeneDoc (26) and JalView (42). A phylogenetic tree was built on the basis of multiple alignments using the maximum-likelihood (ML) method as implemented in the software package MEGA4 (33), which was also used for bootstrap analysis (500 replicates) and graphical representation of the resulting tree.
Cells and viruses.
Spodoptera frugiperda cells (Sf9; Invitrogen) were maintained in SF900-II serum-free medium (Invitrogen) at 27°C under standard conditions. All AcMNPV recombinant bacmids and viruses were derived from the commercially available bacmid bMON14272 (19) (Invitrogen) or its modified version, the Ac-Δvp80 bacmid (23), both propagated in the Escherichia coli strain DH10β. The Ac-Δvp80 bacmid repaired with a full-length vp80 ORF containing a Flag tag at its N terminus under the control of the vp80 promoter was designated Ac-Δvp80-vp80 (23) and was used as a control. This repaired construct also carried an egfp reporter gene under p10 promoter control. Both the vp80 and the egfp ORFs were inserted in the polyhedrin (polh) locus).
DNA constructions.
In order to generate a set of AcMNPV bacmids encoding different vp80 gene truncations, the bacmid Ac-Δvp80 (23) was modified. At first, an egfp reporter was amplified from plasmid pEGFP-N3 (Clontech) with the primers egfp-F and egfp-R (23) and cloned downstream of the p10 promoter in the pFB-Pvp80 (22) plasmid, to create pFB-Pvp80-egfp. Flag-tagged vp80 ORFs encoding four N-truncated VP80 protein variants designated VP80-ΔN1 (amino acids [aa] 47 to 691), VP80-ΔN2 (aa 262 to 691), VP80-ΔN3 (aa 424 to 691), and VP80-ΔN4 (aa 500 to 691) and one C-truncated VP80 named VP80-ΔC (aa 1 to 580) were amplified from the bMON14272 (19) bacmid. To this aim the vp80 gene-specific primers vp80-3-F or vp80-1-R (23) were used in combination with newly designed vp80 gene truncation-specific primers (Table 1). The amplified genes were cloned between SacI and XbaI sites downstream of the vp80 promoter in the donor pFB-Pvp80-egfp plasmid. The developed expression cassettes were finally transposed into the polh locus of the Ac-Δvp80 (23) bacmid according to the Bac-to-Bac protocol (Invitrogen). The final recombinant bacmids/viruses were analogously named Ac-Δvp80-vp80-ΔN1, Ac-Δvp80-vp80-ΔN2, Ac-Δvp80-vp80-ΔN3, Ac-Δvp80-vp80-ΔN4, and Ac-Δvp80-vp80-ΔC.
Table 1.
Primers used for PCR
| Primer | Sensea | Sequence (5′–3′) |
|---|---|---|
| vp80-N1-F | F | TTATCTTGAGCTCAATATGGATTACAAGGATGACGACGATAAGTTGGCTGTAAATAGCCCGC |
| vp80-N2-F | F | TTATCTTGAGCTCAATATGGATTACAAGGATGACGACGATAAGGTCAAGACGGTTCTTTTGCG |
| vp80-N3-F | F | TTATCTTGAGCTCAATATGGATTACAAGGATGACGACGATAAGAAGAGATCTGCGGAAGACGA |
| vp80-N3h-F | F | TTATCTTGAGCTCAATATGCATCATCACCATCACCATAAGAGATCTGCGGAAGACGA |
| vp80-N4-F | F | TTATCTTGAGCTCAATATGGATTACAAGGATGACGACGATAAGATTGTGGTCACCGACGGTATG |
| vp80-N4h-F | F | TTATCTTGAGCTCAATATGCATCATCACCATCACCATAAGATTGTGGTCACCGACGGTATG |
| vp80-C-R | R | AAGGTTCTCTAGATTAACTGGCCAAAAATATGTAATTTA |
F, forward; R, reverse.
N-terminally His-tagged, vp80 truncated ORFs encoding His-VP80-ΔN3 and His-VP80-ΔN4 proteins were amplified with the primers vp80-1-R (23) and vp80-N3h-F or vp80-N4h-F, respectively (Table 1). The PCR products were cloned in a similar way between SacI and XbaI in the donor plasmid pFB-Pvp80-egfp, and recombinant bacmids designated Ac-Δvp80-His:vp80-ΔN3 and Ac-Δvp80-His:vp80-ΔN4 were generated.
Functional analysis of VP80 truncations.
Five micrograms of DNA of the bacmids described above were used to transfect 106 Sf9 cells with Cellfectin II (Invitrogen). At 5 days posttransfection (p.t.) the BV-enriched culture supernatant was centrifuged for 5 min at 2,000 × g and used to infect 1.5 × 106 Sf9 cells. Viral propagation was followed by fluorescence microscopy at indicated time points p.t. or postinfection (p.i.). In addition, production of BV virions was checked by negative staining electron microscopy (EM) as previously described (23). The Ac-Δvp80-vp80 (23) bacmid/virus served as a positive control.
To monitor the BV release kinetics, one million cells were transfected with 5 μg bacmid DNA, and cell culture media were harvested at various time points and analyzed for the presence of infectious BV by endpoint dilution. The averages of the infectious titers from three independent transfections/infections were calculated and plotted into graphs.
DNA replication assay.
To determine the viral DNA replication capacity of the AcMNPV Δvp80 (23) genome, a quantitative real-time PCR (Q-PCR)-based assay was carried out as previously described (37) with minor modifications. Briefly, Sf9 cells (106 cells/35-mm dish) were transfected in an independent triplicate assay with 5 μg of either Ac-Δvp80 (23), Ac-Δvp39 (25), or Ac-Δgp64 (20) bacmid DNA. At the indicated time points p.t., total DNA was purified from the cells using a genomic DNA from blood kit (Macherey-Nagel, Germany). The original bacmid inputs were eliminated from the DNA samples by DpnI treatment at 37°C for 4 h. An 8-μl aliquot (10 ng of DNA) was taken and mixed with a 2-μl mixture of the 65972F and 66072R (37) primers (both primers at 5 pmol/μl) and then with a 10-μl volume of the Quantitect SYBR green master mix (Qiagen). Q-PCRs were performed in a RotorGene 2000 thermal cycler (Corbett Research), and fluorescence was recorded on the FAM channel. The program used was as follows: 15 min at 95°C; followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s; followed in turn by a melting curve (60 to 99°C). To determine calibration standard curves, purified bMON14272 bacmid (19) was serially diluted (10-fold intervals) and amplified with the 65972F and 66072R (37) primers (independent duplicate assay). Nontemplate controls were also included. The percentage of viral DNA synthesis was calculated according to the following formula: % viral DNA synthesis = [(test DNAnh − test DNA0 h)/(Ac-Δgp64 DNA96 h − Ac-Δgp64 DNA0 h)] × 100, where nh represents indicated time point p.t.
EM and immunolabeling.
Sf9 cells (3.5 × 106 cells/flask) were infected with the Ac-Δvp80-Flag:vp80 (23) virus (MOI = 10). At 24 h p.i., the cells were harvested and processed for transmission EM as described previously (35). For immunolabeling, we used primary mouse anti-Flag antibody (Stratagene) diluted 1:500, followed by colloidal-gold (5-nm particle)-conjugated secondary goat antibody (Aurion). Specimens were observed in a Philips CM12 electron microscope.
Production and purification of His-tagged VP80 variants.
To produce His-tagged VP80-ΔN3 and VP80-ΔN4 proteins, Sf9 cells (3 × 108 cells) were transfected with 100 μg of either the Ac-Δvp80-His:vp80-ΔN3 or Ac-Δvp80-His:vp80-ΔN4 bacmid. At 72 h p.t., the cells were harvested and washed once with 1× phosphate-buffered saline (PBS). The cell pellets were sonicated in buffer A (20 mM Tris-HCl [pH 7.2], 100 mM KCl, 5 mM imidazole, 1 mM phenylmethylsulfonyl fluoride) on ice and then centrifuged (15,000 × g, 30 min at 4°C). The supernatants were loaded onto Ni-NTA agarose affinity columns (Qiagen), and bound proteins were eluted with 250 mM imidazole in buffer A and stored in this solution at 4°C or flash frozen at −80°C for long storage. The concentrations of purified proteins were determined by the standard Bradford method.
To study dimerization capacities of both VP80-ΔN3 and VP80-ΔN4, the purified proteins were selectively processed for protein electrophoresis in the presence or absence of either β-mercaptoethanol or with or without thermal denaturation (95°C, 5 min) or both. The protein samples were then electrophoretically separated in a 4 to 20% gradient polyacrylamide gel followed by Western blot analysis with anti-His antibody (Molecular Probes).
Immunoprecipitation.
To study the possible self-association of VP80-ΔN3, Sf9 cells (3.0 × 107 cells) were cotransfected with Ac-Δvp80-Flag:vp80-ΔN3 (25 μg) and Ac-Δvp80-His:vp80-ΔN3 (100 μg) bacmids. At 3 days p.t., the cells were harvested and washed once in 1× PBS. A Flag-tagged protein immunoprecipitation kit (Sigma-Aldrich) was used to fish out protein complexes according to the manufacturer's protocol. Bound proteins were analyzed on Western blots with mouse anti-Flag (Stratagene) or anti-His (Molecular Probes) antibodies. Sf9 cells transfected with Ac-Δvp80-His:vp80-ΔN3 bacmid (100 μg) were used as a negative control for immunoprecipitation reactions.
Electrophoretic mobility shift assay (EMSA).
A 204-bp DNA probe encompassing a partial sequence of the AcMPNV vp80 promoter, which contains an E-box-like sequence, was amplified from the Ac-Δvp80 (23) bacmid DNA with vp80L-F and vp80L-R (23) primers. The DNA probe was then terminally labeled with [γ-32P]ATP using polynucleotide kinase (Fermentas). DNA-binding reactions (20 μl) were carried out in 20 mM HEPES (pH 8.0), 100 mM KCl, and 2 mM dithiothreitol where the labeled DNA probe (∼100 nmol) was mixed with various amounts of VP80-ΔN3 and VP80-ΔN4, respectively. To check the specificity, the tested proteins were presaturated with a 500-fold molar excess of unlabeled DNA probe, designated inhibitor DNA, prior to DNA-binding reactions. The assembled reactions were incubated for 30 min at 25°C. One-half of reaction volume was then run on 6% polyacrylamide gels in standard 1× Tris-borate-EDTA (pH 8.3) buffer at room temperature. The gels were autoradiographed using a PhosphorImager (Molecular Dynamics).
To study the DNA-binding specificity of the VP80-ΔN3 protein, two 18-bp DNA probes were generated. The first probe, designated as E-box DNA, contained the consensual E-box motif (5′-GGAAGGCACGTGAAGAAG-3′; the E-box motif is denoted by italics), while the second probe was designated as the control DNA, in which the E-box is replaced with a random, nonpalindromic sequence (5′-GGAAGGGTTTTCAAGAAG-3′). These probes were labeled and used for comparative EMSA experiments according to the protocol described above. The EMSA results were quantified by densitometric analysis of the bands corresponding to the unbound (free) DNA probes using Quantity One software (Bio-Rad). The significance of the observed differences was analyzed by a Student t test.
Structure prediction and molecular modeling.
An initial search for suitable threading templates for the putative bHLH domains in AcMNPV (Asp457 to Lys515) and Choristoneura fumiferana MNPV (CfMNPV) (Glu398 to Ile458) VP80 performed with the help of the automated mode of Swiss-Model (2) yielded no reliable templates. Therefore, 3D models of the AcMNPV and CfMNPV VP80-bHLH domains were generated by a homology modeling method using the crystal structures of human MyoD (Protein Data Bank [PDB]: 1MDY [21]), and mouse E47 and NeuroD1 (PDB: 2QL2 [17]). Briefly, sequence alignments were constructed with ClustalX (11) and structural models were generated using the automodel function in Modeller 9v6 (7). The models with the lowest values of the Modeller objective function were selected for further evaluation.
In parallel, the alignment mode of Swiss-Model (2) and I-TASSER (31) web-based workspaces were used to build analogous VP80-bHLH models in order to compare resulting structures with the Modeller prediction. The models were finally evaluated by PROCHECK (12) and ANOLEA (24) to check for backbone problems and Ramachandran outliers. The predicted VP80-bHLH structures were in silico complexed with an 14-bp DNA molecule (5′-TCAACAGCTGTTGA-′3) that was borrowed from the crystal structure of human MyoD-DNA complex [PDB: 1MDY [21]). Final adjustments and graphical visualization of 3D models were performed with Chimera (30) software.
RESULTS
Evolutionary history of baculoviruses coding for VP80 protein.
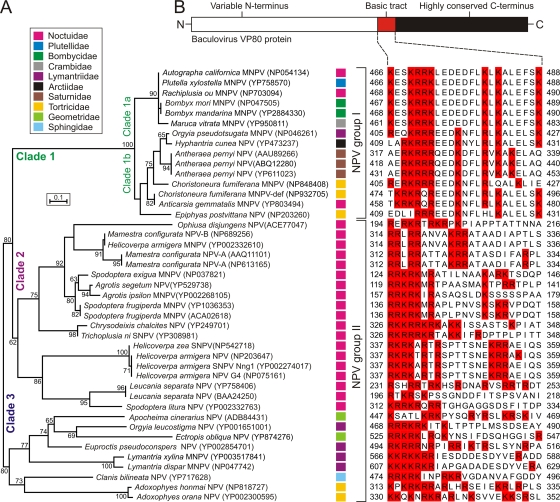
To obtain a broader view of the diversity of VP80, we scanned the GenBank nonredundant database using AcMNPV VP80 as a query. As a result, we have collected a set of 44 protein sequences showing significant similarity (homology) to AcMNPV VP80. All identified VP80 homologues are found in viruses belonging to the genus Alphabaculovirus (lepidopteran NPVs). No VP80-related sequences were found in members of other genera (Betabaculovirus, Gammabaculovirus, and Deltabaculovirus) or in other arthropods-infecting viruses. Based on the available VP80 sequences, we have inferred a detailed phylogenetic ML-tree illustrating a grouping of the VP80 protein family into three distinct clades 1 to 3 (Fig. 1A). A similar tree topology was also obtained using the neighbor-joining method (data not shown). Clade 1 includes the VP80 sequences of group I NPVs, where two monophyletic subclades (1a and 1b) have been formed (Fig. 1A). Subclade 1a includes AcMNPV and its variants infecting lepidopteran species of the families Noctuidae, Plutellidae, Bombycidae, and Crambidae. Subclade 1b constitutes viruses causing infections in hosts from the lepidopteran families Lymantriidae, Arctiidae, Saturnidae, Tortricidae, and Noctuidae. The VP80 proteins of group II NPVs are disjointing into two monophyletic clusters (clades 2 and 3). Clade 2, which interestingly shares common ancestry (bootstrap value 80) with group I NPVs (clade 1), represents a uniform group of viruses infecting only insects in the family Noctuidae. On the other hand, viruses belonging to clade 3 infect species of the families Geometridae, Lymantriidae, Sphingidae, or Tortricidae (Fig. 1A). The revealed phylogenetic linkage between VP80 of group I NPVs (clade 1) and clade 2, with noctuid-infecting group II NPVs could be explained by a recombination event, in which an ancestral group II NPV obtained the vp80 gene from a group I NPV coinfecting the same host. This is possible since some group I NPVs (e.g., AcMNPV, Rachiplusia ou MNPV and Anticarsia gemmatalis MNPV) also infect noctuids.
Fig 1.
Overview of VP80 phylogeny and sequence conservation. (A) Unrooted ML-tree built up from VP80 protein sequence data. Forty-four VP80 homologous sequences were aligned with the help of ClustalX. All positions containing gaps and missing data were eliminated from the data set, and phylogenetic analysis was conducted in MEGA4. Numbers at branches denote percent bootstrap values (out of 500 replicates), and numbers in the brackets indicate the GenBank accession number. Color squares at each baculovirus strain indicate host lepidopteran family as explained in legend (top frame). (B) All VP80 proteins contain a tract of basic amino acid residues. Schematic representation of VP80 sequence conservation (top scheme) with a list of aligned VP80 basic tracts. Basic amino acid residues are highlighted in red boxes. The alignment was constructed using ClustalX, manually edited in GeneDoc, and coloring was performed in JalView.
Careful inspections of the aligned VP80 sequences revealed a remarkable evolutionary conservation of the VP80 carboxy-terminal ends (aa 457 to 691 in AcMNPV VP80), while their amino-terminal ends are extremely variable. Importantly, all VP80 proteins contain a tract of basic amino acids (Fig. 1B) localized nearly upstream of the conserved C terminus. Interestingly, this basic amino acid-rich region is situated downstream from the putative nuclear localization signal (NLS) of the VP80 protein (22), indicative of a different function than nuclear targeting. Hence, the role of these blocks of positively charged amino acid residues in VP80 biology is unclear, but their presence suggest a possible interaction between VP80 and nucleic acids (e.g., DNA or RNA).
Highly conserved C-terminal region of VP80 plays a crucial role in baculovirus infection.
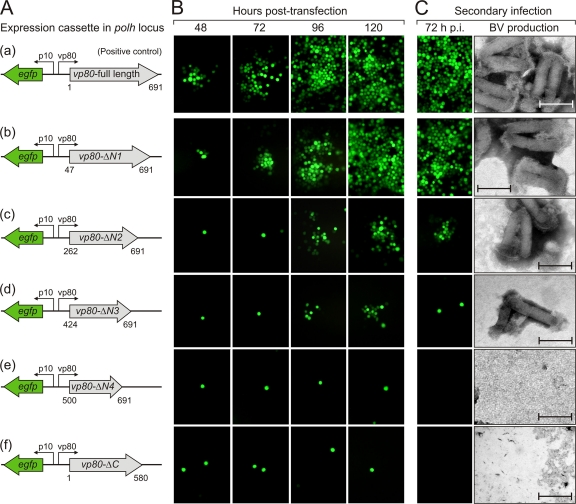
The AcMNPV vp80 gene encodes a polypeptide of 691 aa residues with a theoretical molecular mass of 80.9 kDa. To define the minimal functional domain of VP80 required for baculovirus infection, a set of vp80 gene truncations was constructed. Four truncations targeted the variable N-terminal end (vp80-ΔN1 to vp80-ΔN4), and one truncation targeted the conserved C-terminal end (vp80-ΔC). The cloned vp80 truncations together with an egfp reporter gene were transposed into the polh locus of the defective Δvp80 (23) AcMNPV genome (Fig. 2A). The abilities of all vp80 truncations to rescue the Δvp80 genotype were examined by transfection-infection assays in Sf9 cells, followed by time course fluorescence microscopy (Fig. 2B). BV production was checked by both negative staining EM (Fig. 2C) and endpoint dilution (Fig. 3). The truncated proteins illustrated in Fig. 2A were all expressed at levels comparable to the level of full-length VP80 protein as detected on Western blots (data not shown). As seen by microscopic observations (Fig. 2B and C), removal of 46 aa residues from the N terminus (VP80-ΔN1) resulted in an almost complete rescue of virus propagation, although rigorous BV-titrations revealed a consequential negative effect on viral replication kinetics (Fig. 3A). More importantly, severe truncations (up to 423 aa residues from the N terminus) still resulted in virus propagation; however, these repaired viruses were strongly handicapped compared to the full-length VP80 rescued virus (Fig. 2B and C and 3A). Finally, the truncation (VP80-ΔN4) missing 499 aa from the N terminus, including both the putative NLS (22) and the identified basic tract (Fig. 1B), completely failed to repair the Δvp80 phenotype. These results indicate that the NLS and likely the conserved basic tract (Fig. 1B) are essential for the function of VP80 during infection.
Fig 2.
Essential role of C-terminal region of the VP80 protein. (A) Schematic representation of expression cassettes encoding vp80 gene truncations along with an egfp reporter gene, which were introduced into the polh locus of the Ac-Δvp80 bacmid. (B) Ability of VP80 truncation mutants to rescue the Ac-Δvp80 replication-deficient bacmid. Sf9 cells were transfected with DNA of each bacmid construct and virus propagation was assessed by fluorescence microscopy at the indicated time points p.t. The bacmid Ac-Δvp80-vp80 coding for full-length VP80 protein was used as a positive control. (C) Secondary infection assay. BV-enriched cell culture supernatants from transfection assays were collected and used to infect a new batch of Sf9 cells. Spread of infection was monitored by fluorescence microscopy at 72 h p.i. Release of BV particles was controlled in cell culture media by negative staining EM. Bar, 200 nm.
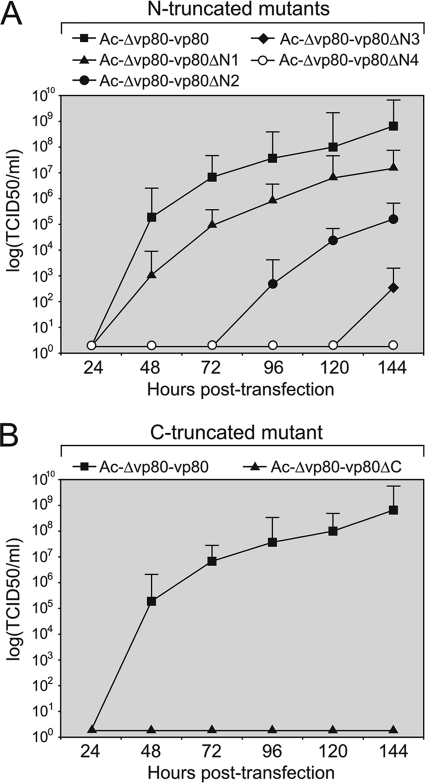
Fig 3.
One-step growth curves of the Ac-Δvp80 genome repaired with different VP80 truncation variants. Sf9 cells were transfected with 5.0 μg of DNA of either N-terminally truncated (Ac-Δvp80-vp80ΔN1, Ac-Δvp80-vp80ΔN2, Ac-Δvp80-vp80ΔN3, and Ac-Δvp80-vp80ΔN4) (A) or C-terminally truncated (Ac-Δvp80-vp80ΔC) (B) repaired bacmids. For all of these constructs the viral propagation was compared to that of Ac-Δvp80-vp80 coding for full-length VP80 protein. Cell culture supernatants were harvested at the indicated times p.t. and analyzed for release of infectious BV by endpoint dilution. Infectivity was determined by monitoring EGFP expression. The points indicate the averages of titers derived from three independent transfections, and the error bars represent the standard deviations (SD).
Consistent with previous observations (23), no viral propagation was observed when 111 aa residues (VP80-ΔC) were removed from the C-terminal end of VP80, confirming its functional importance in the virus infection cycle (Fig. 2B and C and 3B).
VP80 does not appear to be involved in viral DNA replication.
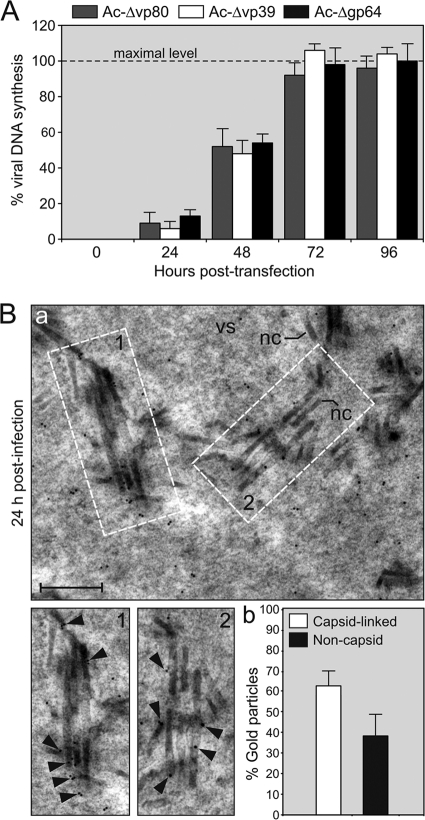
Our previous study revealed that AcMNPV VP80 colocalized with DNA during viral infection (23), suggesting its involvement in a yet-to-be-determined process, in which DNA plays a central role (e.g., virus genome replication/processing, viral transcription, or genome packaging into progeny capsids). To assess whether the VP80 protein is involved in the replication of the viral genome, a comparative DNA-replication assay was performed. Sf9 cells were transfected with the Ac-Δvp80 (23) bacmid, and the level of viral DNA was monitored at different time points as indicated in Fig. 4A using Q-PCR. In order to compare DNA-replication kinetics of the Δvp80 knockout, two control AcMNPV mutants, Ac-Δgp64 (20) and Ac-Δvp39 (25), were used. Analogous to the Ac-Δvp80 bacmid, these mutants are not able to initiate viral cell-to-cell transmission. It is supposed that deletion of either the gp64 (37) or vp39 gene does not affect the viral DNA replication capacity, allowing their application in comparative replication assays. The cells transfected with the Ac-Δvp80 bacmid showed a similar, if not identical profile of viral DNA replication kinetics as both control bacmids (Fig. 4A). The maximal abundance of viral DNA was reached at 72 h p.t. and started to plateau thereafter, probably because of an inability to spread the infection to surrounding cells. The results thus indicate that deletion of the vp80 gene neither affects DNA-replication nor events antecedent to genome replication.
Fig 4.
VP80 is not required for viral DNA replication but appears to assist during nucleocapsid maturation. (A) Virus genome replication assay. Sf9 cells transfected with Ac-Δvp80 bacmid were harvested and total DNA was purified at indicated time points. Two control bacmids, Ac-Δgp64 and Ac-Δvp39, were used as prototypes showing a “single cell infection phenotype.” The data are expressed as the fold of viral DNA (%) relative to level of synthesized DNA by the Ac-Δgp64 bacmid determined at 96 h p.t. (means ± the SD) and represent the means from three separate experiments. (B) Immunogold labeling of VP80 in baculovirus-infected cells. (a) Representative view of the virogenic stroma showing VP80 localization. Sf9 cells were infected with the Ac-Δvp80-Flag:vp80 virus (MOI = 10). At 24 h p.i., the cells were harvested and processed for EM. Ultrathin sections were adsorbed on EM grids and immunostained by mouse anti-Flag antibody, followed by goat anti-mouse immunoglobulin antibody conjugated with 5-nm gold particles. Insets 1 and 2 show typical labeling of progeny nucleocapsids with gold particles. The black arrowheads point to localizations of gold particles. Abbreviations: nc, nucleocapsid; vs, virogenic stroma. Bar, 200 nm. (b) Quantitative analysis of number of gold particles in the nuclear virogenic stroma of baculovirus-infected Sf9 cells. Twenty-three cells (n = 23) were inspected, and the data are expressed as percentage (%) numbers (means ± the SD) of gold particles associated either with progeny nucleocapsids (capsid-linked) or freely distributed in the virogenic stroma (noncapsid).
Recently, we showed that VP80 protein associates with nucleocapsid fractions of both BV and ODV virions, specifically with one end of the nucleocapsids (22). Immunogold labeling of VP80 in virus-infected cells (24 h p.i.) resulted in specific staining of cell nuclei with minimal numbers of gold particles present in the cytoplasm. Within the cell nuclei, most gold particles localized in the virogenic stroma, and a smaller amount was observed in the peristromal compartment, the so-called ring zone (data not show). Comprehensive inspection of the virogenic stroma revealed that gold particles were preferentially situated in less-condensed areas of the stroma, typically localizing to de novo-assembled capsids (Fig. 4Ba). Statistical analysis provided information that the majority of gold particles (62% ± 7%) was localized in close proximity to progeny nucleocapsids. The remainder (38% ± 10%) was homogenously distributed through the virogenic stroma without any apparent pattern (Fig. 4Bb). The fact that VP80 comes into contact with DNA in vivo (23), together with its localization to progeny nucleocapsids in the virogenic stroma, strongly supports the hypothesis that VP80 is involved in nucleocapsid maturation and/or DNA encapsidation.
VP80 contains a putative bHLH domain, a DNA-binding module typical for transcription factors.
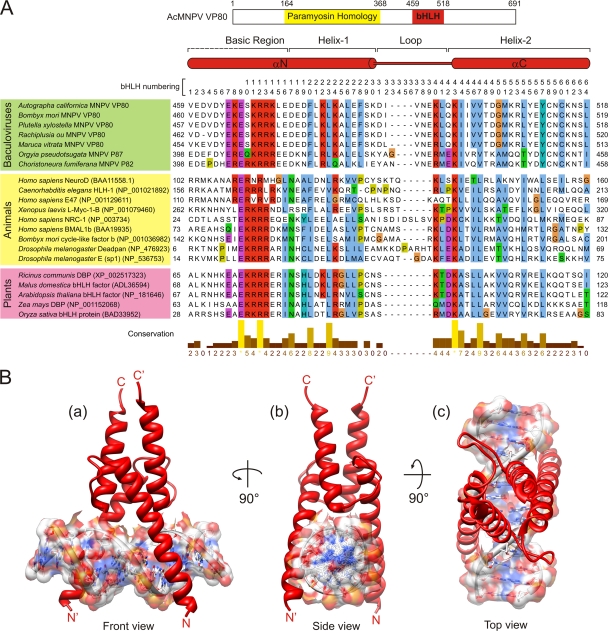
To explain the putative in vivo interaction between VP80 and DNA (22), the protein sequence of AcMNPV VP80 was searched against protein databases to identify similarity with currently known DNA-interacting domains. No highly significant hit was revealed using most of the prediction tools. However, when we extracted the AcMNPV VP80 sequence encompassing the basic region (aa 459 to 518) and used it to search the SMART (14) server, a similarity with bHLH protein domains was identified. Interestingly, use of the homologous CfMNPV VP80 sequence (aa 398 to 458) as a query resulted in a much more apparent relationship with the bHLH protein family. The sequences of putative VP80-bHLH domains from selected members of group I NPVs aligned with a representative set of experimentally verified animal and plant bHLH domains are illustrated in Fig. 5A. In general, DNA-binding bHLH domains are composed of 50 to 60 aa that form two distinct parts: a tract of 10 to 15 mainly basic aa (the basic region) and a segment of approximately 40 aa folding into two amphipathic α-helices (helices 1 and 2) separated by a variable loop (the helix-loop-helix region). The basic region mediates main contacts with DNA, while the α-helices ensure the formation of homo- or heterodimeric complexes between bHLH proteins, a critical requirement for DNA binding (see references 13 and 40 for reviews).
Fig 5.
Structure of baculovirus VP80-bHLH. (A) Alignment of representative baculovirus VP80-bHLH protein domains with several well-defined bHLH domains present in animal and plant proteins. The top scheme shows topology identified functional domains in AcMNPV VP80. Secondary structure elements are indicated over the aligned sequences; the consensual numbering system of bHLH domain is indicated over the alignment. The number in parentheses at each protein sequence represents GenBank accession number. The baculovirus VP80 GenBank accession numbers are identical to those in Fig. 1. (B) Ribbon representations of the structure solution (homodimer) of AcMNPV VP80-bHLH sequence Asp457-Lys515 in complex with a 14-bp DNA molecule containing E-box core sequence (5′-CACGTG-3′). The structure is rendered in front (a), side (b), and top (c) views.
Structural model of baculovirus VP80-bHLH.
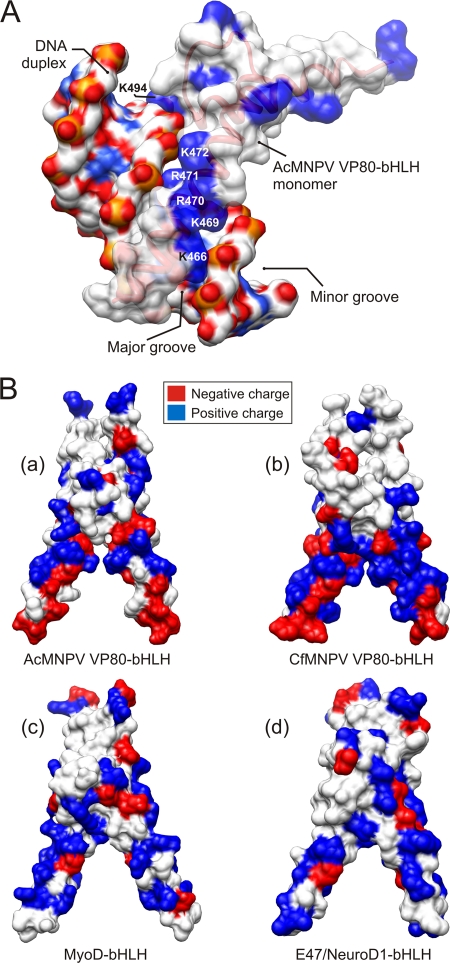
Using molecular modeling approaches, we generated a structural model of the putative AcMNPV VP80-bHLH domain in complex with DNA (Fig. 5B). The alignment (Fig. 5A) shows a conserved tract of basic residues (position 466 and positions 469 to 472) similar to those present in animal and plant bHLH proteins and demonstrates that these residues perfectly fit into the major groove of the DNA double helix as demonstrated by a 3D surface model (Fig. 6A). In contrast to animal and plant bHLH domains, the tract of basic residues within the basic region of the VP80 bHLH is flanked on both sides with a number of negatively charged residues (Fig. 5A), as can clearly be seen on surface 3D models (Fig. 6B). Importantly, within the basic region, the presence of a highly conserved glutamic acid (E) at position 467 in AcMNPV VP80 suggests that the VP80-bHLH may belong to E-box-binding bHLH proteins recognizing the hexanucleotide sequences 5′-CANNTG-3′, known as E-boxes (4). Moreover, a number of highly conserved hydrophobic residues (AcMNPV positions: F478, L481, L484, I498, V501, M505, and L508) can be found within the bHLH helices 1 and 2 (Fig. 5A). These hydrophobic residues putatively stabilize the dimeric bHLH structure (Fig. 5B).
Fig 6.
Surface models of VP80-bHLH domains. (A) Surface model of AcMNPV VP80-bHLH (monomer) in complex with a double-stranded DNA molecule. The distribution of positively (blue) charged residues within the basic region (K466, K469, R470, R471, K472, and K494) of bHLH perfectly matches the major groove of DNA duplex flanked by negatively (red) charged sugar-phosphate backbone. (B) Gallery of bHLH dimer structures for AcMNPV-VP80 (a), CfMNPV-VP80 (b), human MyoD (c), and E47/NeuroD1 (d). Surface models show the distribution of positive (blue) and negative (red) charged residues. Typical for baculovirus VP80-bHLH domains is the presence of negatively (red) charged residues at the proximal domain ends.
Evidence for self-association and DNA-binding activity of VP80.
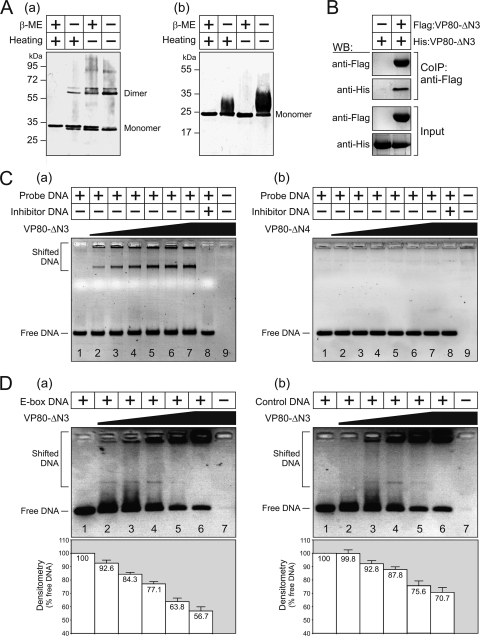
Biochemical assays were performed to verify the dimerization and DNA-binding properties of VP80 (Fig. 5). For this purpose, two His-tagged VP80 truncated proteins, VP80-ΔN3 (aa 424 to 691) and VP80-ΔN4 (aa 500 to 691), were affinity purified from baculovirus-infected Sf9 cells (data not shown). Electrophoretic separation of the purified proteins processed in the absence of a reducing agent (β-mercaptoethanol) and/or thermal denaturation (95°C) revealed that the VP80-ΔN3 protein (theoretically 32.9 kDa) migrates as two bands of ∼33 and ∼65 kDa, respectively. The latter band likely corresponds to the dimeric form. This effect was most obvious when both reducing condition and thermal denaturation were omitted (Fig. 7Aa), signifying the self-association tendency of VP80. On the other hand, the VP80-ΔN4 (∼24 kDa), a protein variant missing the putative bHLH domain, always migrated as a single band corresponding to the monomeric form, although a certain band shift could be observed under nondenatured conditions (Fig. 7Ab). Self-association of the VP80-ΔN3 was subsequently confirmed by coimmunoprecipitation. Herein, Sf9 cells were simultaneously coinfected with two baculovirus seeds, the first inoculum expressed Flag:VP80-ΔN3, while the second seed produced the His-tagged equivalent (His:VP80-ΔN3). The latter His:VP80-ΔN3 specifically copurified with the Flag:VP80-ΔN3 when anti-Flag antibody coated resin was used to fish out the protein complexes (Fig. 7B).
Fig 7.
Dimerization and DNA-binding activity of truncated VP80-ΔN3 and VP80-ΔN4 protein variants. (A) Purified VP80-ΔN3 (a) and VP80-ΔN4 (b) proteins were processed in the presence (+) or absence (−) of β-mercaptoethanol (β-ME), with or without heating to 95°C, and analyzed by protein electrophoresis. (B) Immunoprecipitated Flag:VP80-ΔN3 with its counterpart, His:VP80-ΔN3. Sf9 cells were transfected with either the Ac-Δvp80-His:vp80-ΔN3 bacmid alone or together with the Ac-Δvp80-Flag:vp80-ΔN3 bacmid. Proteins bound to the anti-Flag antibody were analyzed on Western blots using anti-Flag and anti-His antibody, respectively. Cell lysates are shown as input. (C) EMSA for VP80-ΔN3 (a) and VP80-ΔN4 (b) proteins. Increasing amounts of the protein (50, 150, 300, 400, 500, and 600 nmol) were loaded in lanes 2 to 7, as indicated by the wedge (0 nmol in lane 1). Proteins (600 nmol) presaturated with a 500-fold molar excess of unlabeled inhibitor DNA were loaded in lane 8, and only proteins (600 nmol) were loaded in lane 9. (D) DNA-binding capacity of VP80-ΔN3 for E-box-containing (a) and control DNA (b). Increasing amounts of the protein (100, 200, 400, 800, and 1,200 nmol) were loaded in lanes 2 to 6 as indicated by the wedge (0 nmol is lane 1), and only protein (1,200 nmol) was loaded in lane 7. The bottom graphs show the results of densitometric analysis, indicating the proportion of unbound (free) DNA relative to the input. The results of three independent measurements, with error bars giving the SD, are shown.
Finally, DNA-binding capacities of both the VP80-ΔN3 and VP80-ΔN4 proteins were compared by EMSAs. The VP80-ΔN3 protein obviously showed an ability to bind a double-stranded DNA probe containing an E-box-like site (5′-CACATG-3′), as demonstrated by the shifted DNA migration in the presence of the VP80-ΔN3 protein (Fig. 7Ca). This DNA retardation was dose dependent and was totally suppressed when the VP80-ΔN3 protein was presaturated with unlabeled DNA probe (Fig. 7Ca). On the other hand, the VP80-ΔN4 protein completely failed to cause a similar DNA shift, indicating its inability to interact with DNA (Fig. 7Cb). Lastly, comparative in vitro DNA-binding assays with E-box-containing and control (random) DNA probes showed that the binding of the VP80-ΔN3 protein does not depend on the presence of an E-box element (Fig. 7D), although densitometric measurements showed a moderate, statistically significant (P < 0.05), preference of VP80-ΔN3 for the E-box-containing DNA sequence (Fig. 7D). Together, our results provide experimental evidence for the DNA-binding activity of the AcMNPV VP80 protein via its C-terminal bHLH-like domain.
DISCUSSION
Our previous studies showed that the baculovirus VP80 protein is required for formation of BV and ODV virions (23). The exact role of VP80 in virion morphogenesis was not clear, but its involvement has been implicated both in the packaging of progeny nucleocapsids (23) and in their transport from the virogenic stroma toward the nuclear periphery by a mechanism, in which the F-actin cytoskeleton also plays an essential role (22).
Protein database mining resulted in 44 proteins with significant homology to AcMNPV VP80. All identified VP80 family members are encoded by lepidopteran NPV genomes (Fig. 1). Interestingly, a similar evolutionary conservation is seen for the P78/83 protein, suggesting a functional linkage between these two F-actin interacting proteins. Comparative analysis of VP80 protein sequences revealed that the C-terminal region (distal third) is highly conserved across all VP80 family members. On the other hand, the N-terminal and middle part (proximal two-thirds) of the AcMNPV VP80, showing sequence similarity to invertebrate paramyosins (22), is poorly conserved between VP80 proteins. Truncation analysis of VP80 definitively confirmed a crucial role of its conserved C-terminal region in the infection cycle (Fig. 2). Unexpectedly, removal of 423 aa from the N terminus of VP80 (full-length 691 aa) still resulted in production of infectious BV virions, although this mutant showed strongly delayed kinetics of virus spread in cell culture (Fig. 2 and 3). The results thus provide information that the paramyosin-like motif identified in AcMNPV VP80 (aa 164 to 368) (22) is not strictly required, but its presence significantly contributes to productive infection. These findings suggest that a common ancestor of the currently known VP80 proteins executed its original, essential function via a domain corresponding to the C-terminal regions of present VP80 proteins. On the other hand, the N-terminal domain of the VP80 proteins could strongly diverge during evolution, probably specializing for different hosts, hand-in-hand with the speciation process of the order Lepidoptera. Host specificity of VP80 can be perfectly documented by unsuccessful attempts to functionally replace BmNPV vp80 gene with its AcMNPV homologue despite their sequence identity of over 96% (34). Importantly, the majority of sequence differences is localized in the variable N-terminal regions of both VP80 protein, strongly supporting our evolutionary hypothesis.
Sequence analysis showed that all VP80 proteins contain a strongly basic tract in the essential C-terminal region (Fig. 1B), suggesting a possible interaction between VP80 and DNA, as was also concluded from previous localization studies (22). The fundamental aspects of this interaction are unclear but point toward the involvement of VP80 in DNA-based operations, such as viral transcription, viral genome replication, or eventually encapsidation into progeny nucleocapsids. Using the vp80-null AcMNPV genome, we subsequently showed that VP80 is not required for viral genome replication (Fig. 4A). Immunolabeling of VP80 in baculovirus-infected cells showed an association of VP80 with the virogenic stroma, where VP80 was preferentially found in close proximity to progeny nucleocapsids (Fig. 4B). Hence, these observations rather demonstrate that VP80 is involved in nucleocapsid maturation (e.g., in the DNA encapsidation process), fully in accordance with previous findings, where we recorded a high number of aberrantly filled progeny nucleocapsids in the absence of VP80 (23). Similar functional involvements were also concluded for the BmNPV homologue (34).
An unexpected finding was obtained using the entire basic region of AcMNPV VP80 protein sequence (aa 459 to 518) as a query. These searches found a putative bHLH domain, a DNA-binding module typical for eukaryotic transcription factors (Fig. 5). Subsequent homology modeling recognized several highly conserved residues within this domain, such as a conserved glutamic acid (E) at position 9 (Fig. 5A). This conserved glutamic acid is a typical feature for E-box-binding bHLH proteins. However, the 3D modeling indicated that VP80 constitutes a novel type of bHLH domain, because its proximal part is formed by a number of negatively charged residues instead of basic residues, as is characteristic for human MyoD or mouse E47/NeuroD transcription factors (Fig. 6B). The biological implications of these structural differences between VP80-bHLH and other currently known bHLH proteins remain unclear. Nevertheless, our in vitro assays clearly demonstrated that the AcMNPV VP80 protein forms homodimers and that VP80 binds DNA (Fig. 7). Both of these attributes are typical for functional bHLH proteins (13). Since there are more than 200 copies of E-box-like sequence motifs homogeneously interspersed over the AcMNPV genome (data not shown), we were interested in the DNA-binding specificity of the VP80 protein. Our comparative DNA retardation assay showed that VP80 efficiently binds DNA in an E-box-independent manner, at least under in vitro conditions. Recently, it was shown for some other bHLH proteins, for instance, Drosophila melanogaster HLH-27 and HLH-29, that they recognize different, E-box-unrelated DNA sequence motifs (4).
Taken together, we demonstrated that the AcMNPV VP80, the fourth F-actin-interacting capsid protein (22), contains a C-terminal domain competent to bind DNA and sharing characteristics with bHLH domains. There are two explanations why VP80 may have a bHLH domain, a feature typical for transcription factors. First, VP80 may somehow be involved in viral transcription. Since vp80 is a viral late gene (18), it is theoretically possible that VP80 is part of a complex machinery that either shuts off early transcription (negative regulations) or starts to express late and very late genes (positive regulations). Both regulations are probably tightly regulated by levels of G- and F-actins (see reference 38) for a review). It is important to note that in the absence of VP80, the virus's very late gene expression still occurs at a level similar to that of the wild type (23). The second scenario includes the fact that VP80 protein associates with nucleocapsids of both BV and ODV, typically with one of the ends (22). Hence, it is possible that VP80 is a part of the nucleocapsid portal, where its DNA-binding activity is essential for proper genome encapsidation. The latter scenario can explain why absence of VP80 resulted in a number of improperly DNA-filled progeny nucleocapsids (23).
Finally, taking into account that VP80 interacts probably via its N-terminal paramyosin-like domain with the virus-triggered nuclear F-actin scaffold, an essential component required for crucial steps in baculovirus morphogenesis, such as nucleocapsid assembly and maturation (28). It is quite possible that nucleocapsid building and nucleocapsid egress are coupled actions, where virus-encoded actin-interacting proteins (VP39, P78/83, BV/ODV-C42, and VP80) and the host actin-myosin complex play pivotal roles.
ACKNOWLEDGMENTS
We are grateful to Jan van Lent (Wageningen University) for help with the immunogold labeling and electron microscopy.
M.M. and O.-W.M. were financed by the project BACULOGENES of the European Union (contract FP6-037541). M.M.V.O. was sponsored partially by the Program Strategic Alliances of the Royal Dutch Academy of Sciences.
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Altschul S, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- 3. Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2011. GenBank. Nucleic Acids Res. 39: D32–D37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Masi F, et al. 2011. Using a structural and logics systems approach to infer bHLH-DNA binding specificity determinants. Nucleic Acids Res. 39: 4553–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dohner K, Sodeik B. 2004. The role of the cytoskeleton during viral infection, Curr. Top. Microbiol. Immunol. 285: 67–108 [DOI] [PubMed] [Google Scholar]
- 6. Dreschers S, Roncarati R, Knebel-Morsdorf D. 2001. Actin rearrangement-inducing factor of baculoviruses is tyrosine phosphorylated and colocalizes to F-actin at the plasma membrane. J. Virol. 75: 3771–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fiser A, Sali A. 2003. MODELLER: generation and refinement of homology-based protein structure models. Methods Enzymol. 374: 461–491 [DOI] [PubMed] [Google Scholar]
- 8. Goley ED, et al. 2006. Dynamic nuclear actin assembly by Arp2/3 complex and a baculovirus WASP-like protein. Science 314: 464–467 [DOI] [PubMed] [Google Scholar]
- 9. Reference deleted.
- 10. Lanier LM, Volkman LE. 1998. Actin binding and nucleation by Autographa californica M nucleopolyhedrovirus. Virology 243: 167–177 [DOI] [PubMed] [Google Scholar]
- 11. Larkin MA, et al. 2007. ClustalW and ClustalX version 2.0. Bioinformatics 23: 2947. [DOI] [PubMed] [Google Scholar]
- 12. Laskowski RA, Macarthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–291 [Google Scholar]
- 13. Ledent V, Vervoort M. 2001. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 11: 754–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Letunic I, Doerks T, Bork P. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37: D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li K, et al. 2010. The putative pocket protein binding site of Autographa californica nucleopolyhedrovirus BV/ODV-C42 is required for virus-induced nuclear actin polymerization. J. Virol. 84: 7857–7868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Long G, Pan X, Kormelink R, Vlak JM. 2006. Functional entry of baculovirus into insect and mammalian cells is dependent on clathrin-mediated endocytosis. J. Virol. 80: 8830–8833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Longo A, Guanga GP, Rose RB. 2007. Crystal structure of E47-NeuroD1/Beta2 bHLH domain-DNA complex: heterodimer selectivity and DNA recognition. Biochemistry 47: 218–229 [DOI] [PubMed] [Google Scholar]
- 18. Lu A, Carstens EB. 1992. Nucleotide sequence and transcriptional analysis of the p80 gene of Autographa californica nuclear polyhedrosis virus: a homologue of the Orgyia pseudotsugata nuclear polyhedrosis virus capsid-associated gene. Virology 190: 201–209 [DOI] [PubMed] [Google Scholar]
- 19. Luckow VA, Lee SC, Barry GF, Olins PO. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67: 4566–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lung O, Westenberg M, Vlak JM, Zuidema D, Blissard GW. 2002. Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J. Virol. 76: 5729–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. MA PCM, Rould MA, Weintraub H, Pabo CO. 1994. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell 77: 451–459 [DOI] [PubMed] [Google Scholar]
- 22. Marek M, Merten O-W, Galibert L, Vlak JM, van Oers MM. 2011. Baculovirus VP80 protein and the F-actin cytoskeleton interact and connect the viral replication factory with the nuclear periphery. J. Virol. 85: 5350–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marek M, van Oers MM, Francis-Devaraj F, Vlak JM, Merten O-W. 2011. Engineering of baculovirus vectors for the manufacture of virion-free biopharmaceuticals. Biotechnol. Bioeng. 108: 1056–1067 [DOI] [PubMed] [Google Scholar]
- 24. Melo F, Feytmans E. 1998. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 277: 1141–1152 [DOI] [PubMed] [Google Scholar]
- 25. Merten O-W, Marek M, van Oers MM. 2011. Baculovirus-based production of biopharmaceuticals free of contaminating baculoviral virions. European patent EP2292781. [Google Scholar]
- 26. Nicholas KB, Nicholas HB, Deerfield DW. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4: 14 [Google Scholar]
- 27. Ohkawa T, Rowe AR, Volkman LE. 2002. Identification of six Autographa californica multicapsid nucleopolyhedrovirus early genes that mediate nuclear localization of G-actin. J. Virol. 76: 12281–12289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ohkawa T, Volkman LE. 1999. Nuclear F-actin is required for AcMNPV nucleocapsid morphogenesis. Virology 264: 1–4 [DOI] [PubMed] [Google Scholar]
- 29. Ohkawa T, Volkman LE, Welch MD. 2010. Actin-based motility drives baculovirus transit to the nucleus and cell surface. J. Cell Biol. 190: 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pettersen EF, et al. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- 31. Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Slack J, Arif BM. 2006. The baculoviruses occlusion-derived virus: virion structure and function. Adv. Virus Res. 69: 99–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- 34. Tang X-D, et al. 2008. Characterization of a Bombyx mori nucleopolyhedrovirus with Bmvp80 disruption. Virus Res. 138: 81–88 [DOI] [PubMed] [Google Scholar]
- 35. van Lent JWM, et al. 1990. Localization of the 34-kDa polyhedron envelope protein in Spodoptera frugiperda cells infected with Autographa californica nuclear polyhedrosis virus. Arch. Virol. 111: 103–114 [DOI] [PubMed] [Google Scholar]
- 36. van Oers MM, Vlak JM. 2007. Baculovirus genomics. Curr. Drug Targets 8: 1051–1068 [DOI] [PubMed] [Google Scholar]
- 37. Vanarsdall AL, Okano K, Rohrmann GF. 2005. Characterization of the replication of a baculovirus mutant lacking the DNA polymerase gene. Virology 331: 175–180 [DOI] [PubMed] [Google Scholar]
- 38. Volkman LE. 2007. Baculovirus infectivity and the actin cytoskeleton. Curr. Drug Targets 8: 1075–1083 [DOI] [PubMed] [Google Scholar]
- 39. Wang Q, Liang C, Song J, Chen X. 2007. HA2 from the Helicoverpa armigera nucleopolyhedrovirus: a WASP-related protein that activates Arp2/3-induced actin filament formation. Virus Res. 127: 81–87 [DOI] [PubMed] [Google Scholar]
- 40. Wang Y, Chen K, Yao Q, Wang W, Zhu Z. 2007. The basic helix-loop-helix transcription factor family in Bombyx mori. Dev. Genes Evol. 217: 715–723 [DOI] [PubMed] [Google Scholar]
- 41. Ward BM. 2011. The taking of the cytoskeleton one two three: how viruses utilize the cytoskeleton during egress. Virology 411: 244–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]