Abstract
Hepatitis C virus (HCV) reorganizes intracellular membranes to establish sites of replication. How viral and cellular proteins target, bind, and rearrange specific membranes into the replication factory remains a mystery. We used a lentivirus-based RNA interference (RNAi) screening approach to identify the potential cellular factors that are involved in HCV replication. A protein with membrane-deforming activity, proline-serine-threonine phosphatase-interacting protein 2 (PSTPIP2), was identified as a potential factor. Knockdown of PSTPIP2 in HCV subgenomic replicon-harboring and HCV-infected cells was associated with the reduction of HCV protein and RNA expression. PSTPIP2 was localized predominantly in detergent-resistant membranes (DRMs), which contain the RNA replication complex. PSTPIP2 knockdown caused a significant reduction of the formation of HCV- and NS4B-induced membranous webs. A PSTPIP2 mutant defective in inducing membrane curvature failed to support HCV replication, confirming that the membrane-deforming ability of PSTPIP2 is essential for HCV replication. Taking these results together, we suggest that PSTPIP2 facilitates membrane alterations and is a key player in the formation of the membranous web, which is the site of the HCV replication complex.
INTRODUCTION
Hepatitis C virus (HCV), like other RNA viruses, can reorganize cellular membranes to form double- or multimembrane vesicles, including autophagosomes (28) and membranous webs (6). Viral nonstructural proteins (NS3–NS5B), which build up RNA replication complexes (9, 22, 26), and viral RNA are both associated with membranous webs (6, 9). Membranous webs are accumulations of heterogeneous vesicles derived mainly from the endoplasmic reticulum (ER) membrane (6, 22). These membrane structures are induced by viral proteins and presumably protect the HCV replication complex (RC) from the attack of host nucleases and proteases (20, 22). Among all HCV viral proteins, NS4B, which is modified by lipids and has polymerization activity (34), is required for membranous web formation (1, 6, 17). However, what cellular factors coordinate with NS4B to induce the formation of membranous webs is still unknown.
The Pombe Cdc15 homology (PCH) family proteins, such as CIP4 (14) and FCHo (12), are a group of proteins which regulate cytoskeletal and membrane dynamics. They can deform membranes into membrane curvatures during the initiation stage of vesicle formation (27). The membrane-deforming activity is mainly attributed to the intrinsic banana-shaped F-BAR-domain homodimer, which binds to the membrane with its concave surface (8, 24). Recent studies also revealed that proteins of the PCH family can interact with lipids, in particular, phosphatidylinositol (PI) (30); for example, FBP17, CIP4, Toca-1, and PSTPIP2 can interact with phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] (31). FBP17 also has binding affinity to phosphatidylinositol 4-phosphate (PI4P) and phosphatidylinositol 3,4,5-triphosphate [PI(3,4,5)P3] (31), and CIP4 can interact with PI3P (14).
PSTPIP2 is a 37-kDa PCH protein that is also known as macrophage actin-associated and tyrosine-phosphorylated protein (MAYP) (4, 33) and contains an F-BAR domain. PSTPIP2 is expressed in macrophages and is an actin-bundling protein that regulates filopodium formation and macrophage motility (33). PSTPIP2 is expressed in mouse liver cells (5); however, the status of its expression and the functional role of PSTPIP2 in human liver cells are still not clear.
In this study, we used lentivirus-based RNA interference (RNAi) screening to identify PSTPIP2 as a cellular factor involved in HCV replication. We showed that knockdown of PSTPIP2 reduced both the formation of HCV-induced membranous webs and HCV replication, whereas the overexpression of PSTPIP2 enhanced HCV replication. The membrane-deforming ability of PSTPIP2 is important for the enhancement of HCV replication. These studies thus identified a novel protein, PSTPIP2, as a player in HCV-induced membrane rearrangement, which leads to the formation of the HCV replication complex.
MATERIALS AND METHODS
Cells, media, and reagents.
Huh-7, Huh-7.5 (2), and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, nonessential amino acids, 100 units/ml of penicillin, and 100 μg/ml of streptomycin at 37°C in a 5% CO2 incubator. Two HCV subgenomic replicons, HCV-EV71I-Luc and HCVrep-HA, were derived from the original HCV replicon 1bneo/delS (11). HCV-EV71I-Luc was generated by modification of 1bneo/delS by insertion of an EV71-internal ribosome entry site (IRES)-driven luciferase gene between the neo gene and encephalomyocarditis virus (EMCV)-IRES (Fig. 1A); HCVrep-HA was generated by insertion of a hemagglutinin (HA) tag in the C-terminal region of NS5A as previously described (21).
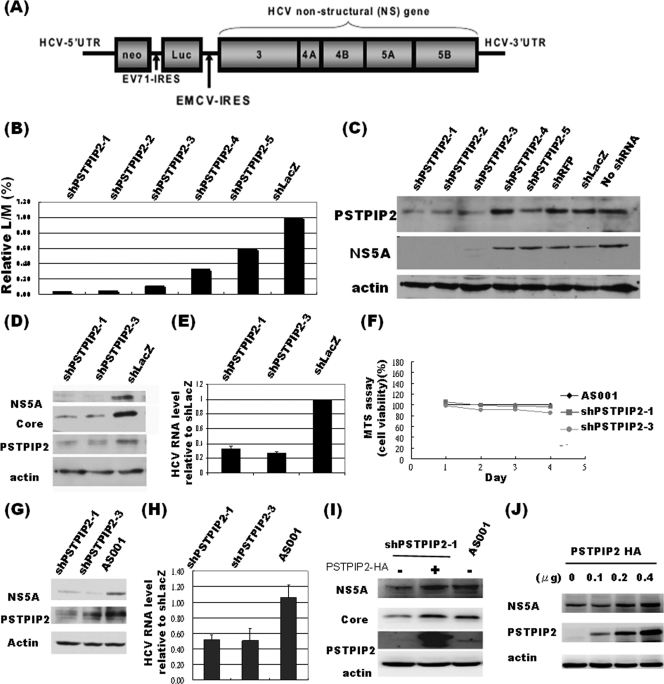
Fig 1.
The expression of PSTPIP2 correlates with HCV replication in replicon and HCV-infected cells. (A) Schematic representation of the configuration of the HCV-EV71I-Luc replicon construct. (B) Relative luciferase activity (L)/cell viability (M) ratio of PSTPIP2 knocked-down cells in the primary screen. (C) Western blot analysis of HCV NS5A and PSTPIP2 proteins in HCV replicon cells transduced with different shPSTPIP2 clones (clone 1 to clone 5), control shRFP, or control shLacZ. Cell lysates were collected on the day 4 postransduction after puromycin selection. The proteins were separated by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and followed by standard Western blot analysis. (D and E) shPSTPIP2-1 and shPSTPIP2-3 were transduced into Huh-7.5 cells and selected with puromycin for 2 days. Cells were then subcultured and infected with JC1 virus (MOI = 0.6). Three days later, cells were collected and analyzed by immunoblotting (D) and quantitative RT-PCR (E) (normalized to GAPDH RNA; data represent means ± standard deviations [SD]). (F) MTS assays of lentivirus-infected and puromycin-selected Huh-7.5 cells were performed on each day after reseeding (days 1 to 4) (data represent means ± SD; n = 3). (G and H) shPSTPIP2-1 and shPSTPIP2-3 were transduced into HCV replicon cells and selected with puromycin for 2 days. The cells were then collected and analyzed by immunoblotting (G) and quantitative RT-PCR (H) (normalized to GAPDH RNA) (data represent means ± SD). (I) The lentiviruses carrying shPSTPIP2-1 or mock AS001 were transduced into Huh-7.5 cells to establish stable cell lines and then infected with JC1 virus. The next day, a plasmid expressing PSTPIP2-HA or empty vector was transfected into the cells described above. Three days after transfection, cell lysates were collected for immunoblot analysis using the various antibodies. (J) Different amounts of wild-type PSTPIP2 were transfected into JC1-infected cells. Three days later, cell lysates were analyzed by Western blotting.
Plasmids and viruses.
All the plasmids required for lentivirus production were provided by the National RNAi Core Facility, Academia Sinica, Taipei, Taiwan. Five pLKO.1-shRNA vectors used for knockdown of PSTPIP2 were as follows: TRCN10000003019 (shPSTPIP2-1), TRCN0000003020 (shPSTPIP2-2), TRCN0000003021 (shPSTPIP2-3), TRCN0000003022 (shPSTPIP2-4), and TRCN0000003023 (shPSTPIP2-5). The pLKO.1-shLacZ control plasmid was TRCN0000072237 (shLacZ). pLKO_AS001, which served as a lentivirus-based RNAi control, was derived from pLKO.1-shLacZ by replacing the shLacZ sequence with a non-short-hairpin-RNA (non-shRNA) sequence, TGG(T)8GCTAGCTTGC. Trans-IT transfection reagent (Mirus Bio.) was used for lentiviral production in 293T cells with a packaging construct (pCMV-_R8.91), an envelope construct (pMD.G), and different shRNA or rescue constructs, according to the protocol specified on the RNAi Core website (http://rnai.genmed.sinica.edu.tw/file/protocol/2_LentivirusProductionV4.pdf). The viral titer was determined in Huh-7 cells by using a cell viability assay (relative infection unit [RIU] method) according to the RNAi Core's instructions (http://rnai.genmed.sinica.edu.tw/file/protocol/4_1_EstimationLentivirusTiterRIUV1.pdf). To generate the plasmid for expression of PSTPIP2, the full-length cDNA of PSTPIP2 was amplified from Huh-7 total cDNA and inserted into XbaI and XmaI sites of pUI vector. The pJC1 plasmid, which encodes the genotype 1a chimera genome of HCV J6CF/JFH1, was constructed as previously described (25). The JC1 viruses were produced and titrated in Huh-7.5 cells based on a method previously described (19). pUI-PSTPIP2-HA and pUI-PSTPIP2-FLAG were constructed by insertion of the PSTPIP2 cDNA sequence into XbaI and XmaI sites of pUI vector, which was derived from pCI (Promega) by replacement of the cytomegalovirus (CMV) promoter with a ubiquitin promoter and a reverse primer which carries an in-frame HA tag or FLAG tag. pCI-HA-NS3/4A, pCI-NS5A-HA, and pCI-HA-GST have been described previously (18, 19). pUI-NS4B-HA was constructed by insertion of the NS4B sequence into the EcoRI and XbaI sites of pUI vector and a reverse primer which carries an in-frame HA tag. pCAG2-HA-NS5B was constructed by insertion of the NS5B sequence into NheI and NotI sites of pCAG2 vector.
Generation of HCV-EV71I-Luc and HCVrep-HA subgenomic replicons.
In vitro-transcribed replicon RNAs were electroporated into Huh-7 cells (975 μF, 220 V) and then selected with G418 solution at concentration of approximately 0.5 to 1 mg/ml for 3 weeks. For HCV-EV71I-Luc replicons, 50 single colonies were picked, expanded, and assayed for luciferase activity. The expanded colony with the highest luciferase activity and HCV protein levels was chosen and maintained in G418 (0.5 mg/ml) for subsequent experiments. For the HCVrep-HA replicon, the G418-resistant cells were pooled and maintained in 0.8 mg/ml of G418.
Screening for cellular factors required for HCV RNA replication by lentiviral shRNAs.
HCV-EV71I-luc replicon cells were plated (1 × 104 cells per well) in 96-well plates 24 h prior to infection. Cells were then infected with KP subset lentiviruses, which target 1,210 human kinase- and phosphatase-related genes, at a multiplicity of infection (MOI) of 3 in the presence of Polybrene (hexadimethrine bromide; Sigma H9268) at a final concentration of 8 μg/ml. Cells were infected with the virus and incubated for 24 h prior to replacement of the media with selective media containing puromycin (3 μg/ml). Luciferase activities (L) and cell viabilities (M) were determined at postransduction day 5. Luciferase activities were determined by using a Bright-Glo luciferase assay system (Promega). Cell viabilities were measured by using a 3-(4-5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt (MTS) assay as described below. The knockdown effects on HCV replication were evaluated by L/M assays. The relative L/M values were normalized to the L/M ratio determined for noninfected control cells without puromycin selection.
By measuring the luciferase activity (L) and cell viability (M), the effects of knockdown on HCV replication in transduced cells were evaluated. Two independent primary screens were performed to ascertain that candidate genes were reproducible and not the results of screening artifacts. Candidates in which each gene hit was shown by at least 2 unique shRNAs were selected, resulting in a more than 50% enhancing or inhibitory effect on HCV replication. Several genes known to be involved in the HCV life cycle, including TYK2, PAK1, and RHO, were among the 40 candidate genes identified. The candidates were further confirmed by quantitative reverse transcription-PCR (qRT-PCR) to quantify the RNA of target genes and the HCV replicon. The candidates were ruled out if their gene knockdown efficiency did not match the change in HCV replication. Finally, four genes were selected for further investigation. Proline-serine-threonine phosphatase-interacting protein 2 (PSTPIP2) was one of these candidates.
Antibodies.
The mouse monoclonal antibodies (MAb) specific for HCV-1b strain NS5A were purchased from Biodesign. The MAb for JC1 NS5A detection was obtained from Austral Biologicals. Chicken polyclonal anti-PSTPIP2 antibody was purchased from Abcam. Mouse MAb against actin was obtained from Millipore. Rat MAb against HA epitopes was purchased from Roche Diagnostics. Rabbit polyclonal anti-HA antibody for immunofluorescence staining was obtained from Santa Cruz. Alexa Fluor 488-conjugated anti-mouse, Alexa Fluor 488-conjugated anti-rabbit, Alexa Fluor 568-conjugated anti-rat, and Alexa Fluor 568-conjugated anti-rabbit secondary Abs were purchased from Invitrogen-Molecular Probes.
Quantitative detection of HCV and PSTPIP2 RNA by qRT-PCR.
For intracellular RNA detection, total RNA was extracted using TRIzol (Invitrogen). The concentration of the extracted RNA was determined by spectrophotometry. cDNA from 1 μg of total RNA was synthesized in a 20-μl total volume by using a SuperScript III first-strand synthesis system (Invitrogen). The primers for reverse transcription were oligo(dT)20 and an HCV-specific RT primer (5′-CACTCGCAAGCACCCTATCA-3′). For real-time PCR, we followed the standard TaqMan method using the Universal ProbeLibrary system and LightCycler (Roche Diagnostics). The primers were as follows: for HCV, the sense primer was 5′-CATGGCGTTAGTATGAGTGTCG-3′ and the antisense primer was 5′-GGTTCCGCAGACCACTATG-3′ (with Universal Probe 75; Roche). As an internal control, we amplified human GAPDH (glyceraldehyde 3-phosphate dehydrogenase) with the following primer set: sense, 5′-AGCCACATCGCTCAGACAC-3′; antisense, 5′-GCCCAATACGACCAAATCC-3′ (with Universal Probe 60; Roche).
Membrane flotation assay.
Cells were first lysed in 500 μl of phosphate-buffered saline (PBS) buffer and passed through a homogenizer 20 times. Total lysate was then centrifuged at 5,000 × g for 60 min in a microcentrifuge at 4°C. Pellet was resuspended in TNE buffer (25 mM Tris-HCl [pH 7.6], 15 mM NaCl, 5 mM EDTA) with or without 1% Triton X-100 and incubated on ice for 15 min. Then, the sample was centrifuged at 5,000 × g for 30 min at 4°C. The pellet was resuspended in 40% OptiPrep solution in TNE buffer and loaded onto ultracentrifuge tubes and then overlaid with 3 ml of 30% OptiPrep and 0.5 ml of 5% OptiPrep. The gradient was centrifuged at 36,000 rpm in a Beckman SW-60 rotor for 18 h at 4°C. Fractions (0.5-ml) were taken from the top of the gradient. The fractions were mixed with sample buffer and analyzed on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel.
Coimmunoprecipitations.
Various plasmids expressing HCV viral proteins were transfected into HEK293T cells by the use of Lipofectamine 2000 transfection reagent (Invitrogen). Cells were collected 48 h after transfection. The preparation of total lysates and immunoprecipitation for FLAG-tagged proteins were performed according to instructions provided with anti-FLAG M2 affinity gel (Sigma-Aldrich). The immunoprecipitated proteins were run on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel, and immunoblot analysis was performed using the antibodies described above.
Immunofluorescence staining.
Huh-7.5 cells infected with HCV and transfected with pUI-PSTPIP2-HA were cultured on glass chamber slides (Lab-Tek II). Cells were washed twice with PBS, fixed with 4% paraformaldehyde–PBS for 20 min, and then permeabilized with 0.2% Triton X-100–PBS. Samples were blocked with 0.25% bovine serum albumin (BSA)–PBS for 1 h. Primary Abs were diluted in blocking buffer and incubated with cells for 1 h. After three washes in PBS, Alexa Fluor 488- and 568-conjugated secondary Abs were diluted together with DAPI (4′,6′diamidino-2-phenylindole; Sigma-Aldrich) in blocking buffer and added to cells for 1 h of incubation. After staining, slides were washed three times with PBS and mounted using a ProLong Antifade kit (Invitrogen-Molecular Probes). All the procedures were carried out at room temperature. Photographs were taken with a confocal microscope (Zeiss LSM 510 and 780 confocal laser-scanning microscope). Image analysis was performed using the standard system operating software provided with the microscope.
Electron microscopy.
All procedures were done at 4°C or on ice. Cells were cultured on Aclar embedding film (Electron Microscopy Sciences) and then rinsed with 0.1 M cacodylate buffer (0.1 M sodium cacodylate, 3.4% sucrose, pH 7.4), fixed with 2.5% glutaraldehyde–0.1 M cacodylate, washed with 0.1 M cacodylate buffer, postfixed in 1% osmium tetroxide, prestained with 1% uranyl acetate, dehydrated in a graded series of ethanol concentrations, and embedded in Spurr's resin. Then, 100-nm-thick sections were cut, stained with 5% uranyl acetate–50% methanol, and viewed on a Tecnai G2 Spirit Twin transmission electron microscope (FEI Company) with a Gatan 794 MultiScan charge-coupled-device (CCD) camera. For immunogold labeling, cells were pelleted, fixed in 4% paraformaldehyde–0.2% glutaraldehyde–0.1 M cacodylate for 30 min at 4°C, washed with 0.1 M cacodylate buffer, dehydrated in ethanol, and infiltrated with LR Gold. Cell pellets were transferred to capsules containing fresh LR Gold and 0.1% benzyl and polymerized at −20°C under UV light. Thin sections were cut, collected, and blocked with 2% BSA–PBS for 1 h. Ultrathin sections (100 nm) were incubated with primary Abs (mouse anti-NS5A and rabbit anti-HA) for 2 h, washed in PBS, and incubated for an additional 1 h in colloidal gold goat anti-mouse IgG (12 nm) and colloidal gold goat anti-rabbit IgG (18 nm). The grids were sequentially washed with PBS and distilled water, stained with uranyl acetate, and viewed on a Tecnai G2 Spirit Twin transmission electron microscope (EM) (FEI Company).
Cell sorting.
Cells infected with shPSTPIP2 or control AS001 lentivirus were transfected using pUI-NS4B-eGFP and Trans-IT reagent (Mirus Bio.). Two days later, cells were trypsinized and resuspended in culture medium. Then, cell sorting was performed on a FACSVantage DiVa flow cytometer.
MTS assay.
A CellTiter96 AQueousOne solution cell proliferation assay kit (Promega) was used to evaluate cell viability. Cells (1 × 104) were seeded onto a 96-well microtiter plate. Cell media were replaced with phenol red-free DMEM (Invitrogen) containing 10% CellTiter96 AQueousOne solution reagent. The plates were incubated for an additional 30 min at 37°C. Light absorbance was measured at 490 nm with a 96-well plate reader. Each experiment was performed in three replicated wells.
Membranous web quantification.
We performed three independent experiments for counting the membranous webs. Monolayers of cells were used for EM analysis by observing the cells at different section levels every 100 nm of depth from the cell attachment surface to the top of the cells. Intact nuclei and membranous webs were seen at approximately 500 to 800 nm from the cell attachment surface. We chose one depth between the approximately 500- to 800-nm sections for counting. Any cell containing at least one membranous web was scored as positive.
RESULTS
PSTPIP2 is involved in the replication of the HCV subgenomic replicon.
To identify cellular factors involved in HCV RNA replication, we performed an RNAi high-throughput screening. A tricistronic HCV replicon, HCV-EV71I-Luc, harboring a subgenomic HCV (genotype 1b) and an EV71-IRES-driven luciferase gene, was introduced into Huh-7 cells to establish a stable cell line (Fig. 1A). A subset of vesicular stomatitis virus-G (VSV-G)-pseudotyped lentiviruses which express distinct shRNA-targeting human kinase- and phosphatase-related genes was then employed for screening (3). PSTPIP2 was hit by three unique shRNAs showing an approximately 80% reduction in the L/M ratio (Fig. 1B).
A bicistronic HCV replicon, HCVrep-HA (21), was used to confirm the result from the shRNA screening. We attempted to use five different shRNA clones to knock down PSTPIP2 expression in HCVrep-HA cells. The immunoblotting result showed that, compared to the shLacZ and shRFP controls, three different clones (clone 1 to clone 3) of shPSTPIP2, one of which (clone 1 [shPSTPIP2-1]) targets the 3′ untranscribed region (UTR) of the endogenous PSTPIP2 transcript, had good specific gene knockdown efficiency and correspondingly decreased HCV NS5A protein and viral RNA expression (Fig. 1C, G, and H). This result confirms that knockdown of PSTPIP2 inhibits HCV replication in replicon cells.
The expression level of PSTPIP2 correlates with the degree of replication of infectious HCV.
Since the HCV replicon RNA used in the previous assay contained several heterologous sequences, we further determined the importance of PSTPIP2 by the use of an HCV infection system. Huh-7.5 cells were first transduced with shPSTPIP2 (clone 1 or clone 3) or control shLacZ lentiviruses. After puromycin selection, cells were reseeded and infected with the HCV JC1 strain. On day 3 postinfection, cells were collected for detection of PSTPIP2 and two representative viral proteins, core and NS5A (Fig. 1D), by immunoblotting and for measurement of intracellular HCV RNA by quantitative RT-PCR (Fig. 1E). The results revealed that the expression of shPSTPIP2 significantly inhibited HCV replication. MTS assays examining shPSTPIP2-1-, shPSTPIP2-3-, and mock-AS001 lentivirus-treated cells (Fig. 1F) showed that the cell viability was not affected. Furthermore, overexpression of PSTPIP2, which is not susceptible to knockdown by shPSTPIP2 in the shPSTPIP2-1-treated cells, reversed the decrease of HCV replication caused by shPSTPIP2, indicating that the effect of shPSTPIP2 was specifically due to the knockdown of PSTPIP2 (Fig. 1I). Finally, we expressed different amounts of PSTPIP2 in JC1-infected cells and found that the amounts of NS5A increased with increasing amounts of PSTPIP2 in a dose-dependent manner (Fig. 1J). This result confirms the positive correlation between PSTPIP2 expression and HCV replication.
These results together indicate that PSTPIP2 is involved in HCV replication not only in the replicon cells but also in the infectious viral system. Since the HCV replicon and JC1 virus used in this study belong to genotypes 1b and 2a, respectively, these results also indicate that the involvement of PSTPIP2 in HCV replication is not limited to a certain viral genotype.
Cofractionation of endogenous PSTPIP2 with HCV replication complexes.
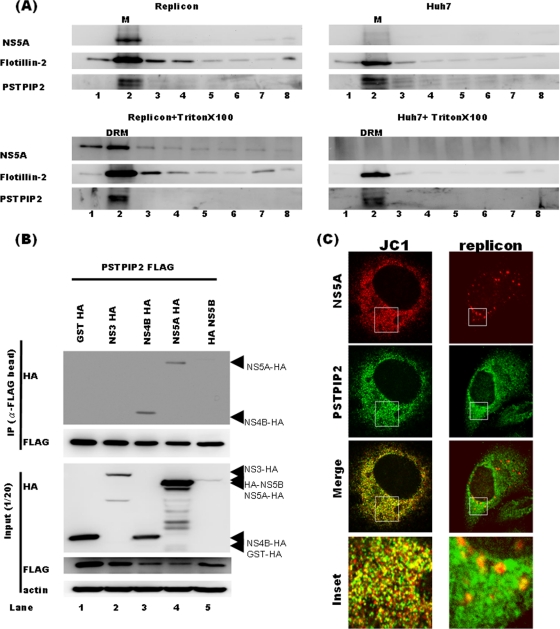
To elucidate how PSTPIP2 is involved in HCV replication, we examined the subcellular localization of the endogenous PSTPIP2 and HCV replication complexes (RCs) by the use of a biochemical fractionation method. HCV RCs are known to be associated with detergent-resistant membranes (DRMs) (26), which have the properties of lipid rafts. Membrane flotation analysis demonstrated that PSTPIP2, flotillin-2 (a DRM marker), and NS5A were present in the DRM fractions both in the presence and in the absence of the HCV replicon (Fig. 2A). The presence of the HCV replicon did not affect the distribution of PSTPIP2. These properties suggest that PSTPIP2 is an endogenous lipid raft-associated protein and is in a position to interact with the components of the HCV replication complexes.
Fig 2.
PSTPIP2 is in the DRM fraction and directly interacts with NS4B and NS5A in JC1-infected cells. (A) Detergent-resistant membranes were prepared from Huh-7 cells with and without HCV replicons by Triton X-100 treatment at 4°C as described in the text. Protein lysates from each fraction were subjected to 10% SDS-PAGE and probed for PSTPIP2, HCV NS5A, or flotillin-2. The membranes without the detergent treatment served as controls (upper panels). M, membrane; DRM, detergent-resistant membrane. (B) 293T cells were cotransfected with PSTPIP2-FLAG and HA-NS3-, NS4B-HA-, NS5A-HA-, HA-NS5B-, or GST-HA-expressing vector individually. At 48 h later, cell lysates were collected and immunoprecipitated with anti-FLAG agarose. The eluate was subjected to Western blotting with anti-HA and anti-FLAG antibodies. (C) Confocal images showing that PSTPIP2 partially colocalizes with NS5A in JC1-infected and replicon cells. The results of immunofluorescence labeling of PSTPIP2-HA and NS5A in JC1-infected Huh-7.5 cells and HCVrep-HA cells are shown. JC1-infected cells were transfected with pUI-PSTPIP2-HA. Two days later, cells were fixed and stained (left panels). HCVrep-HA replicon cells were fixed 2 days after seeding for immunofluorescence labeling of endogenous PSTPIP2 and NS5A (right panels). The “Inset” panels are zoomed images from the boxed areas in the top panels, showing regions of partial colocalization. Yellow areas indicate colocalization between PSTPIP2 and the viral proteins.
Direct interaction and partial colocalization between PSTPIP2 and viral proteins.
Since PSTPIP2 cofractionated with HCV RC in the DRM, we next studied whether PSTPIP2 interacts with any viral protein. For this purpose, we cotransfected 293T cells with PSTPIP2-FLAG and glutathione transferase-HA (GST-HA)-, NS3/4A-HA-, NS4B-HA-, NS5A-HA-, or NS5B-HA-expressing vectors individually and performed coimmunoprecipitation experiments using anti-FLAG M2 affinity gel. The HCV proteins were detected using anti-HA antibody. We used 293T rather than Huh-7 cells because of the higher transfection efficiency (>80%) in the former. We found that PSTPIP2 coprecipitated with NS4B and NS5A but not with NS3/4A, NS5B, or GST (Fig. 2B). We then investigated subcellular localization of PSTPIP2 and viral proteins in HCV-infected cells by the use of confocal microscopy. For this purpose, PSTPIP2-HA was expressed in JC1-infected cells. Immunofluorescence labeling showed that the overexpressed PSTPIP2 and NS5A were partially colocalized in the cytoplasm (Fig. 2C, left panels). We also studied the possible colocalization of the endogenous PSTPIP2 and NS5A in HCV replicon cells (Fig. 2C, right panels). The confocal microscopy showed that they were also partially colocalized. These results together indicate that PSTPIP2 is closely associated with the HCV replication complex. It should be noted that NS5A is involved in both HCV RNA replication and virion assembly, the latter of which does not take place in the HCV replicon cells. Therefore, the NS5A distribution is much more limited in the replicon cells. Nevertheless, the NS5A colocalized with PSTPIP2 in both cell systems.
PSTPIP2 has a potential to deform cellular membranes.
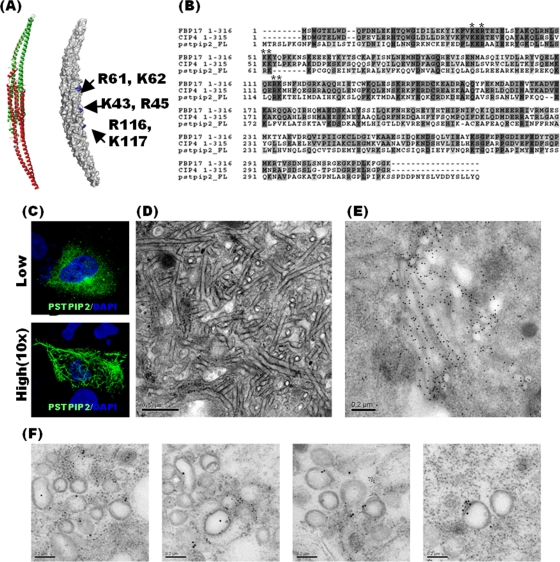
To elucidate how PSTPIP2 is involved in HCV replication, biochemical characteristics of PSTPIP2 were investigated. PSTPIP2 belongs to the PCH protein family, which has an F-BAR domain with the ability to deform cellular membranes in vivo and in vitro (31). Since the structure of PSTPIP2 is currently not known, we used the Protein Homology/analogY Recognition Engine (Phyre) program to perform structural modeling based on amino acid sequences. The model generated for the PSTPIP2 sequence based on the crystal structure of formin-binding protein 17 (FBP17) (27) has a very low E-value (1.5e-33), indicating that the model is reliable. The predicted structure of PSTPIP2 shows a crescent-shaped dimer with a positively charged concave surface (Fig. 3A). The positively charged surface of the domain in FBP17 is thought to bind to the negatively charged inner surface of the membrane (27, 31).
Fig 3.
PSTPIP2 has the potential to deform cellular membranes. (A) Structural modeling of the PSTPIP2 F-BAR domain generated by the Phyre program based on the crystal structure of FBP17 (27). The amino acids proposed to be responsible for lipid binding are shown in blue (right). One monomer of each F-BAR module is shown in green and the other in red (left). (B) Sequence alignment of PSTPIP2 and F-BAR domains of other PCH family proteins. Residues that are identical or similar among family members are shaded in dark gray and light gray, respectively. (C) Immunofluorescence staining of PSTPIP2-HA in Huh-7.5 cells transfected with PSTPIP2-expressing vector. The amount of pUI-PSTPIP2-HA used for transfection in the upper panel was 1/10 of that used in the lower panel. (D) EM of Huh-7.5 cells transfected with PSTPIP2-HA-expressing vector. (E) Gold-labeled anti-HA antibody was observed on the tubule in the cells transfected with pUI-PSTPIP2-HA. Gold grains, 18 nm. (F) Double immunogold staining shows NS5A (smaller gold grains [12 nm]) and PSTPIP2 (larger gold grains [18 nm]) on the membranous web. Huh-7.5 cells were infected by JC1 virus. On day 7 postinfection, cells were reseeded and transfected with pUI-PSTPIP2-HA. Two days later, cells were fixed for sample preparation. Scale bar, 200 nm.
PSTPIP2 has high similarity to other proteins of the PCH family (Fig. 3B). Most of the key residues (K43, R45, R61, K62, R116, and K117) that are essential for lipid binding (31) of the PCH proteins are consensus sequences and are located in the concave area of the modeled structure of PSTPIP2 (Fig. 3A and B). We found that when PSTPIP2 was expressed at a low level, it exhibited small-spot-like morphology (Fig. 3C, top panel), which is the typical distribution pattern of PSTPIP2 in normal cells (10); however, when it was expressed at a high level, its distribution showed tubular morphology similar to that exhibited by overexpression of other proteins of the PCH family (Fig. 3C, bottom panel). The PSTPIP2-induced tubular structure was found to be composed of tubule-shaped membranes under electron microscopy (Fig. 3D) and may represent the accumulation of membrane curvature caused by the imbalance of vesicle processing (15, 31). Since the ability of PSTPIP2 to form a membrane curvature is difficult to quantify, we used the tubular morphology, induced by the overexpressed PSTPIP2, to assay the membrane-deforming activity of PSTPIP2. Under immunoelectron microscopy, PSTPIP2 was found to concentrate on the surface of many tubules (Fig. 3E), suggesting that PSTPIP2 is associated with tubule formation or its maintenance. These data suggest that PSTPIP2 has the potential to deform cellular membranes (29).
The ability of PSTPIP2 to deform membranes is essential for HCV replication.
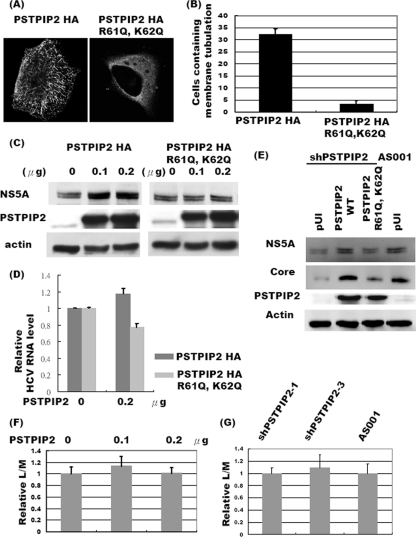
We further investigated whether the ability of PSTPIP2 to deform membranes is important for HCV replication. Since the positively charged residues of the F-BAR domain are essential for binding and deforming of lipids (16, 31), we mutated two basic amino acid residues (R61Q and K62Q) which are located at the concave side of the molecule and consensus sequence among members of the PCH family (Fig. 3B). The R61Q/K62Q mutant (Fig. 4A, right panel), when overexpressed, exhibited primarily homogeneous staining patterns in most of the cells, in contrast to the tubulation patterns induced by wild-type (WT) PSTPIP2 (32 of 50 WT cells versus 3 of 50 mutant cells had the tubulation pattern) (Fig. 4B). The expression level of wild-type PSTPIP2 was similar to that of the PSTPIP2 mutant as judged by immunoblotting (data not shown); thus, the difference in the degrees of membrane tubulation was not caused by a difference in protein expression levels. These results indicated that the PSTPIP2 mutant has reduced membrane-deforming activity. When the WT or mutant PSTPIP2 was transfected into JC1-infected cells, the amounts of NS5A increased with increased amounts of wild-type PSTPIP2 but not with increased amounts of the PSTPIP2 mutant (Fig. 4C). Correspondingly, the HCV RNA level increased with the expression of wild-type PSTPIP2. In contrast, the expression of the PSTPIP2 mutant caused a decrease in the HCV RNA level (Fig. 4D), suggesting that the PSTPIP2 mutant, which does not deform membranes, may serve as a dominant-negative inhibitor of HCV replication. We further found that in the rescue experiment, the expression of wild-type PSTPIP2, which is not susceptible to knockdown by shPSTPIP2, in the shPSTPIP2-1-treated cells reversed the decrease of HCV replication as judged by the amounts of NS5A and core proteins, but the expression of the PSTPIP2 mutant enhanced HCV replication to only a small extent (Fig. 4E). These results demonstrated that the membrane-deforming activity of PSTPIP2 is important for HCV replication. We also showed that the knockdown or the enhancement of PSTPIP2 did not significantly affect the efficiency of HCV entry by using luciferase-encoding HIV-1 particles pseudotyped with HCV envelope glycoproteins (HCVpp) (Fig. 4F and G) (7). Therefore, PSTPIP2, with its membrane-deforming activity, is specifically involved in HCV replication but not virus infection.
Fig 4.
The membrane-deforming ability of PSTPIP2 is essential for HCV replication. (A) Immunofluorescence staining of wild-type (left) or R61Q K62Q mutant (right) PSTPIP2-HA in Huh-7.5 cells. Gray shading indicates anti-HA (PSTPIP2). (B) The numbers of cells showing the tubulation pattern. Fifty cells transfected with the HA-tagged wild-type strain or the K61Q/K62Q PSTPIP2 mutant were examined for the appearance of a tubulation pattern. Three independent experiments were performed. Cells containing a tubule more than 2 μm in length were scored as positive. The results are presented as numbers of positive cells among 50 cells examined (data represent means ± SD). (C and D) The wild-type strain or the R61Q K62Q mutant of PSTPIP2 was transfected into JC1-infected cells. Three days later, cell lysates were analyzed by Western blotting and quantitative RT-PCR (normalized to GAPDH RNA) (data represent means ± SD). (E) The lentiviruses carrying shPSTPIP2-1 or mock AS001 were transduced into Huh-7.5 cells to establish stable cell lines and then infected with JC1 virus. The next day, a plasmid expressing PSTPIP2-HA or mutant PSTPIP2-HA was transfected into the cells described above. Three days after transfection, cell lysates were collected for immunoblot analysis using the various antibodies. (F) Huh-7.5 cells were transfected with different amounts of plasmid expressing PSTPIP2-HA. The next day, cells were infected by luciferase-encoding HIV-1 particles pseudotyped with HCV envelope glycoproteins (HCVpp) for 12 h. Three days later, cells were collected for luciferase and MTS assays. (G) shPSTPIP2-1 and shPSTPIP2-3 lentivirus-transduced Huh-7.5 cells were infected by HCVpp, and cells were collected for luciferase and MTS assays after 3 days.
PSTPIP2 is involved in HCV-induced membranous web formation.
On the basis of the membrane-deforming characteristics of PSTPIP2, we hypothesize that PSTPIP2 is involved in the membrane rearrangement associated with HCV replication. To investigate this hypothesis, we first investigated the relationship between PSTPIP2 and a viral protein (NS5A) by immunoelectron microscopy and found that NS5A and PSTPIP2-HA were often observed to be colocalized on the same or nearby membranous web structures in JC1-infected cells (Fig. 3F). This result is consistent with that of the immunofluorescence studies (Fig. 2C).
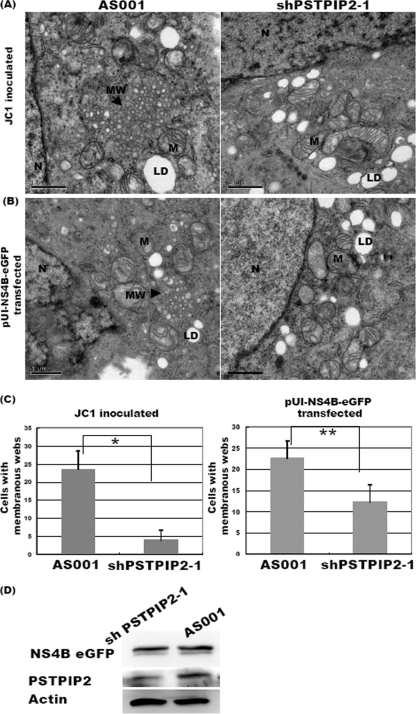
We then examined the effects of PSTPIP2 knockdown on endogenous membrane formation in uninfected cells. We transduced shPSTPIP2 or mock lentivirus into Huh-7.5 cells; however, there was no obvious difference in cellular morphology, including the number and shape of vesicles or other membrane structures, between these cells (data not shown). One possible explanation for the lack of effects of PSTPIP2 knockdown is that the formation of normal membranous substructures may require only a very small amount of PSTPIP2. We next investigated the effect of silencing PSTPIP2 expression on HCV-infected cells. Huh-7.5 cells were transduced with shPSTPIP2-1 or mock lentiviruses (AS001) and then infected with HCV JC1. In the HCV-infected cells transduced with the mock lentivirus, patches of vesicles of heterogeneous sizes and shapes, resembling membranous webs (6), were seen in the perinuclear region; in contrast, very few membranous webs were seen in the HCV-infected cells transduced with shPSTPIP2 (Fig. 5A). We performed three independent experiments and examined 50 cells per experiment for the presence of the membranous web structure; an average of three counts showed that 23 of 50 JC1-infected cells transduced with mock lentivirus had membranous webs but that only 4 of 50 shPSTPIP2-transfected cells had this structure (Fig. 5A and C). These results indicate that knockdown of PSTPIP2 reduced the HCV-induced membranous web formation.
Fig 5.
Silencing PSTPIP2 reduces membranous web formation. (A) Cells were transduced with shPSTPIP2-1 (right panels) or mock AS001 (left panels) lentivirus and infected with HCV for 3 days (MOI = 0.6). Three independent experiments were performed, and 50 cells per experiment were examined. Cells having membranous webs were scored as positive. (B) The shPSTPIP2- or control AS001 lentivirus-transduced cells were reseeded and transfected with pUI-NS4B-eGPF for 2 days. Cells expressing or not expressing NS4B were separated by FACS and processed for EM observation. Three independent experiments were performed, and 100 cells per experiment were examined (scale bar, 1 μm.) N, nucleus; M, mitochondria; LD, lipid droplet; MW, membranous web. (C) The numbers of cells containing membranous webs as determined from panels A and B. A single asterisk (*) represents P < 0.05; a double asterisk (**) represents P < 0.01. (D) Cells prepared in the experiment described for panel B were collected for Western blot analysis.
It has been reported that the presence of NS4B alone is sufficient for the induction of membranous web formation (6, 17). We therefore tested the effect of PSTPIP2 knockdown on the NS4B-induced formation of membranous webs. The shPSTPIP2-transduced cells were transfected with a plasmid expressing enhanced green fluorescent protein (eGFP)-tagged NS4B. The NS4B-expressing cells were sorted by fluorescence-activated cell sorter (FACS) analysis and examined for the presence of membranous webs. Three independent experiments were performed, and 100 cells per experiment were observed. We observed that 22 of 100 cells (Fig. 5B and C) transduced with mock lentivirus but only 12 of 100 cells transduced with shPSTPIP2 lentivirus contained membranous webs (Fig. 5B and C). The expression levels of NS4B, as determined by immunoblotting, in the two cell populations were comparable (Fig. 5D). The result showed that knockdown of PSTPIP2 reduced the NS4B-induced formation of membranous webs, similar to its effects on HCV-induced membranous webs. The finding that the inhibitory effect of shPSTPIP2 in NS4B-transfected cells was not as pronounced as in HCV-infected cells may have been the result of the high expression level of NS4B, which is the primary inducer of membranous webs.
DISCUSSION
In this study, we demonstrated that PSTPIP2 expression is important for HCV replication in both replicons and HCV infection systems: its knockdown decreased viral replication, while its overexpression enhanced viral replication. We showed also that the knockdown or the enhancement of PSTPIP2 did not significantly affect the efficiency of HCV entry (Fig. 4F and G). Furthermore, PSTPIP2 is distributed in the DRM fraction, which is the site of the viral replication complex. Further analysis by immunofluorescence and immuno-EM studies indicated that PSTPIP2 was partially colocalized with NS5A, which is a component of the RC. Immunoprecipitation results indicated that PSTPIP2 has the potential to interact with NS4B and NS5A directly. In addition, we found that the membrane-deforming ability of PSTPIP2 is important for the enhancement of HCV replication and that knockdown of PSTPIP2 reduced the formation of JC1- or NS4B-induced membranous webs. Therefore, we conclude that PSTPIP2 could deform the cellular membrane surrounding the HCV replication complex by interacting with viral proteins, thereby enhancing the formation of membranous webs to facilitate HCV RNA replication.
Our studies provided opportunities to investigate the relationship between membranous webs and HCV replication. We found that knockdown of PSTPIP2 reduced both HCV replication and the formation of membranous webs, whereas overexpression of PSTPIP2 enhanced HCV replication. Interestingly, the expression of a mutant PSTPIP2 reduced viral replication. These results indicate that membranous webs induced by HCV support and enhance HCV replication. The membranous webs may enhance local concentration of the viral nonstructural proteins to increase the probability of formation of the viral replication complex. Furthermore, such a structure may protect the replication complex from the attack of the host antiviral system.
How PSTPIP2 induces membrane rearrangement to form membrane curvature and tubules is an interesting issue. The major structural lipids in eukaryotic membranes are the glycerophospholipids, such as phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidylinositol (PI), and phosphatidic acid (PA) (32). Among these lipids, phosphorylated derivatives of PI are important in participating in signaling and recognition (30), defining organelle identity, and recruiting both soluble and membrane proteins to specific membranes. Very recent findings demonstrated that the metabolic product of PI4K-IIIα, PI4P, which is significantly enhanced in HCV-infected cells and provides lipid microenvironment in the membrane, is an important regulator of flaviviral RNA replication (13). The F-BAR domain of PSTPIP2 can bind to phosphatidylinositide lipids (14, 31) and may thereby target them to intracellular membranes to induce the formation of membrane curvature, thus initiating membranous web formation.
Another member of the PCH family, Bin1, has been reported to be involved in the HCV life cycle (23). Bin1 regulates the phosphorylation of NS5A and participates in virus assembly (23). The ability of bin1 to deform intracellular membranes and induce the membrane curvature to form membranous organelles may explain its ability to participate in the process of virus assembly or budding. Interestingly, PSTPIP2 also interacts with NS5A; thus, whether NS5A also participates in the formation of membranous web and whether PSTPIP2 also participates in virus assembly are interesting issues.
ACKNOWLEDGMENTS
We thank members of M. M. C. Lai's laboratory for helpful discussions, for the technical support from the EM facilities, and for the shRNA constructs from the National RNAi Core platform in Academia Sinica in Taiwan.
Footnotes
Published ahead of print 30 November 2011
REFERENCES
- 1. Aligo J, Jia S, Manna D, Konan KV. 2009. Formation and function of hepatitis C virus replication complexes require residues in the carboxy-terminal domain of NS4B protein. Virology 393: 68–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76: 13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen YC, et al. 2010. Polo-like kinase 1 is involved in hepatitis C virus replication by hyperphosphorylating NS5A. J. Virol. 84: 7983–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chitu V, et al. 2005. The PCH family member MAYP/PSTPIP2 directly regulates F-actin bundling and enhances filopodia formation and motility in macrophages. Mol. Biol. Cell 16: 2947–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chitu V, Stanley ER. 2007. Pombe Cdc15 homology (PCH) proteins: coordinators of membrane-cytoskeletal interactions. Trends Cell Biol. 17: 145–156 [DOI] [PubMed] [Google Scholar]
- 6. Egger D, et al. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76: 5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evans MJ, et al. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446: 801–805 [DOI] [PubMed] [Google Scholar]
- 8. Frost A, et al. 2008. Structural basis of membrane invagination by F-BAR domains. Cell 132: 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gosert R, et al. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77: 5487–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grosse J, et al. 2006. Mutation of mouse Mayp/Pstpip2 causes a macrophage autoinflammatory disease. Blood 107: 3350–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guo JT, Bichko VV, Seeger C. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75: 8516–8523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Henne WM, et al. 2010. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science 328: 1281–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hsu NY, et al. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141: 799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu J, et al. 2009. F-BAR-containing adaptor CIP4 localizes to early endosomes and regulates epidermal growth factor receptor trafficking and downregulation. Cell Signal. 21: 1686–1697 [DOI] [PubMed] [Google Scholar]
- 15. Itoh T, et al. 2005. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev. Cell 9: 791–804 [DOI] [PubMed] [Google Scholar]
- 16. Itoh T, et al. 2001. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science 291: 1047–1051 [DOI] [PubMed] [Google Scholar]
- 17. Konan KV, et al. 2003. Nonstructural protein precursor NS4A/B from hepatitis C virus alters function and ultrastructure of host secretory apparatus. J. Virol. 77: 7843–7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lai CK, Jeng KS, Machida K, Cheng YS, Lai MM. 2008. Hepatitis C virus NS3/4A protein interacts with ATM, impairs DNA repair and enhances sensitivity to ionizing radiation. Virology 370: 295–309 [DOI] [PubMed] [Google Scholar]
- 19. Lai CK, Jeng KS, Machida K, Lai MM. 2008. Association of hepatitis C virus replication complexes with microtubules and actin filaments is dependent on the interaction of NS3 and NS5A. J. Virol. 82: 8838–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miyanari Y, et al. 2003. Hepatitis C virus non-structural proteins in the probable membranous compartment function in viral genome replication. J. Biol. Chem. 278: 50301–50308 [DOI] [PubMed] [Google Scholar]
- 21. Moradpour D, et al. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78: 7400–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moradpour D, et al. 2003. Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex. Antiviral Res. 60: 103–109 [DOI] [PubMed] [Google Scholar]
- 23. Nanda SK, Herion D, Liang TJ. 2006. The SH3 binding motif of HCV [corrected] NS5A protein interacts with Bin1 and is important for apoptosis and infectivity. Gastroenterology 130: 794–809 [DOI] [PubMed] [Google Scholar]
- 24. Peter BJ, et al. 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303: 495–499 [DOI] [PubMed] [Google Scholar]
- 25. Pietschmann T, et al. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. U. S. A. 103: 7408–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MM. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77: 4160–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shimada A, et al. 2007. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell 129: 761–772 [DOI] [PubMed] [Google Scholar]
- 28. Sir D, et al. 2008. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 48: 1054–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suetsugu S. 2009. The direction of actin polymerization for vesicle fission suggested from membranes tubulated by the EFC/F-BAR domain protein FBP17. FEBS Lett. 583: 3401–3404 [DOI] [PubMed] [Google Scholar]
- 30. Takenawa T. 2010. Phosphoinositide-binding interface proteins involved in shaping cell membranes. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsujita K, et al. 2006. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J. Cell Biol. 172: 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Meer G, Voelker DR, Feigenson GW. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9: 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yeung YG, Soldera S, Stanley ER. 1998. A novel macrophage actin-associated protein (MAYP) is tyrosine-phosphorylated following colony stimulating factor-1 stimulation. J. Biol. Chem. 273: 30638–30642 [DOI] [PubMed] [Google Scholar]
- 34. Yu GY, Lee KJ, Gao L, Lai MM. 2006. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J. Virol. 80: 6013–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]