Abstract
The latent nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus (KSHV) is required for the replication and partitioning of latent viral genomes. It contains an extended internal repeat (IR) region whose function is only incompletely understood. We constructed KSHV genomes lacking either LANA (KSHV-ΔLANA) or the IR region of LANA (KSHV-LANAΔ329-931). Although still capable of replicating a plasmid containing a latent origin of replication, LANAΔ329-931 does not support the establishment of stable cell lines containing a KSHV genome. These findings suggest a role for the LANA IR in KSHV episomal maintenance without its being required for replication.
TEXT
Kaposi's sarcoma-associated herpesvirus (KSHV) establishes a latent infection in primary cells and established cell lines (11, 13), which is characterized by an episomal replication of the circular KSHV genome and the expression of a set of latent viral transcripts that encodes the latent nuclear antigen (LANA), vCYC, vFLIP, kaposin/K12, a cluster of microRNAs (miRNAs), and in B cells additionally, vIRF3/K10.5 (7, 8, 16–18). LANA is essential for KSHV infection. It tethers, through its N- and C-terminal domains, the KSHV episomes to mitotic chromosomes and chromatin (1–3). In addition to the N- and C-terminal domains, LANA has an internal repeat (IR) domain. The IR consists of three segments, a segment (amino acids [aa] 340 to 431) containing multiple repeats of the amino acids DEED or DEEED; a segment (aa 440 to 756) containing multiple repeats of the motifs QQQEP, QQREP, and QQQDE; and a segment (aa 760 to 931) containing multiple repeats of the motifs QEQELEE and QELEVEE with L, V, and Q spaced in a leucine zipper-like pattern (19). The length of this IR varies between different isolates (17). The function of this IR region is not completely resolved. In a transient transfection assay LANA deletion mutants lacking the IR region were capable of replicating a plasmid containing the binding site in the terminal repeat (TR) subunits of the KSHV genome, either at levels comparable to those of KSHV-LANAwt or at slightly reduced levels (9, 15, 20). In addition, such mutants were able to repress transcription from the KSHV TR region (20). Two studies have suggested an immune evasion role for the IR similar to that of Epstein-Barr virus nuclear antigen 1 (12, 21).
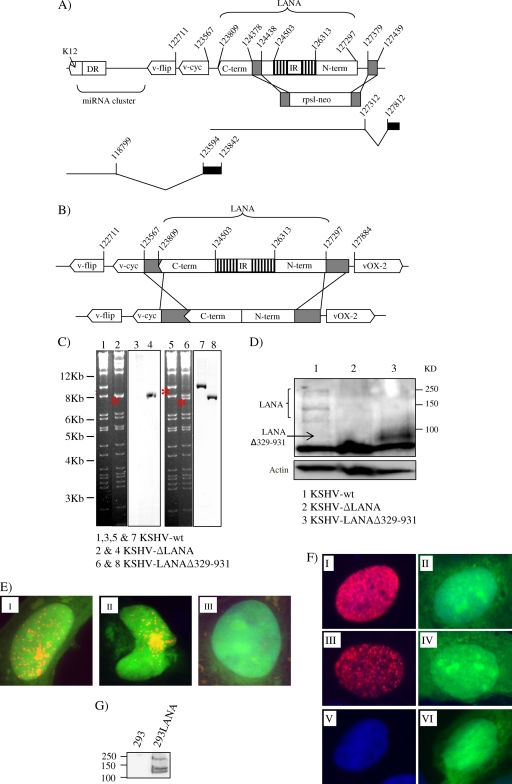
Here we used a reverse genetic approach on a KSHV genome cloned into a bacterial artificial chromosome (BAC36) (22) also containing green fluorescent protein (GFP) and hygromycin resistance genes to investigate the role of the LANA IR region in the context of the entire KSHV genome. We constructed two LANA mutants lacking either the gene for LANA, orf73, or its IR. To delete the coding region for LANA, we inserted a kanamycin resistance gene instead of most of the LANA gene using an ET cloning strategy (Gene Bridges) (Fig. 1A). As a result of the cassette insertion, KSHV-ΔLANA lacks the whole N-terminal domain, the IR, and a small part of the C-terminal domain of orf73, corresponding to nucleotide position 127379 upstream of the LANA ATG and nucleotide position 124438 downstream of the IR region (equivalent to aa 1 to 954 of LANA) (Fig. 1A). The rationale for this design was not to disturb a latent promoter reported to exist in the 3′ end of orf73 (5, 14) (Fig. 1A). Additionally, the upstream splice donor for LANA transcription at position 127812 was left intact (17) (Fig. 1A). To delete the IR region only, we used the shuttle mutagenesis strategy (Fig. 1B) (4) and generated KSHV-LANAΔ329-931. The deleted sequence of the IR corresponds to aa 329 to 931 of LANA. The integrity of the two KSHV LANA mutants was confirmed by restriction digestion and Southern blotting (Fig. 1C). Immunoblot assays of lysates from HEK293 cells transiently transfected with the KSHV genome containing wild-type or mutant LANA and sorted with a fluorescence-activated cell sorter (FACS) for GFP expression showed the presence of the expected 150- to 250-kDa LANA protein in the case of KSHV-LANAwt and a band of approximately 90 kDa for KSHV-LANAΔ329-931, while no specific band was seen in KSHV-ΔLANA-transfected cells (Fig. 1D). An immunofluorescence assay of HeLa cells transiently transfected with wild-type KSHV (KSHV-wt), KSHV-ΔLANA, or KSHV-LANAΔ329-931 confirmed the expression of the LANAΔ329-931 protein, which localizes to the nucleus and shows a speckled pattern similar to that of full-length LANA in the context of the entire genome (Fig. 1E).
Fig 1.
Construction and characterization of KSHV-ΔLANA and KSHV-LANAΔ329-931. (A) KSHV-ΔLANA was generated by ET cloning. A DNA fragment carrying the kanamycin resistance cassette (rpsl-neo) flanked by 60 bp homologous to the area to be knocked out was generated by PCR and inserted instead of aa 1 to 954 of LANA-1. The schematic transcript below the genome on the right side represents the LANA transcript and its upstream splice donor, which was left intact. The transcript encoding vcyc and vFLIP (left side) starts from a reported latent promoter in the 3′ end of orf73 (see text). term, terminus. (B) The KSHV-LANAΔ329-931 construct was generated by shuttle mutagenesis. LANA lacking aa 329 to 931 flanked by extra homologous regions (in gray) was cloned into the shuttle vector (pST76-NSR), electroporated into Escherichia coli harboring the KSHV-wt genome, and then passed through several steps of selection and counterselection. Numbers indicate positions on the genome according to the sequence with GenBank accession no. NC_009333. (C) Restriction digestion analysis of the KSHV LANA mutants with the KpnI enzyme. The 9.6-kb band in the KSHV-wt is shifted to around 8.0 kb in the case of KSHV-ΔLANA (lanes 1 and 2). The same band was shifted to 7.8 kb in the case of KSHV-LANAΔ329-931 (lanes 5 and 6). To confirm the identities of these bands, Southern blot analysis was performed with the rpsl-neo cassette in the case of KSHV-ΔLANA (lanes 3 and 4) and the amino-terminal domain of LANA (aa 1 to 328) as a probe in the case of KSHV-LANAΔ329-931 (lanes 7 and 8). (D) Expression of LANA in the context of the whole KSHV-BAC genome. HEK293 cells were transfected with KSHV-wt, KSHV-ΔLANA, or KSHV-LANAΔ329-931 and then sorted with a FACS for GFP expression and lysed with lysis buffer for Western blot analysis. (E) Immunofluorescence assay of HeLa cells transfected with KSHV-wt (I), KSHV-LANAΔ329-931 (II), or KSHV-ΔLANA (III) DNA showing the intracellular localization of LANA and LANAΔ329-931. (F) Immunofluorescence assay showing the expression and localization of LANA and LANAΔ329-931 from the pGTR4 plasmid constructs. I and II, pGTR4:LANA; III and IV, pGTR4:LANAΔ329-931; V and VI, PGTR4. (G) Immunoblot assay showing the expression of LANA in the 293LANA cell line. Human serum from a patient with Kaposi's sarcoma was used at a dilution of 1:500 to detect the expression of LANA and LANAΔ329-931.
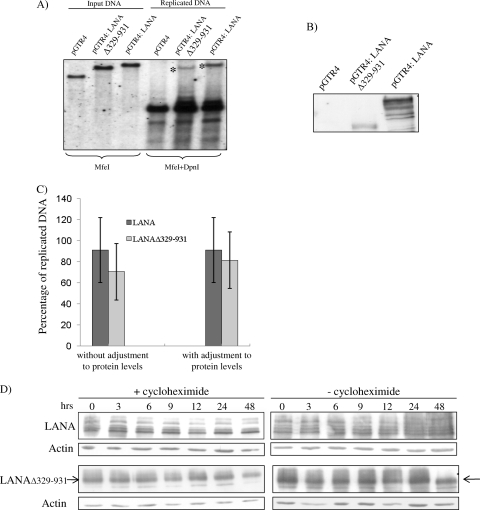
We first evaluated the ability of KSHV-wt and the two KSHV LANA mutants to establish stable clones under selection. HEK293 cells were transfected with KSHV-wt or the KSHV-LANA mutants. Hygromycin selection using different concentrations of hygromycin in independent cultures was applied after 24 h. Cells transfected with KSHV-wt formed hygromycin-resistant colonies at all concentrations, and they could be isolated and propagated to form stable clones (Table 1). As expected, the KSHV-ΔLANA genome failed to produce stable clones under selection, supporting an important role for LANA in KSHV episomal persistence (Table 1). Interestingly, cells transfected with KSHV-LANAΔ329-931 also failed to establish stable clones in the presence of all of the hygromycin concentrations tested (Table 1). The lack of stable transfectants observed with KSHV-LANAΔ329-931 is not due to a lack of LANAΔ329-931 or its reduced expression, as shown by immunoblotting and immunofluorescence (Fig. 1D and E). Furthermore, LANAΔ329-931 does not appear to be degraded more quickly than wild-type LANA in the presence of cycloheximide (see Fig. 3D) and increased turnover is therefore not the explanation for the failure of LANAΔ329-931 to support the establishment of stable transfectants carrying the entire KSHV genome (Table 1).
Table 1.
Numbers of stable clones produced by KSHV-ΔLANA and KSHV-LANAΔ329-931 in comparison with KSHV-wt under selection with hygromycin in HEK293 and 293LANA cells
| Strain | No. of stable clones produceda |
||||
|---|---|---|---|---|---|
| HEK293 cells |
293LANA cells (150)b | ||||
| 50b | 75b | 100b | 150b | ||
| KSHV-wt | 17 | 34 | 39 | 28 | 98 |
| KSHV-ΔLANA | 0 | 0 | 0 | 0 | 86 |
| KSHV-LANAΔ329-931 | 0 | 0 | 0 | 0 | 92 |
Data from one representative experiment out of three are shown.
Hygromycin concentration (μg/ml).
Fig 3.
Replication competence of the LANAΔ329-931 protein. (A) KSHV minireplicons carrying four copies of the KSHV TR region (which contains the LANA binding sites) and LANA (pGTR4:LANA), LANAΔ329-931 (pGTR4:LANAΔ329-931), or no LANA (pGTR4) were transfected into HEK293 cells. Extrachromosomal DNA was extracted from transfected cells by Hirt extraction on day 3 after transfection and digested with MfeI (input DNA) or MfeI and DpnI (unmethylated, replicated DNA). A TR DNA fragment was used as the probe for Southern blotting. pGTR4:LANA and pGTR4:LANAΔ329-931 replicate the minireplicon (asterisks), while pGTR4 does not. (B) Immunoblot assay showing the expression of the LANA and LANAΔ329-931 proteins in these cultures. (C) Quantification of the replication efficiency of LANA and LANAΔ329-931 from panel A. After the bands were measured with the ImageJ program, the quantity of the replicated DNA (represented by the band after digestion with MfeI and DpnI) was divided by the quantity of the input DNA (represented by the band after digestion with MfeI only) for each construct separately and multiplied by 100 to calculate the percentage of replicated DNA. The average percentage of four experiments, without adjustment for LANA protein levels, is shown on the left side of the panel. The right side of the panel presents replication efficiency results adjusted for LANA protein expression levels. (D) Stability of LANA and LANAΔ329-931 in the presence of cycloheximide. At 24 h after the transfection of 293 cells with pGTR4 expressing LANA or LANAΔ329-931, cells were treated with 100 μg/ml cycloheximide or left untreated. Cells were then lysed at the indicated time points after the addition of cycloheximide, and the lysates were immunoblotted for LANA and LANAΔ329-931 with human serum from a patient with Kaposi's sarcoma at a dilution of 1:500.
The phenotype of KSHV-LANAΔ329-931 and KSHV-ΔLANA (lack of stable transfectants) could be complemented by transfecting these genomes into HEK293 cells stably expressing LANA (Table 1; Fig. 1G) and selecting the transfectants with 150 μg hygromycin. This result excludes the possibility of an inadvertent alteration in the remainder of the KSHV genome or the bacterial artificial chromosome backbone being responsible for the observed phenotype.
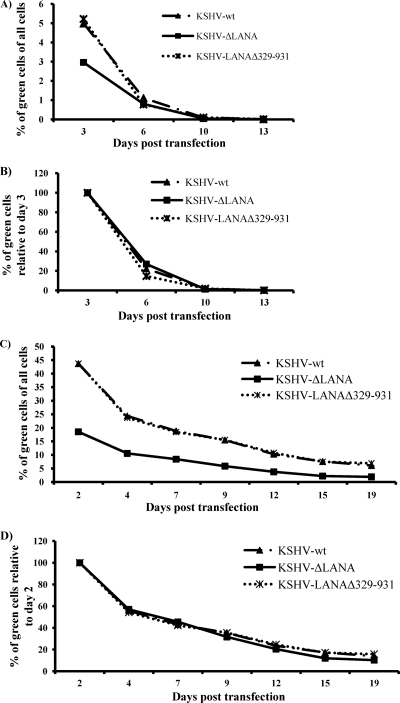
To study the role of the LANA IR region in the retention of KSHV episomes in cells after transient transfection without selection, HEK293 cells were transfected with equal amounts of DNA of the genomes of KSHV-wt, KSHV-LANAΔ329-931, and KSHV-ΔLANA. Early after transfection, wells transfected with KSHV-wt and KSHV-LANAΔ329-931 consistently showed a higher number of GFP-expressing cells than wells transfected with KSHV-ΔLANA (Fig. 2A). This may reflect the transient replication of the KSHV episome in cells transfected with KSHV-wt and KSHV-LANAΔ329-931 but not in KSHV-ΔLANA. The transfected cells were split regularly, and GFP expression was monitored by FACS. To compare the rates of loss of GFP-positive cells in the three constructs, the proportion of these cells on day 3 in each culture was normalized to 100 and the decrease in GFP-positive cells over 2 weeks was plotted relative to this value. As shown in Fig. 2B, we observed comparable rates of GFP-positive cell loss in cultures transfected with KSHV-wt, KSHV-LANAΔ329-931, and KSHV-ΔLANA, indicating that the presence of LANA does not improve the retention of the KSHV genome in HEK293 cells in the absence of selection. We performed the same experiment with the 293LANA cell line. The number of GFP-positive cells was much higher in this experiment (Fig. 2C), indicating that the presence of LANA in trans enhanced the replication of transfected KSHV genomes and thereby the expression of the GFP marker in the backbone of the bacterial artificial chromosome. As in the absence of LANA provided in trans, the number of GFP-positive cells (as a proportion of the total number of cells in the culture) was consistently higher in cells transfected with the KSHV-wt and KSHV-LANAΔ329-931 genomes than in those transfected with KSHV-ΔLANA (Fig. 2C). The fact that the cultures transfected with KSHV-wt and KSHV-LANAΔ329-931 showed a proportion of GFP-positive cells higher than that of KSHV-ΔLANA-transfected cells suggests an additional role for LANA provided in cis and that LANAΔ329-931 behaves like wild-type LANA-1 in this context. As shown in Fig. 2D, the presence or absence of LANA-1 or its IR in cis, however, did not have an impact on the rate of loss of viral episomes, even in the presence of excess LANA-1 provided in trans. KSHV latent episomes are also lost quickly from endothelial cells, the natural host of KSHV infection (10). HEK293 cells were shown to retain KSHV episomes in a small percentage of originally infected cells, slightly more than several other cell lines (10). Therefore, in this regard, HEK293 cells are, although not the natural host of KSHV infection, as good a model for studying KSHV persistence in culture as any other cell line susceptible to KSHV infection.
Fig 2.
KSHV genomes with mutant forms of LANA are lost from HEK293 and 293LANA cells at the same rate as KSHV genomes with wild-type LANA. All three KSHV constructs were transfected into HEK293 cells (A and B) or the 293LANA cell line (C and D), and the loss of GFP-positive cells was monitored over time by FACS. (A and C) Fractions of GFP-positive cells as percentages of all of the cells in the culture plotted over time. (B and D) The percentage of GFP-positive cells on days 3 and 2, respectively, was normalized to 100 for each transfection, and the decrease in GFP-positive cells over time was plotted for each culture.
We employed the minireplicon system to study the ability of LANAΔ329-931 to replicate viral DNA (10). The minireplicon (pGTR4:LANA) replicates spontaneously because it carries four copies of the KSHV TR, which contains the latent origin of replication, in addition to LANA. LANAΔ329-931 was inserted into pGTR4:LANA instead of LANA. A short-term replication assay (9, 20) was employed to study the ability of LANAΔ329-931 to replicate this plasmid. In five independent experiments, pGTR4:LANAΔ329-931 replicated the minireplicon either at a level comparable to that of pGTR4:LANA or at slightly reduced levels, which were, however, comparable to those of pGTR4:LANA when the different levels of protein expression of LANA and LANAΔ329-931 in minireplicon experiments were taken into account (Fig. 3A, B, and C).
A recent study (6) found that KSHV with extensive internal deletions of LANA removing most of the N-terminal domain and the entire IR region (e.g., LANAΔ33-929) retained the ability to replicate viral DNA in the transient replication assay shown in Fig. 3 but was not capable of mediating the persistence of viral episomes in BJAB B cells under selection. As an extension of and in accordance with this study, we show here that, in the context of the entire viral genome, deleting only the IR region (LANAΔ329-931) leads to a loss of persistence of viral DNA under selection, while the ability to replicate viral DNA is retained. We conclude that the LANA IR region plays an important role in supporting the persistence of latent viral genomes.
ACKNOWLEDGMENTS
We thank Martin Messerele, Adam Grundhoff, and Matthias Ballmaier.
This work was supported by grants Schu1668-2/3 and SFB 900 TPC1 from the Deutsche Forschungsgemeinschaft, as well as grant EU Integrated Project INCA (LSHC-CT-2005-018704).
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Ballestas ME, Chatis PA, Kaye KM. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641–644 [DOI] [PubMed] [Google Scholar]
- 2. Ballestas ME, Kaye KM. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barbera AJ, et al. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311:856–861 [DOI] [PubMed] [Google Scholar]
- 4. Borst EM, Posfai G, Pogoda F, Messerle M. 2004. Mutagenesis of herpesvirus BACs by allele replacement. Methods Mol. Biol. 256:269–279 [DOI] [PubMed] [Google Scholar]
- 5. Cai X, Cullen BR. 2006. Transcriptional origin of Kaposi's sarcoma-associated herpesvirus microRNAs. J. Virol. 80:2234–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De León Vázquez E, Kaye KM. 2011. The internal Kaposi's sarcoma-associated herpesvirus LANA regions exert a critical role on episome persistence. J. Virol. 85:7622–7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dittmer D, et al. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dupin N, et al. 1998. Prevalence of human herpesvirus 8 infection measured by antibodies to a latent nuclear antigen in patients with various dermatologic diseases. Arch. Dermatol. 134:700–702 [DOI] [PubMed] [Google Scholar]
- 9. Garber AC, Hu J, Renne R. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277:27401–27411 [DOI] [PubMed] [Google Scholar]
- 10. Grundhoff A, Ganem D. 2004. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J. Clin. Invest. 113:124–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krishnan HH, et al. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J. Virol. 78:3601–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kwun HJ, et al. 2007. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mimics Epstein-Barr virus EBNA1 immune evasion through central repeat domain effects on protein processing. J. Virol. 81:8225–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lagunoff M, et al. 2002. De novo infection and serial transmission of Kaposi's sarcoma-associated herpesvirus in cultured endothelial cells. J. Virol. 76:2440–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H, Komatsu T, Dezube BJ, Kaye KM. 2002. The Kaposi's sarcoma-associated herpesvirus K12 transcript from a primary effusion lymphoma contains complex repeat elements, is spliced, and initiates from a novel promoter. J. Virol. 76:11880–11888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lim C, Seo T, Jung J, Choe J. 2004. Identification of a virus trans-acting regulatory element on the latent DNA replication of Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 85:843–855 [DOI] [PubMed] [Google Scholar]
- 16. Muralidhar S, et al. 1998. Identification of kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) transforming gene. J. Virol. 72:4980–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rainbow L, et al. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rivas C, Thlick AE, Parravicini C, Moore PS, Chang Y. 2001. Kaposi's sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J. Virol. 75:429–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Russo JJ, et al. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. U. S. A. 93:14862–14867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Viejo-Borbolla A, et al. 2005. Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J. Virol. 79:13618–13629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zaldumbide A, Ossevoort M, Wiertz EJ, Hoeben RC. 2007. In cis inhibition of antigen processing by the latency-associated nuclear antigen I of Kaposi sarcoma herpes virus. Mol. Immunol. 44:1352–1360 [DOI] [PubMed] [Google Scholar]
- 22. Zhou FC, et al. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]