Abstract
For retroviruses such as HIV-1 and murine leukemia virus (MLV), active receptor recruitment and trafficking occur during viral entry. However, the underlying mechanisms and cellular factors involved in the process are largely uncharacterized. The viral receptor for ecotropic MLV (eMLV), a classical model for retrovirus infection mechanisms and pathogenesis, is mouse cationic amino acid transporter 1 (mCAT-1). Growth factor receptor-bound protein 2 (GRB2) is an adaptor protein that has been shown to couple cell surface receptors, such as epidermal growth factor receptor (EGFR) and hepatocyte growth factor receptor, to intracellular signaling events. Here we examined if GRB2 could also play a role in controlling infection by retroviruses by affecting receptor function. The GRB2 RNA interference (RNAi)-mediated suppression of endogenous GRB2 resulted in a consistent and significant reduction of virus binding and membrane fusion. The binding between eMLV and cells promoted increased GRB2–mCAT-1 interactions, as detected by immunoprecipitation. Consistently, the increased colocalization of GRB2 and mCAT-1 signals was detected by confocal microscopy. This association was time dependent and paralleled the kinetics of cell-virus membrane fusion. Interestingly, unlike the canonical binding pattern seen for GRB2 and growth factor receptors, GRB2–mCAT-1 binding does not depend on the GRB2-SH2 domain-mediated recognition of tyrosine phosphorylation on the receptor. The inhibition of endogenous GRB2 led to a reduction in surface levels of mCAT-1, which was detected by immunoprecipitation and by a direct binding assay using a recombinant MLV envelope protein receptor binding domain (RBD). Consistent with this observation, the expression of a dominant negative GRB2 mutant (R86K) resulted in the sequestration of mCAT-1 from the cell surface into intracellular vesicles. Taken together, these findings suggest a novel role for GRB2 in ecotropic MLV entry and infection by facilitating mCAT-1 trafficking.
INTRODUCTION
As obligatory parasites, viruses have evolved to exploit host cellular mechanisms to facilitate viral replication and infection. Cell entry is the first step in viral infection. Viral entry involves receptor binding and movement, either into the cell or across the cell membrane, followed by the penetration of the cell membrane. In the case of enveloped viruses, this step involves membrane fusion between the virus and cell membranes (15). For many retroviruses, active receptor recruitment and trafficking occur during entry. For example, receptor trafficking is indispensable for HIV infection. The binding of HIV to CD4, which resides in lipid rafts (membrane microdomains enriched in cholesterol, glycosphingolipids, and signaling phospholipids), results in the subsequent recruitment of the coreceptors CXCR4 and CCR5 to the lipid raft (44). For ecotropic murine leukemia virus (MLV) (eMLV), a distantly related retrovirus receptor, trafficking is also important. Soon after cell contact, eMLV appears to “surf” along cell filopodia toward the cell body (24). Moreover, eMLV is able to establish filopodium bridges between infected and uninfected cells to facilitate cell-to-cell transmission. Both processes are highly dependent on virus envelope glycoprotein-receptor interactions (42). However, the cellular factors that trigger and mediate the movement of the virus-receptor complexes on the surface and into cells are not well understood. After contact with the cell body, the virus is thought to either fuse with the plasma membrane or be taken up by clathrin-independent endocytosis and enters the cell cytoplasm (18, 23).
The principal receptor for eMLV is mouse cationic amino acid transporter 1 (mCAT-1) (3, 19, 50). mCAT-1 is a single polypeptide of 622 amino acids with 14 transmembrane domains and intracellular N and C termini (3). It is a member of the SLC7A amino acid transporter family, and its mammalian homologs share >80% amino acid identity along their entire lengths. Amino acid differences in the third extracellular loop control eMLV tropism, with the human protein being converted to a functional receptor by the exchange of residues in this loop (2). The remainder of the protein shares 89% amino acid identity between human and mouse homologs. Under physiological conditions, CAT-1 functions to transport cationic amino acids across the plasma membrane by facilitated diffusion. In resting cells, CAT-1 is distributed predominantly on the plasma membrane and resides in lipid rafts. Raft disruption by methyl-beta-cyclodextrin (a drug that extracts cholesterol) reduces syncytium formation and infection by eMLV without decreasing surface mCAT-1 levels (28). Consistent with its localization in lipid rafts and the role of caveolae in infection, mCAT-1 colocalizes with caveolin in different cell lines (33) and is internalized independently of clathrin-coated pits (23). Beyond the primary receptor, few other proteins have been shown to be important for eMLV infection. Earlier work demonstrated the importance of cytoskeletal integrity, a requirement for microtubule function, and actin polymerization in infection (24). However, the cellular proteins and molecular mechanism(s) that mediate eMLV–mCAT-1 trafficking are largely unknown.
GRB2 is a signaling adaptor protein ubiquitously expressed in all embryonic and adult tissues (14) and has been shown to couple epidermal growth factor (EGF) receptor (EGFR) and hepatocyte growth factor receptor activation to downstream signaling and receptor internalization (25, 27). Here we show that GRB2 is also important for eMLV entry and infection. The RNA interference (RNAi)-mediated suppression of endogenous GRB2 led to a specific reduction of eMLV infection, while vesicular stomatitis virus G protein (VSV-G)-pseudotyped particles showed no drop in the signal. The reduction corresponded to changes in virus binding and entry into cells, which was consistent with decreases seen in cell surface expression levels of mCAT-1. Using confocal microscopy and immunoprecipitation, a direct interaction of mCAT-1 and GRB2 was seen in resting cells. Interestingly, this interaction increased after eMLV binding to the cell surface. Moreover, the increase in mCAT-1–GRB2 binding correlated with the kinetics of virus-cell membrane fusion. The overexpression of the GRB2 R86K dominant negative mutant resulted in both the sequestration of mCAT-1 in the cytoplasm and the inhibition of eMLV infection. In contrast to the binding of GRB2-ligand-activated signaling receptors, such as EGFR or Met, the recruitment of GRB2 to eMLV-bound mCAT-1 does not require the phosphotyrosine binding site on the GRB2-Src homology 2 (SH2) domain. This study is the first to indicate a novel role for GRB2 in the early steps leading to eMLV infection, through the altered trafficking of the viral receptor mCAT-1.
MATERIALS AND METHODS
Reagents, cell culture, and antibodies.
All general reagents were obtained from Sigma-Aldrich or Fisher unless indicated otherwise. Plasmids pEYFP-N1-GRB2, pEYFP-N1-GRB2R86K (51), pGEX-4T-GRB2, and pGEX-4T-SH2 (55) were described elsewhere previously. All cell lines except for XC cells were maintained in high-glucose Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) containing 10% fetal bovine serum (FBS) plus penicillin-streptomycin (Pen-Strep). HEK293 cells and Moloney MLV-producing cells (CL-1) were obtained from the ATCC. XC cells were maintained in minimal essential medium (MEM) (Invitrogen) containing 5% FBS and Pen-Strep. The antibodies used were mouse anti-influenza virus hemagglutinin (HA) (12CA5; Roche), mouse anti-glutathione S-transferase (GST) (catalog number 13-6700; Zymed Laboratories), mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GADPH) (AM4300; Ambion), rabbit anti-GRB2 (sc-255; Santa Cruz), mouse anti-human transferrin receptor (catalog number 13-6800; Invitrogen), Alexa Fluor 647-conjugated mouse anti-human CD4 (BD Pharmingen), and goat anti-GP70 (ATCC) antibodies.
siRNA transfection.
The small interfering RNAs (siRNAs) used were human GRB2-1 (siGENOME no. 4; Dharmacon), human GRB2-2 (FlexiTube siRNA, Hs-GRB2-5; Qiagen), Hs-GAPDH-FlexiTube siRNA (Qiagen), and Silencer negative-control siRNA 1 (Ambion). HEK293 cells expressing mCAT-1–HA were transfected with siRNA in 96-well plates by using a reverse transfection method. First, 6 pmol siRNA rehydrated with Dharmafect transfection medium was added to each well, and the mixtures were incubated for 5 min at room temperature. Lipofectamine 2000 transfection reagent (Invitrogen) was then added, followed by 30 min of incubation. Aliquots (200 μl) of freshly suspended cells were added in DMEM to give 105 cells per well. The plate was incubated overnight at 37°C in a CO2 incubator. The next day, media were replaced with complete culture medium, and the plate was incubated for up to 72 h posttransfection. Controls were cells treated with transfection reagent alone or with nontargeting siRNAs.
Production of pseudotyped MLV and purification of wild-type Moloney MLV.
293FT cells (Invitrogen) were transfected with plasmids by calcium phosphate precipitation according to our previously reported method (21). Luciferase- or green fluorescent protein (GFP)-encoding pseudotyped MLV used for infection assays was generated with pGAG-POL, Friend MLV (pFr-MLV) Env or pVSV-G, and pΨ Luciferase or GFP (encoding firefly luciferase or GFP under the control of the MLV long terminal repeat [LTR] and packaging sequence). Limiting dilution of the luciferase-encoding virus indicated that 1 CFU corresponded to 100 cps of luciferase activity. Recombinant retroviruses containing firefly luciferase for use in the content-mixing assay were generated by using the same plasmids, with the addition of a Nef-luciferase construct (41). GFP-tagged eMLV- and GFP-tagged VSV-G-pseudotyped MLVs were generated by the same method with the same plasmids but with addition of an enhanced GFP-matrix fusion protein (43). The Env− MLV particles were generated by the same method but omitting the plasmid encoding Env. After an overnight incubation of the transfection mixture and cells, the medium was replaced. Supernatants were collected and filtered through a 0.45-μm cellulose acetate filter at 48 h and 60 h to remove cell debris. Moloney MLVs were collected from the culture supernatant from CL-1 cells. All medium was filtered through a 0.45-μm cellulose acetate filter. Virus particles were pelleted through a 20% (wt/vol) sucrose cushion for 4 h at 20,000 × g. The virus pellet was resuspended in DMEM and stored frozen at −80°C. GFP-tagged eMLV- and GFP-tagged VSV-G-pseudotyped MLV were titrated on HEK293 cells expressing mCAT-1–HA, and titers were matched for all work. The titers of eMLV on HEK293 and mouse NIH 3T3 cells (naturally expressing mCAT-1) were also compared and were found to be within 2-fold of each other. MLV was titrated with an XC plaque assay described previously (10).
Virus binding assay.
HEK293 cells expressing mCAT-1–HA seeded in 8-well chamber slides were reverse transfected with human GRB2-1 siRNA (siGENOME no. 4; Dharmacon), Allstars negative-control siRNA (Qiagen), and Ambion negative-control siRNA. At 72 h posttransfection, cells were washed once with ice-cold medium and then chilled to 14°C. eMLV-, VSV-G-, or Env−-pseudotyped EGFP-tagged MLV particles were incubated with cells at 14°C for 1 h. The incubation at 14°C allowed virus binding while preventing endocytosis so that only surface interactions of particles with cells were examined. Unbound virus was removed by rinsing cells twice with phosphate-buffered saline (PBS) and then fixed with 4% paraformaldehyde in PBS. To visualize the outline of cells, F-actin close to the plasma membrane was stained with Alexa Fluor 594-conjugated phalloidin without permeabilization. Under these conditions, some phalloidin penetrated through the cell membrane and bound cortical actin. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Enhanced yellow fluorescent protein (YFP) (EYFP)-tagged viruses on cells were imaged by using a Zeiss LSM 510 confocal microscope with a 100× oil (1.4-numerical-aperture [NA]) immersion objective. A total of 100 to 150 cells were examined under each condition.
Biotinylation and analysis of protein surface levels.
Before biotinylation, HEK293 cells expressing mCAT-1–HA, seeded in 96-well plates, were transfected with 2 independent specific siRNAs against GRB2 (for GRB2-1, siGENOME no. 4 [Dharmacon]; for GRB2-2, FlexTube siRNA, Hs-GRB2-5 [Qiagen]) or with nontargeting siRNA from Ambion, GAPDH siRNA, or transfection reagent only. At 72 h posttransfection, cells were rinsed twice with ice-cold PBS. An EZ-Link sulfo-N-hydroxysuccinimide (NHS)-LC-biotin (Pierce) stock solution (200 mg/ml) was freshly prepared and diluted to 0.5 mg/ml with ice-cold PBS prior to use. Next, 50 μl of biotin solution was added to each well, and the plate was then incubated at 4°C for 1 h with gentle agitation. The reaction was quenched with 1 volume of ice-cold 0.3 M glycine (pH 7.0), and the cells were washed once again with PBS. Cell lysates were prepared and then precipitated with streptavidin-agarose resin (Pierce) according to the manufacturer's instructions. The amounts of surface mCAT-1 and transferrin receptor were analyzed by Western blotting. One-tenth of the lysate was used to determine the total mCAT-1 or transferrin receptor content. All results were quantified by image densitometry using ImageJ software (1) of autoradiographs scanned at a 16-bit image depth. Amounts of surface mCAT-1 and transferrin receptor were compared to total amounts of mCAT-1 and transferrin receptor, respectively, to reveal the percentage of protein on the plasma membrane.
eMLV RBD binding assay.
A GFP-tagged receptor binding domain (RBD) (RBD-GFP) was made for the surface localization analysis of mCAT-1. The construct consisted of a synthetic gene, codon optimized for expression in human cells, flanked by BamHI and NotI restriction endonuclease sites. The gene was a fusion between the RBD (residues 1 to 271) of ecotropic murine leukemia virus, Friend 57, gp85, and GFP at its C terminus. The RBD portion was previously crystallized, and its structure was determined (11). In-frame XhoI and EcoRV sites followed in the nucleotide sequence. These were then followed by a sequence encoding a chitin binding domain (NEB, MA), a factor Xa site (IEGR), and six histidine residues. The open reading frame (ORF) ended in a TAA stop codon. This fragment was inserted into pcDNA 3.1 (Invitrogen), and EGFP was inserted in frame between the XhoI and EcoRV sites.
Protein production was performed with HEK293 cells by selecting G418-resistant colonies and choosing those with the highest level of protein production by Western blot analysis, probing for both GFP and the chitin binding domain by using commercially available antibodies. One clone that produced the largest amount of protein was cultivated by using a Fibercell system (medium cartridge; Fibercell, Frederick, MD) in 293 SFM culture medium (Invitrogen) supplemented with 1% FBS. The culture fluid surrounding the cells was collected on days 4 and 5 after the start of culture, when the cell density had reached 108 cells per ml. The secreted fusion protein was purified over a nickel-chelating Sepharose column (GE Biosciences, Piscataway, NJ) and eluted by using 0.3 M imidazole (pH 6.4). The protein purity was >80%, and the protein was fluorescent by fluorimetry.
For binding assays, cells were removed from plates using 2 mM EDTA in PBS (without calcium or magnesium) and then resuspended in 1 ml of ice-cold DMEM with 10% FBS in tubes. For each assay point, 104 cells were incubated at 14°C (to prevent endocytosis) in the presence of the indicated amount of protein for 1.5 h. The cells were then washed twice with PBS and analyzed for bound fluorescent protein by fluorescence-activated cell sorter (FACS) analysis (Accuri, Ann Arbor, MI). Control experiments using purified GFP gave signals that were 40-fold lower than peak signals seen with the recombinant RBD protein. Also, cells lacking the mCAT-1 protein showed similar background signals.
Content-mixing assay to measure virus entry signal and kinetics.
HEK293 cells expressing mCAT-1–HA (105 cells/sample) were incubated for 45 min with Env-Luc-containing virus at a multiplicity of infection (MOI) of 0.1 to 0.5. Excess virus was washed free of cells by pelleting at 200 × g for 5 min, and cells were resuspended in DMEM. The cells were pelleted again and resuspended in 0.1 ml of luciferase assay buffer (Promega) lacking detergent. Luciferase activity was measured after 1 min with a Turner Designs TD 20/20 luminometer and expressed as counts per second. Virus entry kinetics were determined by incubating 5 × 106 cells with virus at an MOI of 10 for 5 min at 37°C. Unbound virus (95%) was removed by rapid washing three times in DMEM and pelleting of the cells at 200 × g for 3 min. Cells were resuspended in 1 ml of DMEM and incubated at 37°C with gentle agitation. Samples of 0.1 ml of cells were removed at 15-min intervals for up to 105 min, and the luciferase activity of each aliquot was measured as described above.
GST pulldown assay and immunoprecipitation.
First, HEK293 cells expressing HA-tagged mCAT-1 were incubated with Moloney MLV at an MOI of 5 at 37°C for 0 min (no virus) and for 5, 10, 15, 30, and 45 min (5 × 106 cells for each time point), and the medium was then removed. Cells were immediately washed with ice-cold PBS 3 times and then lysed on ice in pulldown buffer (30 mM octyl-β-d-glucopyroside, 50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2 mM EDTA, 10% glycerol, 4 mM Na3VO4, 20 mM NaF, and EMD protease inhibitor cocktail). This buffer was used for all immunoprecipitation experiments. Cell lysates were cleared by centrifugation at 12,000 × g at 4°C for 15 min, and the supernatant was collected. Next, 0.5 to 1 mg of total protein from the supernatant was added to GST-GRB2 or GST-SH2 fusion protein-coated glutathione-agarose, incubated overnight at 4°C, and then washed 3 times with ice-cold lysis buffer, followed by washing with 50 mM Tris-HCl (pH 7.5). The bound protein was then eluted by the addition of 8 M urea-containing SDS-PAGE buffer (2% SDS, 8 M urea, 100 mM Tris-HCl, 4 mM Na3VO4, EMD protease inhibitor cocktail) at room temperature. The supernatant was collected by centrifugation at 12,000 × g for 5 min and then subjected to SDS-PAGE. After the detection of mCAT-1–HA by use of monoclonal antibody 12CA5 (Roche), the same membrane was stripped and blotted with antibody against GST (Invitrogen). For immunoprecipitation, HEK293 cells expressing HA-tagged mCAT-1 were stimulated with Moloney MLV, and cell lysates were made by the method described above for the GST pulldown assay. Anti-HA antibody 12CA5 was prebound to NHS-activated Sepharose (GE Biosciences, Piscataway, NJ). One milligram of each lysate was incubated with the antibody-coated beads at 4°C overnight. The beads were then washed, and the protein was eluted by using urea-containing SDS-PAGE buffer as described above. Immunocomplexes were analyzed by SDS-PAGE and detected by blotting with antibody against GRB2 (Santa Cruz Biotechnology, Santa Cruz, CA). Goat anti-eMLV gp70 antibody (6) was used to detect the eMLV envelope protein SU.
Infection assay and flow cytometry analysis.
HEK293 cells expressing mCAT-1–HA were transfected with pcDNA3-YFP, pEYFP-N1-GRB2, pEYFP-GRB2(R86K), or pEYFP-GRB2(P49L+G203R) by calcium phosphate precipitation. After 30 h, cells were infected with recombinant eMLV encoding truncated CD4 at an MOI of 0.1. The virus was left on the cells overnight at 37°C in a CO2 incubator and then removed the next morning. At 48 h postinoculation, cells were harvested in 5 mM EDTA in PBS and then fixed in freshly made 4% paraformaldehyde (pH 7.4) in PBS for 5 min. The infection marker was detected by surface staining with anti-CD4 antibody conjugated with Alexa Fluor 647 antibody (Becton Dickinson) according to the manufacturer's protocol. Stained cells were analyzed with a FACSCanto instrument (Becton Dickinson). Three independent experiments were performed, and at least 4 × 105 cells were collected each time. The cells were gated based on forward and side light scatter. Debris or aggregates were excluded, and only the individual cells were counted. The normalized percentage of infection within the transfected population was calculated as the proportion of cells that were positive for both Alexa Fluor 647 and YFP divided by the proportion of cells expressing YFP, which is the sum of the proportion of uninfected YFP-expressing cells and the proportion of infected YFP-expressing cells.
Confocal microscopy and colocalization analysis.
HeLa–mCAT-1–mStrawberry cells were seeded in 8-well chamber slides and then transfected with YFP-GRB2 or the YFP-GRB2(R86K) mutant with FuGene 6 (Roche) transfection reagent. After 30 h, the medium was changed to DMEM, and cells were returned to a 37°C incubator overnight. Cells were then incubated with Moloney MLV cells at an MOI of 10 for 45 min at 37°C. Cells were washed twice with PBS and fixed with freshly made 4% formaldehyde in PBS for 10 min. Cells were also stained with phalloidin conjugated with Alexa Fluor 633 (Invitrogen) at room temperature for 30 min. Cells were washed 3 times with PBS and then mounted with Slow Fade reagent with DAPI (Invitrogen). Chamber slides were observed with a Zeiss LSM 510 confocal microscope with a 100× oil (1.4-NA) immersion objective. Colocalization analysis was performed with AutoQuant X 2.2.1 software (Media Cybernetics, Bethesda, MD). GraphPad Prism software (GraphPad, La Jolla, CA) was used for statistical analysis using one-way analysis of variance (ANOVA) with Tukey's posttest or Student's t test.
RESULTS
RNAi suppression of endogenous GRB2 expression inhibits eMLV infection.
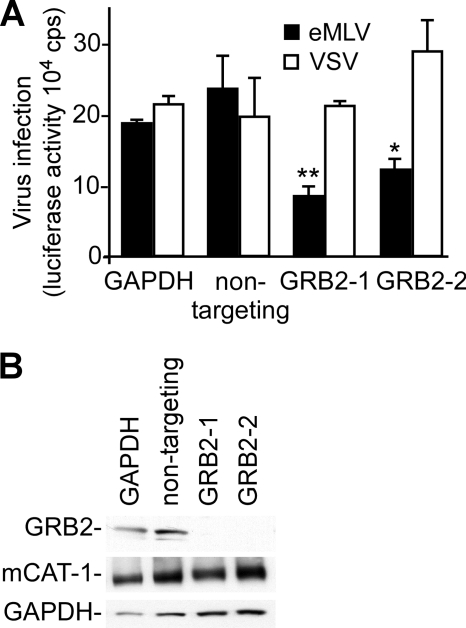
To determine if GRB2 played a role in MLV infection, we treated HEK293 cells expressing HA epitope-tagged mCAT-1 with siRNA against GRB2. The tagged protein retains wild-type virus receptor and amino acid transport properties and was characterized in previous work (8). The clone of HEK293 cells that was used in this work was selected to have levels of virus binding similar to that seen for mouse NIH 3T3 cells, indicating similar levels of surface-exposed receptor protein (not shown). For infection analysis, we used pseudotyped MLV bearing glycoproteins of either ecotropic murine leukemia virus (eMLV) or vesicular stomatitis virus (VSV). The eMLV glycoprotein gp85 mediates entry into the cell by a pH-independent mechanism either at the cell surface or from within caveolae (18, 32). In contrast, the G protein of VSV mediates entry by a clathrin- and pH-dependent mechanism (30). The VSV-G-pseudotyped virus thus served to show that the impact of each treatment was specific for the MLV envelope glycoprotein and not downstream of the receptor interaction. A pool of 4 siRNAs targeting GRB2 was found to significantly inhibit eMLV infection by 50% relative to that of control transfected cells (data not shown). This finding was validated with two distinct siRNAs against GRB2 (Fig. 1A). The VSV-G-pseudotyped virus was unaffected by this treatment, indicating that GRB2 suppression specifically impacted eMLV glycoprotein function and was not a downstream component of the retroviral infection mechanism. Western blots confirmed that GRB2 expression was suppressed by the GRB2-targeted siRNA (Fig. 1B). Conversely, GRB2 expression and eMLV infection were unaffected by transfection with 2 different nontargeting siRNAs.
Fig 1.
siRNA-mediated reduction of GRB2 expression inhibits eMLV infection. (A) Impact of siRNA on infection. Two siRNAs targeting distinct regions of the GRB2 mRNA were introduced into HEK293 cells stably expressing mCAT-1. siRNAs targeting GAPDH and a nontargeting siRNA (Ambion) were used as nonspecific siRNA controls. At 72 h posttransfection, cells were infected with eMLV gp85- or VSV-G-pseudotyped MLV encoding firefly luciferase at an MOI of 0.1. Infection levels were determined by measuring the luciferase activity of cells at 36 h postinoculation. The data represent 3 independent experiments. (B) Determination of RNAi-mediated reduction in GRB2 expression levels. Cells transfected with siRNA were collected at 48 h, and the levels of GRB2, mCAT-1, and GAPDH were examined by Western blotting using specific antibodies against GRB2 and GAPDH. The expression of mCAT-1 was detected with an anti-HA (12CA5) monoclonal antibody recognizing an HA epitope tag at the C terminus of mCAT-1. ∗, P < 0.05; ∗∗, P < 0.01 (determined by one-way ANOVA).
Impact of GRB2 on eMLV binding.
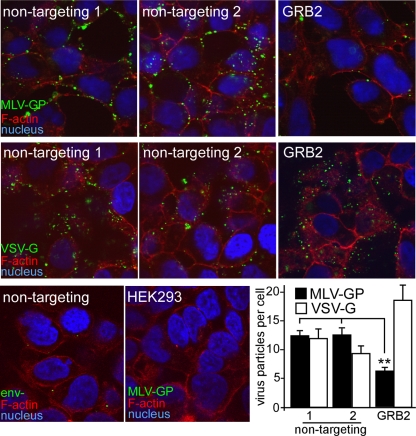
The only difference between the eMLV- and VSV-pseudotyped retroviruses is in the glycoproteins present on the virion surface. Since the VSV-G-pseudotyped virus was not inhibited by reduced GRB2 expression levels, it was likely that GRB2 played a specific role in eMLV glycoprotein-mediated entry and infection. The binding of virus to cells is the first step in entry, so we first investigated whether GRB2 plays a role in eMLV binding. eMLV particles were made fluorescent by the incorporation of a matrix-GFP fusion protein (43). These particles were visualized to bind to cells by conventional microscopy as well as confocal epifluorescence microscopy. Image analysis revealed that the number of eMLV particles associated with cells treated with GRB2 siRNA was <50% (P < 0.01) (Fig. 2) of that seen for cells transfected with nontargeting siRNA (Fig. 2, top panels), while VSV-G-pseudotyped particles showed no similar drop (Fig. 2, middle panels), consistent with what was seen in the infection assay. For this experiment, cells were incubated with virus at 14°C to prevent endocytosis (47), and Polybrene was not used. Under these conditions, nonspecific interactions were minimized, with particles lacking an envelope protein (Env−) or cells lacking a receptor showing few eMLV-pseudotyped particles bound (Fig. 2, bottom panels). The results of this binding assay indicated that GRB2 suppression directly affects virus-receptor interactions with cells.
Fig 2.
Suppression of endogenous GRB2 inhibits eMLV binding. Fluorescent eMLV particles were generated by producing virus particles containing a fusion of the MLV matrix protein coupled to GFP. Virus particles were purified by pelleting through a 20% sucrose cushion and titrated on HEK293 cells expressing mCAT-1–HA. siRNA against GRB2 or nontargeting siRNA (Ambion, Qiagen) was transfected into cells, and virus binding was measured after 72 h. Cells were first chilled to 14°C to prevent endocytosis, and GFP-tagged eMLV gp85 (top panels)- or VSV-G (middle panels)-pseudotyped particles were added for 1 h. Unbound virus was removed by rinsing with DMEM, and cells were fixed in paraformaldehyde. In order to visualize the cell outline, the nonpermeabilized cells were treated with Alexa Fluor 594-conjugated phalloidin. Under these conditions, some stain permeated through the cell membrane and stained cortical actin. Cell nuclei were stained with DAPI. Samples were then examined by using confocal microscopy. As a control for the nonspecific binding of particles, particles lacking an envelope glycoprotein (Env−) or eMLV g85-pseudotyped particles were incubated with mCAT-1-expressing cells or HEK293 cells lacking a receptor, respectively (bottom panels). The number of virus particles bound to the cells was determined by using CellProfiler image analysis software to count virus particles associated with each cell. The average numbers of viral particles per cell (with standard deviations) are shown and represent results obtained from 10 random images, each containing >20 cells for each individual treatment (bottom right). ∗∗, P < 0.01, determined by one-way ANOVA.
RNAi-mediated suppression of endogenous GRB2 reduces mCAT-1 expression on the cell surface.
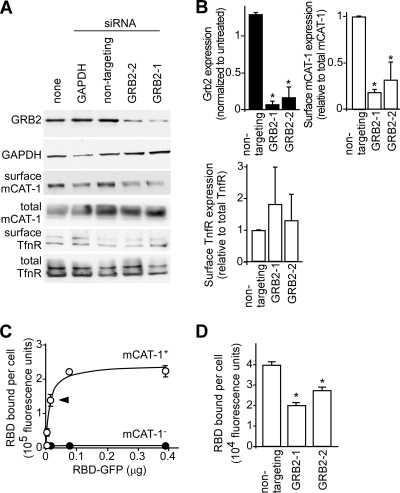
One mechanism for the siRNA suppression of GRB2 in reducing virus particle binding is the alteration of mCAT-1 surface expression. Cells were treated with control or GRB2 siRNAs, and cell surface proteins were labeled with the membrane-impermeable biotinylation reagent sulfo-NHS-LC-biotin. Cell lysates were prepared, biotin-labeled surface proteins were precipitated by using streptavidin-agarose resin, and mCAT-1 or the cell surface protein transferrin was detected by Western blotting (Fig. 3A). The intensities of the bands were quantified by densitometry (Fig. 3B). Both siRNAs targeting GRB2 (GRB2-1 and GRB2-1) reduced GRB2 expression levels to <20% of those in untreated cells or nontargeting-siRNA-treated cells. In these cells, the ratio of surface levels of mCAT-1 to total mCAT-1 levels was reduced to <30% of that of untreated or nontargeting-siRNA-treated cells, while the total mCAT-1 level remained relatively constant (Fig. 3A, third panel, and B, right). The surface localization of transferrin receptor, another integral membrane protein with a single transmembrane segment, was on average not reduced.
Fig 3.
RNAi suppression of endogenous GRB2 reduces surface mCAT-1 localization. (A) HEK293 cells stably expressing mCAT-1–HA were transfected with siRNAs against GAPDH, nontargeting siRNA (Ambion), or two siRNAs targeting GRB2. At 48 h posttransfection, cells were either lysed in SDS-PAGE buffer or surface labeled with membrane-impermeable sulfo-NHS-LC-biotin, and labeled proteins were then immunoprecipitated with streptavidin-agarose resin. Proteins present in cell lysates or bound to beads were detected after Western blotting using anti-GRB2 (top panel), anti-GAPDH (second panel), anti-HA (third and fourth panels), or anti-human transferrin receptor (TfnR) (fifth and sixth panels) antibodies. (B) Densitometry and analysis of band intensities from 3 independent Western blots. GRB2 levels were normalized to those of untreated samples (top left). Surface-labeled mCAT-1 (top right) and transferrin receptor (bottom left) were normalized to total mCAT-1 or transferrin receptor levels in cell lysates for nontargeting and GRB2-specific siRNAs, averaged between 2 independent experiments. (C) Binding curve for the MLV receptor binding domain (RBD)-GFP fusion protein binding to cells. Protein purified over a nickel chelating Sepharose column was incubated with cells at 14°C for 1.5 h. After washing, the mean fluorescence intensity of the cells was determined by FACS analysis. Data for signals from HEK293 cells expressing mCAT-1 (open circles) or the parental cell line lacking a receptor (solid circles) are shown. The arrowhead indicates the amount of protein (15 ng per sample) used in subsequent assays. (D) Binding of the RBD-GFP fusion protein to cells transfected with the indicated siRNAs. The subsaturating amount of RBD-GFP indicated in panel C was used, and the incubation time was 1 h at 14°C. ∗, P < 0.05, determined by one-way ANOVA.
As an additional test, surface levels of mCAT-1 were measured by using a fluorescently tagged MLV receptor binding domain (11). This protein was generated in HEK293 cells to ensure correct folding and was purified by a 6×His tag at its C terminus. The protein demonstrated saturable binding kinetics on HEK293 cells expressing the mCAT-1 receptor but showed no specific interaction with HEK293 cells lacking the receptor (Fig. 3C). As described above, cells were treated with siRNA against GRB2, and the binding of the RBD-GFP fusion was measured by FACS analysis. Compared to cells treated with nontargeting siRNA, the GRB2-targeting siRNA reduced the mean fluorescent intensity of cells by 50% and 30%, respectively (Fig. 3D). While not as great as that seen in the biotinylation experiment, both surface labeling experiments were consistent with GRB2 being a necessary factor in modulating surface levels of mCAT-1.
Impact of GRB2 on eMLV entry.
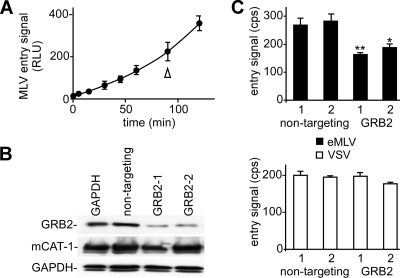
We have shown that endogenous GRB2 plays a role in eMLV binding by modulating the expression of surface mCAT-1. If GRB2 is controlling this surface receptor access, then a similar drop in the virus entry efficiency should be seen. A content-mixing assay used by ourselves and others (21, 41, 52) was used to measure the movement of virion contents into the cell cytosol that occurs shortly after glycoprotein-induced membrane fusion. Luciferase is encapsulated into the lumen of fully infectious, intact viral particles. When the virus envelope fuses with the cell membrane, the encapsulated luciferase can access its substrates, ATP and luciferin, the latter of which is readily taken into the cells by facilitated diffusion.
The kinetics of eMLV entry were measured by using the luciferase-containing virus. Instead of cell suspensions, the assay was modified to work with adherent cells on 96-well plates. The kinetics were measured, and the signal was found to increase semilinearly up to 2 h (Fig. 4A). In further work, the signal plateaued after this time (not shown). We therefore chose a 90-min time point for subsequent measurements. Both siRNAs targeting GRB2 again reduced GRB2 expression levels (Fig. 4B) and reduced the eMLV entry signal significantly (P < 0.05) (Fig. 4C) but did not affect the signal from VSV-G-pseudotyped particles. This result indicated that the suppression of GRB2 impacts virus entry, as would be expected if receptor function had been affected.
Fig 4.
Suppression of endogenous GRB2 inhibits eMLV entry. A content-mixing assay was used to assess the impact of the reduction of GRB2 expression levels on virus entry. (A) The assay was adapted to 96-well plates, and the entry kinetics of eMLV particles were measured. In other assays, the entry signal reached a plateau after 120 min, and so a 90-min time point was chosen for subsequent assays. RLU, relative light units. (B) The impact of the GRB2-targeting siRNA was confirmed with cells used in the entry assay and showed a reduction in GRB2 levels but no change in either mCAT-1 or GAPDH levels in cells. (C) The impact of the indicated siRNA on eMLV entry into cells was then measured. Cells were transfected with siRNA and then challenged with eMLV gp85 (top)- or VSV-G (bottom)-pseudotyped, luciferase-containing MLV 72 h later. After 90 min, the luciferase activity in intact cells was measured by using luciferin-containing buffer lacking detergent. ∗, P < 0.05; ∗∗, P < 0.001 (determined by one-way ANOVA).
mCAT-1 binds to GRB2 in vitro.
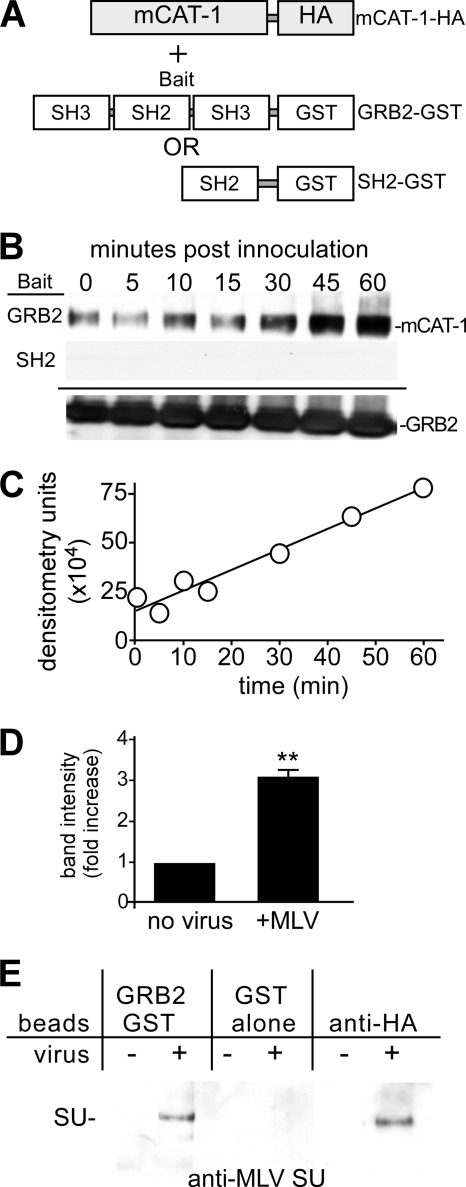
To gain further insight into the molecular role of GRB2 in eMLV entry, we tested whether mCAT-1 and GRB2 interact. Glutathione S-transferase (GST)-GRB2 fusion proteins (wild-type GRB2 and portions of different functional domains) were used previously to identify and characterize proteins that interact with GRB2 (4, 25). A nonionic detergent (n-octyl-β-d-glucopyranoside)-containing buffer was modified from that previously demonstrated to preserve the interaction between mCAT-1 and MLV gp85 (8) and was used for pulldown assays. Cell lysates from HEK293 cells expressing HA-tagged mCAT-1 before or after incubation with virus were applied onto glutathione beads coupled to GST-GRB2 (Fig. 5A). In resting cells, mCAT-1 was found to bind GRB2 at a basal level (Fig. 5B, top, lane 1). Interestingly, in the presence of virus, a time-dependent increase in the association between mCAT-1 and GRB2 was observed. Binding increased up to 60 min (Fig. 5B, top, lanes 2 to 7). In other work, a plateau was seen after this time. This increase followed kinetics similar to those seen with the eMLV content-mixing assay.
Fig 5.
In vitro binding of mCAT-1 to GRB2. (A) Schematic representation of the configuration of the pulldown assay using GST-GRB2 and GST-GRB2-SH2 fusion proteins as bait. (B) GST pulldown assay. HEK293 cells stably expressing HA-tagged mCAT-1 were incubated with eMLV for the indicated times. Cell lysates were then made, and the protein concentration was determined by a bicinchoninic acid (BCA) assay. The same amount of total protein from each sample was incubated with glutathione-agarose beads that had been coated with either GST-GRB2 (top) or GST-GRB2-SH2 (middle). The amount of HA-tagged mCAT-1 retained on the beads was analyzed by Western blotting using an anti-HA monoclonal antibody. The GST-tagged GRB2 present in each sample was blotted with an anti-GST antibody as a loading control (bottom). (C) Densitometry analysis of HA–mCAT-1 precipitated with GST-GRB2 shown in panel B. (D) Relative fold change of mCAT-1 binding to GST-GRB2 in untreated cells after a 45-min exposure to eMLV. Data represent 3 independent experiments. ∗∗, P < 0.01 (determined by Student's t test). (E) The surface domain of MLV gp85 (SU) coprecipitates with GRB2. Untreated (−) or virus-treated (+) cells were prepared as described above and applied onto GST-GRB2-, GST-, or anti-HA-coated beads. The bound protein on Western blots was then analyzed by probing with an anti-SU antibody.
GRB2 binds to receptor tyrosine kinases (RTKs) through specific phosphotyrosine residues in its Src homology 2 (SH2) domain, and the isolated SH2 domain retains binding activity (29). However, for mCAT-1, a GST-tagged SH2 domain did not bind mCAT-1 (Fig. 5B, middle). This indicated that binding between GST-GRB2 and mCAT-1 is specific but that the SH2 domain is insufficient in isolation to elicit the GRB2–mCAT-1 association. Compared to that in resting cells, quantitative analysis of multiple blots showed a consistent 3-fold increase in the mCAT-1 binding to GRB2 at 45 min after incubation with virus (P < 0.01) (Fig. 5D). As a control for receptor integrity in the buffer and the specificity of the beads, blots were probed for MLV gp85 by using an antibody against the surface domain (SU). With either GST-GRB2- or anti-HA-primed beads, SU was observed but not when GST alone was used as the bait (Fig. 5E).
Immunoprecipitation reveals that mCAT-1 binds to endogenous GRB2.
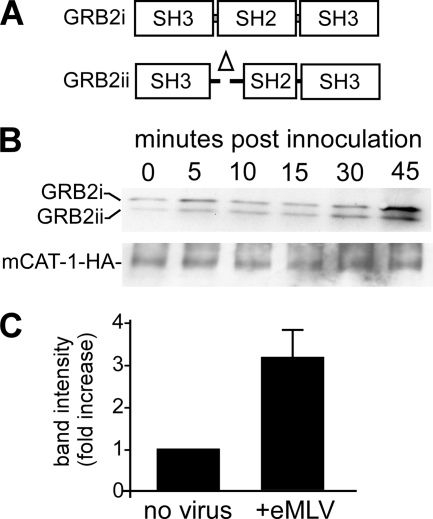
GRB2 is composed of one SH2 domain flanked by two Src homology 3 (SH3) domains (Fig. 6A, top) (27). The SH2 domain binds to specific phosphorylated tyrosine peptide sequences and protein ligands (31), whereas the N-terminal SH3 domain binds the guanine nucleotide exchange factor SOS1, an important mediator that couples activated growth factor RTKs to Ras signaling (16). The N-terminal SH3 domain was also reported previously to play an important role in receptor endocytosis, by interacting with the E3 ubiquitin ligase Cbl and the GTPase dynamin 1 (4). The C-terminal SH3 domain of GRB2 also functions to link activated signaling receptors to the phosphatidylinositol 3-kinase (PI3K)/Akt pathway by binding to GRB2-associated binding protein 1 (Gab1) (26). GRB2 mRNA exists as two splice variants (12). Full-length GRB2 mRNA gives a protein of 25 kDa, while the smaller mRNA gives a 20-kDa protein, with a deletion in the N-terminal portion of the SH2 domain from amino acids 59 to 100 (Fig. 6A). This portion harbors the phosphotyrosine recognition site used to bind EGFR and other receptor tyrosine kinases, rendering it unable to bind via this motif. Accordingly, the smaller product behaves as a dominant negative protein in EGFR trafficking (12).
Fig 6.
Association of endogenous GRB2 with mCAT-1 increases in a time-dependent manner after eMLV exposure. (A) Schematic showing predominant 25-kDa and 20-kDa forms of GRB2 seen in cells due to the alternative splicing of pre-mRNA. The smaller protein differs by a deletion at the N-terminal side of the SH2 domain. (B) mCAT-1 immunoprecipitates GRB2 from cells. HEK293 cells stably expressing HA-tagged mCAT-1 were incubated with eMLV, and cell lysates were made as described in the legend of Fig. 5. Protein was then immunoprecipitated by using anti-HA covalently coupled Sepharose beads. Endogenous GRB2 bound to mCAT-1 was detected by Western blotting with an anti-GRB2 antibody. The membrane was stripped and then reprobed with anti-HA antibody to determine the amount of mCAT-1 present in each sample. (C) Quantitative analysis of the GRB2 binding level determined by densitometry. The relative fold change in endogenous GRB2 binding to mCAT-1 after a 45-min exposure to eMLV was calculated relative to that of cells not incubated with virus. Data are the averages and standard errors from 3 independent experiments.
The in vitro binding assay (Fig. 5) showed that mCAT-1 associated with GST-tagged GRB2 in a time-dependent manner upon eMLV stimulation. To determine if GRB2 and mCAT-1 associate in cells, lysates were prepared from HEK293 cells expressing mCAT-1–HA and were incubated with beads covalently coupled to anti-HA monoclonal antibody. The bound protein was then eluted by the addition of buffer containing SDS and urea and by applying the eluate onto SDS-PAGE gels. Western blotting showed that endogenous GRB2 coprecipitated with mCAT-1 (Fig. 6B). Interestingly, both forms of GRB2 were present. At this point, while we do not know whether GRB2 interacts with mCAT-1 directly or indirectly through other cellular factors, this finding indicated that the mCAT-1 interaction with GRB2 does not require the SH2 phosphotyrosine recognition site of GRB2 used by EGFR and some other receptor tyrosine kinases. Similar to data from the in vitro binding studies, quantitative analysis of multiple blots showed a 3-fold increase in binding between endogenous GRB2 and mCAT-1 after a 45-min incubation with virus (Fig. 6C).
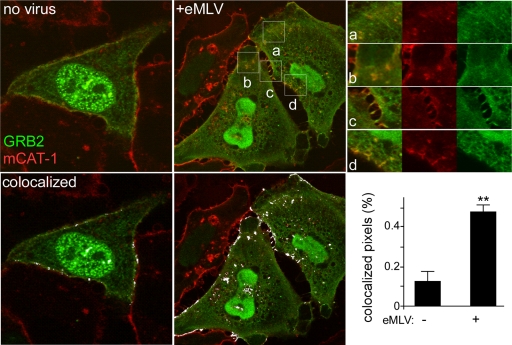
eMLV stimulation results in the subcellular redistribution of GRB2.
To determine the subcellular localization of the GRB2–mCAT-1 complex, HeLa cells stably expressing mStrawberry-tagged mCAT-1 were transfected with YFP-tagged GRB2. Consistent with previous reports of GRB2 localization, YFP-GRB2 was seen diffusely distributed in the cytoplasm and in nuclei, and some was close to the plasma membrane (46). As expected, mCAT-1 was primarily on or close to the plasma membrane, with some protein present in the perinuclear region consistent with the endoplasmic reticulum (ER) or Golgi apparatus (Fig. 7, left). Importantly, some of the YFP-tagged GRB2 overlapped with the mStrawberry-tagged mCAT-1 signal on the plasma membrane (Fig. 7, bottom left) and is consistent with the basal-level binding that was detected by immunoprecipitation (Fig. 5 and 6). As with the immunoprecipitation, after 45 min of incubation with Moloney MLV, higher levels of GRB2 appeared colocalized with the mCAT-1 signal (Fig. 7, center and top right); colocalization analysis revealed a 3-fold increase in the level of the GRB2 signal overlapping that of mCAT-1 (Fig. 7, bottom right). The mCAT-1 protein was also visible within the cell cytoplasm in punctate clusters together with GRB2, which could represent endocytic vesicles. Interestingly, a similar redistribution of GRB2 with EGFR was reported previously after the treatment of cells with EGF for 20 min (46). The level of colocalization of mCAT-1 with GRB2 also appeared to be increased at contact points between cells (Fig. 7, top right).
Fig 7.
Confocal microscopy analysis shows increased colocalization of GRB2 and mCAT-1 signals during eMLV infection. HeLa cells stably expressing mStrawberry-tagged mCAT-1 were seeded into 8-well chamber slides and then transfected with YFP-tagged GRB2. At 48 h posttransfection, cells were untreated (no virus) or incubated with eMLV for 45 min at 37°C at an MOI of 10. Unbound virus was then removed, and cells were fixed and examined with a confocal microscope using a 100× objective lens. Images at the bottom show highlighting (white) of overlapping mCAT-1 and GRB2 signals, as determined by using AutoQuant X v.2.2.1 software. Panels at the top right show magnified views and each fluorescent channel of the indicated areas labeled a to d. The bar graph (bottom right) shows averages and standard errors of the means (SEM) of data obtained from 9 images from 3 independent experiments including more than 45 cells. ∗∗, P < 0.001, determined by Student's t test.
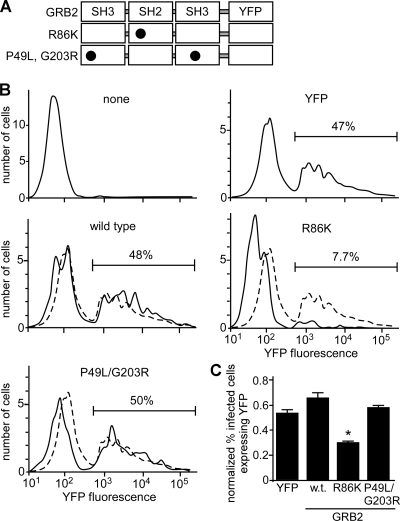
Overexpression of the dominant negative GRB2 R86K mutant reduces eMLV infectivity.
After the binding of EGF to EGFR, a rapid recruitment of GRB2 occurs at the plasma membrane. The association of EGFR with GRB2 occurs through the interaction of the GRB2 SH2 domain with phospho-Tyr1068 in the activated EGFR (39). The mutation of R86 to K in the GRB2 SH2 domain (Fig. 8A) results in a dominant negative protein that is unable to recognize and bind to phospho-Tyr1068 on activated EGFR and the subsequent inhibition of downstream Ras signaling (5, 29). Consistent with this, the GRB2 R86K mutant is unable to translocate to the membrane upon EGF stimulation. Conversely, the mutations P49L (N-terminal SH3 domain) and G203R (C-terminal SH3 domain) block the interactions of dynamin 1, Cbl, and Gab1 with GRB2 (51).
Fig 8.
FACS analysis reveals that overexpression of the dominant negative GRB2 R86K mutant inhibits eMLV infection. (A) Schematic of the YFP-tagged GRB2 wild type, SH2 domain R86K mutant, and SH3 domain P49L G203R mutant. Circles indicate sites of amino acid substitutions. (B) HEK293 cells expressing mCAT-1–HA were untreated (none) or transfected with plasmids encoding YFP, YFP-GRB2 (wild type), YFP-GRB2(R86K), and YFP-GRB2(P49L G203R). Two days after transfection, cells were challenged with recombinant eMLV encoding a truncated CD4 protein as an infection marker. Two days after infection, cells were stained with Alexa Fluor 647-labeled anti-CD4 antibody and analyzed by FACS. At least 5 × 105 cells were analyzed. The horizontal bar shows the gate containing infected cells, with percentages of infected cells indicated. (C) To compensate for differences in expression levels of each YFP-tagged protein, data were normalized by calculating the percentage of infected cells within the transfected population (number of cells infected and expressing YFP/total number of YFP-expressing cells). Data are the averages and standard deviations from 3 independent experiments. ∗, P < 0.01, determined by one-way ANOVA.
To gain a further understanding of the mechanism of action of GRB2 for eMLV infection, we expressed wild-type GRB2 or the dominant negative GRB2 mutants in mCAT-1-expressing HEK293 cells, and virus infection was measured by FACS analysis. The GRB2 constructs were tagged with EYFP, a modification that was previously shown not to affect the phenotypes of wild-type GRB2 and its mutants (22). A recombinant eMLV encoding a truncated form of CD4 was used as an infection marker. This marker can be stained with anti-CD4 antibody, but the truncation prevents signaling (38). Cells expressing the YFP-tagged GRB2 R86K construct showed a 6-fold reduction in infection compared to cells expressing YFP alone or wild-type GRB2-YFP (Fig. 8B). In contrast, the SH3 mutant form did not alter infection (Fig. 8B, bottom left). To compensate for differences in transfection and expression efficiencies, the data were normalized by comparing the number of infected cells with a strong YFP signal to the number of infected cells lacking a YFP signal (Fig. 8C). While this analysis reduced the overall impact of the R86K mutant to a 2-fold drop, this was significant (P < 0.01). An examination of the expression intensity of the R86K form (data not shown) suggested that it may have a higher turnover rate than the wild-type protein, and so the normalized data may be an underestimate of its potency. All three constructs did not affect cell viability, as determined by a trypan blue exclusion assay.
Overexpression of the GRB2 R86K mutant depletes mCAT-1 from the cell surface.
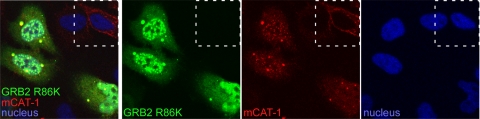
The expression of the dominant negative GRB2 R86K mutant inhibited eMLV infection. However, the canonical recognition site for phosphotyrosine residues present in the SH2 domain of GRB2 is apparently dispensable for the binding of GRB2 to mCAT-1 during virus entry. To further elucidate the mechanism of the GRB2–mCAT-1 interaction in infection, the YFP-tagged GRB2 R86K mutant was expressed in HeLa cells expressing mStrawberry-tagged mCAT-1 and examined by confocal microscopy. The R86K form showed a very distinct colocalization with mCAT-1. However, the expression of the GRB2 R86K mutant resulted in a prominent redistribution of plasma membrane-associated mCAT-1 into intracellular puncta containing the GRB2 R86K form (Fig. 9). This was in striking contrast to the pattern seen in neighboring cells lacking GRB2 R86K expression (Fig. 9, top right). This finding indicated that the GRB2 R86K mutant inhibits eMLV infection by the intracellular sequestration of mCAT-1, making it inaccessible to virus. In addition, while the SH2 domain may be dispensable for the association of mCAT-1 and GRB2, it must play a role in the transportation to or retention of mCAT-1 in the plasma membrane, presumably through SH2-mediated interactions with an as-yet-unidentified protein.
Fig 9.
Overexpression of dominant negative GRB2(R86K) causes a loss of mCAT-1 from the cell surface and accumulation inside the cell. HeLa cells stably expressing mStrawberry-tagged mCAT-1 were transfected with a plasmid encoding the YFP-tagged GRB2 R86K mutant. At 48 h posttransfection, cells were fixed, and nuclei were stained with DAPI and then observed by confocal microscopy. Composite images as well as each fluorescence channel are shown. The square on the top corner shows a cell not expressing the GRB2 R86K mutant.
DISCUSSION
Viruses have developed complex interactions with host cellular machinery to promote viral invasion, replication, and persistence. Ecotropic MLV, a gammaretrovirus, utilizes mouse or rat cationic amino acid transporter 1 as its principal receptor (3). In this study, we report that GRB2, a major cellular adaptor protein, is important for eMLV entry and infection by interacting with the virus receptor mCAT-1. We first showed that the RNAi-mediated suppression of endogenous GRB2 resulted in a 50% reduction in eMLV infection (Fig. 1), an observation that was later substantiated by the overexpression of a dominant negative form of GRB2 (Fig. 8). Since cell entry is the first step for the virus to establish infection, the role of GRB2 in eMLV entry was next examined. Consistent with the reduction in infection observed, the inhibition of endogenous GRB2 also resulted in a similar reduction in levels of eMLV binding and membrane fusion, while the infection and entry of a VSV-G-pseudotyped MLV were not affected (Fig. 2 and 4, respectively). Because the only differences between the MLV- and VSV-pseudotyped viruses are the envelope glycoproteins that they bear and receptors with which they interact, we investigated the impact of GRB2 on the surface expression of mCAT-1. Both the chemical labeling of cell surface protein and an RBD (receptor binding domain of gp85) binding assay indicated that surface mCAT-1 levels were significantly reduced as a result of the GRB2 depletion, while the total mCAT-1 levels remained comparable to those of the controls. A combination of biochemical and imaging approaches was then used to dissect the molecular mechanism of the mCAT-1–GRB2 interaction. These studies revealed that mCAT-1 associated with GRB2 in uninfected cells. However, this interaction was enhanced by the incubation of cells with purified MLV (Fig. 5, 6, and 7). Furthermore, the interaction between GRB2 and mCAT-1 increased in a time-dependent manner that correlated with the entry kinetics of eMLV (Fig. 5 and 6).
GRB2 has been shown to bind a variety of receptor and nonreceptor tyrosine kinases, such as the EGFR (14). In response to EGF binding to its receptor, GRB2 is recruited to plasma membrane-localized EGFR through its SH2 domain and subsequently cointernalizes with EGFR into early endosomes in an SH3 domain-dependent manner (51). In contrast to the EGFR, our studies revealed that the binding between GRB2 and mCAT-1 did not depend on the recognition of phosphotyrosine resides through the SH2 domain. Two isoforms of endogenous GRB2 bound to mCAT-1 were detected by immunoprecipitation in HEK293 cells. Both isoforms were previously identified in different cell types, and they are produced by splice variation in the GRB2 pre-mRNA (12). The shorter form differs from the full-length protein by lacking part of the SH2 domain (residues 59 to 100), which is responsible for binding phosphotyrosine residues in the receptor tyrosine kinase (45). Arginine 86 is a key residue important for this function and is within the portion missing in the smaller isoform. The finding that this isoform binds to and responds by increased binding to mCAT-1 in the presence of eMLV, like the full-length protein, indicates that this part of the SH2 domain is not necessary to bind mCAT-1 (Fig. 6). Consistent with this observation, confocal imaging also showed that the SH2-defective GRB2 R86K mutant still bound mCAT-1 (Fig. 9). The in vitro binding work showed that the GRB2-SH2 domain, by itself, did not bind mCAT-1 (Fig. 5). This result showed that the mCAT-1–GRB2 interaction is distinct from growth factor receptors like EGFR and must utilize a different strategy to engage GRB2. In recent years, numerous studies have reported that some SH2 domain interactions do not require the canonical phosphotyrosine-dependent mechanisms. For example, the in vivo association of V-Abl and GRB2 was found previously to involve a non-phosphotyrosine-dependent mechanism (37). Similarly, the HIV Nef protein interacts with the Lck-SH2 domain in a phosphotyrosine-independent manner (9).
Both RNAi and the overexpression of dominant negative GRB2 imply that GRB2 plays a significant role in the plasma membrane distribution of mCAT-1. Two possible mechanisms that could contribute to the reduced level of surface mCAT-1 are that the inhibition of the GRB2 function either promotes the endocytosis of mCAT-1 or decreases its anterograde trafficking to the plasma membrane. The expression of the GRB2 R86K protein resulted in the localization of most of the mCAT-1 protein to the cytosol, rendering it inaccessible to MLV and resulting in reduced viral infection. Unfortunately, our data do not discriminate if the intracellular protein was newly synthesized or cycling from the cell surface. Since GRB2 does not act alone in trafficking proteins, it is likely that protein binding partners could also be affected by the change in the SH2 domain. This activity would perturb downstream signaling events that depend on GRB2-SH2 domain interactions. Since the GRB2 R86K mutant retains two functional SH3 domains, it remains competent for binding to mCAT-1 but is unable to interact with downstream factors that are necessary for mCAT-1 surface localization. This finding suggests that mCAT-1's functional trafficking relies on the integrity of GRB2.
The only other cellular factor that is known to affect CAT-1 surface expression is protein kinase C-α (PKC-α). The activation of PKC indirectly promotes the endocytosis of human CAT-1 (40). Recently, it was reported that human CAT-1 undergoes clathrin-dependent endocytosis as a result of PKC activation (49). However, eMLV uses a clathrin-independent endocytic pathway to infect cells (18, 23). This suggests that the PKC-mediated downregulation of CAT-1 may not play a major role in MLV infection. Further work will be required to determine if GRB2 has a role in PKC activation as well as how the reduction in GRB2 levels impacts the anterograde trafficking of mCAT-1.
Previous studies on the trafficking of insulin receptor (IR) showed that ligand stimulation (insulin-like growth factor 1 or insulin) resulted in the recruitment of GRB2 to caveolae, and GRB2 has been implicated in the bidirectional trafficking of the caveosome: the retrograde transport of cargo from the caveosome to the ER or the anterograde transport back to the surface by transient fusion between the caveosome and the early or recycling endosome (35). The caveosome is an endosomal compartment distinct from the “conventional” early endosome (EEA1+/Rab5+). Caveosomes have a neutral pH and stain negative for the early endosome marker EEA1 but are positive for caveolin-1 (36). Endocytosis to caveosomes is a clathrin-independent mechanism that can be blocked by either cholesterol extraction or the overexpression of the dominant negative dynamin II K44A mutant (36). Another study showed that cholesterol depletion led to a reduced amount of GRB2 in the caveolar fraction (13). Together with data from the insulin receptor studies, this suggests a possible role of GRB2 in mediating caveosome trafficking.
Interestingly, mCAT-1 has been shown to associate with caveolin in a number of cell lines (28), and Moloney MLV was reported to use the clathrin-independent endocytic pathway to enter cells (23). We speculate that GRB2 may play a role in the recycling of mCAT-1 through the regulation of caveosome trafficking, but further investigation will be required to examine this possibility.
It remains unclear whether the interaction between mCAT-1 and GRB2 is direct or involves additional proteins. Zharikov and Block reported previously that the association with the cytoskeleton binding protein fodrin is important for the full activity of CAT-1 (53). The actin-disrupting toxin swinholide leads to a 25% reduction of l-arginine transport in pulmonary artery endothelial cells, and conversely, the actin-stabilizing toxin jasplakinolide increases l-arginine transport without an alteration of the total CAT-1 expression level (54). Likewise, the disruption of the cell actin network by cytochalasin D, a drug that depolymerizes filamentous actin, inhibits eMLV entry into rat cells without downregulating the surface expression of mCAT-1 (20). Several proteins important for actin remodeling and cellular compartmentalization contain SH3 binding motifs that can interact with GRB2 (17). For example, the N-terminal SH3 domain of GRB2 binds to the guanine nucleotide exchange factor SOS1, the E3 ubiquitin ligase Cbl, and dynamin. Dynamin is a GTPase that is essential for coated-vesicle formation and receptor-mediated endocytosis. The C-terminal SH3 domain binds to GRB2-associated binding protein 1 (Gab1) to relay signals to PI3K pathways (34). Of note, several viral proteins have been reported to interact directly with SH3 domain-containing proteins. HIV Nef and feline immunodeficiency virus (FIV) Nef bind to a variety of SH3 domains (7), the hepatitis C virus (HCV) NS5 protein interacts directly with the N-terminal GRB2 SH3 domain (48), and the ORF3 protein of hepatitis E virus (HEV) activates mitogen-activated protein kinase (MAPK) through the binding of the SH3 domain (22). However, these are all interactions that occur between GRB2 and virus proteins in the cell cytoplasm. To date, our data demonstrating that GRB2 interacts with a viral receptor, mCAT-1, and that this interaction is stimulated by the virus are novel.
We propose a model in which GRB2 regulates mCAT-1 trafficking. Normally, GRB2 binds to mCAT-1 at a basal level in resting cells, mainly on the plasma membrane, which accounts for the basal binding that we observed in the immunoprecipitation, pulldown, and confocal imaging experiments. The physiological level of endogenous GRB2 is important for regulating the turnover of mCAT-1, which may include the anterograde transport of mCAT-1-recycling endosomes from the caveosome and/or from the trans-Golgi network to the plasma membrane. The binding of eMLV recruits more GRB2 to associate with mCAT-1. We speculate that GRB2 binding to mCAT-1 triggers the movement of the receptor on the plasma membrane and/or the internalization of the virus–mCAT-1–GRB2 complex into intracellular compartments. In this way, GRB2 may also play a role in the surface availability of mCAT-1.
In conclusion, we identified the signaling adaptor protein GRB2 as an important factor for facilitating mCAT-1 trafficking and, hence, ecotropic MLV entry and infection. The knowledge that we gained from this study not only elucidates an important part of the molecular mechanism of receptor trafficking during ecotropic retrovirus entry but also reveals the previously unknown mechanism of mCAT-1 participation in signaling pathways by its interaction with GRB2. Additional work needs to be done to investigate if these mechanisms apply to other CAT-1 proteins and to other retrovirus receptors.
ACKNOWLEDGMENTS
This work was supported by grant R01AI063513-02 awarded to R.A.D. and a McLaughlin predoctoral fellowship awarded to Z.C.
We thank Mardelle Susman and Kyle DeShazo for assistance with the editing of the manuscript.
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Abramoff MD, Magelhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int. 11:36–42 [Google Scholar]
- 2. Albritton LM, Kim JW, Tseng L, Cunningham JM. 1993. Envelope-binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J. Virol. 67:2091–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Albritton LM, Tseng L, Scadden D, Cunningham JM. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659–666 [DOI] [PubMed] [Google Scholar]
- 4. Ando A, et al. 1994. A complex of GRB2-dynamin binds to tyrosine-phosphorylated insulin receptor substrate-1 after insulin treatment. EMBO J. 13:3033–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baughn LB, Rosenberg N. 2005. Disruption of the Shc/Grb2 complex during Abelson virus transformation affects proliferation, but not apoptosis. J. Virol. 79:2325–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chesebro B, et al. 1983. Characterization of monoclonal antibodies reactive with murine leukemia viruses: use in analysis of strains of Friend MCF and Friend ecotropic murine leukemia virus. Virology 127:134–148 [DOI] [PubMed] [Google Scholar]
- 7. Collette Y, et al. 2000. HIV-2 and SIV nef proteins target different Src family SH3 domains than does HIV-1 Nef because of a triple amino acid substitution. J. Biol. Chem. 275:4171–4176 [DOI] [PubMed] [Google Scholar]
- 8. Davey RA, Hamson CA, Healey JJ, Cunningham JM. 1997. In vitro binding of purified murine ecotropic retrovirus envelope surface protein to its receptor, MCAT-1. J. Virol. 71:8096–8102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dutartre H, Harris M, Olive D, Collette Y. 1998. The human immunodeficiency virus type 1 Nef protein binds the Src-related tyrosine kinase Lck SH2 domain through a novel phosphotyrosine independent mechanism. Virology 247:200–211 [DOI] [PubMed] [Google Scholar]
- 10. Fan H, Paskind M. 1974. Measurement of the sequence complexity of cloned Moloney murine leukemia virus 60 to 70S RNA: evidence for a haploid genome. J. Virol. 14:421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fass D, et al. 1997. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science 277:1662–1666 [DOI] [PubMed] [Google Scholar]
- 12. Fath I, et al. 1994. Cloning of a Grb2 isoform with apoptotic properties. Science 264:971–974 [DOI] [PubMed] [Google Scholar]
- 13. Furuchi T, Anderson RG. 1998. Cholesterol depletion of caveolae causes hyperactivation of extracellular signal-related kinase (ERK). J. Biol. Chem. 273:21099–21104 [DOI] [PubMed] [Google Scholar]
- 14. Giubellino A, Burke TR, Jr, Bottaro DP. 2008. Grb2 signaling in cell motility and cancer. Expert Opin. Ther. Targets 12:1021–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greber UF. 2002. Signalling in viral entry. Cell. Mol. Life Sci. 59:608–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huebner K, et al. 1994. Chromosome locations of genes encoding human signal transduction adapter proteins, Nck (NCK), Shc (SHC1), and Grb2 (GRB2). Genomics 22:281–287 [DOI] [PubMed] [Google Scholar]
- 17. Kaneko T, Li L, Li SS. 2008. The SH3 domain—a family of versatile peptide- and protein-recognition module. Front. Biosci. 13:4938–4952 [DOI] [PubMed] [Google Scholar]
- 18. Katen LJ, Januszeski MM, Anderson WF, Hasenkrug KJ, Evans LH. 2001. Infectious entry by amphotropic as well as ecotropic murine leukemia viruses occurs through an endocytic pathway. J. Virol. 75:5018–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim JW, Closs EI, Albritton LM, Cunningham JM. 1991. Transport of cationic amino acids by the mouse ecotropic retrovirus receptor. Nature 352:725–728 [DOI] [PubMed] [Google Scholar]
- 20. Kizhatil K, Albritton LM. 1997. Requirements for different components of the host cell cytoskeleton distinguish ecotropic murine leukemia virus entry via endocytosis from entry via surface fusion. J. Virol. 71:7145–7156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kolokoltsov AA, Davey RA. 2004. Rapid and sensitive detection of retrovirus entry by using a novel luciferase-based content-mixing assay. J. Virol. 78:5124–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Korkaya H, et al. 2001. The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J. Biol. Chem. 276:42389–42400 [DOI] [PubMed] [Google Scholar]
- 23. Lee S, Zhao Y, Anderson WF. 1999. Receptor-mediated Moloney murine leukemia virus entry can occur independently of the clathrin-coated-pit-mediated endocytic pathway. J. Virol. 73:5994–6005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lehmann MJ, Sherer NM, Marks CB, Pypaert M, Mothes W. 2005. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J. Cell Biol. 170:317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li N, Lorinczi M, Ireton K, Elferink LA. 2007. Specific Grb2-mediated interactions regulate clathrin-dependent endocytosis of the cMet-tyrosine kinase. J. Biol. Chem. 282:16764–16775 [DOI] [PubMed] [Google Scholar]
- 26. Lock LS, Royal I, Naujokas MA, Park M. 2000. Identification of an atypical Grb2 carboxyl-terminal SH3 domain binding site in Gab docking proteins reveals Grb2-dependent and -independent recruitment of Gab1 to receptor tyrosine kinases. J. Biol. Chem. 275:31536–31545 [DOI] [PubMed] [Google Scholar]
- 27. Lowenstein EJ, et al. 1992. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell 70:431–442 [DOI] [PubMed] [Google Scholar]
- 28. Lu X, Silver J. 2000. Ecotropic murine leukemia virus receptor is physically associated with caveolin and membrane rafts. Virology 276:251–258 [DOI] [PubMed] [Google Scholar]
- 29. Lung FD, Tsai JY. 2003. Grb2 SH2 domain-binding peptide analogs as potential anticancer agents. Biopolymers 71:132–140 [DOI] [PubMed] [Google Scholar]
- 30. Marsh M. 1984. The entry of enveloped viruses into cells by endocytosis. Biochem. J. 218:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matsuda M, Mayer BJ, Fukui Y, Hanafusa H. 1990. Binding of transforming protein, P47gag-crk, to a broad range of phosphotyrosine-containing proteins. Science 248:1537–1539 [DOI] [PubMed] [Google Scholar]
- 32. McClure MO, Sommerfelt MA, Marsh M, Weiss RA. 1990. The pH independence of mammalian retrovirus infection. J. Gen. Virol. 71(Pt. 4):767–773 [DOI] [PubMed] [Google Scholar]
- 33. McDonald KK, Zharikov S, Block ER, Kilberg MS. 1997. A caveolar complex between the cationic amino acid transporter 1 and endothelial nitric-oxide synthase may explain the “arginine paradox.” J. Biol. Chem. 272:31213–31216 [DOI] [PubMed] [Google Scholar]
- 34. Meisner H, Conway BR, Hartley D, Czech MP. 1995. Interactions of Cbl with Grb2 and phosphatidylinositol 3′-kinase in activated Jurkat cells. Mol. Cell. Biol. 15:3571–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nichols B. 2003. Caveosomes and endocytosis of lipid rafts. J. Cell Sci. 116:4707–4714 [DOI] [PubMed] [Google Scholar]
- 36. Pelkmans L, Kartenbeck J, Helenius A. 2001. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 3:473–483 [DOI] [PubMed] [Google Scholar]
- 37. Raffel GD, Parmar K, Rosenberg N. 1996. In vivo association of v-Abl with Shc mediated by a non-phosphotyrosine-dependent SH2 interaction. J. Biol. Chem. 271:4640–4645 [DOI] [PubMed] [Google Scholar]
- 38. Roberts MR, et al. 1994. Targeting of human immunodeficiency virus-infected cells by CD8+ T lymphocytes armed with universal T-cell receptors. Blood 84:2878–2889 [PubMed] [Google Scholar]
- 39. Rojas M, Yao S, Lin YZ. 1996. Controlling epidermal growth factor (EGF)-stimulated Ras activation in intact cells by a cell-permeable peptide mimicking phosphorylated EGF receptor. J. Biol. Chem. 271:27456–27461 [DOI] [PubMed] [Google Scholar]
- 40. Rotmann A, Strand D, Martine U, Closs EI. 2004. Protein kinase C activation promotes the internalization of the human cationic amino acid transporter hCAT-1. A new regulatory mechanism for hCAT-1 activity. J. Biol. Chem. 279:54185–54192 [DOI] [PubMed] [Google Scholar]
- 41. Saeed MF, Kolokoltsov AA, Davey RA. 2006. Novel, rapid assay for measuring entry of diverse enveloped viruses, including HIV and rabies. J. Virol. Methods 135:143–150 [DOI] [PubMed] [Google Scholar]
- 42. Sherer NM, et al. 2007. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat. Cell Biol. 9:310–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sherer NM, et al. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4:785–801 [DOI] [PubMed] [Google Scholar]
- 44. Singer II, et al. 2001. CCR5, CXCR4, and CD4 are clustered and closely apposed on microvilli of human macrophages and T cells. J. Virol. 75:3779–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Songyang Z, et al. 1994. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol. Cell. Biol. 14:2777–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sorkin A, McClure M, Huang F, Carter R. 2000. Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr. Biol. 10:1395–1398 [DOI] [PubMed] [Google Scholar]
- 47. Sullivan PC, Ferris AL, Storrie B. 1987. Effects of temperature, pH elevators, and energy production inhibitors on horseradish peroxidase transport through endocytic vesicles. J. Cell. Physiol. 131:58–63 [DOI] [PubMed] [Google Scholar]
- 48. Tan SL, et al. 1999. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc. Natl. Acad. Sci. U. S. A. 96:5533–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vina-Vilaseca A, Bender-Sigel J, Sorkina T, Closs EI, Sorkin A. 2011. Protein kinase C-dependent ubiquitination and clathrin-mediated endocytosis of the cationic amino acid transporter CAT-1. J. Biol. Chem. 286:8697–8706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang H, Kavanaugh MP, North RA, Kabat D. 1991. Cell-surface receptor for ecotropic murine retroviruses is a basic amino-acid transporter. Nature 352:729–731 [DOI] [PubMed] [Google Scholar]
- 51. Yamazaki T, et al. 2002. Role of Grb2 in EGF-stimulated EGFR internalization. J. Cell Sci. 115:1791–1802 [DOI] [PubMed] [Google Scholar]
- 52. Yu D, Wang W, Yoder A, Spear M, Wu Y. 2009. The HIV envelope but not VSV glycoprotein is capable of mediating HIV latent infection of resting CD4 T cells. PLoS Pathog. 5:e1000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zharikov SI, Block ER. 2000. Association of L-arginine transporters with fodrin: implications for hypoxic inhibition of arginine uptake. Am. J. Physiol. Lung Cell. Mol. Physiol. 278:L111–L117 [DOI] [PubMed] [Google Scholar]
- 54. Zharikov SI, Sigova AA, Chen S, Bubb MR, Block ER. 2001. Cytoskeletal regulation of the L-arginine/NO pathway in pulmonary artery endothelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 280:L465–L473 [DOI] [PubMed] [Google Scholar]
- 55. Zhou D, Noviello C, D'Ambrosio C, Scaloni A, D'Adamio L. 2004. Growth factor receptor-bound protein 2 interaction with the tyrosine-phosphorylated tail of amyloid beta precursor protein is mediated by its Src homology 2 domain. J. Biol. Chem. 279:25374–25380 [DOI] [PubMed] [Google Scholar]