Abstract
The genome of bat adenovirus 2 was sequenced and analyzed. It is similar in size (31,616 bp) to the genomes of bat adenovirus 3 and canine adenoviruses 1 and 2. These four viruses are monophyletic and share an identical genome organization, with one E3 gene and four E4 genes unique to this group among the mastadenoviruses. These findings suggest that canine adenoviruses may have originated by interspecies transfer of a vespertilionid bat adenovirus.
TEXT
Adenoviruses (AdVs) have been described in every vertebrate class and in general exhibit a strong species specificity. Nevertheless, a study published recently (2011) by Chen et al. described the zoonotic transmission potential of AdVs from New World monkeys to humans (2). Also, bats have been recognized particularly as potential reservoir hosts of numerous zoonotic viruses (9).
The first bat AdV was isolated from a fruit bat in the suborder Megachiroptera (12). In 2008, we isolated the first AdV (bat adenovirus 2 Pipistrellus pipistrellus virus 1 [BtAdV-2 PPV1]) from a microchiropteran bat in Germany (13). A second microchiropteran bat AdV (BtAdV-3 strain TJM) was isolated from an Asian bat (Myotis ricketti), and an almost complete genome sequence has been reported (11). Twenty-nine additional bat AdVs have since been detected in China, Hungary, and Germany (5, 7, 15), and, by metagenomic study of bat guano specimens, the United States (10). In an initial study of BtAdV-2, a close phylogenetic relationship to canine adenoviruses (CAdVs) was observed (13). Although most AdVs are strictly host specific, CAdVs have been detected in an unusually broad range of carnivores (e.g., bears, wolves, raccoons, and sea lions) (1). Moreover, in their respective hosts, CAdVs are more severely pathogenic than the majority of adenoviruses. These observations and the close phylogenetic relationship led us to hypothesize an interspecies transmission event of a bat AdV to a carnivore. To explore this hypothesis, the whole genome of BtAdV-2 PPV1 was determined and analyzed.
In 2008, we detected a novel adenovirus in 3 of 55 free-ranging bats from Germany (13). We have now increased the number of animals examined to 330 European bats, with 116 belonging to two species of pipistrelles (Pipistrellus pipistrellus and P. nathusii). A full necropsy was performed on each bat, followed by histopathological examination.
For detection of BtAdV-2 DNA, a real-time PCR targeting the DNA polymerase gene was utilized (13). In addition to the initial three bats carrying BtAdV-2 (13), we detected the virus (sequence confirmed) by real-time PCR in an additional nine bats from Germany. Molecular biological examinations indicated distinct organ tropisms for intestine, kidney, and liver (Table 1), whereas histopathological changes were mostly limited to lung and spleen.
Table 1.
Molecular biological and histopathological results for pipistrelle bats found positive for BtAdV-2
| Bat host | Species | Age | Sex | Origin | Organ tropism (PCR results) |
Histopathological resultsb |
||
|---|---|---|---|---|---|---|---|---|
| Organ(s) PCR positive | No. of viral DNA copiesa | Histopathological change(s) | Autolysis | |||||
| 198/07 | P. pipistrellus | Adult | Male | Bavaria, Germany | Intestine | >1.0 × 106 | Lung, nonsuppurative interstitial | +++ |
| Liver | 1.4 × 103 | pneumonia (+) | ||||||
| Kidney | 3.1 × 105 | |||||||
| 199/07 | P. pipistrellus | Adult | Female | Bavaria, Germany | Intestine | >1.0 × 106 | Lung, nonsuppurative interstitial | ++ |
| Liver | 2.0 × 103 | pneumonia (++) and | ||||||
| Kidney | 2.0 × 103 | leucocytostasis of blood vessels (+++) | ||||||
| 200/07 | P. pipistrellus | Adult | Male | Bavaria, Germany | Intestine | >1.0 × 106 | Lung, nonsuppurative interstitial | ++ |
| Spleen | 1.4 × 103 | pneumonia (++); spleen, | ||||||
| Kidney | 2.6 × 102 | follicular hyperplasia (+++) | ||||||
| 279/08 | P. pipistrellus | Adult | Male | Lower Saxony, Germany | Intestine | 7.9 × 103 | Lung, nonsuppurative interstitial pneumonia (++) and leucocytostasis of blood vessels (+++) | + |
| 332/08 | P. nathusii | Adult | Male | Bavaria, Germany | Intestine | 2.0 × 103 | Lung, leucocytostasis of blood | − |
| Kidney | 3.2 × 101 | vessels (+); spleen, follicular hyperplasia (+) | ||||||
| 348/08 | P. pipistrellus | Subadult | Female | Bavaria, Germany | Intestine | 3.2 × 101 | Lung, neutrophilic infiltration of | + |
| Salivary glands | 6.9 × 101 | alveolar septa (+); spleen, follicular hyperplasia (+++) and colliquative necrosis (+++) | ||||||
| 097/09 | P. pipistrellus | Subadult | Male | Bavaria, Germany | Intestine | 4.4 × 105 | Skin, focal purulent ulcerative dermatitis (+++) | +++ |
| 142/09 | P. pipistrellus | Subadult | Male | Bavaria, Germany | Intestine | 7.9 × 103 | Lung, nonsuppurative interstitial pneumonia (++) | − |
| 173/09 | P. pipistrellus | Adult | Male | Berlin, Germany | Intestine | 3.2 × 101 | Spleen, follicular hyperplasia (+) | ++ |
| 198/09 | P. pipistrellus | Juvenile | Male | Berlin, Germany | Intestine | 6.7 × 102 | Lung, nonsuppurative interstitial pneumonia (+++) | − |
| 199/09 | P. pipistrellus | Adult | Male | Berlin, Germany | Intestine | 2.9 × 102 | Spleen, follicular hyperplasia (++) | − |
| 228/09 | P. pipistrellus | Adult | Male | Berlin, Germany | Intestine | 1.6 × 101 | Spleen, follicular hyperplasia (+++) | + |
Number of copies of viral DNA per ml of homogenized organ tissue (average sample size, 8 mm3 per ml).
Degree of severity: −, none; +, mild; ++, moderate; +++, severe.
Shotgun pyrosequencing was carried out using a 454 Genome Sequencer FLX (454 Life Sciences, Branford, CT) in accordance with the manufacturer's protocol. A total of 15,565 reads (3,651,529 bp) were used for the Newbler and MIRA assembly, with an average coverage of 115.8 (3). Protein-coding open reading frames (ORFs) and splice sites were identified by comparative genomics with other mastadenoviruses.
The genome sequence (GenBank accession no. JN252129) consists of 31,616 bp, with an average G+C content of 53.5%, has an inverted terminal repeat (ITR) of 146 bp, and contains 31 predicted genes (Table 2).
Table 2.
Characteristics of the genes of the two bat and two canine adenovirus types, with human adenovirus 2 included for comparison
| Gene name, type, or product | Coding sequence positionsa | Size of gene (bp or aa) from virus of speciesb: |
||||
|---|---|---|---|---|---|---|
| BtAdV-Bc (virus BtAdV-2 [31,616 bp]) | BtAdV-Ac (virus BtAdV-3 [>31,680 bp])d | CAdV |
HAdV-C (virus HAdV-2 [35,937 bp]) | |||
| Virus CAdV-1 (30,288 bp) | Virus CAdV-2 (31,323 bp) | |||||
| ITR | 1–146, 31471–31616 | 146 | NDe | 199 | 198 | 102 |
| E1A | 490–1000, 1085–1305 | 243 | 217 | 230 | 232 | 289 |
| E1B 19K | 1474–2016 | 180 | 182 | 169 | 169 | 175 |
| E1B 55K | 1836–3170 | 444 | 459 | 444 | 444 | 495 |
| IX | 3237–3548 | 103 | 106 | 103 | 103 | 140 |
| IVa2 | 3545–4881, 5160–5172 c | 449 | 442 | 446 | 446 | 449 |
| pol | 4654–8061, 12628–12636 c | 1,138 | 1,142 | 1,149 | 1,150 | 1,198 |
| pTP | 7917–7740, 12628–12636 c | 610 | 609 | 608 | 610 | 671 |
| 52K | 9777–10970 | 397 | 438 | 389 | 388 | 415 |
| pIIIa | 10843–12591 | 582 | 574 | 563 | 567 | 585 |
| III | 12660–14093 | 477 | 525 | 477 | 477 | 571 |
| pVII | 14125–14508 | 127 | 134 | 170 | 172 | 198 |
| V | 14582–15895 | 437 | 433 | 421 | 428 | 369 |
| pX | 15852–16133 | 93 | 69 | 68 | 69 | 80 |
| pVI | 16186–16932 | 248 | 273 | 238 | 249 | 250 |
| Hexon | 17000–19723 | 907 | 908 | 905 | 905 | 968 |
| Protease | 19735–20355 | 206 | 206 | 206 | 206 | 204 |
| DBP | 20399–21783 c | 461 | 472 | 454 | 454 | 529 |
| 100K | 21796–23895 | 699 | 682 | 689 | 689 | 805 |
| 33K | 23732–23860, 24099–24401 | 143 | 164 | 149 | 149 | 228 |
| 22K | 23732–24262 | 176 | 155 | 128 | 128 | 195 |
| pVIII | 24405–25094 | 229 | 222 | 224 | 224 | 227 |
| E3 12.5K | 25081–25431 | 116 | 118f | 117 | 119 | 107 |
| E3 ORF1 | 25454–26626 | 390 | 382 | 216 | 364 | |
| U exon | 26658–26821 c | 55 | 67 | 55 | 55 | 54 |
| Fiber | 26820–28493 | 557 | 555 | 543 | 542 | 582 |
| E4 ORF6/7 | 28508–28725, 29512–29533 c | 79 | 96 | 86 | 86 | 150 |
| E4 34K | 28748–29533 c | 261 | 260 | 265 | 259 | 294 |
| E4 ORFD | 29534–29986 c | 150 | 130 | 124 | 124 | |
| E4 ORFC | 29862–30275 | 137 | 124 | 132 | 123 | |
| E4 ORFB | 30302–30661 | 119 | 119 | 128 | 134 | |
| E4 ORFA | 30746–31141 | 131 | 187 | 153 | 131 | |
The letter “c” indicates the product is coded on the complementary strand.
The sizes of the ITR are given in base pairs (bp). All other sizes are given in numbers of amino acids (aa).
Proposal is waiting for final approval by ICTV.
The genome ends have not been determined.
ND, not determined.
Not determined in the original submission.
Several features of the BtAdV-2 genome (Fig. 1) are worthy of comment. The BtAdV-2 genome is actually the first full genome of a bat AdV as the ITRs of BtAdV-3 were not determined. The first 40 bp (in the ITR) of BtAdV-2 are identical to those of CAdV-1and CAdV-2.
Fig 1.
Genomic organization of BtAdV-2. The genome is represented by a black horizontal line marked at 2,000-bp intervals. ORFs assumed to encode proteins are shown as arrows. Coding regions in the first exons of spliced genes are represented by rectangles.
Each of the early regions E1, E3, and E4 contains at least one conserved genus-specific gene or ORF.
All three E1 genes (E1A, E1B 19K, and E1B 55K) are present in BtAdV-2, and their ORFs have typical lengths (Table 2).
In the middle part of the genome, 18 genes are conserved among all mastadenoviruses, including BtAdV-2 (4). The spliced nature of some of these genes is also evident. Each of the IVa2, pTP, DNA polymerase, and 33K genes consists of two exons, as in other mastadenoviruses.
The E3 region of BtAdV-2, BtAdV-3, and the CAdVs contains two genes. The first is the 12.5K gene, which is present in the majority of mastadenoviruses. Although this gene was not mentioned in the original description of the BtAdV-3 genome, it is clearly present (11). The second gene is E3 ORF1 and is present in BtAdV-3 and the CAdVs but not in other AdVs studied to date.
Also, the U exon, which is present in almost all AdVs (8, 14), is present in BtAdV-2 between E3 ORF1 and the fiber gene. Sequence conservation in this gene can be detected within but not among AdV genera, and downstream exons have been detected thus far only in members of the mastadenovirus species Human adenovirus C.
The protein playing the most crucial role for virus attachment is the antenna-like projection called “fiber.” All nonprimate mastadenoviruses have a single fiber gene, and BtAdV-2 and BtAdV-3 are no exceptions.
The E4 region is the second most variable region in mastadenovirus genomes (after the E3 region) both in its length and in its genetic contents (14). Adjacent to the fiber gene is the spliced ORF6/7 gene, which occurs in many mastadenoviruses. The adjacent 34K gene is the most conserved gene in the E4 region and consists of the first exon of ORF6/7 extended into the intron. In some mastadenoviruses (e.g., bovine AdV-3 and porcine AdV-5) and seemingly in all atadenoviruses, the 34K gene is duplicated (6, 14), whereas BtAdV-2, BtAdV-3, and CAdVs have only a single instance. To the right of the 34K gene, four novel putative genes (ORFA to -D) have been described in CAdVs and BtAdV-3, although the functions of their predicted protein products are not known (11). Homologous ORFs are present in BtAdV-2.
Similarities between bat and canine AdVs are apparent not only in their genome organization (including unique arrangements of the E3 and E4 genes), but also in their phylogenetic relationship.
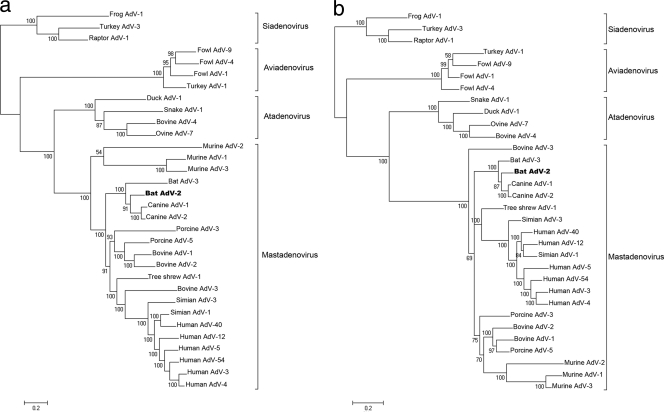
Multiple alignments of predicted amino acid sequences were prepared with the MultAlin 5.4.1 program, and phylogenetic tree reconstructions were performed on six large viral proteins (DNA-dependent RNA polymerase, terminal protein precursor, pIIIa, penton base, hexon, and 100K) by three different methods (distance matrix, maximum likelihood, and Bayesian). In each Bayesian (Fig. 2), maximum likelihood, and distance matrix analysis tree, BtAdV-2, BtAdV-3, and the CAdVs grouped monophyletically, thus strongly supporting the origin of these viruses from a common ancestor.
Fig 2.
Phylogenetic (Bayesian) analysis of BtAdV-2 using amino acid sequences of a nonstructural protein and a structural protein of the four AdV genera containing multiple species. (a) DNA-dependent DNA polymerase; (b) hexon. Posterior probability values are depicted. BtAdV-2 is shown in boldface. The scale bar shows the evolutionary distance of 0.2 amino acid substitution per position. The calculations were unrooted, but for visualization, members of the genus Siadenovirus were used as the outgroup. Accession numbers: BtAdV-2, JN252129; BtAdV-3, GU226970; bovine AdV-1 (BAdV-1), NC_006324; BAdV-2, AC_000001; BAdV-3, AC_000002; BAdV-4, AF036092; CAdV-1, AC_000003; CAdV-2, AC_000020; duck AdV-1 (DAdV-1), AC_000004; fowl AdV-1 (FAdV-1), AC_000014; FAdV-4, AJ431719; FAdV-9, AC_000013; frog AdV-1 (FrAdV-1), AF224336; human AdV-3 (HAdV-3), DQ086466; HAdV-4, AY487947; HAdV-5, AC_000008; HAdV-12, X73487; HAdV-40, L19443; HAdV-54, AB333801; murine AdV-1 (MAdV-1), NC_000942; MAdV-2, HM049560; MAdV-3, EU835513; ovine AdV-7 (OAdV-7), U40839; porcine AdV-3 (PAdV-3), AF083132; PAdV-5, AF289262; raptor AdV-1 (RAdV-1), EU715130; simian AdV-1 (SAdV-1), AY771780; SAdV-3, AY598782; snake AdV-1 (SnAdV-1), DQ106414; tree shrew AdV-1 (TSAdV-1), NC_004453; turkey AdV-1 (TAdV-1), GU936707; and TAdV-3, AC000016.
The overall genetic distance between BtAdV-2 and BtAdV-3 (and also their distance to the cluster of CAdVs) exceeds 5%, which is a prerequisite for assigning AdVs into separate species. As the first bat AdV for which extensive sequence data were derived, BtAdV-3 has recently been proposed as the founding member of a new species called Bat adenovirus A. We now propose BtAdV-2 as the founding member of a new species called Bat adenovirus B. Official approval of these two species is currently awaiting voting by the International Committee on Taxonomy of Viruses (ICTV).
In summary, the results of histopathological and molecular biological investigations indicate that BtAdV-2 may be associated in bats with either an enteric course of infection or a mild to inapparent infection of other organs and strongly suggest that BtAdV-2, BtAdV-3, CAdV-1, and CAdV-2 have descended from a common ancestor. As there are several other vespertilionid AdVs similar to BtAdV-2 and BtAdV-3, the most obvious hypothesis is that the CAdVs originated from a bat AdV by host switching at some point in the past. Incomplete adaptation to the new host is consistent with the high pathogenicity of CAdV-1 and the ease with which it can cross the host species barrier between different carnivore hosts.
Nucleotide sequence accession number.
The GenBank accession number of the sequence reported in this paper is JN252129.
ACKNOWLEDGMENTS
We are grateful to Jule Tesch, Michael Sonntag, Patrycja Machnowska, René Lesnik, Annika Brinkmann, and Arnt Ebinger for outstanding technical assistance, Ursula Erikli for copyediting, and one of the reviewers for extensive help with the final wording.
This study was partially financed by grants provided by the Konrad Adenauer Foundation (fellowship to C.K.), by the Adolf and Hildegard Isler-Stiftung, Klara Samariter-Stiftung, and FAZIT-Stiftung (K.M.), and by the Hungarian Scientific Research Fund (OTKA K72484).
Footnotes
Published ahead of print 30 November 2011
REFERENCES
- 1. Buonavoglia C, Martella V. 2007. Canine respiratory viruses. Vet. Res. 38:355–373. [DOI] [PubMed] [Google Scholar]
- 2. Chen EC, et al. 2011. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony. PLoS Pathog. 7:e1002155 doi: 10.1371/journal.ppat.1002155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chevreux B, et al. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davison AJ, Benkő M, Harrach B. 2003. Genetic content and evolution of adenoviruses. J. Gen. Virol. 84:2895–2908 [DOI] [PubMed] [Google Scholar]
- 5. Drexler JF, et al. 2011. Amplification of emerging viruses in a bat colony. Emerg. Infect. Dis. 17:449–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farkas SL, Harrach B, Benkő M. 2008. Completion of the genome analysis of snake adenovirus type 1, a representative of the reptilian lineage within the novel genus Atadenovirus. Virus Res. 132:132–139 [DOI] [PubMed] [Google Scholar]
- 7. Jánoska M, et al. 2011. Novel adenoviruses and herpesviruses detected in bats. Vet. J. 189:118–121 [DOI] [PubMed] [Google Scholar]
- 8. Klempa B, et al. 2009. A novel cardiotropic murine adenovirus representing a distinct species of mastadenoviruses. J. Virol. 83:5749–5759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kuzmin IV, et al. 2011. Bats emerging infectious diseases, and the rabies paradigm revisited. Emerg. Health Threats 4:7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li L, et al. 2010. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 84:6955–6965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y, et al. 2010. Host range, prevalence, and genetic diversity of adenoviruses in bats. J. Virol. 84:3889–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maeda K, et al. 2008. Isolation of novel adenovirus from fruit bat (Pteropus dasymallus yayeyamae). Emerg. Infect. Dis. 14:347–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sonntag M, Mühldorfer K, Speck S, Wibbelt G, Kurth A. 2009. New adenovirus in bats, Germany. Emerg. Infect. Dis. 15:2052–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ursu K, Harrach B, Matiz K, Benkő M. 2004. DNA sequencing and analysis of the right-hand part of the genome of the unique bovine adenovirus type 10. J. Gen. Virol. 85:593–601. [DOI] [PubMed] [Google Scholar]
- 15. Vidovszky MZ, Boldogh S. Detection of adenoviruses in the Northern Hungarian bat fauna. Magy. Allatorv. 133:747–753 (In Hungarian.) [Google Scholar]