Abstract
Roseolovirus, or human herpesvirus 6 (HHV-6), is a ubiquitous human pathogen infecting over 95% of the population by the age of 2 years. As with other herpesviruses, reactivation of HHV-6 can present with severe complications in immunocompromised individuals. Recent studies have highlighted the importance of herpesvirus-derived microRNAs (miRNAs) in modulating both cellular and viral gene expression. An initial report which computed the likelihood of various viruses to encode miRNAs did not predict HHV-6 miRNAs. To experimentally screen for small HHV-6-encoded RNAs, we conducted large-scale sequencing of Sup-T-1 cells lytically infected with a laboratory strain of HHV-6B. This revealed an abundant, 60- to 65-nucleotide RNA of unknown function derived from the lytic origin of replication (OriLyt) that gave rise to smaller RNA species of 18 or 19 nucleotides. In addition, we identified four pre-miRNAs whose mature forms accumulated in Argonaute 2. In contrast to the case for other betaherpesviruses, HHV-6B miRNAs are expressed from direct repeat regions (DRL and DRR) located at either side of the genome. All miRNAs are conserved in the closely related HHV-6A variant, and one of them is a seed ortholog of the human miRNA miR-582-5p. Similar to alphaherpesvirus miRNAs, they are expressed in antisense orientation relative to immediate-early open reading frames (ORFs) and thus have the potential to regulate key viral genes.
INTRODUCTION
Human herpesvirus 6 (HHV-6) is one of eight herpesviruses known to infect humans and was first isolated in 1986 from immunocompromised patients suffering from lymphoproliferative disorders (43). HHV-6 is a ubiquitous human pathogen with seroprevalence rates exceeding 95% in industrialized nations. Together with HHV-7, it belongs to the Roseolovirus genus within the betaherpesvirus subfamily. It exists as two variants, HHV-6A and HHV-6B, which differ with respect to their biological properties, tropisms, and clinical manifestations (14). In 1988, HHV-6B was identified as the causative agent of the childhood disease exanthem subitum (roseola or 3-day fever) (57). Primary infection typically presents as an acute febrile illness, sometimes followed by a red rash. Additional symptoms may include otitis, meningitis, and seizures (42). While severe complications resulting from primary infection, such as encephalitis and encephalopathy, are rare, reactivation of HHV-6 in transplant patients, particularly those receiving hematopoietic stem cells, can present with life-threatening complications, most notably due to encephalitis (31, 58). In addition, HHV-6 may facilitate the progression of other viral infections, trans-activating HHV-7, human cytomegalovirus (HCMV), and HIV-1, which generally results in a poorer prognosis (reviewed in reference 14). Finally, HHV-6 has been implicated in a range of neurological diseases, most prominently multiple sclerosis (MS), although a causative role of HHV-6 in MS pathology remains controversial (36).
The HHV-6 genome consists of a linear, double-stranded DNA molecule of 160 to 162 kb, comprising a unique region (U) of 145 kb flanked by terminal direct repeats (DRL and DRR) of 8 to 9 kb. HHV-6 has a broad cellular tropism, as its receptor (CD46) is expressed on all nucleated cells. The sites of latency are not precisely defined yet but could consist of CD4+ T cells, monocytes, and early bone marrow progenitor cells (14, 32, 49).
HHV-6, like HCMV (although it is less well studied), uses a multitude of approaches to modulate the host-pathogen environment and establish latency. Previous studies have focused on the role of viral proteins that modulate major histocompatibility complex (MHC) class I gene expression and of viral chemokines and receptors that interact with the host (34). Recently, many members of the herpesvirus family have been shown to express their own microRNAs (miRNAs) (11, 19, 40, 41). Since the seminal discovery of miRNAs in Epstein-Barr virus (EBV) (41), more than 235 miRNAs of viral origin have been identified and listed in the miRNA repository miRBase (v.17) (25, 26). Conserved throughout metazoan evolution, miRNAs derive from a large primary transcript (pri-miRNA) which is sequentially processed by the nuclear and cytoplasmic RNase III enzymes Drosha and Dicer to generate the mature, ∼22-nucleotide (nt) miRNA. This miRNA becomes functional only after its incorporation into an Argonaute (Ago)-containing RNA-induced silencing complex (RISC), where it will mediate the regulation of target messenger RNAs via binding to partially complementary sites, usually located in the 3′-untranslated regions (3′-UTRs) (3).
The vast majority of virus-encoded miRNAs derive from herpesviruses, and so far HHV-3, HHV-6, and HHV-7 are the only herpesviruses for which miRNAs have not been characterized. For HHV-3, deep sequencing of latently infected trigeminal ganglia failed to identify any miRNAs (55). To resolve the question of whether HHV-6 encodes miRNAs, we performed small RNA cloning followed by massively parallel sequencing to identify the small RNAs deriving from the virus during lytic infection.
We cloned small RNAs both from total RNA isolated from a CD4+ T lymphocyte cell line (Sup-T-1) infected with a laboratory strain of HHV-6B and from RNAs coimmunoprecipitated with Ago2. Here we report that HHV-6B encodes at least four pre-miRNAs. The respective miRNAs are all conserved in the closely related virus HHV-6A, although they display intervariant and interstrain sequence variations. These viral miRNAs are encoded at two positions within the genome (in both DRL and DRR) and are encoded in antisense orientation relative to predicted HHV-6B-specific genes. One HHV-6 miRNA is a seed ortholog of the human miRNA miR-582-5p, which is not expressed in Sup-T-1 T cells. Interestingly, 5′ RNA ligase-mediated rapid amplification of cDNA ends (5′ RLM-RACE) to detect viral miRNA-induced cleavage products enabled us to identify a locus expressing miRNAs from both strands of the genome. In addition, we identified a novel viral transcript of 60 to 65 nt deriving from the minimal efficient origin of lytic replication (OriLyt) that gives rise to abundant RNA species of 18 and 19 nt that may represent a new class of origin-derived viral small RNAs.
MATERIALS AND METHODS
Cell culture and virus infection.
Sup-T-1 cells (CRL-1942; ATCC) were maintained in RPMI 1640 medium containing 7% fetal bovine serum (FBS). A total of 3 × 105 cells/ml were infected with HHV-6B (using a strain that is routinely used for immunofluorescence-based antibody detection in the laboratory) by cocultivation of uninfected Sup-T-1 cells with infected cells at a ratio of 1:1,000. Samples were harvested at 0, 24, 48, 72, and 96 h postinfection (hpi), and total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations.
Immunofluorescence staining.
For immunofluorescence analysis to determine the rate of HHV-6 infection, 2 × 105 cells in 150 μl phosphate-buffered saline (PBS) per slide were pelleted on a glass slide (by cytospin centrifugation) at 800 rpm for 10 min. Slides were air dried, fixed, and permeabilized with fixative solution for a CINAkit (Argene, Sherley, NY) and permeabilization solution for a CINAkit (Argene, Sherley, NY). The primary antibody (sc-65447; Santa Cruz) for fluorescence staining was applied at a concentration of 0.5% for 1 h, followed by 1 h of incubation with the Alexa Fluor 488-conjugated donkey anti-mouse secondary antibody (Invitrogen, Darmstadt, Germany). Evans blue (1%) was used for counterstaining. Approximately 80% of the cells used for deep sequencing were positive for HHV-6B p41 staining.
IP of hAgo2 complexes.
For RISC immunoprecipitation (IP), 1 × 108 cells were used for each replicate and were washed twice in PBS before lysis in 10 ml of lysis buffer containing 25 mM Tris-HCl, pH 7.5, 150 mM KCl, 2 mM EDTA, 0.5% NP-40, 0.5 mM dithiothreitol (DTT), and Complete protease inhibitor (Roche). DTT and protease inhibitors were always prepared freshly and added immediately before use. Lysates were incubated for 30 min at 4°C and cleared by centrifugation at 20,000 × g for 30 min at 4°C. Total RNA was prepared from 100 μl of cell lysate by using an miRNeasy kit (Qiagen) following the manufacturer's instructions. Ago2 immunoprecipitation was performed as previously described (16). Briefly, 6 μg of purified monoclonal hAgo2 antibody (anti-hAgo2; 11A9) or control monoclonal bromodeoxyuridine (BrdU) antibody (Abcam) was added to 5 ml of RPMI medium and incubated with 30 μl of protein G-Sepharose beads (GE Healthcare) in Pierce centrifuge columns (Thermo Scientific) with constant rotation at 4°C overnight. Columns were drained by gravity flow and washed once with lysis buffer. Beads were subsequently incubated with 5 ml of cell lysate for 2.5 h with constant rotation at 4°C. After incubation, the beads were washed four times with IP wash buffer (300 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 0.1% NP-40, 1 mM NaF) and once with PBS to remove residual detergents. RNA was recovered from the beads by adding 700 μl of Qiazol to the columns. After 5 min, the Qiazol lysates were collected from the columns. This step was repeated once, and the Qiazol lysates were combined. RNA was prepared using an miRNeasy kit (Qiagen) according to the manufacturer's instructions. RNA samples were eluted in 30 μl H2O and stored at −80°C until subjected to further analyses.
cDNA synthesis and quantitative PCR on a LightCycler instrument.
To monitor the efficiency of Ago2 IP and to check for selective enrichment of HHV-6B miRNA candidates, cDNAs were prepared from 2.5 μl Ago2 IP, BrdU IP, and total RNAs (1:10) in a single-step reaction mixture by use of a miScript reverse transcription (RT) kit (Qiagen). LightCycler quantitative RT-PCR (qRT-PCR) was performed for the cellular miRNA let-7a, using the forward primer let-7a for (5′-TGAGGTAGTAGGTTGTATAGTT), or with the respective sequences of the mature HHV-6 miRNA candidates by using a miScript SYBR green PCR kit (Qiagen) following the manufacturer's instructions.
Small RNA cloning and sequencing.
RNAs were extracted from noninfected and HHV-6B-infected (7 days postinfection [dpi]) Sup-T-1 cells by using TRI reagent (MRC, Inc.) per the manufacturer's instructions. Small RNA cloning was conducted with 30 μg of total RNA as previously described (40), except that PCR products were not concatenated and instead were sent directly for large-scale sequencing. Small RNA sequencing was performed at the Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Illkirch, France, using an Illumina Genome Analyzer IIx instrument with a read length of 36 bp. Similarly, for deep sequencing of small RNAs incorporated into Ago2-containing RISCs, a small RNA library was created from hAgo2 IP samples from Sup-T-1 cells infected with HHV-6B at 4 days postinfection, using a read length of 54 bases.
Processing and annotation of small RNA sequences.
An in-house Perl analysis pipeline was used to analyze the vast amount of data produced during next-generation small RNA sequencing. After 3′ adaptor removal and size selection (exclusion of trimmed reads shorter than 15 nt), nonredundant sequences were mapped to the genomes from which they may have been derived and to other already annotated RNAs by using Nexalign (http://genome.gsc.riken.jp/osc/english/software/src/nexalign-1.3.5.tgz), permitting up to 2 mismatches. The Homo sapiens and HHV-6B genome sequences were downloaded from the UCSC repository (assembly version hg19) and the RefSeq or GenBank database (accession number NC_000898.1 for the Z29 strain and AB021506.1 for the HST strain), respectively. The following sources of annotated transcripts were used: miRBase v.16 for miRNAs, GenBank v.180 for Homo sapiens rRNA, tRNA, small nuclear-small nucleolar RNA (sn-snoRNA), small cytoplasmic RNA (scRNA), and Piwi-interacting RNA (piRNA) and Repbase v.16.01 for Homo sapiens and common ancestral repeats. By doing so, small RNAs that mapped unambiguously to sequences from one single functional category were easily classified, while the others were identified by applying the following annotation rule based on the abundances of various types of sequences in the cell: rRNA > tRNA > sn-snoRNA > miRNA > piRNA > repeat > pathogen genome > host genome > unknown. Sequences annotated as HHV-6B sequences were represented graphically on the HHV-6B (strain Z29) genome by using the R/Bioconductor package girafe (52).
Northern blotting.
Total RNA was extracted from Sup-T-1 cells infected with HHV-6B (laboratory strain) by using TRI reagent (MRC, Inc.) following 1, 2, 3, and 4 days of infection. Total RNA (10 μg) or 5 μl of BrdU IP or Ago2 IP sample was mixed with an equal volume of formamide loading dye and heated at 95°C for 30 s prior to separation in a 17.5% urea-acrylamide gel. RNA was transferred to a HybondNX membrane (Amersham Biosciences) in Milli-Q water and chemically cross-linked at 60°C by using 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) (Sigma) for 1 h 30 min, followed by extensive washing in Milli-Q water prior to prehybridization. Membranes were prehybridized for 2 h in PerfectHyb Plus solution (Sigma) at 42°C. For each of the 8 candidate miRNAs, antisense DNA oligonucleotides containing 3 locked nucleic acid (LNA) bases (Eurogentec) or standard antisense DNA oligonucleotides (for hhv6b-miR-Ro6-3-5p, hhv6b-miR-Ro6-4, and sequences antisense to positions 69121 to 69141 and 69141 to 69161 of HHV-6B [strain Z29]) were 5′ end labeled with 25 μCi of [γ-32P]dATP by using T4 polynucleotide kinase (Fermentas). The labeled probes were hybridized to the blot overnight at 42°C. The blot was then washed at 55°C twice for 10 min with low-stringency buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]–0.1% SDS), followed by a single wash in a higher-stringency buffer (1× SSC–0.1% SDS) for 5 min. Northern blots were exposed overnight to phosphorimager plates (Fuji) and scanned using an FLA-5000 series phosphorimager (Fuji).
5′ RACE.
To determine whether hhv6b-miR-Ro6-3 could direct cleavage of the viral transcript (B1) expressed antisense to the pre-miRNA, we performed 5′ RACE by using non-tobacco alkaline phosphatase (TAP)-, non-calf intestine phosphatase (CIP)-treated RNA following 3 days of infection with HHV-6B. RNA from Sup-T-1 cells served as an internal control for the specificity of the RACE product. RNA (5 μg) was treated with DNase twice according to the manufacturer's instructions (Fermentas) and was purified by phenol-chloroform extraction. The RNA pellet was precipitated and resuspended in water. An RNA oligonucleotide (200 pmol; 5′-GUUCAGAGUUCUACAGUCCGACGAUC) was added and denatured at 90°C for 2 min. Ligation was performed for 1.5 h at 37°C by using T4 RNA ligase 1 (NEB). RNA was purified as previously described, and one quarter of the product was used for the RT step. An oligonucleotide specific to B1 (5′-GCCCAGGGAGGACGGATAGCAGGAGTCG) was used for RT at 55°C, using Superscript III polymerase (Invitrogen) following the manufacturer's instructions. No-RT controls served as negative controls. The resulting product(s) was purified and amplified in an initial PCR using Phusion high-fidelity DNA polymerase (Finnzymes), using the adaptor-specific 5′ oligonucleotide 30.300 (5′-CAGCCAACAGGCACCGTTCAGAGTTCTACA) and the RT primer with the following PCR program: 98°C for 30 s followed by 40 cycles of 98°C for 10 s and 72°C for 25 s, with a final step of 72°C for 2 min. A second round of PCR was used to amplify B1-specific products, using an internal primer for B1 (5′-CGCAGACTGGCGTGTGGACCGT) and the 5′ oligonucleotide (30.300) under the same PCR conditions. The PCR products were purified, an A residue was added by performing 20 cycles of two-step PCR with DreamTaq polymerase (Fermentas), and the product was repurified and cloned into pDRIVE following the manufacturer's recommendations. Eleven positive clones were obtained and sequenced using the M13Rev primer; sequences were mapped to the HHV-6B genome by using BLAST.
Microarray data accession number.
The data discussed in this paper have been deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE34196.
RESULTS AND DISCUSSION
Identification of HHV-6B-derived small RNAs.
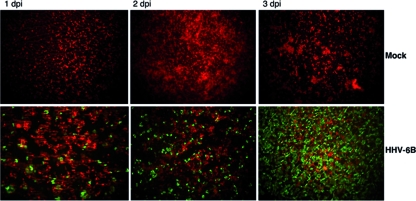
To identify viral miRNAs encoded by HHV-6B, we performed small RNA cloning and deep sequencing of Sup-T-1 cells lytically infected for 7 days with a laboratory strain of HHV-6B. Sup-T-1 is a CD4+ T lymphocyte cell line that is permissive to HHV-6, HHV-7, and HIV-1 infection (7, 13). We confirmed the permissibility of Sup-T-1 cells to HHV-6 by immunofluorescence staining against the HHV-6 p41 early antigen, which showed that 80% of the cells were infected at 3 dpi (Fig. 1). Total RNA from HHV-6B-infected cells was size fractionated in a polyacrylamide gel, and RNA species of between 19 and 24 nt were cloned as previously described (39, 40). In total, 24,380,617 reads were obtained, among which 93.9% could be mapped to either the human or HHV-6B genome, and were categorized into RNA classes based on sequence annotation (Table 1). An unusually large number of reads were annotated as rRNA (71.58%). Most likely, this was due to nonsynchronized, prolonged infection, i.e., infected cells were added to uninfected cells at 1:1,000 and thus reduced the RNA quality. Similar findings were previously noted for small RNA libraries generated from HCMV-infected cells (40). Allowing two mismatches for the alignment to the viral genome, only a limited number of reads (12,350 [0.05%]) could be assigned as viral sequences. Of these, 9,138 sequences mapped to the HHV-6B Z29 genome.
Fig 1.
Visualization of HHV-6B infection. Infection of HHV-6B was verified and monitored by immunofluorescence staining of the HHV-6 p41 early antigen in Sup-T-1 cells infected with a laboratory strain of HHV-6B. The percentage of HHV-6B-positive cells increased throughout the 3-day time course.
Table 1.
Distribution of small RNA reads from both librariesa
| RNA classification | % of total clones |
|
|---|---|---|
| HHV-6B total RNA | HHV-6B Ago2 IP RNA | |
| rRNA | 71.58 | 0.29 |
| tRNA | 1.12 | 0.25 |
| sn-snoRNA | 1.00 | 0.45 |
| scRNA | 0.14 | 0.09 |
| miRNA | 11.97 | 94.96 |
| Repeats | 0.93 | 0.13 |
| Host genome | 6.82 | 1.03 |
| HHV-6B (Z29) genome | 0.05 | 0.69 |
| Unknown | 6.38 | 2.11 |
| Total | 100.00 | 100.00 |
sn-snoRNA, small nuclear-small nucleolar RNA; scRNA, small cytoplasmic RNA. Shading indicates the species examined in this study.
To enhance the sensitivity and boost the degree of confidence in our analysis, we performed small RNA cloning and deep sequencing analysis of RNAs obtained after Ago2 immunoprecipitation (Ago2 IP) of Sup-T-1 cells lytically infected with HHV-6B. In total, 30,225,650 reads were obtained, of which 97.93% were mappable. We observed that ∼95% of all reads were annotated as miRNAs. Due to the prevalence of miRNAs within the Ago2 sample, we could be confident that the majority of sequences deriving from the virus (207,394 reads [0.69%]) were likely to represent genuine viral miRNAs (Table 1). Such a limited number of viral sequences is intriguing compared to results for other herpesviruses, particularly those for the betaherpesvirus family (17, 40), where viral miRNAs often constitute 10 to 60% of the cellular miRNA pool late in infection. However, it was previously noted for mouse CMV (MCMV) that the proportion of viral sequences present in a given library is dependent on the efficiency of viral infection, multiplicity of infection (MOI), and cell type. For instance, in a small-scale library created from bone marrow macrophages infected with MCMV, only ∼0.5% of sequences represented viral sequences, compared to ∼30% for mouse embryonic fibroblasts (MEFs) (8). Similarly, a recent report showed that less than 2% of small RNA sequences cloned from marmoset T cells infected with herpesvirus saimiri mapped to the viral genome (12). Similar to the case for HHV-6B, miRNAs characterized during lytic infection of the integrating, oncogenic Marek's disease virus (MDV) contributed only ∼1% of cloned sequences (9).
Distribution of HHV-6B small RNA sequences across the genome.
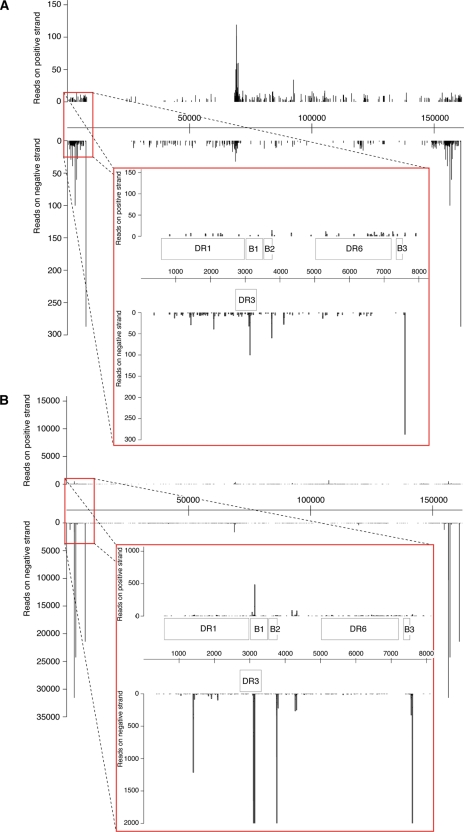
To simplify the analysis and to reduce noise, only sequences with zero mismatches to HHV-6B (strain Z29) and between 19 and 24 nt (with no corresponding human genomic hits) were used to search for putative miRNA candidates. Sequences were mapped to the HHV-6B Z29 reference genome (GenBank accession no. NC_000898.1) to look for hot spots across the genome. The HHV-6B genome consists of a unique long region (U) of 145 kb flanked by two identical repetitive regions of 8 to 9 kb that contain direct repeats (DRL and DRR). The distribution of HHV-6B small RNA sequences across the genome was quite striking. Mapping of HHV-6B small RNA sequences revealed that the majority of viral sequences derived from either the direct repeat regions or the region between open reading frames (ORFs) U41 and U42 that contains the OriLyt (15). Entire genomic representations of cloned sequences from both total RNA (Fig. 2A) and RNA obtained from Ago2 IP (Fig. 2B) are given with reference to the Z29 strain. Comparing whole genome reads between total RNA and Ago2 IP RNA, we observed a distinct enrichment in certain sequences deriving from the direct repeat regions (Fig. 2B). Reads mapped from small RNA cloning of total RNA clearly defined a central peak which correlated with the OriLyt (Fig. 2A). Sequences deriving from in or around the OriLyt of both rat and mouse cytomegaloviruses (RCMV and MCMV, respectively), as well as some other herpesviruses, such as herpes simplex virus 1 (HSV-1) and HSV-2, have previously been reported to encode miRNAs or postulated to act as RNA primers for the replication of the viral genome (8, 17, 30, 35).
Fig 2.
Genomic distribution of HHV-6B small RNA sequences by mapping of small RNA reads to the Z29 strain of HHV-6B. (A) Small RNA cloning from total RNA of Sup-T-1 cells infected with a laboratory strain of HHV-6B (7 dpi). The inset zooms into the left direct repeat (DRL), indicating the positions of DR1, DR3, and DR6, along with the HHV-6B-specific ORFs (B1, B2, and B3), with respect to the cloned small RNA sequences. (B) Representation of small RNA sequences cloned from the Ago2 IP RNA (4 dpi) mapped to the HHV-6B genome. Three peaks deriving from DRL and DRR are clearly visible; the inset highlights sequences deriving from DRL, using a cutoff of 2,000 reads.
Identification of HHV-6 miRNAs.
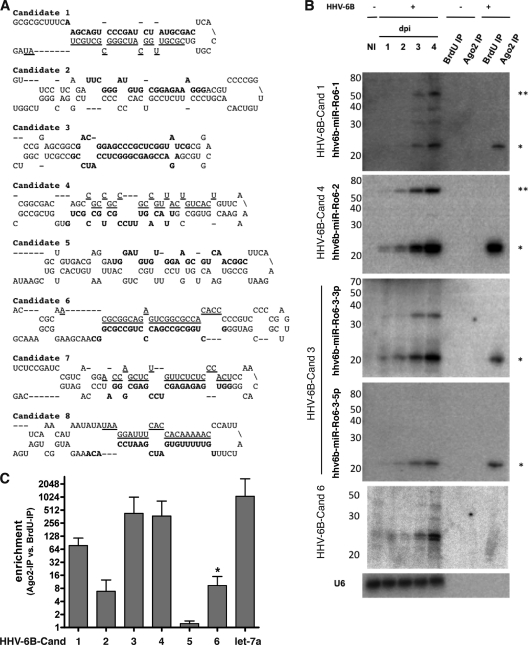
Those sequences that were cloned multiple times and had characteristic 5′ homogeneity were assessed manually for the ability to fold into typical stem-loop precursor structures by using Mfold (59). Eight sequences (named candidates 1 to 8) deriving from eight putative classical pre-miRNA stem-loop structures that matched perfectly to the HHV-6B Z29 genome were initially selected for validation (Fig. 3A). To validate these candidates, we performed Northern blot analysis of RNA taken from a time course of HHV-6B infection from 1 to 4 dpi; noninfected Sup-T-1 cells were used as a control. In addition, to verify if these were bona fide miRNAs, we loaded RNA samples from Ago2 IPs (and from BrdU IPs as controls) from both noninfected and infected cells (4 dpi). Three of these sequences (candidates 1, 3, and 4) were validated as novel HHV-6B miRNAs by their detection in both total RNA and Ago2 IP RNA by Northern blotting; both the 3p and 5p arms of candidate 3 were detected by Northern blotting of total RNA and RNA isolated from Ago2 IP samples (Fig. 3B).
Fig 3.
Predicted structures and validation of HHV-6B miRNAs. (A) Predicted fold-back structures of 8 HHV-6B miRNA candidates; in each case, the putative mature miRNA that was cloned is highlighted in bold, and putative star sequences are underlined. (B) Northern blots of candidates 1, 3, 4, and 6 against total RNA (10 μg) obtained from a time course of HHV-6B infection (1 to 4 dpi) of Sup-T-1 cells. RNAs isolated from immunoprecipitation (IP) of Ago2 complexes and from a control IP using an antibody to BrdU for both mock- and HHV-6B-infected Sup-T-1 cells (4 dpi) were loaded to determine whether the candidate sequences could be found in Ago2 RNA. (C) Fold enrichment of let-7a (control) and viral miRNA candidates for comparison of miRNA levels in the Ago2 IP sample and the BrdU IP control by qPCR (n = 2). A strong enrichment was observed for all four mature miRNA candidates also confirmed by Northern blotting, validating them as bona fide viral miRNAs.
Although sequences representing candidates 2, 6, 7, and 8 were cloned from RNA isolated after Ago2 IP (237, 289, 1,214, and 1,639 times, respectively), we were unable to formally validate them as miRNAs. While candidates 2, 7, and 8 remained undetectable in both total RNA and Ago2 IP RNA by Northern blotting, candidate 6 strongly accumulated during the time course of infection, as two distinct RNA species, but interestingly, it remained undetectable in the Ago2 IP samples. In addition, a small nonspecific signal was detected in the control (Fig. 3B). These results indicate that this small RNA most probably represents a degradation product or as yet unclassified small RNA species.
We further verified these results by using quantitative real-time PCR to identify enrichment of the small viral RNAs in Ago2 versus BrdU control samples; consistent with the Northern blot data, qPCR revealed significant enrichment of candidates 1, 3, and 4 in the Ago2 IP samples (Fig. 3C). Candidates 2 and 6 were significantly enriched in the Ago2 IP samples compared to BrdU IP controls, although the signal for candidate 6 was weak and nonspecific, reflecting the Northern blot data (Fig. 3A and C). Candidate 5 was cloned from total RNA and was not present in the Ago2 IP RNA sequence data, but it was selected as a candidate because it derived from a nonrepetitive genomic region and folded into a hairpin structure. No enrichment in Ago2 IP RNA was observed for candidate 5 by qPCR, confirming the sequencing data; no signal was observable by Northern blot analysis (Fig. 3C and data not shown).
To distinguish HHV-6 from the related HHV-7, HHV-6B miRNAs are named hhv6b-miR-Ro6-1 (candidate 1), hhv6b-miR-Ro6-2 (candidate 4), and hhv6b-miR-Ro6-3-3p and hhv6b-miR-Ro6-3-5p (candidates 3-3p and 3-5p, respectively). We refer to these miRNAs by their official names hereinafter. The sequences of the mature miRNAs and their star sequences are given in Table 2, along with the numbers of times they were cloned from both total RNA and Ago2 IP RNA libraries. The sequences of the predicted pre-miRNAs shown in Fig. 3A are given in Table 3, with reference to the Z29 strain of HHV-6B.
Table 2.
Mature HHV-6B miRNA sequences, genomic positions, and distribution in the two librariesa
| miRNA | Cloned sequence (major form) | Strand | Size (nt) | No. of reads for total RNA | No. of reads for Ago2 IP RNA | Genomic location in DRL | Genomic location in DRR |
|---|---|---|---|---|---|---|---|
| hhv6b-miR-Ro6-1 | AAGCAGUCCCGAUCUAUGCGAC | − | 22 | 287 | 25,449 | 7590–7611 | 160911–160932 |
| hhv6b-miR-Ro6-1* | CGCGUUGGCAUCGGGCGCUGCUAU | 24 | 1 | 946 | 7557–7580 | 160878–160901 | |
| hhv6b-miR-Ro6-2 | UUACAGUUUCCGCUGCCGCUGU | − | 22 | 110 | 55,321 | 3754–3775 | 157076–157096 |
| hhv6b-miR-Ro6-2* | CGCCGCCGCCGUUACUGUCACU | 22 | 2 | 612 | 3794–3815 | 157115–157136 | |
| hhv6b-miR-Ro6-3-5p | GACGGAGUCCGCUCGGUAUCG | − | 21 | 100 | 21,460 | 3128–3148 (1) | 156449–156469 (1) |
| hhv6b-miR-Ro6-3-3p | AGACCGAGCGGGCUCCAUCCG | − | 21 | 32 | 61,900 | 3097–3117 | 156418–156438 |
| hhv6b-miR-Ro6-4 | AUACCGAGCGGACUCCGUCGCU | + | 22 | 2 | 947 | 3130–3151 (2) | 156451–156472 |
| hhv6b-miR-Ro6-4* | CGGAUGGAGCCCGCUCGGUCUCG | + | 23 | 0 | 20 | 3097–3119 | 156418–156440 |
The predominantly cloned sequences are indicated. 5p, 5′ arm of the hairpin precursor; 3p, 3′ arm of the hairpin precursor; *, nonfunctional star sequence. Positions are given relative to the published genomic sequence of the Z29 strain of HHV-6B. Nucleotides in bold denote mismatches to the Z29 genome but matches to the HST strain, nontemplate nucleotides are underlined, and numbers of mismatches are indicated in parentheses.
Table 3.
HHV-6B pre-miRNA sequences and their genomic positions
| Pre-miRNA | Sequencea | Strand | Genomic location in DRL | Genomic location in DRR |
|---|---|---|---|---|
| hhv6b-miR-Ro6-1 | GCGCGCUUUCAAGCAGUCCCGAUCUAUGCGACUCACCGUGUCGCGUUGGCAUCGGGCGCUGCUAUAG | − | 7555–7621 | 160876–160942 |
| hhv6b-miR-Ro6-2 | CGGCGACAGCCGCCGCCGCCGUUACUGUCACUGUUCAAAGAACGUGGCCGUUACAGUUUCCGCUGCCGCUGUGGUCGCCGC | − | 3745–3825 | 157066–157146 |
| hhv6b-miR-Ro6-3 | CCGGAGCGGCGACGGAGCCCGCUCGGUAUCGCGGACGUGCGAGACCGAGCGGGCUCCAUCCGCCGCUCGGUC | − | 3087–3158 | 156408–156479 |
| hhv6b-miR-Ro6-4 | GACCGAGCGGCGGAUGGAGCCCGCUCGGUCUCGCACGUCCGCGAUACCGAGCGGGCUCCGUCGCCGCUCCGG | + | 3087–3158 | 156408–156479 |
The indicated sequences correspond to the sequences used to make the predicted fold-back structures shown in Fig. 3A and in Fig. S1C in the supplemental material. Nucleotides highlighted in bold indicate the mature miRNA sequences. Positions are given relative to the published genomic sequence of the Z29 strain of HHV-6B.
Characteristics of HHV-6 miRNAs.
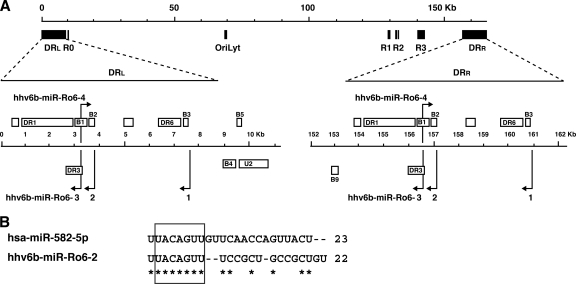
A schematic representation of HHV-6B miRNAs with respect to their genomic localization in the direct repeats is given in Fig. 4A. Note that hhv6b-miR-Ro6-3 lies antisense to the predicted B1 ORF and is encoded within DR3, while hhv6b-miR-Ro6-2 lies antisense to the predicted B2 ORF. B1 and B2 are unique to HHV-6B among all herpesviruses and were recently identified as 2 of the 8 immediate-early (IE) genes expressed by HHV-6B via transcriptome analyses (53). The functions of B1 and B2 are unknown, but it is thought that due to their classification as IE genes and their specificity to HHV-6B, they may play important roles in HHV-6B-specific infection (53). The identification of viral miRNAs that are transcribed antisense to viral genes is now a well-described feature of known viral miRNAs: proven regulation of antisense targets has been defined for EBV (4), HCMV (24), HSV-1 and -2 (50, 51, 54, 56), and polyomaviruses (47, 48). Interestingly, such viral miRNAs do not always regulate their antisense targets by cleavage, as is the case for the simian virus 40 (SV40) miR SV40-miR-S1 and its target, the large T antigen (47), but they may inhibit their targets by the classical translational inhibition model, as is the case for HCMV-miR-UL112-1 and its target, UL114 (24), and putatively the case for HSV-1-miR-H2-3p and its target, ICP0 (54). We therefore hypothesize from these previous observations that HHV-6B could use hhv6b-miR-Ro6-2 and hhv6b-miR-Ro6-3 to downregulate B2 and B1, respectively. Note that hhv6b-miR-Ro6-2 bridges the 3′ coding region and the presumed 3′-UTR of B2 with the 3p and 5p arms of its precursor. To our knowledge, such a feature has not been described previously for any viral or cellular miRNA. As for hhv6b-miR-Ro6-2, hhv6b-miR-Ro6-3 also lies antisense to an immediate-early transcript (B1) but is encoded within DR3. DR3 is a gene of unknown function which is expressed as early as 6 h following infection and is blocked by cycloheximide (CHX) treatment (53). Since DR3 has not been classified in terms of its kinetics, we presume that DR3 and hhv6b-miR-Ro6-3 share similar kinetics, although it is possible that both of these miRNAs are transcribed by an upstream promoter, as they share the same expression pattern for both the mature and precursor miRNA forms (Fig. 3B). Finally, hhv6b-miR-Ro6-1, which was the most abundant miRNA identified from sequencing of total RNA (7 dpi) (Table 2), was detectable only following 3 dpi (2 days later than the other HHV-6B miRNAs). Therefore, we assume that all three of these miRNAs are not produced from one large transcript but that at least hhv6b-miR-Ro6-1 is transcribed from a separate promoter. Like the miRNAs located antisense to B1 and B2, hhv6b-miR-Ro6-1 is encoded antisense to the potential 3′-UTR of B3, an HHV-6B-specific gene of unknown function that has been classified as either an IE (38) or early (53) gene. In general, all HHV-6B miRNAs have the potential to regulate HHV-6B-specific genes, which is an unusual feature of these miRNAs. Currently, no antibodies are available against B1 to B3. In addition to these particular features of HHV-6B miRNAs, hhv6b-miR-Ro6-2 is a seed ortholog (bases 2 to 8) of the human miRNA miR-582-5p (Fig. 4B). Northern blots to detect the poorly conserved miR-582-5p from noninfected and infected cells failed to detect the expression of this cellular miRNA in Sup-T-1 cells (data not shown). This is in agreement with the sequencing data, in which miR-582-5p was absent. This miRNA is upregulated in certain pituitary adenomas and has provisionally been linked with targeting SMAD3 to downregulate transforming growth factor beta (TGF-β) (10); however, further validation is required. We hypothesize that similar to the Kaposi's sarcoma-associated herpesvirus (KSHV) miR-155 ortholog miR-K12-11 (44), hhv6b-miR-Ro6-2 may thus share a broad range of targets with miR-582-5p and function in cells that do not normally express this cellular miRNA to modulate the host-pathogen environment.
Fig 4.
Genomic localization and features of HHV-6B miRNAs. (A) Genomic architecture of HHV-6B indicating the relative positions of the repetitive regions (DRL, R0, R1, R2 [R2A and R2B], R3, and DRR) and OriLyt, with a zoom-in to each DR region. The direction of transcription is given by placement above (positive) or below (negative) the scale bar. All HHV-6B miRNAs are expressed from both repeat regions. All HHV-6B miRNAs are expressed from both repeat regions; miRNAs are indicated with arrows and labeled hhv6b-miR-Ro6-1, -Ro6-2, -Ro6-3, and -Ro6-4. DR3 encodes miR-Ro6-3, and each miRNA is located antisense (either to the coding region or to the putative 3′-UTR) to an HHV-6B-specific ORF that has no homolog with HHV-6A (see the text for details). (B) hhv6b-miR-Ro6-2 is a perfect seed ortholog of a human miRNA that is not expressed in Sup-T-1 cells (data not shown). A sequence alignment of miR-Ro6-3 with its seed ortholog, miR-582-5p, is shown. The seed is boxed, and homology (ClustalW) is indicated by asterisks.
5′ RACE identifies the presence of an antisense miRNA.
To determine whether hhv6b-miR-Ro6-3 could regulate the B1 ORF carried antisense to the miRNA sequence, 5′ RLM-RACE was performed to assess whether this RNA could induce cleavage of B1. A cleavage product was identified that resided exactly at the 5′ terminus of the predicted pre-miRNA on the opposite strand, not at the expected cleavage site if the transcript were cleaved by the miRNA within Ago2 (see Fig. S1A in the supplemental material). These 5′ RACE results were initially disappointing, although it remained possible that this miRNA regulated B1 via translational inhibition or that the cleavage product is extremely rare or rapidly degraded by exonucleases to generate the observed cleavage product. While the observed cleavage product from 5′ RACE could result from RNA cleavage of B1, we noted that small RNA reads with a distinct miRNA-like signature (Fig. 2B; see Fig. S1B in the supplemental material) were located antisense to both the 5p and 3p arms of hhv6b-miR-Ro6-3, indicating a potential additional miRNA not originally considered a candidate. Antisense miRNAs (miR-AS) have previously been described for three other members of the betaherpesvirus family, namely, human CMV (46), murine CMV (8, 17), and rat CMV (35), as well as for animals (45). We therefore decided to perform Northern blot analysis to detect this miRNA, as the 5′ RACE results together with sequencing data were indicative of a cleavage site within the B1 transcript, mediated not by hhv6b-miR-Ro6-3 but by Drosha. Although the number of sequence reads for this antisense miRNA was small, a miRNA derived from a classical pre-miRNA hairpin structure (see Fig. S1C in the supplemental material) was detectable following 4 days of infection and was enriched in Ago2 IP samples (see Fig. S1D in the supplemental material). This miRNA has a unique seed, and as such it was named hhv6b-miR-Ro6-4. We also identified the presence of sequencing reads located on the opposite strand for the two other miRNAs, albeit at extremely low levels (Fig. 2B).
Conservation of HHV-6 miRNAs.
Because a laboratory strain was used for infection, we performed an additional analysis by reannotating the small RNA libraries against the HST strain of HHV-6B to look for potential interstrain variations in the validated miRNA sequences. hhv6b-miR-Ro6-1 and hhv6b-miR-Ro6-2 share identical pre-miRNA sequences in both strains, while there are notable interstrain variations of hhv6b-miR-Ro6-3 (see Fig. S2 in the supplemental material). hhv6b-miR-Ro6-3-5p (not an original candidate due to having a mismatch with respect to the Z29 genome) matches perfectly to the HST sequence but bears one mismatch (C to U), at position 7 (position 6 of the seed), to the Z29 sequence. Inversely, hhv6b-miR-Ro6-3-3p matches the Z29 sequence perfectly but is altered (G to A) at position 2 (seed position 1) with respect to the HST sequence; hairpin structures are unaffected by these differences. In addition, the genome sequences were obtained from plasmid clones, which would not have represented the genetic diversity present in virus populations. The relevance of these interstrain variations is unknown and would require careful sequencing analysis of all known strains of HHV-6B. However, variations in miRNA sequence, particularly in the seed region, may contribute to functional differences with respect to cellular targets. In effect, with the predicted targeting of their antisense transcripts, interstrain variations would not affect a potential regulation of the relevant viral antisense transcripts. In terms of intervariant variation, HHV-6B and HHV-6A share over 90% sequence homology, although they demonstrate greater diversity within genes encoding IE-1 and within the terminal direct repeats, which results in the B1 to B3 ORFs being specific to HHV-6B (18). All HHV-6B miRNAs are conserved within HHV-6A at both the structural and sequence levels, although they display minor variations in their sequences (see Fig. S2 in the supplemental material). In addition, we cloned several distinct isoforms of each miRNA that demonstrate 5′ and 3′ heterogeneity (see Table S1 in the supplemental material). In contrast, hhv6b-miR-Ro6-2 sequence is perfectly conserved but presents with significant differences within the predicted pre-miRNA. These differences may have functional consequences with respect to the different biological properties of these two viruses. However, HHV-6A isolates with natural deletions of a large part of the direct repeat region (positions 60 to 5545; U1102 strain) that encompasses the HHV-6A homologues of hhv6b-miR-Ro6-2 and hhv6b-miR-Ro6-3 do not show in vitro replication defects but in fact possess enhanced replication capabilities in vitro (6). Proper functional analysis of these miRNAs in an in vitro setting will require the construction of a bacterial artificial chromosome (BAC) for HHV-6B, as a BAC is currently available only for HHV-6A (5).
Other small RNAs identified in HHV-6.
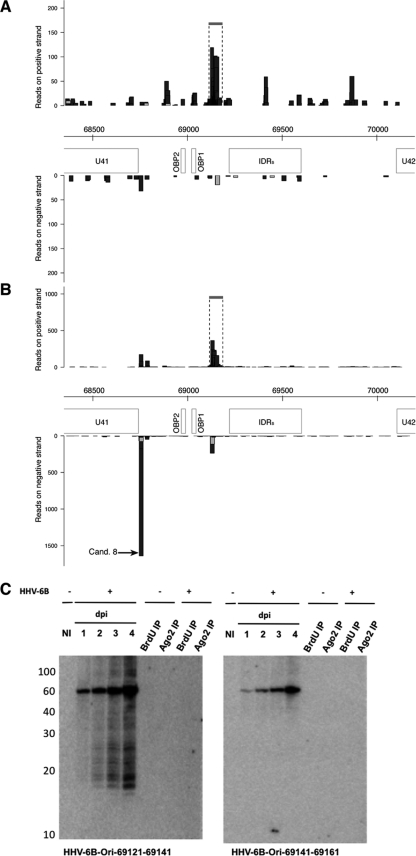
An overall analysis of small viral RNAs in the Ago2 RNA sample indicated that the four miRNA hairpins verified to give rise to HHV-6B miRNAs comprise ∼80% of all viral sequences. Note that candidates 7 and 8 form fold-back hairpins (Fig. 3A) with characteristic 5′ homogeneity and were enriched in libraries generated from Ago2 RNA compared to total RNA (Fig. 2A and B and 5A and B). These candidates were tested by both qPCR and Northern blotting; however, we were unable to verify these as miRNAs, likely owing to their low abundance. Considering that ∼95% of all cellular RNAs found in the Ago2 IP samples were classifiable as miRNAs, it is interesting to speculate that these sequences may simply derive from rare HHV-6B miRNAs not detectable under the conditions used. Experiments to assess the role of HHV-6B miRNAs in latency and in vivo will assess whether these sequences represent miRNAs. Sequences and genomic positions of all the candidates can be found within Table S2 in the supplemental material. A breakdown of the distribution of sequence reads across the genome identifies ∼85% as deriving from the direct repeats (DR), ∼7.5% as deriving from the OriLyt, and ∼7.5% as deriving from elsewhere. As mentioned, miRNAs have been verified from in or around the OriLyt regions for other herpesviruses, namely, MCMV (8, 17), RCMV (35), and HSV-1 (30). As such, Northern blots were performed for the three most abundant sequences deriving from the origin: candidate 8 (not defined as a miRNA), on the negative strand (positions c68744 to c68765), and two contiguous sequences that do not form a typical hairpin, located on the positive strand (positions 69121 to 69141 and 69141 to 69161) (Fig. 5A and B). Interestingly, probes against the contiguous sequences identified a novel viral transcript of ∼62 nt spanning this region, located near sites for the origin binding protein (OBP) (29) (Fig. 5C). Note that sequencing data from both libraries indicate multiple RNA species deriving from this locus (sequences in Ago2 RNA ranging from 17 to 32 nt), and this is clearly visible as a broad set of vertical bars in the graphical representation of OriLyt sequences and by a smear on the Northern blot (Fig. 5A, B, and C). Notably, abundant RNA species of 18 and 19 nt were identified from only one arm of the precursor; the Northern blot signal indicates that these were not present in Ago2 RNA at comparable levels to those in total RNA samples (as seen for viral miRNAs in Fig. 3B). As such, their presence in Ago2 cloning almost certainly represents a nonspecific binding to Ago2 due to their abundance within the cell, highlighting the benefit of including RNA from Ago2 IP on Northern blots to distinguish RNA species within the Ago2 sequence from other small RNAs or breakdown products. Interestingly, Tsao et al. recently characterized the expression of lytic genes in HHV-6B and identified a number of intergenic noncoding transcripts (53). Among these transcripts, they identified a 316-nt RNA located such that it could contain the ∼60-nt RNA we detected. It appears reasonable to hypothesize that these small RNAs derive from this novel viral RNA being degraded specifically to generate small RNA species that may represent RNA primers to assist in viral replication. Alternatively, these small RNAs could represent a new class of small RNAs that may be incorporated into another member of the Argonaute family. Performing immunoprecipitations with the three remaining Argonaute proteins will resolve this interesting hypothesis. Further work is required to elucidate their processing and potential functions in HHV-6 biology.
Fig 5.
Identification of an abundant small RNA mapping to the OriLyt. (A) Representation of small RNA reads mapping to the region encompassing the origin of lytic replication from total RNA of Sup-T-1 cells infected with HHV-6B (7 dpi). Sites for OBP are given, along with the positions of the indirect repeat regions (IDRs) and the neighboring genes U41 and U42. The horizontal gray bar indicates the position of the sequence that was analyzed by Northern blotting. (B) Sequences mapping to the OriLyt obtained following sequencing of RNAs within the Ago2 IP sample. A clear enrichment of two sets of antisense sequences was visible following IP. The leftmost sequence (candidate 8, indicated by an arrow) was enriched ∼50 times (compared to that in panel A). The horizontal gray bar indicates the position of the sequence that was analyzed by Northern blotting. (C) Northern blots for two contiguous sequences enriched in Ago2 RNA and deriving from a GC-rich region between OBP1 and the IDR. A novel viral transcript encompassing positions 69121 to 69161 was clearly detectable and increased in abundance throughout the time course of infection. One arm of this transcript generates a range of abundant small RNAs which are not detectable by Northern blotting of Ago2 RNA and may represent a novel class of origin-derived small RNAs.
Conclusions.
Here we showed by using a deep sequencing approach that HHV-6B encodes at least four pre-miRNAs, two of which lie antisense to each other at the same genomic locus. These miRNAs are expressed during lytic infection and are all conserved in HHV-6A. The use of small RNA cloning from Ago2 IP samples greatly simplified the analysis by removing the majority of RNA breakdown products, providing a clean data set to aid in identifying novel viral miRNAs. HHV-6B miRNAs accumulate during HHV-6B infection and, similar to HSV-1/2 miRNAs, are encoded antisense to IE HHV-6B-specific ORFs (B1, B2, B3, and DR3), in line with the paradigm that viral miRNAs can regulate IE genes to modulate their own transcription. Like the case for KSHV-miR-K12-11 and miR-155, hhv6b-miR-Ro6-2 is a seed ortholog of a cellular miRNA (miR-582-5p) that is not expressed in the main target cell of the virus, in this case CD4+ T cells. These data provide good starting points for identifying the cellular and viral targets of these miRNAs. Similar to HCMV, HHV-6 is a betaherpesvirus, and its miRNAs were identified in cells undergoing active lytic infection (19, 23, 40).
This work opens up questions regarding the cloning bias observed in deep sequencing data sets for certain miRNAs; the viral miRNAs reported in this study were present at low abundances in the sequencing data sets. Nevertheless, the miRNA candidates all conform to standard rules governing miRNA classification (1). This was confirmed by their Northern blot detection in both total RNA and Ago2 IP RNA. The inclusion of Ago2 IP RNA in Northern blots aided in our discrimination of miRNAs from non-miRNA-like small RNAs or breakdown products, such as candidate 6.
In terms of the use of bioinformatics to predict a given miRNA, caution is warranted. In the original study by Pfeffer et al., no pre-miRNA hairpins were predicted for HHV-6, although the likelihood that HHV-6 encodes miRNAs was stated at 84% (40). More recently, an open access large database (Vir-miR) was published that predicted viral miRNAs in over 2,000 viral genomes (33), but none of our experimentally validated miRNAs were predicted in that study. These reports open up speculation as to the efficacy of bioinformatic predictions when miRNAs derive from complex genomic loci such as direct repeats. An improved approach relies on the combination of prediction tools and deep sequencing data sets to scan for putative pre-miRNA hairpins, as in miRCat (37), miRDeep (20, 21), and miRAnalyzer (27, 28) software. At least one strand of each HHV-6B pre-miRNA was predicted by one or more of these programs for either the total RNA library, the Ago2 IP library, or both (see Table S3 in the supplemental material), which provides further support regarding their miRNA status.
Unlike all other human herpesviruses, HHV-6A has been shown to integrate into host telomeres as a method of latency in individuals with chromosomally integrated HHV-6 (CIHHV-6), rather than residing as an episome. Importantly, the virus can be reactivated from this state and passed on through generations of such individuals (2). It will be interesting to see if these miRNAs are expressed in latently infected cells as well as in CIHHV-6 patients. The distribution of HHV-6 miRNAs is somewhat different from what has been found to date in other betaherpesviruses but resembles the genomic organization of the miRNAs of the chicken Marek's disease virus, another integrating herpesvirus (9).
The identification of an abundant transcript of 60 to 65 nt expressed from the OriLyt could be reminiscent of other viral pre-miRNAs recently identified to be expressed from the OriLyt regions of MCMV (8, 17), RCMV (35), bovine herpesvirus (BoHV) (22), and HSV-1/2 (30). However, similar to our results, none of the miRNAs reported in these papers were reported conclusively to be in Ago2 RNA; hence, herpesvirus origin-derived small RNAs represent an area of research that clearly deserves further investigation.
As with all viral miRNAs, a thorough decoding of cellular and viral targets is essential to understand the potential therapeutic uses of targeting the viral miRNAs of a given virus in the context of human disease. The identification of miRNAs from HHV-6 will provide researchers with new avenues of research concerning the host-pathogen relationship during HHV-6 infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of our laboratories for critically reviewing the manuscript prior to publication. We thank Timo Lassmann for providing us with an early version of Nexalign.
This work was supported by a grant from the German Bundesministerium für Bildung und Forschung (NGFN-Plus 01GS0801) to L.D. and by the European Research Council (ERC starting grant ncRNAVIR 260767) and a starting grant from the Centre National de la Recherche Scientifique (ATIP) to S.P.
Footnotes
Published ahead of print 23 November 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Ambros V, et al. 2003. A uniform system for microRNA annotation. RNA 9: 277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arbuckle JH, et al. 2010. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc. Natl. Acad. Sci. U. S. A. 107: 5563–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- 4. Barth S, et al. 2008. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 36: 666–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borenstein R, Frenkel N. 2009. Cloning human herpes virus 6A genome into bacterial artificial chromosomes and study of DNA replication intermediates. Proc. Natl. Acad. Sci. U. S. A. 106: 19138–19143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Borenstein R, Zeigerman H, Frenkel N. 2010. The DR1 and DR6 first exons of human herpesvirus 6A are not required for virus replication in culture and are deleted in virus stocks that replicate well in T-cell lines. J. Virol. 84: 2648–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boyd MT, Simpson GR, Cann AJ, Johnson MA, Weiss RA. 1993. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J. Virol. 67: 3649–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buck AH, et al. 2007. Discrete clusters of virus-encoded microRNAs are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J. Virol. 81: 13761–13770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burnside J, et al. 2006. Marek's disease virus encodes microRNAs that map to meq and the latency-associated transcript. J. Virol. 80: 8778–8786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Butz H, et al. 2011. MicroRNA profile indicates downregulation of the TGFbeta pathway in sporadic non-functioning pituitary adenomas. Pituitary 14: 112–124 [DOI] [PubMed] [Google Scholar]
- 11. Cai X, et al. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. U. S. A. 102: 5570–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cazalla D, Xie M, Steitz JA. 2011. A primate herpesvirus uses the integrator complex to generate viral microRNAs. Mol. Cell 43: 982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cermelli C, et al. 1997. SupT-1: a cell system suitable for an efficient propagation of both HHV-7 and HHV-6 variants A and B. New Microbiol. 20: 187–196 [PubMed] [Google Scholar]
- 14. De Bolle L, Naesens L, De Clercq E. 2005. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin. Microbiol. Rev. 18: 217–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dewhurst S, Dollard SC, Pellett PE, Dambaugh TR. 1993. Identification of a lytic-phase origin of DNA replication in human herpesvirus 6B strain Z29. J. Virol. 67: 7680–7683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dolken L, et al. 2010. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe 7: 324–334 [DOI] [PubMed] [Google Scholar]
- 17. Dolken L, et al. 2007. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J. Virol. 81: 13771–13782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dominguez G, et al. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73: 8040–8052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dunn W, et al. 2005. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell. Microbiol. 7: 1684–1695 [DOI] [PubMed] [Google Scholar]
- 20. Friedlander MR, et al. 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 26: 407–415 [DOI] [PubMed] [Google Scholar]
- 21. Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Glazov EA, et al. 2010. Characterization of microRNAs encoded by the bovine herpesvirus 1 genome. J. Gen. Virol. 91: 32–41 [DOI] [PubMed] [Google Scholar]
- 23. Grey F, et al. 2005. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 79: 12095–12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grey F, Nelson J. 2008. Identification and function of human cytomegalovirus microRNAs. J. Clin. Virol. 41: 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Griffiths-Jones S. 2006. miRBase: the microRNA sequence database. Methods Mol. Biol. 342: 129–138 [DOI] [PubMed] [Google Scholar]
- 26. Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hackenberg M, Rodriguez-Ezpeleta N, Aransay AM. 2011. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res. 39: W132–W138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hackenberg M, Sturm M, Langenberger D, Falcon-Perez JM, Aransay AM. 2009. miRanalyzer: a microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 37: W68–W76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Inoue N, Dambaugh TR, Rapp JC, Pellett PE. 1994. Alphaherpesvirus origin-binding protein homolog encoded by human herpesvirus 6B, a betaherpesvirus, binds to nucleotide sequences that are similar to ori regions of alphaherpesviruses. J. Virol. 68: 4126–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jurak I, et al. 2010. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J. Virol. 84: 4659–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim YJ, et al. 2002. Human herpesvirus-6 as a possible cause of encephalitis and hemorrhagic cystitis after allogeneic hematopoietic stem cell transplantation. Leukemia 16: 958–959 [DOI] [PubMed] [Google Scholar]
- 32. Kondo K, et al. 2002. Strong interaction between human herpesvirus 6 and peripheral blood monocytes/macrophages during acute infection. J. Med. Virol. 67: 364–369 [DOI] [PubMed] [Google Scholar]
- 33. Li SC, Shiau CK, Lin WC. 2008. Vir-Mir db: prediction of viral microRNA candidate hairpins. Nucleic Acids Res. 36: D184–D189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lusso P. 2006. HHV-6 and the immune system: mechanisms of immunomodulation and viral escape. J. Clin. Virol. 37(Suppl 1): S4–S10 [DOI] [PubMed] [Google Scholar]
- 35. Meyer C, et al. 2011. Cytomegalovirus microRNA expression is tissue specific and is associated with persistence. J. Virol. 85: 378–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moore FG, Wolfson C. 2002. Human herpes virus 6 and multiple sclerosis. Acta Neurol. Scand. 106: 63–83 [DOI] [PubMed] [Google Scholar]
- 37. Moxon S, et al. 2008. A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics 24: 2252–2253 [DOI] [PubMed] [Google Scholar]
- 38. Oster B, Hollsberg P. 2002. Viral gene expression patterns in human herpesvirus 6B-infected T cells. J. Virol. 76: 7578–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pfeffer S. 2007. Identification of virally encoded microRNAs. Methods Enzymol. 427: 51–63 [DOI] [PubMed] [Google Scholar]
- 40. Pfeffer S, et al. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2: 269–276 [DOI] [PubMed] [Google Scholar]
- 41. Pfeffer S, et al. 2004. Identification of virus-encoded microRNAs. Science 304: 734–736 [DOI] [PubMed] [Google Scholar]
- 42. Pruksananonda P, et al. 1992. Primary human herpesvirus 6 infection in young children. N. Engl. J. Med. 326: 1445–1450 [DOI] [PubMed] [Google Scholar]
- 43. Salahuddin SZ, et al. 1986. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science 234: 596–601 [DOI] [PubMed] [Google Scholar]
- 44. Skalsky RL, et al. 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 81: 12836–12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stark A, et al. 2008. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 22: 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stark TJ, Arnold JD, Spector DH, Yeo GW. 2012. High-resolution profiling and analysis of viral and host small RNAs during human cytomegalovirus infection. J. Virol. 86: 226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435: 682–686 [DOI] [PubMed] [Google Scholar]
- 48. Sullivan CS, et al. 2009. Murine polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology 387: 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takahashi K, et al. 1989. Predominant CD4 T-lymphocyte tropism of human herpesvirus 6-related virus. J. Virol. 63: 3161–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tang S, et al. 2008. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc. Natl. Acad. Sci. U. S. A. 105: 10931–10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tang S, Patel A, Krause PR. 2009. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J. Virol. 83: 1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Toedling J, Ciaudo C, Voinnet O, Heard E, Barillot E. 2010. Girafe—an R/Bioconductor package for functional exploration of aligned next-generation sequencing reads. Bioinformatics 26: 2902–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tsao EH, et al. 2009. Microarray-based determination of the lytic cascade of human herpesvirus 6B. J. Gen. Virol. 90: 2581–2591 [DOI] [PubMed] [Google Scholar]
- 54. Umbach JL, et al. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454: 780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR. 2009. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J. Virol. 83: 10677–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Umbach JL, et al. 2010. Identification of viral microRNAs expressed in human sacral ganglia latently infected with herpes simplex virus 2. J. Virol. 84: 1189–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yamanishi K, et al. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet i: 1065–1067 [DOI] [PubMed] [Google Scholar]
- 58. Zerr DM, Gupta D, Huang ML, Carter R, Corey L. 2002. Effect of antivirals on human herpesvirus 6 replication in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 34: 309–317 [DOI] [PubMed] [Google Scholar]
- 59. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.