Abstract
The broadly neutralizing monoclonal antibodies (MAbs) 4E10, 2F5, and Z13e1 target membrane-proximal external region (MPER) epitopes of HIV-1 gp41 in a manner that remains controversial. The requirements for initial lipid bilayer binding and/or CD4 ligation have been proposed. To further investigate these issues, we probed for binding of these MAbs to human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus (SIV) virions with protein A-conjugated gold (PAG) nanoparticles using negative-stain electron microscopy. We found moderate levels of PAG associated with unliganded HIV-1 and SIV virions incubated with the three MAbs. Significantly higher levels of PAG were associated with CD4-liganded HIV-1 (epitope-positive) but not SIV (epitope-negative) virions. A chimeric SIV virion displaying the HIV-1 4E10 epitope also showed significantly higher PAG association after CD4 ligation and incubation with 4E10. MAbs accumulated rapidly on CD4-liganded virions and slowly on unliganded virions, although both reached similar levels in time. Anti-MPER epitope-specific binding was stable to washout. Virions incubated with an irrelevant MAb or CD4-only (no MAb) showed negligible PAG association, as did a vesicle-rich fraction devoid of virions. Preincubation with Fab 4E10 inhibited both specific and nonspecific 4E10 IgG binding. Our data provide evidence for moderate association of anti-MPER MAbs to viral surfaces but not lipid vesicles, even in the absence of cognate epitopes. Significantly greater MAb interaction occurs in epitope-positive virions following long incubation or CD4 ligation. These findings are consistent with a two-stage binding model where these anti-MPER MAbs bind first to the viral lipid bilayer and then to the MPER epitopes following spontaneous or induced exposure.
INTRODUCTION
The typical human immunodeficiency virus (HIV) or simian immunodeficiency virus (SIV) virion has about 5 to 15 envelope spikes on its surface (15, 83, 84), each of which is a trimer with a gp41 transmembrane stalk and a gp120 head region (21, 63). Each gp41 protomer may be further subdivided into a fusion peptide, polar region, N-terminal heptad repeat, connecting loop region, C-terminal heptad repeat, membrane-proximal external region (MPER), transmembrane domain, and cytoplasmic tail (reviewed in reference 49).
The MPER is rarely targeted by effective neutralizing antibodies and might therefore seem to be an unattractive target for vaccine production (20, 26, 28, 72, 80, 81). However, because the region is highly conserved across clades and, though rare, some patient-derived antibodies with broadly neutralizing activity targeting this region have been described (7, 12, 26, 29, 43, 68, 74, 80), considerable effort has gone into attempts to characterize the few available MPER-specific monoclonal antibodies (MAbs) and into developing methods for enhancing the immunogenicity of this region for purposes of vaccine development (9, 49).
Of the three broadly neutralizing anti-MPER MAbs (4E10, 2F5, and Z13e1) (12, 53, 54, 86), 2F5 is the most potent and 4E10 has the broadest cross-clade neutralization capacity (8, 48). These MAbs are effective in protecting against infection in a SHIV model (33) and have shown considerable therapeutic potential (33, 45). Together, the 4E10 and 2F5 epitopes cover most of the MPER, with the 4E10 epitope occupying the C-terminal half and the 2F5 epitope occupying the N-terminal half. The Z13 epitope is situated in an intermediate position and partially overlaps the two neighboring epitopes (54, 60). Although the structure of the MPER in its native form in the context of the trimer has not been resolved, all three anti-MPER MAbs have been crystallized in complex with their cognate MPER peptides (11, 13, 37, 57). For 4E10 and 2F5, one face of the associated peptide is predominantly hydrophilic and the other is hydrophobic with the hydrophilic surface making contact with a grooved paratope on the cognate MAb. All three MAbs possess qualities suitable for interaction with membrane-associated epitopes. 4E10 and 2F5 MAbs share the unusual property of having long VH-H3 loops tipped with hydrophobic residues which likely play no role in direct peptide contact (2) but rather appear to interact with the viral membrane (49).
4E10, in particular, has affinity for lipids in addition to its peptide binding capacity (2, 4, 32, 66, 69, 75, 77). The affinity of 2F5 for lipids is controversial (38, 46, 66, 70, 77, 85). Both MAbs bind with higher affinities to their respective MPER peptides when presented in a lipid environment (2, 77). These observations have led to the suggestion that the 4E10 peptide region is closely associated with, and likely partially submerged within, the lipid bilayer (1, 10, 13, 30, 57, 66). Recent biophysical data (2, 23, 73) provide strong evidence that this is indeed the case and supports a model in which the 4E10 MAb is specially adapted to partially insert (via the hydrophobic tips) into the lipid bilayer as a prelude to successful engagement of the epitope within the recessed hydrophilic portion of its CDR (73). The 2F5 epitope, in contrast, may reside just above the plane of the membrane but close enough for the hydrophobic elements of the CDR to engage the membrane (2, 23). The Z13e1 MAb appears to exert its neutralizing effect by binding to and immobilizing a small hinge-like region in the MPER (73).
As originally described by Haynes et al., the hydrophobic nature of the 4E10 and 2F5 paratopes are sufficient to bind lipids directly, even in the absence of the MPER peptide, suggesting that these MAbs are polyreactive and perhaps function as, or derive from, autoantibodies (26, 27, 32, 52, 66). Each of these assertions has been questioned (46, 70, 75, 76). Nevertheless, a variety of evidence supports the lipid-binding nature of these MAbs (1, 46, 57, 75, 77, 78).
Whether the anti-MPER MAbs bind to their respective gp41 epitopes prior to CD4 ligation or require ligation for binding is also controversial (reviewed in reference 49). Various evidences have been interpreted to indicate that the MPER epitopes are exposed or formed in the CD4-liganded Env spike but are not present, or are only minimally present, on the unliganded spike, leaving a relatively narrow window of opportunity for MAb binding and neutralization (2, 6, 16, 24, 31, 82). A two-step model has been proposed in which 4E10 and 2F5 MAbs bind to, and diffuse within, the plane of the membrane prepositioning these MAbs for efficient interaction with the membrane-associated exposure/creation of the cognate epitopes upon CD4-induced Env spike triggering (1, 2). In contrast, a recent study has provided evidence that both 4E10 and 2F5 neutralize HIV-1 and induce gp120 shedding in the absence of CD4 ligation, but only after prolonged incubation (64). Still others find minimal recognition by these MAbs before or after CD4 ligation compared to nonneutralizing anti-MPER antibodies (14).
We have addressed these and related issues by assaying both virions and vesicles incubated with anti-MPER MAbs under a variety of conditions using negative stain electron microscopy (EM) coupled with protein-A gold (PAG) probes. The results show that anti-MPER MAbs moderately associate with virions, including those devoid of MPER epitopes, and that this interaction is strong enough to resist washout. MPER epitope-bearing virions liganded with CD4 show a much higher association of anti-MPER antibodies compared to the unliganded virions. These results suggest that CD4 liganding enhances the exposure of the MPER epitopes but also are consistent with a lipid-binding capacity.
MATERIALS AND METHODS
Reagents.
The following reagents were obtained through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Disease: MAbs 4E10 (catalog no. 10091) from Hermann Katinger, 2F5 (catalog no. 1475) from Hermann Katinger, Z13e1 (catalog no. 11557) from Michael Zwick, and 2G12 (catalog no. 1476) from Hermann Katinger and soluble 4-domain CD4. The AIDS Vaccine Program (SAIC Frederick, National Cancer Institute [NCI], Frederick, MD) supplied the following highly purified aldrithiol-2-treated viruses: HIV-1 (mn) CL.4/SUPT1, lot P3806 (HIV-1 MN); HIV-1 BaL/SUPT1, lot P3955 (HIV-1 BaL); SIVmac239/CEMX 174(T1), lot P3660 (SIVmac239); SIV 239/251 TAIL/SUPT1-CCR5 CL.30, lot P3978 (SIV short-tailed); SIV*4E10/SUPT1-CCR5 CL.30, lot P4131 (4E10 epitope-grafted SIV or SIV-4E10); HIV-1SF162/SupT1-R5, lot P3916 (HIV-1 SF162); and SUPT1-CCR5 CL.30 microvesicles, lot P4138 (microvesicles). The data on the estimated number of Env spikes per virion (see Table S1 in the supplemental material) were provided by Elena Chertova, AIDS and Cancer Virus Program, NCI-Frederick, using previously described methods (15). Genetically engineered 4E10 Fab (40) was generously provided by Pamela Bjorkman, and MAb 2909 was generously provided by Susan Zolla-Pazner. Uranyl formate was obtained from SPI Supplies (West Chester, PA). Protein A-conjugated 5-nm gold nanoparticles (PAG) were purchased from the Cell Microscopy Center (University Medical Center, Utrecht, Netherlands). Trehalose was obtained from Sigma (St. Louis, MO). Thin-bar hexagonal 600-mesh copper grids were purchased from Electron Microscopy Sciences (Hatfield, PA).
MAb binding and gold labeling assay.
Original virus stocks (1.5 to 2.5 mg/ml protein) were 1:1 diluted with phosphate-buffered saline (PBS). Portions (10 μl) of diluted virions were incubated with 1.0 μl of CD4 (1.0 mg/ml stock) or 1.0 μl of PBS (control) at 37°C for 15 min. The molar ratio of soluble CD4 to the gp120 monomer was about 3:1. After incubation, CD4-liganded and -unliganded viruses were incubated with 2 μl of MAb (1 mg/ml stock) at 37°C for additional 30 min. The MAb/monomeric gp120 molar ratio was 15:1 to 30:1. After incubation, excess MAbs and CD4 were removed by the addition of 127 μl of PBS, pelleting using a Beckman Airfuge (A-100/18 rotor; catalog no. 347593) at ∼140,000 × g for 10 min, and resuspended in 10 μl of PBS on ice. To gold label the bound MAb, 1.0 μl of PAG (freshly prepared 1:1 dilution of the original PAG stock with PBS) was added to the virus suspension, followed by incubation with rotation at room temperature for 30 min. After incubation, the virions were fixed by adding an equal volume (11 μl) 5% glutaraldehyde, incubation at 4°C for 30 min, and washing with 118 μl of 20% BSB (20 mM H3BO3, 5 mM Na2B4O7·10H2O, 15 mM NaCl buffer [pH 8.2]) at ∼140,000 × g for 10 min prior to preparation of the EM-negative staining grids.
Fab inhibition assay.
Portions (10 μl) of HIV-1 or SIV-4E10, diluted 1:1 in PBS, to which 1.0 μl of PBS (control) or 1.0 μl of CD4 (1.0 mg/ml) in PBS were added, were incubated at 37°C for 15 min. Then, 6 μl of Fab 4E10 (1.0 mg/ml in PBS) or PBS (for control) was then added, followed by incubation at 37°C for an additional 30 min. The Fab 4E10/monomeric gp41 molar ratio was 40:1 to 60:1. Immediately after the Fab incubation, the virus mixtures were incubated with 2 μl of MAb 4E10 (1.0 mg/ml stock) at 37°C for 30 min. Excess CD4, Fab 4E10, and MAb 4E10 was removed by dilution with 121 μl of PBS, pelleting at ∼140,000 × g for 10 min, and resuspension in 10 μl of PBS on ice. After washing, the bound MAb was labeled with PAG as described above.
Washout assay.
Portions (10 μl) of HIV-1, SIV short-tailed, SIV-4E10, or microvesicles (1:1 diluted original virus stock with PBS) were incubated with 2 μl of anti-MPER MAb (1.0 mg/ml stock) at 37°C for 30 min. The molar ratio of MAbs to gp41 monomer was 15:1 to 30:1. Excess MAb was washed out as described above, and the virions/microvesicles were resuspended in 10 μl of PBS on ice. Virions and/or microvesicles with bound MAbs were additionally incubated at 37°C for 30 min and then washed again as described above with PBS prior to labeling with PAG.
Negative staining and EM.
Gold-labeled, glutaraldehyde-fixed, and washed virions were resuspended in 9 μl of BSB previously diluted 1:5 with H2O. Virions were affixed to thin carbon membranes supported by thin bar hexagonal 600-mesh copper grids and stained with 1% uranyl formate containing 2% ethanol and 0.5% trehalose as follows: 4-μl virion samples were applied to the carbon substrate, incubated at room temperature for 60 s, and blotted from the bottom with filter paper. Immediately after the blotting, 4 μl of stain solution was applied, incubated for 30 to 60 s, and removed by blotting. Air-dried grids were imaged at 100 keV in a JEOL 1200EX or FEI CM120 transmission electron microscope at 30K or 50K magnification.
Scoring and statistical analysis.
PAG particles bound to the surface of the virions were counted visually from the EM negatives. Gold particles within close proximity (∼20 nm) to the virions were considered as associated. For each virion/reagent combination, two to four independent experiments were performed. Statistical calculations were made using Origin 7.5 software package (OriginLab, Northampton, MA).
RESULTS
Negative-stain EM analysis of MAb association with the viruses.
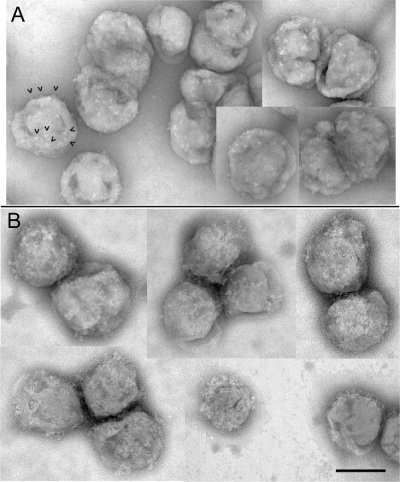
An initial attempt to visualize the binding of an anti-MPER MAb to epitope-bearing virions was conducted with 4E10 in complex with a chimeric form of SIV displaying the HIV-1 4E10 epitope (SIV-4E10). This target construct was chosen because it expresses higher levels of Env than wild-type HIV and has been shown to be readily neutralizable by 4E10 (80). Figure 1A shows a collage of virions displaying well-delineated Env spikes. When reacted with MAb 4E10, considerable additional surface features are evident as a reticular meshwork over much of the surface (Fig. 1B). However, the level of detail was insufficient to distinguish between spike elements and MAbs on the crowded viral surface, precluding further analysis by this method. Attempts to analyze 4E10-liganded virions by cryo-EM were similarly inconclusive (data not shown).
Fig 1.
Negative-stain EM montage of SIV-4E10. (A) Virions showing Env spikes (arrowheads show some examples). (B) Virions incubated with MAb 4E10 showing multiple complexes (protein meshwork). Bar, 100 nm.
As an alternative approach, the association of broadly neutralizing MAbs with individual virions was measured by a gold-labeling assay in which the presence of virion-associated MAbs was detected and quantified by counting the number of protein A-labeled gold particles (PAG) associated with each virion as visualized by negative-staining EM. Virions or control vesicles were first incubated with MAb, washed, and then incubated with 5-nm PAG. In some cases, CD4 was also added and incubated with the virions prior to PAG labeling. The conjugated virions were then fixed with glutaraldehyde to stabilize the structures and allowed to adhere to carbon-coated EM grids, followed by negative staining and TEM analysis.
Negative controls show minimal background PAG binding.
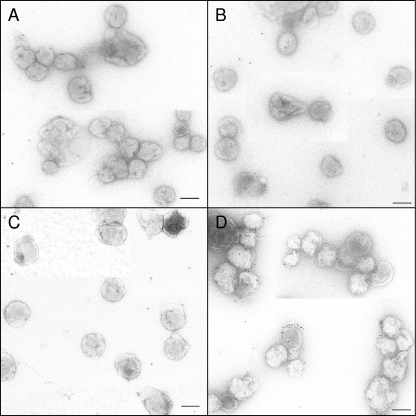
HIV-1 MN virions were initially assayed. Negative controls include HIV-1 MN reacted with no antibody, an irrelevant anti-Env antibody (antibody 2909), or CD4. MAb 2909 recognizes a strain (SF162)-specific V2/V3 conformational epitope not expressed on HIV-1 MN (25, 34). For these controls, less than 1 PAG/virion (PAG/v), on average, was detected (Table 1). A short-tailed (high Env-expressing) mutant version of SIV also tested negative for reactivity when incubated with CD4 or MAb 2909, as did a second mutant form of SIV expressing a grafted 4E10 epitope in the MPER (SIV-4E10) (Table 1). Figure 2 shows collages of HIV-1 MN virions reacted with PAG alone (Fig. 2A), CD4 + PAG (Fig. 2B), and irrelevant MAb 2909 + PAG (Fig. 2C). Only occasional scattered gold particles are observed.
Table 1.
Statistical data from PAG binding control assays
| Reactant(s) | PAG/virion |
No. of viruses assayed | ||||
|---|---|---|---|---|---|---|
| Mean | SD | SE | Range | Median | ||
| HIV-1 MN | 0.20 | 0.52 | 0.03 | 0–3 | 0 | 373 |
| HIV-1 MN+ CD4 | 0.25 | 0.55 | 0.04 | 0–3 | 0 | 187 |
| HIV-1 MN + MAb 2909 | 0.62 | 1.00 | 0.09 | 0–5 | 0 | 121 |
| HIV-1 MN + MAb 2G12 | 7.81 | 6.68 | 0.50 | 0–34 | 6 | 176 |
| HIV-1 SF162 + MAb 2909 | 4.28 | 4.17 | 0.44 | 0–18 | 3 | 92 |
| SIV short-tailed + CD4 | 0.22 | 0.54 | 0.04 | 0–3 | 0 | 156 |
| SIV short-tailed + MAb 2909 | 0.36 | 0.69 | 0.08 | 0–3 | 0 | 75 |
| SIV-4E10 + CD4 | 0.57 | 1.07 | 0.10 | 0–3 | 0 | 115 |
| SIV-4E10 + MAb 2909 | 0.55 | 1.08 | 0.10 | 0–2 | 0 | 110 |
Fig 2.
EM images of HIV-1 MN samples. (A) HIV-1 MN alone incubated with PAG (negative control). (B) HIV-1 MN liganded with soluble CD4 and then incubated with PAG (negative control). (C) HIV-1 MN treated with irrelevant MAb 2909 and incubated with PAG (negative control). (D) HIV-1 MN treated with MAb 2G12 (anti-gp120) and labeled with PAG (positive control). Scale bar, 100 nm.
Positive controls show PAG binding to virions.
When reacted with HIV-1 SF162 (3 to 6 spikes/virion, see Table S1 in the supplemental material), 2909 + PAG averaged 4.3 PAG/v (Table 1). Figure 2D shows another positive control for binding consisting of HIV-1 MN (8 to 10 spikes/virion) incubated with the anti-gp120 MAb 2G12, an MAb which binds to high mannose glycan epitopes on HIV-1 gp120 (67). Note the numerous gold particles associated with some of the virions (7.8 PAG/v, Table 1).
In summary, a combination of gold labeling and negative-stain EM techniques appears to be an effective tool for the analysis of MAb association with these viruses yielding, in these examples, ∼1 PAG/Env spike.
Binding of MPER-specific MAbs to virions with or without CD4 liganding.
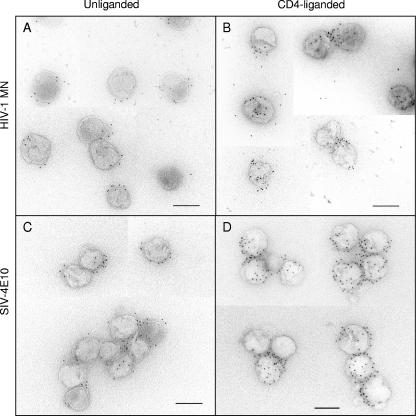
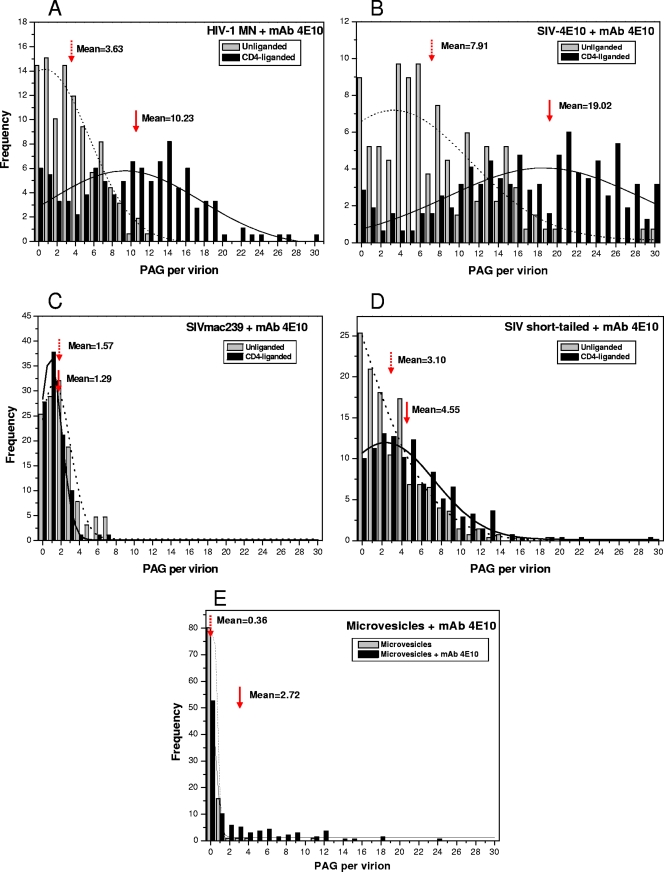
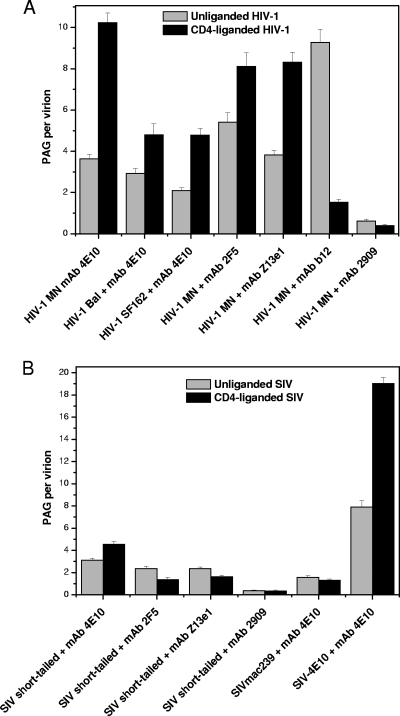
Whether anti-MPER antibodies bind to unliganded virions or require CD4 liganding for epitope expression is controversial (49). Our initial focus was on MAb 4E10. The virions used in the present study have been previously shown not to shed gp120 upon CD4 binding or prolonged incubation at 37°C (15). Figure 3A and B show EM collages of 4E10 reacted with unliganded and CD4-liganded HIV-1 MN virions, respectively. We observed a moderate, though well above background, level of PAG associated with the unliganded virions and a significantly greater level of binding to the CD4-liganded virions. A direct count shows 3.6 PAG/v for the unliganded form and 10.2 PAG/v for the liganded form (Fig. 4A; see Table S2 in the supplemental material for statistical data on all binding assays). A frequency plot reveals a wide range of PAG binding values, especially for the CD4-liganded virions (Fig. 4A). 4E10 was also reacted with a mutant form of SIV in which the 4E10 epitope was orthotopically grafted into the MPER (SIV-4E10) (80). This mutant expresses a higher level of Env (34 to 58 Env spikes/virion; see Table S1 in the supplemental material) than its wild-type counterpart, SIVmac239 (7 to 14 Env spikes per virion). Interestingly, the PAG association values for SIV-4E10 were about twice those observed for HIV-1 MN for both unliganded (7.9 PAG/v) and liganded (19 PAG/v) virions (Fig. 4B), likely reflecting the greater number of epitope-bearing spikes on SIV-4E10.
Fig 3.
EM images of MAb 4E10 and PAG association with the unliganded HIV-1 MN (A), CD4-liganded HIV-1 MN (B), unliganded SIV-4E10 (C), and CD4-liganded SIV-4E10 (D). Scale bar, 100 nm.
Fig 4.
PAG frequency analysis for MAb 4E10 bound to unliganded and CD4-preliganded virions and to microvesicles. The arrows indicate mean PAG/virion values. Gaussian curve fitting is used to represent the PAG distribution. The dotted and solid lines represent the unliganded and CD4-preliganded virions, respectively.
It would seem that although CD4 liganding clearly enhances binding, in its absence, a significant level of 4E10 binding occurs, and this binding may be influenced by the amount of Env present, possibly by direct association with the virus lipid bilayer. To explore this issue further, we incubated 4E10 and PAG with wild-type SIV (low number of spikes, lacking the 4E10 epitope), as well as the SIV short-tail virions (i.e., high number of spikes, lacking the 4E10 epitope), the parent strain from which SIV-4E10 was derived. The high-spike SIV short-tail virions bound more PAG (3.1 PAG/v) than wild-type SIVmac239 (1.6 PAG/v) (Fig. 4D and C, respectively). Also, CD4 liganding did not appreciably affect PAG binding in 4E10 epitope-negative virions.
In comparing the distribution patterns among the four viruses, the following patterns emerge: (i) in the absence of CD4 liganding, the presence of the 4E10 epitope enhances 4E10 binding (compare Fig. 4A and B to Fig. 4C and D); (ii) virions with greater numbers of spikes bind more 4E10 even in the absence of the 4E10 epitope (compare Fig. 4B and D to Fig. 4A and C); and (iii) CD4 liganding greatly enhances 4E10 binding on epitope-positive virions but not on 4E10 epitope-negative virions (compare Fig. 4A and B to Fig. 4C and D).
To determine whether 4E10 binds to lipid bilayers in general or has a preference for virion membranes, we probed a microvesicle-rich fraction obtained from noninfected cell of the same line that was used to generate the virions. Although some vesicles displayed associated PAG, the vast majority did not, indicating that a lipid bilayer, per se, is not sufficient for appreciable 4E10 interaction. This assay also serves as an important control since most virus preparations contain a certain level of contaminating microvesicles that copurify with the virion-rich fraction.
HIV-1 MN was also tested for reactivity with anti-MPER MAbs 2F5 and Z13e1, without and with CD4 liganding (Fig. 5A and see Fig. S1A and B in the supplemental material). As with 4E10, both MAbs bound to unliganded viruses (ca. 4 to 5 PAG/v) and PAG binding increased when CD4 liganded (8 PAG/v). The irrelevant anti-gp120 MAb, 2909, did not appreciably bind to HIV-1 MN (Table 1, Fig. 5A, and see Fig. S1C in the supplemental material) in the absence or presence of CD4 preliganding (Fig. 5A and see Fig. S1C in the supplemental material).
Fig 5.
Mean PAG binding for MAbs 4E10, 2F5, Z13e1, and 2909 bound to the unliganded and CD4-liganded HIV-1 (A) and SIV (B) virions.
HIV-1 MN virions reacted with MAb b12 were used as a control to assess the maximum PAG binding for this HIV-1 strain in the absence of the sort of steric restrictions that likely affect MPER-targeting MAbs (Fig. 5A and see Fig. S1D in the supplemental material). MAb b12 is a broadly neutralizing antibody that binds to and blocks the CD4 binding site on gp120 and has been shown by cryo-EM tomography and negative-stain EM to project radially from the Env spike head well above the membrane (39, 44). In this position, steric interference from neighboring gp120 elements or the viral membrane, as could be the case for the anti-MPER MAbs, are highly unlikely. An average of 9.3 PAG/v was detected for the b12-treated HIV-1, which is comparable to the levels of 4E10-, 2F5-, and Z13e1-mediated PAG binding to CD4-liganded HIV-1 (Fig. 5A). These data indicate that once triggered by CD4, the cognate MPER epitopes are readily accessible. When HIV-1 is preliganded with CD4, b12 binding to the virus is reduced to 1.5 PAG/v (Fig. 5A), demonstrating the efficiency of CD4 binding in our assays.
The HIV-1 BaL and HIV-1 SF162 isolates were also tested for MAb 4E10 binding to unliganded and CD4 liganded virions (Fig. 5A and see Fig. S1E and F in the supplemental material). Unliganded HIV-1 BaL and HIV-1 SF162 showed 2 to 3 PAG/v, on average, whereas CD4 liganded virions showed ∼5 PAG/v. Like HIV-1 MN, these other tier 1 isolates showed increased PAG binding after CD4 liganding.
When the same set of anti-MPER MAbs was reacted with SIV virions lacking the three epitopes, all showed 2 to 3 PAG/v on average (versus <1 for anti-gp120 MAb 2909) (Fig. 5B and see Fig. S2 in the supplemental material). With the possible exception of 4E10 (which was unexpectedly high), CD4 ligation yielded comparably low levels of PAG binding.
Fab 4E10 can block both MPER epitope-specific and nonspecific IgG 4E10 interactions with the viruses.
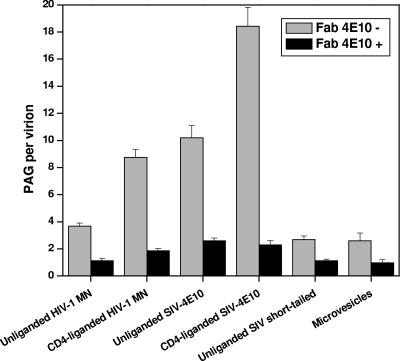
To determine whether Fc interactions or MAb bivalency plays an important role in 4E10 IgG binding, we preincubated virions with Fab 4E10 and assayed for blockage of both specific and nonspecific interactions (Fig. 6 and see Fig. S3 in the supplemental material; see Table S3 in the supplemental material for statistical data on all Fab inhibition assays). HIV-1 unliganded and CD4-liganded virions not treated with Fab 4E10 have an average of 3.7 and 8.7 PAG/v, respectively. When pretreated with Fab 4E10, PAG binding dropped to 1.1 and 1.9 PAG/v, respectively. Similarly, whereas 4E10-liganded SIV-4E10 virions showed 11.2 and 18.4 PAG/v in the absence and presence of CD4, respectively, preincubation with 4E10 Fab reduced binding to 1.4 and 3.6 PAG/v, respectively. When short-tailed SIV virions and microvesicles were reacted with MAb 4E10, a modest level of PAG binding was observed (∼2.6 PAG/v) as indicated above. Preincubation with 4E10 Fab reduced this level of MAb binding (∼1.0, PAG/v).
Fig 6.
Fab 4E10 inhibition assay. Fab 4E10 is used to block the binding of MAb 4E10 to virions and microvesicles.
Together, these data indicate that MAb 4E10 binds to virions both through specific epitope-mediated interactions and nonspecific presumptive membrane interactions and that both modes of binding are mediated by the Fab arms of 4E10.
The strength of the association of anti-MPER MAbs with the viral lipid membrane is substantial.
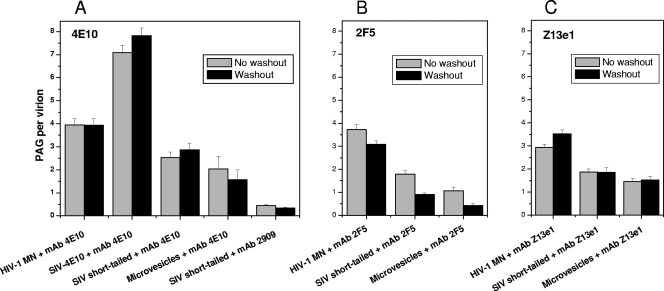
To gain a measure of the persistence of anti-MPER MAb association with the viral membrane, we performed a washout study that incorporated a second 30-min incubation at 37°C in the absence of MAb and a wash to remove any dissociated MAb prior to labeled with PAG. For HIV-1 MN, washout resulted in an average of 3.9 PAG/v, which was virtually the same as without washout (Fig. 7A and see Fig. S4A in the supplemental material; see Table S4 in the supplemental material for statistical data on all washout assays). Similarly, SIV-4E10, short-tailed SIV, and microvesicles showed no significant difference in 4E10 binding when comparing the before and after washing data (Fig. 7A and see Fig. S4B to D in the supplemental material). These results indicate a rather stable interaction between MAb 4E10 and membrane components. Similarly, Z13e1 appears stably associated with virions and microvesicles in the presence or absence of the cognate epitope (Fig. 7C and see Fig. S6 in the supplemental material). Only 2F5 showed a statistically significant loss of MAb/PAG binding following washout, although the drop-off in binding was small (Fig. 7B and see Fig. S5 in the supplemental material).
Fig 7.
MAb washout assay. Virions were incubated with anti-MPER MAbs, washed to remove unreacted MAbs, incubated an additional 30 min, rewashed, and labeled with PAG.
4E10 binding kinetics.
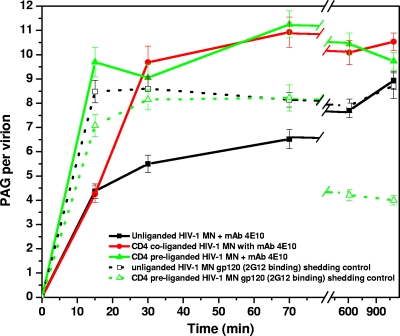
It has been proposed that 4E10 first binds to the lipid membrane and then to the gp41 fusion intermediate form (following CD4 ligation), suggesting a two-stage binding mechanism (2). To test this hypothesis, we assayed 4E10 binding to unliganded, CD4-preliganded, and CD4-coliganded (CD4 and MAb added together) HIV-1 at 15, 30, and 70 min and at 10 and 18 h of incubation (at 37°C) with MAb. As can be seen in Fig. 8, in the absence of CD4 about half of the maximum 4E10 binding occurs within the first 15 min. Over the next ∼18 h, additional 4E10 becomes associated at a decelerating rate (see Table S5 in the supplemental material). For HIV-1 liganded with CD4, as previously observed, considerably more PAG/v is detected. Here, too, most of the 4E10 association occurs within the first 15 min. Interestingly, under coliganding conditions where soluble CD4 and 4E10 are added together, there was a reduced level of PAG/v at 15 min of incubation, but by 30 min the amount of PAG bound equaled that of the preliganded samples. Thus, during coliganding conditions, 4E10 may initially bind to the viral membrane quicker than CD4 can convert Env into fully receptive 4E10 epitope-expressing spikes, a process which is, however, completed within 30 min. Note that the level of 4E10 binding to unliganded virions eventually (after 18 h) reaches a level approaching the maximum observed following CD4-induced binding, whereas the maximum for the CD4 bound virions is reached within minutes of ligation.
Fig 8.
Kinetics of MAb 4E10 and 2G12 binding to unliganded and CD4-liganded HIV-1 MN. MAb 4E10 or 2G12 were added to virions preassociated with CD4 (CD4-preliganded) or added simultaneously with CD4 (CD4-coliganded) and incubated for the indicated time prior to PAG labeling.
Spontaneous and CD4-induced gp120 shedding has been reported for a variety of HIV-1 isolates (15, 22, 50, 61, 64). To determine whether either form of shedding is occurring and, if so, correlates with the binding of 4E10 to the virions, we probed HIV-1 MN with the anti-gp120 MAb, 2G12, over time (18 h) in the absence and presence of CD4. The level of binding for the unliganded virions remained constant at 8 to 10 PAG/v indicating no spontaneous gp120 shedding. For the CD4 preliganded virions, the level of gp120 detected was comparable to that of the unliganded virions through 3 h (see Fig. S5 in the supplemental material) but drops by half by 10 h, suggesting that gp120 is shed only after extended incubation with CD4 for this isolate. Consequently, long-term stability of gp120 in unliganded virions and the long delay in gp120 shedding in CD4-liganded virions would have minimal influence in our findings.
DISCUSSION
The broadly neutralizing MAbs 4E10, 2F5, and Z13e1 target the MPER. Because the MPER is highly conserved and has been demonstrated to be the target of these MAbs and by serum antibodies in some patients, including those with broadly neutralizing antibody activity, extensive efforts have been directed toward determining the nature of both the antibodies and the targeted structural element of this region (reviewed in reference 49).
Despite extensive analyses of the MPER peptides (5, 36, 47, 49, 79), the in vivo/in situ structure of this region of the Env spike is unknown for any stage of the prefusion/fusion sequence. The hydrophobic nature of the MPER suggests a significant association with the lipid bilayer (30, 42, 57, 77). The presence of extended hydrophobic V loops on 4E10 and 2F5 suggests a mechanism for binding wherein the tips of the MAbs can penetrate the lipid bilayer to facilitate engagement with their cognate epitopes (13, 38, 57, 69). This exceptional HV loop hydrophobicity has been used to explain the intriguing though still controversial observation that 4E10 and 2F5 have demonstrable affinity for peptide-free lipid bilayers in addition to their peptide epitope affinities (18). It has also been suggested that the polyreactivity of such antibodies is broader than simple lipid affinity (1, 52). Also controversial is whether CD4-Env liganding is a prerequisite for anti-MPER MAb binding and neutralization (2, 6, 8, 14, 19, 24, 64). In an effort to further understand the nature of the interaction between the anti-MPER MAbs and the virus, we analyzed intact virions in complexed with 4E10, 2F5, and Z13e1 MAbs by EM.
Using negative-stain EM, clear evidence of considerable 4E10 deposition on the virion surfaces was observed; however, the resultant high density of surface protein prevented clear distinction between spike structures and virion-associated MAb (Fig. 1). As an alternative, we developed a negative-stain EM assay in which protein A-gold (PAG) was used to provide a quantifiable visual indication of the relative amounts of anti-MPER MAbs binding to HIV-1 and related virions. A variety of negative controls demonstrated that less than one PAG particle bound, on average, to antibody-free virions or to virions incubated with CD4 or an irrelevant MAb. As a positive control, HIV-1 MN virions were shown to react with the well-characterized anti-gp120 MAbs 2G12 and b12 to yield an average of ca. 8 to 10 PAG/v, which is about one PAG/Env spike (Table 1 and see Table S1 in the supplemental material). The PAG distribution range for these and other spike targeted MAbs is rather broad and consistent with the wide range of Env spike expression on HIV-1 directly counted in our previous cryo-EM study (84).
A comparison of the binding of 4E10, 2F5, and Z13e1 to un(CD4)liganded HIV-1 MN virions showed ca. 3.5 to 4 PAG/v for 4E10 and Z13e1 and 5.5 PAG/v for 2F5 (Fig. 5A). Prior ligation with CD4 markedly enhanced PAG binding for all three MAbs. Similar, although less-pronounced, patterns of binding with 4E10 were observed with two other isolates, HIV-1 BaL and SF162 (Fig. 5A), which have previously been shown to be less susceptible to neutralization by 4E10 than MN (9). These data support previous indications that CD4 ligation is not strictly required for anti-MPER MAb binding (8, 14, 16, 19, 64) but also lend support to the hypothesis that triggering of the spikes by CD4 ligation enhances MPER epitope expression (24, 59). For example, Peachman et al. (59) used a virus capture enzyme-linked immunosorbent assay (ELISA) and surface plasmon resonance to show that 2F5 and 4E10 bind virions before and after CD4 ligation. In their study, for many but not all virus strains tested, binding was stronger after CD4 engagement. For some strains, CD4 ligation was a prerequisite for any anti-MPER MAb binding. These data may explain some of the apparent discrepancies in the literature on this point.
Part of the controversy surrounding the role of CD4 binding and spike triggering in the expression of the MPER epitopes relates to possible artifacts characteristic of the assays used to measure binding. ELISA capture assays, which are often used, have been interpreted as indicating significant anti-MPER binding to unliganded virions (39, 55, 56). However, at high density, anti-MPER antibodies can capture virions even when devoid of Env. At a low antibody density, virions may not be captured even when Env was present (41). The likely mode of binding is through low affinity-high avidity hydrophobic (anti-membrane) forces associated with the solid-phase high-density MAb arrays used in such assays (62). However, Leaman et al. found that if the virions were first incubated with soluble anti-MPER MAbs, Env-positive, but not Env-negative, virions could be readily captured by anti-isotype second antibody on the plate solid phase (41). Because PAG was reacted with virions only after excess MAb was washed out in our assays, PAG detects only MAbs that bound to the virions directly from the solution phase, thus avoiding the issue of artificially high-avidity interactions associated with solid-phase detection.
An important issue is the nature of the binding of anti-MPER MAbs to epitope-negative virions. Some previous studies have failed to detect such interactions or at best showed a weak interaction with viral membranes (35, 41, 49). Others have detected autoimmune-like binding to lipids and lipid membranes (2, 10, 18, 23, 65, 71). To test for direct membrane binding, we reacted our anti-MPER MAbs with virions expressing Env spikes devoid of the cognate epitopes (short-tailed SIV) and found low but significant PAG binding (ca. 2.4 to 2.8 PAG/v) for each of the three anti-MPER MAbs (Fig. 5B). In contrast, the reaction of virions with the irrelevant MAb, 2909, yielded an average of just 0.4 PAG/v (Fig. 5B, Table 1).
The stability of the anti-MPER MAb binding was assessed using a washout assay. The additional incubation and wash step failed to reduce the level of PAG binding for any of the three anti-MPER MAbs to epitope-positive or epitope-negative virions or to microvesicles (see Fig. S4, S5, and S6 in the supplemental material). The anti-MPER MAbs therefore appear to be firmly associated with the virions even in the absence of cognate peptide epitopes (e.g., with SIV short-tailed virions), at least over the timescale tested. These observations support those of Franquelim et al. in which atomic force microscopy was used to reveal that 2F5 and 4E10 bind to, and form aggregates on, artificial lipid bilayers that mimic the composition of HIV-1 membranes (23). In contrast, others have found that 4E10 has considerably higher affinity for lipids than does 2F5 (32, 66, 77).
To further investigate the nature of the association between the anti-MPER MAbs and the virions, especially in instances where either the peptide epitope was not present (e.g., SIV short-tailed, microvesicles) or it is not optimally expressed, e.g., in un(CD4)liganded epitope-positive virions, we performed a series of inhibition assays using Fab 4E10 as the inhibitor and intact MAb 4E10 as the probe. The resultant data demonstrate that 4E10 epitope-specific and nonspecific binding is saturable and that 4E10 Fab arm binding is not strictly dependent upon MAb bivalency or the presence of Fc.
Another pattern that has emerged from the data is the apparent relationship between the number of epitope-positive Env and the level of 4E10 binding in the absence of CD4 triggering (Fig. 4A and B). One possible explanation is that even in the absence of CD4 ligation, there is some 4E10 epitope expression (8). In support of this interpretation is the recent observations of Ruprecht et al. (64) that 4E10 (and 2F5) can neutralize un(CD4)liganded HIV-1 virions and concomitantly induce the shedding of gp120, although the process proceeded slowly over many hours. We suggest that the binding of 4E10 to the virions in our short-term (20- to 60-min) assays demonstrates interaction with what may be transiently accessible 4E10 epitopes and that this binding serves as a prelude to the ultimate spike functional disruption and gp120 release associated with virus neutralization as described by Ruprecht et al.
With regard to the enhanced binding of 4E10 to SIV-4E10, compared to HIV-1 MN, we cannot rule out a completely lipophilic non-epitope-specific model of 4E10 binding in the absence of CD4 triggering. For example, the lipid composition of the HIV-1 MN and SIV-4E10 virions may differ as an indirect effect of the latter having acquired a greater number of Env spikes or through differences in the structure and composition of the gp41 lipid binding/recruitment elements between the two virion forms (3, 78). However, the observation that unliganded short-tailed SIV, which also expresses high numbers of Env spikes, binds considerably less 4E10/PAG than the unliganded SIV-4E10 construct argues against lipid-mediated binding as the prime mechanism for 4E10 association in epitope-positive virions.
An alternative and much more rapid route to neutralization by these anti-MPER MAbs enlists CD4 to induce the conformational changes necessary to expose the MPER epitopes. Haynes and associates, as well as others, have proposed a two-stage binding model in which anti-MPER MAbs initially bind to and diffuse within the plane of the membrane and subsequently bind to the MPER epitopes when exposed (2, 18, 71).
The emerging picture of anti-MPER binding and neutralization is that there are two distinct pathways to viral neutralization, one CD4 dependent and the other CD4 independent. In the CD4-dependent pathway, the MPER epitopes are rapidly exposed as a consequence of the structural rearrangements associated with spike triggering. Anti-MPER antibodies previously associated with the membrane would then be in a position to latch on to their respective epitopes. This mode would not preclude direct binding of the epitopes from the fluid phase but, under steady-state in vivo conditions, the viral membranes would already be preloaded with antibodies making this the primary source for MPER epitope engagement. In the CD4-independent pathway, the data indicate that there is a moderate level of epitope expression likely preexisting in unliganded spikes since we see ca. 50% of maximum (i.e., when CD4-liganded) binding after a 30-min incubation with anti-MPER MAbs. This is not simply lipid bilayer binding since the level of binding to virions is significantly greater in the presence of the cognate epitope. The level of binding then rises over the course of hours, eventually approaching the maximum level observed for CD4-liganded virions. Since Ruprecht et al. (64) observe significant viral neutralization only after many hours of incubation with negligible neutralization following short incubations, simply binding to the MPER epitopes, per se, may not be sufficient to induce neutralization.
This comparison presupposes that our virions are behaving similarly to the pseudovirions used by Ruprecht et al. However, others have shown considerable isolate-specific variations in anti-MPER binding and neutralization (8, 58). One possible explanation for a disconnect between the hypothesized rapid binding of the MAbs to some MPER epitopes and the slow rate of neutralization is that the anti-MPER MAbs may target preexposed epitopes on defective viral spikes, spike fragments, or uncleaved gp160, i.e., forms that are not involved in virion-target cell fusion (14, 17, 51). Although we cannot completely rule out all of these modes of binding in our system, we can exclude binding to gp160, a molecular form that was not detected on our virions. An alternative explanation for the different time scales for binding versus neutralization is that epitope saturation, which may be a slow process, could be required for efficient neutralization. In any case, it would appear that CD4-independent neutralization depends on spontaneous low-frequency/intermittent MPER epitope exposure to produce the necessary degree of epitope saturation for neutralization.
Supplementary Material
ACKNOWLEDGMENTS
We thank Elena Chertova, Julian Bess, Jr., and James D. Roser of the AIDS and Cancer Virus Program, NCI-Frederick, for preparing and providing the viruses and virus compositional analyses.
This study was supported by grants from the National Institutes of Health (AI055461[K.H.R.] and AI057039 [W.E.J.]).
Footnotes
Published ahead of print 16 November 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Alam SM, et al. 2007. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 178: 4424–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alam SM, et al. 2009. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc. Natl. Acad. Sci. U. S. A. 106: 20234–20239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alfsen A, Bomsel M. 2002. HIV-1 gp41 envelope residues 650–685 exposed on native virus act as a lectin to bind epithelial cell galactosyl ceramide. J. Biol. Chem. 277: 25649–25659 [DOI] [PubMed] [Google Scholar]
- 4. Alving CR. 2008. 4E10 and 2F5 monoclonal antibodies: binding specificities to phospholipids, tolerance, and clinical safety issues. AIDS 22: 649–651 [DOI] [PubMed] [Google Scholar]
- 5. Apellániz B, Nir S, Nieva JL. 2009. Distinct mechanisms of lipid bilayer perturbation induced by peptides derived from the membrane-proximal external region of HIV-1 gp41. Biochemistry 48: 5320–5331 [DOI] [PubMed] [Google Scholar]
- 6. Binley JM, et al. 2003. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J. Virol. 77: 5678–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Binley JM, et al. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82: 11651–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Binley JM, et al. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78: 13232–13252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bomsel M, et al. 2011. Immunization with HIV-1 gp41 subunit virosomes induces mucosal antibodies protecting nonhuman primates against vaginal SHIV challenges. Immunity 34: 269–280 [DOI] [PubMed] [Google Scholar]
- 10. Brown BK, et al. 2007. Monoclonal antibodies to phosphatidylinositol phosphate neutralize human immunodeficiency virus type 1: role of phosphate-binding subsites. J. Virol. 81: 2087–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bryson S, Julien JP, Hynes RC, Pai EF. 2009. Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications. J. Virol. 83: 11862–11875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buchacher A, et al. 1994. Generation of human monoclonal antibodies against HIV-1 proteins: electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10: 359–369 [DOI] [PubMed] [Google Scholar]
- 13. Cardoso RM, et al. 2005. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22: 163–173 [DOI] [PubMed] [Google Scholar]
- 14. Chakrabarti BK, et al. 2011. HIV type 1 env precursor cleavage state affects recognition by both neutralizing and nonneutralizing gp41 antibodies. AIDS Res. Hum. Retrovir. 27: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chertova E, Jr, et al. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 76: 5315–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crooks ET, et al. 2005. Characterizing anti-HIV monoclonal antibodies and immune sera by defining the mechanism of neutralization. Hum. Antibodies 14: 101–113 [PMC free article] [PubMed] [Google Scholar]
- 17. Crooks ET, Tong T, Osawa K, Binley JM. 2011. Enzyme digests eliminate non-functional env from HIV-1 particle surfaces leaving native env trimers intact and viral infectivity unaffected. J. Virol. 85: 5825–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dennison SM, et al. 2009. Stable docking of neutralizing human immunodeficiency virus type 1 gp41 membrane-proximal external region monoclonal antibodies 2F5 and 4E10 is dependent on the membrane immersion depth of their epitope regions. J. Virol. 83: 10211–10223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Rosny E, Vassell R, Jiang S, Kunert R, Weiss CD. 2004. Binding of the 2F5 monoclonal antibody to native and fusion-intermediate forms of human immunodeficiency virus type 1 gp41: implications for fusion-inducing conformational changes. J. Virol. 78: 2627–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dhillon AK, et al. 2007. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J. Virol. 81: 6548–6562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eckert D, Kim P. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70: 777–810 [DOI] [PubMed] [Google Scholar]
- 22. Finnegan CM, Berg W, Lewis GK, DeVico AL. 2002. Antigenic properties of the human immunodeficiency virus transmembrane glycoprotein during cell-cell fusion. J. Virol. 76: 12123–12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Franquelim HG, et al. 2011. Anti-HIV-1 antibodies 2F5 and 4E10 interact differently with lipids to bind their epitopes. AIDS 25: 419–428 [DOI] [PubMed] [Google Scholar]
- 24. Frey G, et al. 2008. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc. Natl. Acad. Sci. U. S. A. 105: 3739–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gorny MK, et al. 2005. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J. Virol. 79: 5232–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gray ES, et al. 2009. Broad neutralization of human immunodeficiency virus type 1 mediated by plasma antibodies against the gp41 membrane proximal external region. J. Virol. 83: 11265–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gray ES, Meyers T, Gray G, Montefiori DC, Morris L. 2006. Insensitivity of paediatric HIV-1 subtype C viruses to broadly neutralizing monoclonal antibodies raised against subtype B. PLoS Med. 3: e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gray ES, et al. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 81: 6187–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gray ES, et al. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J. Virol. 83: 8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grundner C, Mirzabekov T, Sodroski J, Wyatt R. 2002. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins. J. Virol. 76: 3511–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gustchina E, Bewley CA, Clore GM. 2008. Sequestering of the prehairpin intermediate of gp41 by peptide N36Mut(e,g) potentiates the human immunodeficiency virus type 1 neutralizing activity of monoclonal antibodies directed against the N-terminal helical repeat of gp41. J. Virol. 82: 10032–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Haynes BF, et al. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308: 1906–1908 [DOI] [PubMed] [Google Scholar]
- 33. Hessell AJ, et al. 2010. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J. Virol. 84: 1302–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Honnen WJ, et al. 2007. Type-specific epitopes targeted by monoclonal antibodies with exceptionally potent neutralizing activities for selected strains of human immunodeficiency virus type 1 map to a common region of the V2 domain of gp120 and differ only at single positions from the clade B consensus sequence. J. Virol. 81: 1424–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huarte N, et al. 2008. The broadly neutralizing anti-human immunodeficiency virus type 1 4E10 monoclonal antibody is better adapted to membrane-bound epitope recognition and blocking than 2F5. J. Virol. 82: 8986–8996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ingale S, Gach JS, Zwick MB, Dawson PE. 2010. Synthesis and analysis of the membrane proximal external region epitopes of HIV-1. J. Pept. Sci. 16: 716–722 [DOI] [PubMed] [Google Scholar]
- 37. Julien JP, Bryson S, Nieva JL, Pai EF. 2008. Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site. J. Mol. Biol. 384: 377–392 [DOI] [PubMed] [Google Scholar]
- 38. Julien JP, et al. 2010. Ablation of the complementarity-determining region H3 apex of the anti-HIV-1 broadly neutralizing antibody 2F5 abrogates neutralizing capacity without affecting core epitope binding. J. Virol. 84: 4136–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kang YK, et al. 2009. Structural and immunogenicity studies of a cleaved, stabilized envelope trimer derived from subtype A HIV-1. Vaccine 27: 5120–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klein JS, et al. 2009. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc. Natl. Acad. Sci. U. S. A. 106: 7385–7390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leaman DP, Kinkead H, Zwick MB. 2010. In-solution virus capture assay helps deconstruct heterogeneous antibody recognition of human immunodeficiency virus type 1. J. Virol. 84: 3382–3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lenz O, et al. 2005. Trimeric membrane-anchored gp41 inhibits HIV membrane fusion. J. Biol. Chem. 280: 4095–4101 [DOI] [PubMed] [Google Scholar]
- 43. Li M, et al. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in southern africa. J. Virol. 80: 11776–11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Manrique A, et al. 2007. In vivo and in vitro escape from neutralizing antibodies 2G12, 2F5, and 4E10. J. Virol. 81: 8793–8808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matyas GR, Beck Z, Karasavvas N, Alving CR. 2009. Lipid binding properties of 4E10, 2F5, and WR304 monoclonal antibodies that neutralize HIV-1. Biochim. Biophys. Acta 1788: 660–665 [DOI] [PubMed] [Google Scholar]
- 47. Matyas GR, et al. 2009. Neutralizing antibodies induced by liposomal HIV-1 glycoprotein 41 peptide simultaneously bind to both the 2F5 or 4E10 epitope and lipid epitopes. AIDS. 23: 2069–2077 [DOI] [PubMed] [Google Scholar]
- 48. Mehandru S, et al. 2004. Neutralization profiles of newly transmitted human immunodeficiency virus type 1 by monoclonal antibodies 2G12, 2F5, and 4E10. J. Virol. 78: 14039–14042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Montero M, van Houten NE, Wang X, Scott JK. 2008. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol. Mol. Biol. Rev. 72: 54–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moore JP, McKeating JA, Weiss RA, Sattentau QJ. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250: 1139–1142 [DOI] [PubMed] [Google Scholar]
- 51. Moore PL, et al. 2006. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 80: 2515–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mouquet H, et al. 2010. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature 467: 591–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Muster T, et al. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67: 6642–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nelson JD, et al. 2007. An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human-immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10. J. Virol. 81: 4033–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nelson JD, et al. 2008. Antibody elicited against the gp41 N-heptad repeat (NHR) coiled-coil can neutralize HIV-1 with modest potency but non-neutralizing antibodies also bind to NHR mimetics. Virology 377: 170–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nyambi PN, et al. 2000. Immunoreactivity of intact virions of human immunodeficiency virus type 1 (HIV-1) reveals the existence of fewer HIV-1 immunotypes than genotypes. J. Virol. 74: 10670–10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ofek G, et al. 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78: 10724–10737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Peachman KK, et al. 2010. The importance of antibody isotype in HIV-1 virus capture assay and in TZM-bl neutralization. Viral Immunol. 23: 627–632 [DOI] [PubMed] [Google Scholar]
- 59. Peachman KK, Wieczorek L, Polonis VR, Alving CR, Rao M. 2010. The effect of sCD4 on the binding and accessibility of HIV-1 gp41 MPER epitopes to human monoclonal antibodies. Virology 408: 213–223 [DOI] [PubMed] [Google Scholar]
- 60. Pejchal R, et al. 2009. A conformational switch in human immunodeficiency virus gp41 revealed by the structures of overlapping epitopes recognized by neutralizing antibodies. J. Virol. 83: 8451–8462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Poignard P, Fouts T, Naniche D, Moore JP, Sattentau QJ. 1996. Neutralizing antibodies to human immunodeficiency virus type-1 gp120 induce envelope glycoprotein subunit dissociation. J. Exp. Med. 183: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Poignard P, et al. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 77: 353–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Roux KH, Taylor KA. 2007. AIDS virus envelope spike structure. Curr. Opin. Struct. Biol. 17: 244–252 [DOI] [PubMed] [Google Scholar]
- 64. Ruprecht CR, et al. 2011. MPER-specific antibodies induce gp120 shedding and irreversibly neutralize HIV-1. J. Exp. Med. 208: 439–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sanchez-Martinez S, et al. 2006. Specific phospholipid recognition by human immunodeficiency virus type-1 neutralizing anti-gp41 2F5 antibody. FEBS Lett. 580: 2395–2399 [DOI] [PubMed] [Google Scholar]
- 66. Sanchez-Martinez S, Lorizate M, Katinger H, Kunert R, Nieva J. 2006. Membrane association and epitope recognition by HIV-1 neutralizing anti-gp41 2F5 and 4E10 antibodies. AIDS Res. and Hum. Retrovir. 22: 998–1006 [DOI] [PubMed] [Google Scholar]
- 67. Sanders RW, et al. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76: 7293–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sather DN, et al. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83: 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Scherer EM, Leaman DP, Zwick MB, McMichael AJ, Burton DR. 2010. Aromatic residues at the edge of the antibody combining site facilitate viral glycoprotein recognition through membrane interactions. Proc. Natl. Acad. Sci. U. S. A. 107: 1529–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Scherer EM, Zwick MB, Teyton L, Burton DR. 2007. Difficulties in eliciting broadly neutralizing anti-HIV antibodies are not explained by cardiolipin autoreactivity. AIDS 21: 2131–2139 [DOI] [PubMed] [Google Scholar]
- 71. Shen X, et al. 2010. Prolonged exposure of the HIV-1 gp41 membrane proximal region with L669S substitution. Proc. Natl. Acad. Sci. U. S. A. 107: 5972–5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shen X, et al. 2009. In vivo gp41 antibodies targeting the 2F5 monoclonal antibody epitope mediate human immunodeficiency virus type 1 neutralization breadth. J. Virol. 83: 3617–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Song L, et al. 2009. Broadly neutralizing anti-HIV-1 antibodies disrupt a hinge-related function of gp41 at the membrane interface. Proc. Natl. Acad. Sci. U. S. A. 106: 9057–9062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tomaras GD, et al. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82: 12449–12463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Vcelar B, et al. 2007. Reassessment of autoreactivity of the broadly neutralizing HIV antibodies 4E10 and 2F5 and retrospective analysis of clinical safety data. AIDS. 21: 2161–2170 [DOI] [PubMed] [Google Scholar]
- 76. Veiga AS, Castanho MA. 2006. The membranes role in the HIV-1 neutralizing monoclonal antibody 2F5 mode of action needs re-evaluation. Antivir. Res. 71: 69–72 [DOI] [PubMed] [Google Scholar]
- 77. Veiga AS, Pattenden LK, Fletcher JM, Castanho MA, Aguilar MI. 2009. Interactions of HIV-1 antibodies 2F5 and 4E10 with a gp41 epitope prebound to host and viral membrane model systems. Chembiochem. 10: 1032–1044 [DOI] [PubMed] [Google Scholar]
- 78. Vincent N, Genin C, Malvoisin E. 2002. Identification of a conserved domain of the HIV-1 transmembrane protein gp41 which interacts with cholesteryl groups. Biochim. Biophys. Acta 1567: 157–164 [DOI] [PubMed] [Google Scholar]
- 79. Vishwanathan SA, Hunter E. 2008. Importance of the membrane-perturbing properties of the membrane-proximal external region of human immunodeficiency virus type 1 gp41 to viral fusion. J. Virol. 82: 5118–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yuste E, et al. 2006. Simian immunodeficiency virus engrafted with human immunodeficiency virus type 1 (HIV-1)-specific epitopes: replication, neutralization, and survey of HIV-1-positive plasma. J. Virol. 80: 3030–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang G, Lu H, Lu Y, Jiang S, Chen YH. 2005. Neutralization of HIV-1 primary isolate by ELDKWA-specific murine monoclonal antibodies. Immunobiology 210: 639–645 [DOI] [PubMed] [Google Scholar]
- 82. Zhou T, et al. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445: 732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhu P, et al. 2003. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl. Acad. Sci. U. S. A. 100: 15812–15817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhu P, et al. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441: 847–852 [DOI] [PubMed] [Google Scholar]
- 85. Zwick MB, et al. 2004. The long third complementarity-determining region of the heavy chain is important in the activity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5. J. Virol. 78: 3155–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zwick MB, et al. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodefeciency virus type 1 glycoprotein gp41. J. Virol. 75: 10892–10905 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.