Abstract
Viral interleukin-6 (vIL-6) specified by human herpesvirus 8 is, unlike its cellular counterpart, secreted very inefficiently and can signal via vIL-62:gp1302 signaling complexes from the endoplasmic reticulum (ER) compartment. Intracellular, autocrine activities of vIL-6 are important for proproliferative and prosurvival activities of the viral cytokine in latently infected primary effusion lymphoma (PEL) cells. However, the molecular determinants of vIL-6 ER localization and function are unclear. Using yeast two-hybrid analysis, we identified the database-documented but uncharacterized splice variant of vitamin K epoxide reductase complex subunit 1 (VKORC1), termed VKORC1 variant 2 (VKORC1v2), as a potential interaction partner of vIL-6. In transfected cells, epitope-tagged VKORC1v2 was found to localize to the ER, to adopt a single-transmembrane (TM) topology placing the C tail in the ER lumen, and to bind vIL-6 via these sequences. Deletion mutagenesis and coprecipitation assays mapped the vIL-6-binding domain (vBD) of VKORC1v2 to TM-proximal residues 31 to 39. However, while sufficient to confer vIL-6 binding to a heterologous protein, vBD was unable to induce vIL-6 secretion when fused to (secreted) hIL-6, suggesting a VKORC1v2-independent mechanism of vIL-6 ER retention. In functional assays, overexpression of ER-directed vBD led to suppression of PEL cell proliferation and viability, effects also mediated by VKORC1v2 depletion and, as reported previously, by vIL-6 suppression. The growth-inhibitory and proapoptotic effects of VKORC1v2 depletion could be rescued by transduced wild-type VKORC1v2 but not by a vIL-6-refractory vBD-altered variant, indicating the functional relevance of the vIL-6–VKORC1v2 interaction. Notably, gp130 signaling was unaffected by VKORC1v2 or vBD overexpression or by VKORC1v2 depletion, suggesting an alternative pathway of vIL-6 activity via VKORC1v2. Combined, our data identify a novel and functionally significant interaction partner of vIL-6 that could potentially be targeted for therapeutic benefit.
INTRODUCTION
Human herpesvirus 8 (HHV-8) was the first virus in which a homologue of interleukin-6 (IL-6) was discovered. This is of particular significance because IL-6 activity has been implicated as a cofactor in the development of HHV-8-associated endothelial tumor Kaposi's sarcoma and the B cell malignancies primary effusion lymphoma (PEL) and multicentric Castleman's disease (9, 13, 15, 17, 25). Viral IL-6 is believed to contribute to these diseases via mitogenic and/or proangiogenic activities. We reported previously that vIL-6 expressed during HHV-8 latent infection in PEL cell lines was necessary to sustain normal growth and viability of these cells in culture and hence could contribute directly to PEL pathogenesis (5). Intracellular activity of vIL-6, mediated from the endoplasmic reticulum (ER) compartment, where the viral cytokine is predominantly localized, is critically important for such progrowth activity.
The mechanism(s) responsible for ER and intracellular retention of vIL-6 has not been resolved, although interaction of vIL-6 with the ER chaperone calnexin appears to contribute, directly or indirectly, to ER localization and stability of the viral cytokine (3). However, the duration of vIL-6 interaction with calnexin is shorter than that of intracellular retention of vIL-6, suggesting that the vIL-6–calnexin association is not the principal determinant of the slow secretion kinetics of vIL-6 (3, 12). In addition to calnexin involvement, gp130 has been reported to promote vIL-6 secretion when introduced into Ba/F3 cells, which do not express endogenous gp130 (12). However, results in transfected HEK293T cells showed that while gp130 overexpression promoted vIL-6 redistribution intracellularly (apparently within pre-Golgi complex compartments) and also stabilized vIL-6 upon calnexin depletion, the signal transducer did not enhance secretion of vIL-6 in this experimental system (3). Regardless of the apparent contradictions of these independent findings, however, the mechanisms of ER localization and function of vIL-6 remain unknown and are important in considering the potentially pathogenically significant contribution of ER signaling by latently expressed vIL-6 to cell proliferation and viability.
Here, we report the identification via yeast two-hybrid screening and subsequent molecular and functional analyses of a novel interaction between vIL-6 and a previously uncharacterized protein predicted from the sequence of a cDNA clone representing a splice variant (variant 2; accession number NM_206824) of vitamin K epoxide reductase complex subunit 1 (VKORC1). While VKORC1 (variant 1) is a well-characterized protein involved in the vitamin K cycle and the target of the anticoagulant warfarin (18), VKORC1 variant 2 (VKORC1v2) shares only N-terminal sequences with its well-known counterpart. This region encompasses the first of three predicted transmembrane (TM) domains and part of the TM1-to-TM2 sequence of variant 1, followed by variant 2-specific “soluble” (non-TM) residues. We show here that variant 2, like variant 1, is localized in the ER but that its membrane topology is reversed, allowing variant 2-specific interaction of vIL-6 in the ER lumen via a sequence motif shared with variant 1. Functional analyses employing binding domain overexpression, VKORC1v2 depletion, and rescue by wild-type versus vIL-6-refractory VKORC1v2, revealed that interaction of vIL-6 with VKORC1v2 contributed to progrowth and antiapoptotic activities of the viral cytokine in HHV-8 latently infected PEL cells, although not significantly to its intracellular retention. The data presented identify a new cellular interaction partner of vIL-6 that is an important contributor to its unique functional properties and which might represent a useful therapeutic drug target.
MATERIALS AND METHODS
Yeast two-hybrid screening.
The vIL-6 open reading frame (ORF) was cloned into the Gal4(1-147) “bait” vector pGBKT7 (catalog number 630303; Clontech, Mountain View, CA) between the NcoI and SalI sites and used to transform Saccharomyces cerevisiae strain AH109 to enable library screening. The library used was the human HeLa Matchmaker cDNA library (catalog number 638862; Clontech). Mating, identification, and confirmation of positive interactions and rescue of prey plasmid were carried out according to the supplier's protocols. Verified positive clones were sequenced, and full-length cDNAs of interest were amplified from BCBL-1 cell mRNA preparations by reverse transcription-PCR (RT-PCR), using appropriate primers to allow subsequent cloning into the eukaryotic expression vector pSG5.CBD (4), allowing eukaryotic expression of the respective proteins fused to the chitin-binding domain (CBD). Binding was confirmed by coprecipitation assays using cell lysates from vIL-6 and “prey” cotransfected cells (see below).
Cell culture and gene transduction.
BCBL-1 and BJAB suspension cultures were maintained in RPMI 1640 supplemented with 10% fetal bovine serum. HEK293T cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. For transfection of HEK293T cells, cultures were passaged 12 to 24 h prior to transfection to produce 60% confluent monolayers; calcium phosphate-DNA coprecipitates were generated by standard methods, using CaCl2 solution and HEPES-buffered saline. Gene transduction into PEL or BJAB cells was mediated via lentiviral vector infection. A total of 1 × 106 cells were mixed with lentivirus in medium containing 5 μg/ml Polybrene and incubated at 37°C. After 5 h the inoculum was replaced with fresh medium, and cells were allowed to rest for 2 days prior to experimental use. For dual lentiviral infections, lentiviruses were mixed and used to cotransduce BCBL-1 cultures.
Measurements of cell growth and apoptosis.
For assays of cell growth, 100 μl of cells at a density of 105 cells/ml was seeded into single wells of a 96-well tissue culture plate, and hemocytometric counting of trypan blue excluding (viable) cells was performed at various time points to generate growth curves. To measure apoptosis, phosphate-buffered saline (PBS)-washed cells were resuspended in 1 ml of binding buffer solution comprising 10 mM HEPES (pH 7.4), 140 mM NaCl, and 2.5 mM CaCl2 to which 1 μl of annexin V-Cy3 stock solution (catalog number 1002-200; BioVision, Mountain View, CA) was added prior to light-occluded incubation at room temperature for 5 min. Cells were then washed with binding buffer, fixed with 4% paraformaldehyde (15 min; room temperature, in the dark), permeabilized with 0.25% Triton X-100, and stained with 4′,6′-diamidino-2-phenylindole (DAPI) for 1 min. Following washing in PBS, cells were placed on a glass slide and visualized by UV microscopy. Several random fields were viewed to calculate average percentages of annexin V-Cy3 staining (apoptotic) cells in total cell populations (DAPI+).
Plasmids and lentiviral vector constructions.
Expression vectors for native, CBD-fused, and KDEL motif-tagged vIL-6 and human IL-6 (hIL-6) have been described previously, as has pSG5-gp130.Fc (4, 5). The vIL-6-binding domain (vBD) of VKORC1v2 was cloned into pSG5-hIL-6 or pSG5-hIL-6-KDEL by ligation of vBD-encoding double-stranded oligomer with appropriate 5′ BglII and 3′ blunt ends between the BamHI and SmaI sites of the respective vectors to generate pSG5-hIL-6-vBD and pSG5-hIL-6-vBD-KDEL. CBD-fused ORFs encoding VKORC1 variants 1 and 2 were generated by cloning of the respective PCR-amplified cDNA sequences as EcoRI-BglII fragments into pSG5.CBD (4). Expression vectors for Flag-tagged VKORC1v1 and VKORC1v2 were made by PCR amplification of the respective ORFs with 3′ primers containing Flag-epitope codons, and equivalent cloning into pSG5. VKORC1v2 C-tail codons 31 to 92 were PCR amplified as a fragment containing terminal BamHI and HindIII sites, used to clone the PCR product between the same restriction sites of vector pET32a (Novagen, Madison, WI) for bacterial expression of the variant 2 C-tail as a fusion with thioredoxin (Trx) and His6 tag. A modified version of the UBC promoter-containing pDUET011 lentiviral vector (catalog number 17627; Addgene, Cambridge, MA) was used for expression in PEL or BJAB cells of VKORC1 proteins (wild type and mutated) and green fluorescent protein (GFP)-KDEL and GFP-vBD-KDEL fusions. PCR-amplified coding sequences were cloned into the vector between the BamHI and XhoI sites, replacing GFP-coding sequences of pDUET011. The lentiviral vector pYNC352, described previously (5), was used for shRNA expression; the vector contains cytomegalovirus immediate-early promoter-driven GFP-coding sequences for identification of transduced cells. Double-stranded oligonucleotides corresponding to shRNA sequences directed to transcripts encoding VKORC1v1 or VKORC1v2 or to a target sequence shared by both mRNAs were cloned between the BamHI and MluI sites of pYNC352. The respective shRNA sequences were CACTACAGCTATTGTTAGG (v1-3), CTTCTCCTCCAGGTTGCCT and TTCTCCTCCAGGTTGCCTG (v2-1 and v2-2), and CGTGGGCACCGCCATCAGC (v1 + 2). Nonsilencing (NS) shRNA sequence (GCGCGCTTTGTAGGATTCG) was also cloned into pYNC352 to provide a negative control.
Lentivirus production.
Lentiviral vectors for shRNA or VKORC1v2 expression (12 μg) were cotransfected with gag-pol and vesicular stomatitis virus G protein expression plasmids (9 μg and 3 μg, respectively) into 60% confluent monolayers of HEK293T cells in 75-cm2 tissue culture flasks. Two days posttransfection, virus was collected from the medium by centrifugation in an SW28 rotor at 24,000 rpm for 1.5 h at 4°C. Virus pellets were resuspended in 3 to 5 ml RPMI 1640 medium supplemented with 10% fetal bovine serum. The infectious titers were established by using GFP-based or immunofluorescence microscopy to identify shRNA and VKORC1v2 vector-transduced cells, respectively, in infected PEL cultures prior to their experimental use in these cells.
ER purification.
For ER purification, cells were washed with cold PBS and centrifuged 5 min at 1,400 × g at 4°C. Pellets were resuspended in 2 ml MTE (270 mM d-mannitol, 10 mM Tris base, 0.1 mM EDTA; pH 7.4) and sonicated on ice three times for 10 s each, separated by 10-s rest intervals on ice. The lysates were centrifuged for 10 min at 1,400 × g at 4°C, and the resulting supernatants were centrifuged for 10 min at 15,000 × g at 4°C. The cleared supernatants were layered onto sucrose gradients (2 ml of 2 M sucrose at the bottom followed by 3 ml of 1.5 M sucrose and overlaid with 3 ml of 1.3 M sucrose) and centrifuged at 152,000 × g at 4°C for 70 min. The ER fraction was extracted at the interface of the 1.3 M sucrose layer, added to 10 ml cold MTE plus 2 mM phenylmethanesulfonylfluoride (PMSF), and centrifuged at 126,000 × g for 45 min at 4°C. The pellets were resuspended in 150 μl MTE without PMSF.
Coprecipitations, immunoprecipitations, and Western blotting.
Coprecipitation assays utilized either CBD or human immunoglobulin Fc fusion proteins, expressed with other (partner) proteins in cotransfected HEK293T cells and precipitated from corresponding cell lysates with chitin beads or protein A-agarose, respectively. The CBD-fused proteins included VKORC1v1, VKORC1v2 (native and derivatives), and vIL-6; CBD tagging of vIL-6 allowed for chitin bead-mediated precipitation and concentration from transfected cell medium as well as its use in coprecipitation assays. Fc-tagged gp130 was used in coprecipitation experiments, and analysis of gp130 activation (phosphorylation) following protein A-agarose-mediated precipitation from transfected cell lysates was undertaken by immunoblotting using phosphotyrosine antibody. For isolation of endogenous gp130 from BCBL-1 cells, the signal transducer was immunoprecipitated from cell lysates prior to phosphotyrosine immunoblotting. Incubation of chitin or protein A beads with cell lysates or medium was undertaken overnight at 4°C; gp130-specific antibody was included for immunoprecipitations. For coprecipitation assays involving vIL-6–CBD and bacterially derived Trx/His6-v2(C tail), vIL-6-bound chitin beads were blocked for 3 h with 1% bovine serum albumin in lysis buffer (20 mM Tris-HCl [pH 8.0], 5 mM EDTA, 50 mM NaCl, 0.2% NP-40) prior to addition of recombinant protein and overnight incubation at 4°C. After washing of precipitates in lysis buffer by repeated suspension and sedimentation, pelleted proteins were denatured and reduced in SDS-PAGE loading buffer, and proteins were resolved by SDS-PAGE and identified by Western blotting. Tricine–SDS-PAGE and electrophoretic transfer to nitrocellulose membranes were carried out by standard procedures. Membranes were blocked in PBS-T buffer (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, 0.1% Tween 20) containing 5% nonfat milk prior to the addition of primary antibody (0.1 to 1.0 μg/ml) and incubation at 4°C overnight. PBS-T-washed membranes were incubated with horseradish peroxidase-conjugated anti-rabbit, anti-goat, or anti-mouse secondary antibodies in PBS-T–5% nonfat milk and washed with PBS-T, and membrane-bound antibody was visualized in a chemiluminescence assay.
Antibodies.
Commercial antibodies used for immunoprecipitation (gp130) and analysis of immunoprecipitates, cell lysates, and subcellular fractions by immunoblotting were as follows: hIL-6 (catalog number AB-206-NA; R&D Systems, Minneapolis, MN); CBD (catalog number E8034S; New England BioLabs, Beverly, MA); gp130 (catalog number 555755; BD Biosciences, Rockville, MD); phosphotyrosine (catalog number 9411; Cell Signaling Technology, Beverly, MA); EEA1 (catalog number 610457), calreticulin (catalog number 612136) and calnexin (catalog number 610523) (all from BD Biosciences); Flag-epitope (catalog number F1804; Sigma, St. Louis, MO); phospho-STAT3 (catalog number 9131; Cell Signaling Technology); STAT3 (catalog number sc-482; Santa Cruz Biotechnology, Santa Cruz, CA); GFP (catalog number 1533-1; Epitomics, Burlingame, CA); His (catalog number SC-803; Santa Cruz Signaling Technology). vIL-6 was immunodetected using custom rabbit polyclonal antiserum (24).
Fluorescence microscopy.
For studies of the subcellular localization of VKORC1v2, HEK293T cells were seeded onto poly-l-lysine-coated chamber slides and transfected with expression plasmids for VKORC1-Flag and ER-directed KDEL motif-tagged GFP. At 24 h after transfection, cells were fixed with 4% paraformaldehyde in PBS and permeabilized with 0.25% Triton X-100 for immunofluorescence assays. Cells were blocked in PBS containing 1% bovine serum albumin and subsequently incubated at room temperature with primary and secondary antibodies (1 h each); PBS was used to rinse cells after antibody incubation. Mouse monoclonal antibody to Flag-epitope was used to identify subcellular localization of Flag-tagged VKORC1v2 relative to ER-directed GFP-KDEL. Fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G (1:100; catalog no. 816511; Zymed Laboratories, San Francisco, CA) was used as the secondary antibody. Cells were counterstained with DAPI and viewed by confocal microscopy.
Endo H and protease digestions.
Recombinant endo-β-N-acetylglucosaminidase H (endo H–maltose-binding protein fusion protein [endo Hf]) was obtained from New England BioLabs (catalog no. P0703S; Ipswich, MA). For endo H analysis of high-mannose glycosylation of VKORC1, vIL-6, and gp130, VKORC1-CBD, vIL-6-CBD, and gp130-Fc were precipitated from transfected cell lysates with chitin or protein A-agarose beads, resuspended in glycoprotein denaturation buffer (0.5% SDS, 40 mM dithiothreitol), and incubated at 100°C for 10 min. A 0.1 volume of 10× endo H reaction buffer (50 mM sodium citrate, pH 5.5) and 1,000 units of endo Hf were added, and the mixture was incubated at 37°C for 1 h. Protease treatment of MTE buffer-suspended sucrose gradient-purified ER membrane preparations was carried out using 50 μg of proteinase K (New England BioLabs) and incubation on ice for 30 min. For both endo H and proteinase K digestions, reactions were stopped by addition of SDS-PAGE loading buffer, and samples of pre- and posttreated products were compared by SDS-PAGE/immunoblotting, along with control proteins known to be resistant or susceptible to digestion.
Recombinant proteins.
Trx/His6 and Trx/His6-fused VKORC1v2 were expressed in Escherichia coli strain ER2566 by overnight culture at 16°C in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were lysed by sonication, and recombinant proteins were affinity purified via Ni-bead column chromatography. After washing, His6-tagged proteins were eluted in 50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole (pH 8.0), and their purity was checked by SDS-PAGE fractionation and Coomassie staining.
RESULTS
Identification of VKORC1 variant 2 as a potential binding partner of vIL-6.
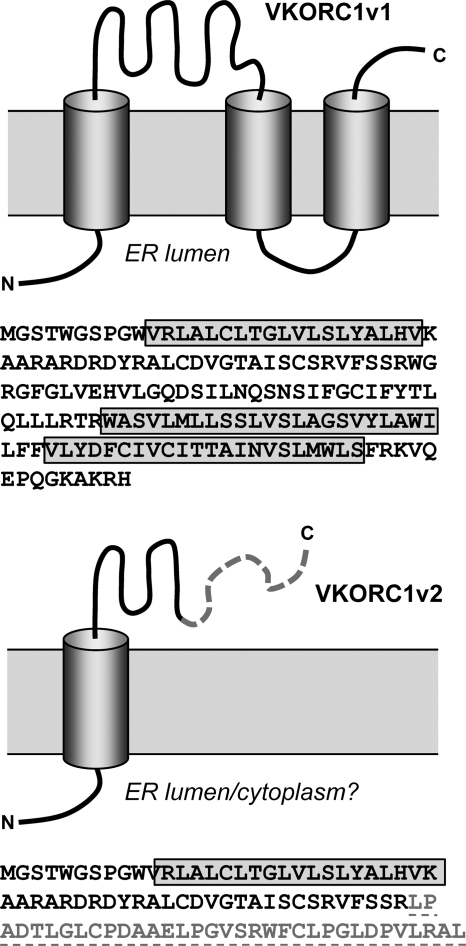
To screen for possible novel interaction partners of vIL-6, we employed the yeast two-hybrid approach, utilizing Gal4 DNA-binding domain-fused vIL-6 (amino acids 1 to 204) to probe a HeLa cell cDNA library cloned into a Gal4 activation domain vector, pGADT7-Rec (see Materials and Methods). From a screen of approximately 3 × 107 clones, with around 2-fold redundancy, 38 clones were obtained upon repeated rounds of selection (Quadruple Dropout; Clontech). Plasmids were prepared from 32 of these clones for sequencing and identification. Of those for which reliable sequence data were obtained and definitive protein matches identified, most corresponded to nuclear, cytoplasmic, or other proteins known not to localize to the ER, the site of vIL-6 intracellular localization, and these were therefore excluded from further consideration (see Table S1 in the supplemental material). Two of the isolates corresponded to independent clones of a splice variant of a transcript encoding VKORC1, which is known to localize to the ER (2, 19, 23). The spliced transcript, referred to as variant 2, was originally identified by sequencing of the corresponding cDNA (accession number NM_206824), but its protein product had not been studied. VKORC1 (variant 1) and VKORC1 variant 2 (VKORC1v2) share the first 58 amino acids but diverge thereafter (Fig. 1). The full length of the VKORC1v2 open reading frame (92 codons from the first ATG) was present in the clones derived from the yeast two-hybrid screen.
Fig 1.
Relationship between VKORC1 variants 1 and 2. Sequences and diagrammatic representations of the predicted structures of VKORC1 variants 1 and 2 are shown. The membrane topology shown for VKORC1v1 corresponds to that deduced by Tie et al. (21). Variant 2 shares the first 58 amino acids with variant 1; residues 59 to 92 are unique.
Interaction between vIL-6 and VKORC1 variant 2.
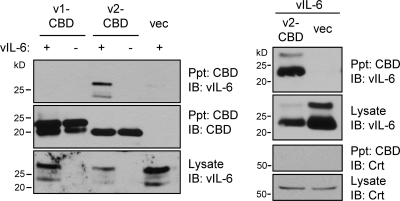
The intracellular interaction between VKORC1v2 and vIL-6 was tested by coprecipitation assay. HEK293T cells were cotransfected with expression vectors for vIL-6 and CBD-tagged VKORC1v2 or VKOCR1v1, allowing chitin bead-mediated precipitation of CBD-VKORC1 proteins. The results demonstrated that vIL-6 could be coprecipitated with VKORC1v2, but VKORC1v1 was unable to associate with the viral cytokine (Fig. 2).
Fig 2.
Interaction between vIL-6 and VKORC1v2. Association of vIL-6 with VKORC1v2 was identified by coprecipitation of vIL-6 with chitin bead-precipitated VKORC1v2-CBD from lysates of transfected HEK293T cells. Precipitated proteins (Ppt) were identified by immunoblotting (IB) for detection of vIL-6 and CBD. The two bands for vIL-6 represent glycosylated and unglycosylated forms of the protein. VKORC1v1-CBD did not precipitate vIL-6 (left). The right panels present the results of an analogous experiment, showing specificity of VKORC1v2 binding to vIL-6; no interaction between VKORC1v2 and ER protein calreticulin (Crt) was detected. vec, empty vector.
ER localization and membrane topology of VKORC1v2.
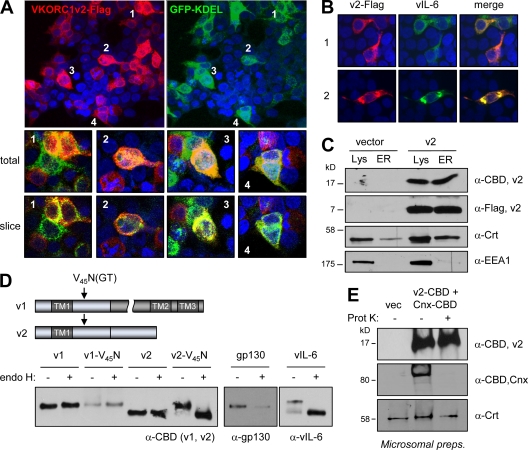
Consistent with the interaction between vIL-6 and VKORC1v2 identified by coprecipitation, confocal immunofluorescence microscopy detected substantial colocalization of ER-directed KDEL motif-fused GFP (an established means of visualizing ER [5, 7]) and Flag-tagged VKORC1v2 in transfected cells (Fig. 3A). Thus, VKORC1v2, like its well-studied variant 1 counterpart, is present in the ER, the major site of vIL-6 localization (3, 23). Indeed, immunofluorescence detection of VKORC1v2-Flag with vIL-6 in cells cotransfected with the respective expression vectors demonstrated a high degree of colocalization of the proteins (Fig. 3B). ER localization of VKORC1v2 was verified independently by detection of VKORC1v2 in sucrose gradient-purified ER membrane preparations from appropriately transfected cells (Fig. 3C).
Fig 3.
ER localization and membrane topology of VKORC1v2. (A) Confocal immunofluorescence assay results, showing colocalization of ER-directed GFP-KDEL and Flag-tagged VKORC1v2 in transfected HEK293T cells. (B) Confocal immunofluorescence-determined colocalization of vIL-6 and VKORC1v2-Flag in transfected HEK293T cells (two fields shown). (C) Immunoblot detection of VKORC1v2 in sucrose gradient-purified ER membrane preparations derived from VKORC1v2-Flag-transfected HEK293T cells, confirming ER localization of VKORC1v2. Calreticulin (Crt; ER localized) and early endosome antigen 1 (EEA1) provided quality control markers for ER preparations. (D) Determination of VKORC1v2 orientation within the ER by N-glycosylation tagging, showing efficient (∼100%) glycosylation of variant 2 upon introduction of an N-glysosylation site (N45GT by V45N mutation) into the post-TM region. These results demonstrate lumenal exposure of this region. In contrast, no such glycosylation was observed for VKORC1v1 containing the identical introduced N-glycosylation site. Controls for endo H digestion and resistance to digestion were provided by vIL-6 (high mannose glycosylated) and gp130 (containing mature glycan moieties), respectively. (E) Protease digestion experiment using C-terminally CBD-tagged VKORC1v2 revealed protection of CBD (and most/all of VKORC1v2) upon exposure of transfected cell-derived ER membrane preparations to proteinase K (Prot. K). The cytoplasmically exposed CBD of C-terminally tagged calnexin (ER transmembrane protein) and lumenally expressed calreticulin were cleaved and protected, respectively, verifying protease activity and membrane integrity.
While this localization of VKORC1v2 was consistent with its detected interaction with vIL-6, the previously reported membrane topology of VKORC1 variant 1 (21) coupled with the common N-terminal (predicted ER-lumenal) sequences of variants 1 and 2 raised the question of how vIL-6 interacts specifically with VKORC1v2; the divergent regions of variants 1 and 2 occur after the first transmembrane domain in a region of variant 1 reportedly exposed to the cytoplasm. Thus, we reasoned that the different abilities of VKORC1v1 and VKORC1v2 to bind vIL-6 were due either to conformational differences or to opposite orientations of the common TM domain of each within the ER membrane. The latter would place the TM downstream shared sequence (residues 30 to 58) and variant 2-specific sequence (residues 59 to 92) of VKORC1v2 within the ER lumen and available for interaction with vIL-6. This was tested in two ways: first, by investigation of N-glycosylation of a target sequence (N45GT) introduced into TM downstream sequences of variants 1 (control) and 2, and second by testing the susceptibility of the C tail of variant 2 to protease digestion. For variant 2, specifically, introduction of the V45N mutation to generate glycosyltransferase-targeted NGT led to glycosylation of the protein, digestible by endonuclease H, which cleaves nascent glycan moieties (Fig. 3C). In contrast, no glycosylation of VKORC1v1-V45N was detected, consistent with published findings noting cytoplasmic exposure of this region (21). Using protease digestion to probe the membrane orientation of variant 2, the C-terminal epitope (CBD) tag of the expressed protein was found to be refractory to proteinase K applied to ER membrane preparations, as determined via its detection by immunoblotting (Fig. 3D). In contrast, the C-terminal epitope tag of calnexin-CBD, coexpressed with VKORC1v2-CBD in the same transfected cells to provide a control, was susceptible to protease digestion, consistent with the known membrane orientation of calnexin (22), resulting in cytoplasmic exposure of the CBD. The ER-lumenal, soluble protein calreticulin (8) was resistant to protease digestion, confirming ER membrane integrity. Thus, data from these two experimental approaches provide robust evidence that the TM-distal tail of variant 2 is within the ER lumen, opposite to the orientation of VKORC1 variant 1 reported by Tie et al. (21) and supported by the resistance of VKORC1v1-V45N (and indeed N77QS of wild-type VKORC1v1) to N-glycosylation. Furthermore, the fact that glycosylated VKORC1v2 is digestible with endo H provides further support for ER localization; endo H-sensitive, immature (high-mannose) glycans are present almost exclusively on pre-Golgi complex-localized glycoproteins (10).
Mapping of the vBD of VKORC1 variant 2.
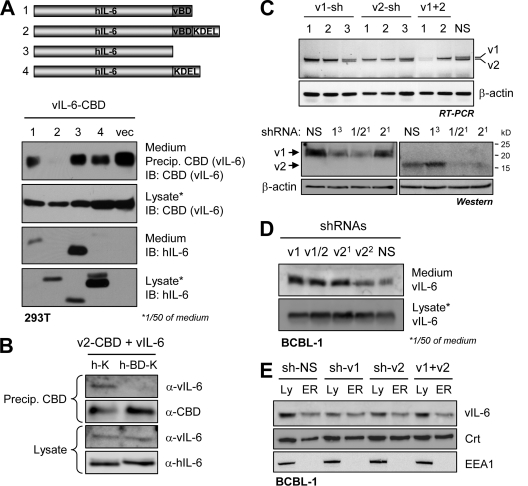
Coprecipitation assays employing transfected vIL-6 and CBD-fused wild-type VKORC1 proteins and deletion and substitution derivatives of variant 2 were undertaken to identify VKORC1 sequences required for interaction with vIL-6. Initially, VKORC1 variants 1 and 2 were used along with variant 2 derivatives from which post-TM sequences common to variants 1 and 2 (residues 31 to 58) or specific to the latter (residues 59 to 92) were deleted. The data from this first experiment identified the TM-proximal amino acids 31 to 58 as required for interaction with vIL-6 (Fig. 4A, top left autoradiographs). Subsequent deletions within this region, in the context of VKORC1v2, further mapped the residues required for vIL-6 interaction to amino acids 31 to 39 (vBD), immediately adjacent to the TM domain (Fig. 4A, right panels). Alanine substitution of vBD residues D38 and Y39 diminished coprecipitation of vIL-6 to background levels, thereby identifying these residues as critically important for vIL-6 binding (Fig. 4A, bottom). That amino acids 31 to 39 of VKORC1v2 did indeed comprise the vIL-6-binding region was confirmed by demonstration that this sequence could confer vIL-6 binding to CBD-fused and KDEL-tagged (ER-retained) hIL-6 (Fig. 4B). Direct binding between vIL-6 and VKORC1v2 was demonstrated by in vitro coprecipitation assays employing CBD-fused vIL-6 isolated from transfected cell medium and recombinant thioredoxin/His6-fused VKORC1v2 C tail (residues 31 to 92). The latter coprecipitated with chitin bead-bound vIL-6–CBD, but not by chitin beads supplemented with hIL-6-CBD (Fig. 4C). Combined, these data identify the amino acid 31 to 39 region of VKORC1v2 as necessary and sufficient for direct interaction with vIL-6.
Fig 4.
Mapping of VKORC1v2 residues interacting with vIL-6. (A) Coprecipitation assays were employed to identify the region(s) of VKORC1v2 required for its interaction with vIL-6; CBD-tagged VKORC1v2 and deleted derivatives were utilized in chitin-mediated precipitations (IP) from transfected cell lysates. The region of VKORC1v2 immediately following the TM domain was found to be required for binding. Dialanine mutations within this region (residues 31 to 39) were introduced into VKORC1v2-CBD and tested for interaction with vIL-6; the DY39AA mutation inhibited binding. IB, immunoblotting; WT, wild type. (B) Residues 31 to 39, constituting the vIL-6 binding domain of VKORC1v2, were transferred to a heterologous protein, KDEL-tagged hIL-6, to test their sufficiency for binding to vIL-6. The nonopeptide sequence was able to confer vIL-6 binding to hIL-6-KDEL in transfected cells. (C) In vitro coprecipitation assay using media-derived vIL-6–CBD and bacterially expressed thioredoxin (Trx)/His6-fused VKORC1v2 C tail (residues 31 to 92), demonstrating direct interaction between the two proteins. Trx-His6 (negative control) was unable to bind vIL-6–CBD, and VKORC1v2 protein could not be coprecipitated with control hIL-6-CBD.
VKORC1v2 and vIL-6 intracellular retention.
To test for a possible contribution of VKORC1v2 to intracellular retention of vIL-6, we first investigated the effects on vIL-6 secretion of vBD-fused secreted and ER-retained (KDEL motif-tagged) hIL-6 constructions in transfected HEK293T cells. Western blotting of media and lysates of transfected cultures confirmed the appropriate secretion of hIL-6–vBD and intracellular retention of hIL-6–vBD-KDEL (Fig. 4A); ER localization of the latter and hIL-6-KDEL (previously demonstrated to localize to the ER [5]) was verified by immunofluorescence assay (see Fig. S1 in the supplemental material). Overexpression of hIL-6–vBD-KDEL led to abrogation of the low levels of vIL-6 secretion, while secreted hIL-6–vBD had no detectable effect on vIL-6 secretion (Fig. 5A). Respectively, these data are consistent with vBD binding of vIL-6 to complement its intracellular retention by another mechanism but inability of vBD to disrupt this mechanism of vIL-6 retention. That overexpression of vBD, as a fusion with hIL-6–KDEL, was indeed able to disrupt the vIL-6–VKORC1v2 interaction (Fig. 5B) provided further evidence that intracellular retention of vIL-6 is mediated independently of VKORC1v2.
Fig 5.
VKORC1v2 in intracellular retention of vIL-6. (A) The influence of VKORC1v2 on intracellular retention of vIL-6 was tested by overexpression of vBD as a fusion with hIL-6 (secreted) or hIL-6-KDEL (ER retained) in vIL-6-cotransfected HEK293T cells. While ER-expressed vBD led to a reduction in the very low levels of secreted vIL-6, consistent with its binding the viral cytokine, vBD in the context of secreted hIL-6 was insufficient to induce, in a dominant fashion, secretion of vIL-6. These data indicate that a mechanism independent of vIL-6–VKORC1v2 association is responsible for ER retention of vIL-6. (B) The ability of hIL-6-fused vBD to disrupt vIL-6–VKORC1v2 binding was verified in a coprecipitation assay utilizing lysates from transfected HEK293T cells. Chitin bead-sedimented VKORC1v2-CBD was able to coprecipitate vIL-6 in the presence of hIL-6-KDEL (control) but not its vBD-containing counterpart. (C) Testing by RT-PCR (top panels) of lentivirus-cloned shRNAs directed to VKORC1 variants 1 and/or 2 transcripts (see Materials and Methods) for their abilities to deplete the respective mRNAs in BCBL-1 cells. Selected, active shRNAs were then tested in VKORC1v1- and VKORC1v2-vector transfected cells to verify by immunoblotting (IB; bottom panels) their activities in respect of VKORC1 protein depletion. (D) shRNA-mediated VKORC1v2 depletion had little effect relative to controls (VKORC1v1 and NS shRNA transduction) on the levels of secreted, medium-derived versus intracellular vIL-6. Secreted vIL-6 was immunoprecipitated from medium, and the proportion loaded onto the gel and immunoblotted represents 50 times the fraction of corresponding cell lysate analyzed. (E) Western analysis of density gradient-purified ER membrane preparations by immunoblotting revealed that there was no influence of VKORC1v2 depletion on ER localization of vIL-6. Calreticulin (Crt; ER marker) and EEA1 (endosomal marker) were detected to check the quality of the ER preparations.
The significance of VKORC1v2 to intracellular retention of endogenously expressed vIL-6 was tested by using a knockdown approach in HHV-8+, latently infected BCBL-1 PEL cells. shRNAs were generated to target sequences specific to variant 1 (variant 2 intron) or variant 2 (splice junction) or that were common to both mRNA transcripts (see Materials and Methods). These were cloned into a lentivirus vector (GFP expressing) for delivery into BCBL-1 cells; their relative effects on VKORC1 transcript levels were tested using RT-PCR analysis (Fig. 5C, top). Selected shRNAs were further tested in VKORC1 expression vector-cotransfected HEK293T cells by immunoblotting to demonstrate effective depletion of the respective epitope-tagged proteins (Fig. 5C, bottom). Neither of the two active VKORC1v2-specific shRNAs nor the functional shRNA targeting variants 1 and 2 led to significant changes in amounts of vIL-6 secreted from BCBL-1 cells relative to levels seen in VKORC1v1 or NS shRNA-transduced control cultures (Fig. 5D). It should be noted that the ratio of intracellular to extracellular vIL-6 was very high (∼50:1) and that the intracellular (functional) levels were essentially unchanged by VKORC1v2 depletion. The possible effect of VKORC1v2 depletion specifically on ER localization of vIL-6 in BCBL-1 cells was investigated by undertaking Western analysis of purified ER membrane preparations, isolated by density gradient fractionation of cell extracts. This analysis showed that there was no change in the level of ER-localized vIL-6 as a consequence of VKORC1v2 depletion (Fig. 5E). Thus, VKORC1v2 appears not to be a determinant of vIL-6 ER retention or otherwise to modulate vIL-6 secretion.
VKORC1v2 contributes to PEL cell growth and survival.
To test the potential relevance of VKORC1v2 to PEL cell proliferation and viability, shown previously to be promoted by vIL-6 from the ER compartment (5), shRNA-mediated depletion was again employed. Lentiviral vectors (GFP+) specifying shRNAs targeting variants 1 and/or 2 or an NS control were used to infect BCBL-1 PEL or control (HHV-8−) BJAB cultures at an efficiency of >90%, as determined by GFP fluorescence (data not shown). Two days after infection, cells were passaged in triplicate, and cell number-normalized cultures were monitored for growth rates by daily cell counting. The results showed clearly that depletion of variant 2 or variants 1 plus 2 led to markedly reduced growth specifically of BCBL-1 cells (Fig. 6A). Variant 1 transcript depletion led to small reductions in growth rates of both BCBL-1 and BJAB cultures. BJAB cells were confirmed by RT-PCR to express both variant 1 and variant 2 VKORC1 transcripts, each of which was suppressed specifically by the respective shRNAs (see Fig. S2 in the supplemental material). Substantial negative effects of VKORC1v2-shRNA on growth of another PEL cell line, JSC-1, were also observed, but growth of control Akata cells (HHV-8−) was unaffected (Fig. 6A). These data indicate that VKORC1v2 plays a positive role in PEL cell growth.
Fig 6.
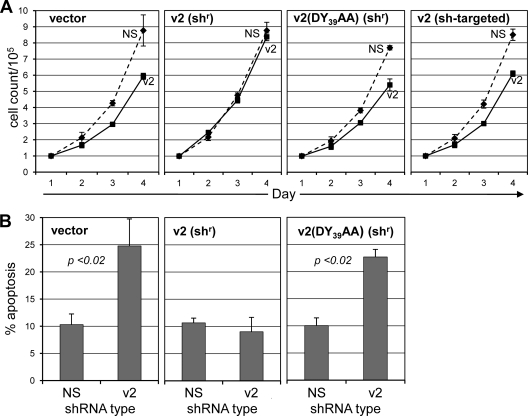
Functional analysis of VKORC1v2 and the vIL-6–VKORC1v2 interaction. (A) Lentivirus-shRNA transduction of PEL cells revealed substantial inhibition of cell growth by VKORC1v2-directed shRNAs (v2 alone or v1+2) relative to NS control and v1-targeted shRNAs. Growth of HHV-8-negative BJAB cells was unaffected or minimally altered by transduction of these VKORC1 shRNAs. JSC-1 PEL cell growth, like that of BCBL-1 cells, was inhibited markedly in response to VKORC1v2-specific shRNA, while control Akata cells (HHV-8 negative) were unaffected. (B) Lentivirus vector-mediated expression in BCBL-1 and JSC-1 cells of ER-directed vBD (GFP-vBD-KDEL), used as a competitor for the VKORC1v2–vIL-6 interaction, also led to growth inhibition. This was not seen in identically treated BJAB and Akata cells. (C) Individual and cotransduction of v2-shRNA and vBD in BCBL-1 cells failed to reveal significant additive effects. These data are consistent with targeting of a common mechanism rather than distinct pathways. (D) Using Annexin V-Cy3 staining for identification of apoptotic cells, both VKORC1v2 depletion and GFP-vBD-KDEL (vBD-K) transduction were found to lead to significantly increased rates of BCBL-1 cell apoptosis relative to those detected in NS-shRNA and GFP-KDEL (K) control cultures (left and right panels, respectively). For all experiments (panels A to D), triplicate cultures were used; error bars represent standard deviations from mean values. In panel D, NS versus v2 P values (t test, unpaired, two tailed) are shown.
To address the significance of the vIL-6–VKORC1v2 interaction specifically, a competition approach was used that employed hIL-6 signal sequence-fused, KDEL motif-tagged (ER-directed) GFP, with or without attachment of the VKORC1v2 vIL-6-binding motif (AARARDRDY [vBD]), in transfected cells to disrupt the vIL-6–VKORC1v2 association (Fig. 5B). Lentiviral vectors were used to enable efficient delivery of the respective coding sequences into PEL cells. Transduction of the vBD-containing fusion protein into BCBL-1 and JSC-1 cells inhibited growth relative to the vBD-deficient GFP construction, indicating that the vIL-6 interaction with VKORC1v2 is important for variant 2 support of BCBL-1 and JSC-1 cell growth (Fig. 6B). Further evidence of the functional relevance of the vIL-6–VKORC1v2 interaction, specifically, was provided by an experiment employing cotransduced shRNA and ER-directed GFP-vBD; each was able to reduce, to equivalent levels, BCBL-1 cell growth, and these effects were greatly diminished in the presence of the reciprocal treatment (Fig. 6C). Alternatively stated, the effects of GFP-vBD and VKORC1v2 shRNA were essentially redundant rather than additive, suggesting that VKORC1v2 and vIL-6 functions are connected and that vBD challenge does not affect vIL-6 activity independently of the vIL-6–VKORC1v2 interaction.
Using the same variant 2 depletion and vBD overexpression approaches, we tested the influence of VKORC1v2 and the variant 2–vIL-6 interaction on BCBL-1 cell viability. In each case, rates of apoptosis (determined based on annexin V-Cy3+ staining) in BCBL-1 cultures were increased significantly (2- to 3-fold) relative to negative controls (Fig. 6D). These data reflect our previous findings from vIL-6 depletion experiments (5) and suggest that VKORC1v2 may enable or enhance vIL-6 prosurvival activity in PEL cells.
Specific role of the VKORC1v2–vIL-6 interaction in PEL growth and survival.
VKORC1v2 depletion, vBD challenge, and vIL-6 depletion all inhibit growth and promote apoptosis of PEL cells, suggesting a link between the vIL-6–VKORC1v2 interaction and vIL-6 activity. To test this further, wild-type VKORC1v2 and a vIL-6-refractory counterpart (DY39AA) (Fig. 4A) were compared for their abilities to rescue growth inhibition and increase apoptosis induced by depletion of endogenous VKORC1v2 in BCBL-1 cells. The constructions for expression of the transduced proteins were engineered to be resistant to shRNA targeted to variant 2 mRNA. As a control, unaltered variant 2 coding sequences, susceptible to shRNA targeting, were also transduced. BCBL-1 cells were coinfected with lentiviral vectors specifying NS (control) or VKORC1v2-specific shRNA together with lentiviral vectors encoding epitope (CBD)-tagged wild-type or DY39AA-mutated VKORC1v2. Viruses were used at a titer sufficient to yield close-to-100% transduction efficiency, as verified by GFP fluorescence (lentivirus-shRNA) or immunofluorescence staining of culture samples (lentivirus–VKORC1v2-CBD) (data not shown). At 48 h after infection, equal cell numbers for each condition (in triplicate) were seeded into fresh media, and cell growth was monitored by daily cell counting. In a parallel experiment, apoptosis was quantified by annexin V-Cy3 staining on day 4. The data (Fig. 7) showed clearly that wild-type VKORC1v2 was fully able to rescue the antigrowth and proapoptotic effects of endogenous VKORC1v2 depletion, but the vIL-6-refractory variant was unable to do so. These data provide strong evidence that the vIL-6 interaction with VKORC1v2 is functionally significant.
Fig 7.
Role of the VKORC1–vIL-6 interaction in PEL cell growth and survival. (A) BCBL-1 cells were coinfected with lentivirus vectors specifying control (NS) or VKORC1v2-specific shRNA and “shRNA-resistant” (shr) wild-type or DY39AA-mutated (vIL-6-refractory) VKORC1v2 proteins, or wild-type VKORC1v2 susceptible to shRNA targeting (control for effective depletion). Cell growth was monitored daily following lentivirus transduction and a 2-day rest by counting of trypan blue-excluding cells. (B) In a parallel experiment, apoptosis was assayed at day 4 based on Annexin V-Cy3 staining and calculated as the fraction of total cells in each of >10 random fields. For growth and apoptosis data, error bars represent deviations from mean values obtained from triplicate cultures; t test values (unpaired, two tailed) are shown in panel B.
Assessment of the VKORC1v2 contribution to vIL-6 signal transduction via gp130.
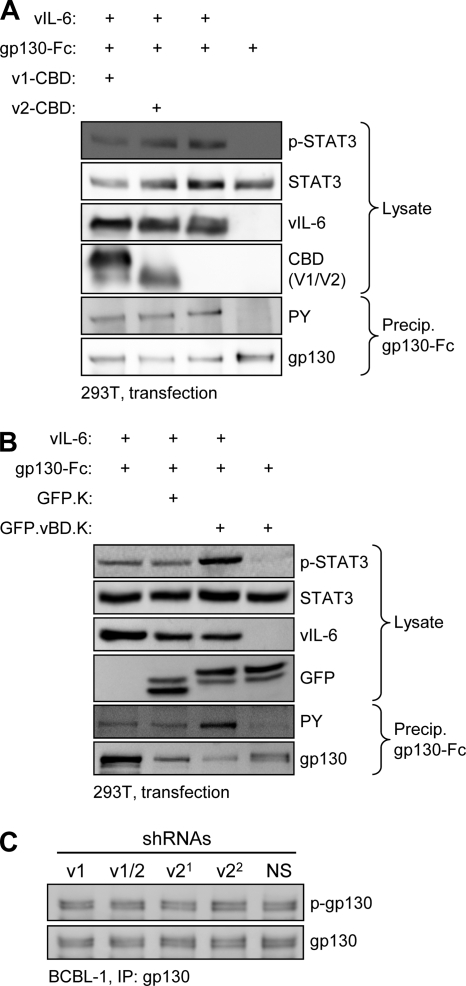
To try to explain the growth- and survival-inhibitory effects of VKORC1v2 depletion and vBD overexpression, we hypothesized that the VKORC1v2–vIL-6 interaction might enhance vIL-6 signal transduction via gp130 and that vBD-mediated competition of such complexing, or the vBD–vIL-6 interaction itself, could be inhibitory to gp130 activation by vIL-6. This was tested initially by overexpressing VKORC1v2 in vIL-6/gp130-cotransfected HEK293T cells and assaying for gp130 and STAT3 activation, detected by phospho-tyrosine-specific or phospho-STAT3-specific immunoblotting of precipitated gp130 or cell extracts, respectively. Both measures of vIL-6-induced signal transduction were essentially unaffected by coexpression of VKORC1v2 (or variant 1, used as a control) (Fig. 8A). Second, we tested the effect of vBD, expressed as a fusion with GFP-KDEL, on vIL-6 signal transduction by using the same readouts. Again, no inhibitory effect was observed; in fact, vIL-6-induced gp130 tyrosine phosphorylation and activated phospho-STAT3 were somewhat increased in GFP-vBD-KDEL-transfected relative to GFP-KDEL- and empty vector-transfected cells (Fig. 8B). Possible explanations for this result are that in this transfection experiment vBD may liberate higher levels of “free” vIL-6 (unbound to VKORC1v2 or other proteins) for interaction with gp130 or otherwise enhance interaction of vIL-6 with the signal transducer, but further investigation is required to resolve this issue. In this experiment, vIL-6 was fused to a KDEL ER retention signal to ensure that all signaling measured was from ER activity of the viral cytokine. Finally, the effect of VKORC1v2 depletion on levels of active (phosphorylated) gp130 in BCBL-1 cells was investigated; neither of the two effective VKORC1v2-specific shRNAs nor v1+2 shRNA had any effect on levels of phospho-gp130, relative to levels in NS or VKORC1v1-specific shRNA-transduced cells (Fig. 8C). Combined, these data indicate that VKORC1v2 and vBD, while respectively enhancing and inhibiting PEL cell growth, do not have corresponding effects on vIL-6/gp130 signal transduction. Coupled with the dissociation of VKORC1v2 and vBD influence on vIL-6 secretion (Fig. 5) and the demonstrated functional significance of the vIL-6 interaction with VKORC1v2 for progrowth and prosurvival activities in BCBL-1 cells (Fig. 7), these results indicate an alternative, novel mechanism of vIL-6 activity via the VKORC1v2 interaction.
Fig 8.
Testing the potential influence of VKORC1v2 on vIL-6 signaling via gp130. (A) Cotransfection assays were undertaken to determine the effect of VKORC1v2 (and VKORC1v1 [control]) overexpression on vIL-6/gp130 signaling, induced by coexpression of vIL-6 and (functional) gp130-Fc. Immunoblotting was used to identify phospho-STAT3 (p-STAT3) in cell lysates and tyrosine-phosphorylated (PY) gp130-Fc following precipitation from cell lysates by protein A-agarose as measures of vIL-6-induced gp130 signaling and activation. VKORC1v2 had no effect on either. (B) Overexpression of vBD, fused to GFP-KDEL, with vIL-6-KDEL (fully ER retained) and gp130-Fc, had no inhibitory effect on STAT3 activation or gp130 phosphorylation but rather seemed to enhance vIL-6 activity. (C) Depletion of VKORC1v2 in BCBL-1 cells did not alter the levels of active (phosphorylated) endogenous gp130, immunoprecipitated from cell lysate prior to immunoblotting for detection of phospho-tyrosine (p-gp130).
DISCUSSION
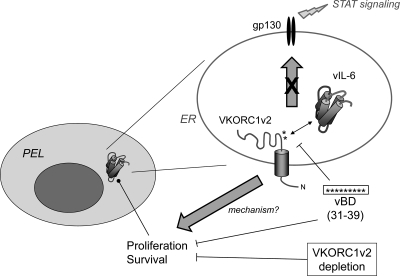
HHV-8 vIL-6 is unusual in its ability to transduce signal directly via gp130 in the absence of the IL-6 receptor α-subunit, gp80, and its accumulation intracellularly, in the ER compartment (1, 5, 6, 11, 12, 14, 16, 24). These properties, together with expression of vIL-6 during latency in PEL cells, enable the viral cytokine to influence growth and viability of PEL cells, implicating a role of vIL-6 in maintenance of latent viral loads (in B cells, at least) in vivo and in PEL pathogenesis (5). The mechanisms of retention and factors regulating the activity of vIL-6 in the ER compartment are as yet poorly understood, although it is known that the vIL-6 interaction with the ER chaperone calnexin is important for vIL-6 stability and may play a role in ER localization and that gp80 independence of vIL-6 is likely to enable vIL-6 activity in this compartment (3, 5). Such mechanistic issues have considerable relevance to the development of potentially therapeutic strategies to inhibit vIL-6 activity. Further addressing this area, the present study reports a new, ER-localized interaction partner of vIL-6, VKORC1 variant 2, which plays a role in vIL-6 function, independently of gp130. The major findings are summarized in Fig. 9 and discussed further below.
Fig 9.
Summary of our main findings and conclusions. Previous studies revealed that vIL-6 is expressed in latently infected PEL cells and functions in the ER compartment to promote cell growth and survival (5). The present study identified VKORC1v2 as an ER-localized binding partner of vIL-6 and mapped the vIL-6-binding region of VKORC1v2 to residues 31 to 39 (vBD), which is capable, when fused to a heterologous protein and targeted to the ER, of interfering with vIL-6 binding to VKORC1v2 and suppressing PEL growth and survival. These outcomes were also seen upon VKORC1v2 depletion and could be rescued by (shRNA-refractory) wild-type VKORC1v2 but not by a dialanine variant (DY39AA) with diminished ability to bind vIL-6. Neither VKORC1v2 depletion nor vBD overexpression had detectable negative effects on gp130 signaling, and VKORC1v2 overexpression did not enhance it. These findings suggest that activities of vIL-6 connected with its binding to VKORC1v2 are mediated independently of g130 signaling, which is known to promote growth and survival of various cell types. However, the mechanism of vIL-6 activity via VKORC1v2 association remains to be determined.
VKORC1 (variant 1) is a 159-amino-acid protein that catalyzes the reduction of vitamin K epoxide in the vitamin K cycle, which is involved in carboxylation of several proteins, including blood coagulation factors (18). The predicted structure of VKORC1 is a triple-transmembrane protein, and its topology deduced experimentally through a combination of glycosylation tagging and protease digestion by Tie and colleagues (21) is consistent with this model and places the N terminus in the ER lumen with the C tail projecting into the cytoplasm. Our data using N-glycosylation site-containing transfected VKORC1 also indicate this topology, as an introduced glycosylation target (NGT) in the TM1-TM2 loop was refractory to glycosylation, in contrast to the identically placed motif in VKORC1 variant 2 (Fig. 3D). However, recently published data from cysteine tagging and maleimide-polyethylene glycol (mal-PEG) modification experiments indicate a possible four-transmembrane structure for VKORC1 with cytoplasmic orientations of the N- and C-terminal regions and ER-lumenal exposure of the TM1-TM2 loop (20). The reason for this discrepancy is unclear, but it is conceivable that the protein can adopt more than one topology. Indeed, the data of Schulman et al. (20) did reveal evidence of mal-PEG modification of native VKORC1, indicating cytoplasmic exposure of TM1-TM2 loop C51 and C85 residues, although this was enhanced by N-terminally introduced cysteine and by ER permeabilization. The C-terminal mal-PEG modification would be expected of either the three- or four-transmembrane structural model and so is not diagnostic. Consistent with ER exposure of the TM1-TM2 loop is interaction via this region of ER-lumenal-localized thioredoxin-related proteins TMX, TMX4, and ERp18 (20), although, again, this could result from only a fraction of total VKORC1 being present in the corresponding orientation. Nonetheless, it is difficult to reconcile these data with the complete absence of glycosylation of our NGT-tagged VKORC1 variant 1, especially as equivalently tagged variant 2 was very efficiently glycosylated (∼100%) (Fig. 3D). Notwithstanding, it is very clear that the topology of VKORC1v2 places the post-TM, C-tail region (experimentally CBD tagged and protease protected [Fig. 3E]) in the ER lumen. The inverse topologies of variants 1 and 2 that our data indicate, while not yet accounted for mechanistically, provide a simple explanation for the divergent abilities of the VKORC1 protein to interact via a shared sequence with ER-lumenal vIL-6.
The sequence of VKORC1v2 interacting with vIL-6 comprises amino acids 31 to 39, this motif being both required and sufficient for vIL-6 binding. Indeed, ER targeting of this motif was able to inhibit growth and viability of HHV-8 latently infected BCBL-1 cells, functions previously shown to be mediated in PEL cells by ER-localized vIL-6 (5). Such inhibitory effects on PEL cells mediated by a 9-mer peptide sequence suggest potential future therapeutic strategies, based on disruption of the vIL-6 interaction with VKORC1v2. However, still to be determined is the mechanism of vIL-6 activity via interaction with variant 2, depletion of which inhibits PEL cell growth and induces apoptosis (reflecting effects of vIL-6 depletion). It is notable that rescue of this depletion phenotype was possible with (shRNA-resistant) wild-type VKORC1v2 but not with a mutated protein containing a vIL-6 interaction-disrupting dialanine substitution in the vIL-6 binding domain. This finding, together with diminished growth and viability induced by overexpression of ER-targeted vBD, indicates strongly that the vIL-6 interaction with VKORC1v2 is functionally relevant and links the common effects on cell growth and survival of vIL-6 and VKORC1v2 depletion and vBD challenge. Importantly, the mechanism of vIL-6 function via VKORC1v2 appears to be independent of gp130 signaling, currently the only known means of vIL-6 signal transduction and activity, as neither overexpression of vBD nor VKORC1v2 depletion had any inhibitory effect on gp130 activation or signaling, and VKORC1v2 overexpression did not affect it. Determining the underlying mechanistic basis of VKORC1v2-mediated vIL-6 activity is the subject of current investigation in our laboratory and may reveal further opportunities for vIL-6-inhibitory strategies and, ultimately, PEL therapy.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant CA76445 from the National Cancer Institute.
We thank Young Bong Choi for helpful discussions throughout the course of this project.
Footnotes
Published ahead of print 30 November 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Adam N, et al. 2009. Unraveling viral interleukin-6 binding to gp130 and activation of STAT-signaling pathways independently of the interleukin-6 receptor. J. Virol. 83:5117–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cain D, Hutson SM, Wallin R. 1997. Assembly of the warfarin-sensitive vitamin K 2,3-epoxide reductase enzyme complex in the endoplasmic reticulum membrane. J. Biol. Chem. 272:29068–29075 [DOI] [PubMed] [Google Scholar]
- 3. Chen D, Choi YB, Sandford G, Nicholas J. 2009. Determinants of secretion and intracellular localization of human herpesvirus 8 interleukin-6. J. Virol. 83:6874–6882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen D, Nicholas J. 2006. Structural requirements for gp80 independence of human herpesvirus 8 interleukin-6 (vIL-6) and evidence for gp80 stabilization of gp130 signaling complexes induced by vIL-6. J. Virol. 80:9811–9821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen D, Sandford G, Nicholas J. 2009. Intracellular signaling mechanisms and activities of human herpesvirus 8 interleukin-6. J. Virol. 83:722–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chow DC, et al. 2002. A structural template for gp130-cytokine signaling assemblies. Biochim. Biophys. Acta 1592:225–235 [DOI] [PubMed] [Google Scholar]
- 7. Dayel MJ, Hom EF, Verkman AS. 1999. Diffusion of green fluorescent protein in the aqueous-phase lumen of endoplasmic reticulum. Biophys. J. 76:2843–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fliegel L, Burns K, MacLennan DH, Reithmeier RA, Michalak M. 1989. Molecular cloning of the high affinity calcium-binding protein (calreticulin) of skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 264:21522–21528 [PubMed] [Google Scholar]
- 9. Gasperini P, Sakakibara S, Tosato G. 2008. Contribution of viral and cellular cytokines to Kaposi's sarcoma-associated herpesvirus pathogenesis. J. Leukoc. Biol. 84:994–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Helenius A, Aebi M. 2001. Intracellular functions of N-linked glycans. Science 291:2364–2369 [DOI] [PubMed] [Google Scholar]
- 11. Li H, Wang H, Nicholas J. 2001. Detection of direct binding of human herpesvirus 8-encoded interleukin-6 (vIL-6) to both gp130 and IL-6 receptor (IL-6R) and identification of amino acid residues of vIL-6 important for IL-6R-dependent and -independent signaling. J. Virol. 75:3325–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meads MB, Medveczky PG. 2004. Kaposi's sarcoma-associated herpesvirus-encoded viral interleukin-6 is secreted and modified differently than human interleukin-6: evidence for a unique autocrine signaling mechanism. J. Biol. Chem. 279:51793–51803 [DOI] [PubMed] [Google Scholar]
- 13. Miles SA, et al. 1990. AIDS Kaposi sarcoma-derived cells produce and respond to interleukin 6. Proc. Natl. Acad. Sci. U. S. A. 87:4068–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Molden J, Chang Y, You Y, Moore PS, Goldsmith MA. 1997. A Kaposi's sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. J. Biol. Chem. 272:19625–19631 [DOI] [PubMed] [Google Scholar]
- 15. Moore PS, Boshoff C, Weiss RA, Chang Y. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739–1744 [DOI] [PubMed] [Google Scholar]
- 16. Mullberg J, et al. 2000. IL-6 receptor independent stimulation of human gp130 by viral IL-6. J. Immunol. 164:4672–4677 [DOI] [PubMed] [Google Scholar]
- 17. Nicholas J, et al. 1997. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat. Med. 3:287–292 [DOI] [PubMed] [Google Scholar]
- 18. Oldenburg J, Bevans CG, Muller CR, Watzka M. 2006. Vitamin K epoxide reductase complex subunit 1 (VKORC1): the key protein of the vitamin K cycle. Antioxid. Redox Signal. 8:347–353 [DOI] [PubMed] [Google Scholar]
- 19. Rost S, et al. 2004. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature 427:537–541 [DOI] [PubMed] [Google Scholar]
- 20. Schulman S, Wang B, Li W, Rapoport TA. 2010. Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc. Natl. Acad. Sci. U. S. A. 107:15027–15032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tie JK, Nicchitta C, von Heijne G, Stafford DW. 2005. Membrane topology mapping of vitamin K epoxide reductase by in vitro translation/cotranslocation. J. Biol. Chem. 280:16410–16416 [DOI] [PubMed] [Google Scholar]
- 22. Wada I, et al. 1991. SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J. Biol. Chem. 266:19599–19610 [PubMed] [Google Scholar]
- 23. Wajih N, Hutson SM, Wallin R. 2007. Disulfide-dependent protein folding is linked to operation of the vitamin K cycle in the endoplasmic reticulum. A protein disulfide isomerase-VKORC1 redox enzyme complex appears to be responsible for vitamin K1 2,3-epoxide reduction. J. Biol. Chem. 282:2626–2635 [DOI] [PubMed] [Google Scholar]
- 24. Wan X, Wang H, Nicholas J. 1999. Human herpesvirus 8 interleukin-6 (vIL-6) signals through gp130 but has structural and receptor-binding properties distinct from those of human IL-6. J. Virol. 73:8268–8278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoshizaki K, et al. 1989. Pathogenic significance of interleukin-6 (IL-6/BSF-2) in Castleman's disease. Blood 74:1360–1367 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.