Abstract
The nonenveloped polyomavirus simian virus 40 (SV40) is taken up into cells by a caveola-mediated endocytic process that delivers the virus to the endoplasmic reticulum (ER). Within the ER lumen, the capsid undergoes partial disassembly, which exposes its internal capsid proteins VP2 and VP3 to immunostaining with antibodies. We demonstrate here that the SV40 genome does not become accessible to detection while the virus is in the ER. Instead, the genome becomes accessible two distinct detection procedures, one using anti-bromodeoxyuridine antibodies and the other using a 5-ethynyl-2-deoxyuridine-based chemical reaction, only after the emergence of partially disassembled SV40 particles in the cytoplasm. These cytoplasmic particles retain some of the SV40 capsid proteins, VP1, VP2, and VP3, in addition to the viral genome. Thus, SV40 particles undergo discrete disassembly steps during entry that are separated temporally and topologically. First, a partial disassembly of the particles occurs in the ER, which exposes internal capsid proteins VP2 and VP3. Then, in the cytoplasm, disassembly progresses further to also make the genomic DNA accessible to immune detection.
INTRODUCTION
The entry pathway of simian virus 40 (SV40) and other polyomaviruses is rather atypical. SV40 entry begins when the virus binds to major histocompatibility class I proteins and ganglioside GM1 at the cell surface (2, 24). SV40 and other polyomaviruses (e.g., MPyV and BKV) are then taken up into the cell by virus-induced, caveola-mediated endocytosis (1, 8, 16, 20). Moreover, SV40, as well as other polyomaviruses, then follows a rather unusual pathway to the nucleus, getting there by first passing through the endoplasmic reticulum (ER) (12, 16, 24).
A key question pertaining to this unique entry pathway concerns the extent of particle uncoating at each stage of the entry pathway. Earlier, we demonstrated that SV40 particles undergo partial disassembly in the ER, as shown by the finding that within that organelle the internal capsid proteins, VP2 and VP3, become accessible to immunostaining with antibodies (19). We sought to determine whether SV40 disassembly in the ER occurs to an extent sufficient to also make the viral genome accessible to an antibody-based detection procedure. Our current experimental findings demonstrate that, whereas the internal SV40 capsid proteins VP2 and VP3 become accessible to immunostaining in the ER, the genomic DNA becomes accessible to each of two independent detection procedures only after the partially disassembled SV40 particles emerge in the cytoplasm. These cytoplasmic particles retain at least some of the three SV40 capsid proteins, as well as the viral genome.
Together, our experimental findings show that SV40 particles undergo discrete disassembly steps during entry that are separated temporally and topologically. First, a partial disassembly of the particles occurs in the ER, which causes the internal capsid proteins VP2 and VP3 to become accessible to detection with antibodies. Then, in the cytoplasm, disassembly progresses further to also make the genomic DNA accessible to immune detection, as well as to an 5-ethynyl-2-deoxyuridine (EdU)-based procedure. These findings are discussed with regard to other recent results that bear on SV40 disassembly and transport out from the ER.
MATERIALS AND METHODS
Reagents.
African green monkey kidney cells (CV-1) were purchased from the American Type Culture Collection. Dulbecco modified Eagle medium (DMEM), 5-bromo-2-deoxyuridine (BrdU), Pen-Strep, and bovine serum albumin (BSA) were purchased from Sigma-Aldrich (St. Louis, MO). Fetal bovine serum (FBS) was purchased from Atlanta Biologicals (Fetal Bovine Serum Premium Select; Lawrenceville, GA). EdU and the Click-iT Alexa Fluor high-throughput imaging (HCS) assay kit were obtained from Invitrogen (Carlsbad, CA). Mounting medium with DAPI (4′,6′-diamidino-2-phenylindole) was obtained from Vector Laboratories (Burlingame, CA), and Fluoromount-G came from Southern Biotech (Birmingham, AL). Rabbit anti-VP2/3 was from A. Oppenheim (Jerusalem, Israel), mouse anti-BrdU was from Invitrogen, monoclonal mouse anti-protein disulfide isomerase (anti-PDI) was from BD Biosciences (San Jose, CA), polyclonal rabbit anti-PDI was from Abcam (Cambridge, MA), and mouse anti-SV40 large-T antigen was from Calbiochem/EMD4 Biosciences (San Diego, CA). PAb597 hybridoma, which produces a monoclonal antibody to SV40 major capsid protein VP1, was provided by Walter Atwood (Brown University). Fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit IgG(H+L), and cyanine 3 (Cy3)-conjugated goat anti-mouse IgG(H+L) were from Jackson Immunoresearch Laboratories (West Grove, PA).
Viral DNA labeling.
SV40 was absorbed onto semiconfluent CV-1 cells growing on 100-mm dishes at 37°C and 5% CO2 in a humidified incubator. At 2 h postattachment, medium containing DMEM plus 10% FBS was added to cells. Because both BrdU and EdU are light sensitive, the rest of the protocol was performed with minimal exposure to light. At 20 h postinfection (hpi) BrdU or EdU was added to the media at a final concentration of 1 μg/ml. The infection was spiked with an additional 1 μg/ml at 5 days postinfection. The infection was allowed to proceed until the cells on the plate were killed (10 to 12 days postinfection), and the cell suspension was then collected. The harvested cell cultures were freeze-thawed three times, the cell debris was removed by centrifugation at 4,000 × g for 45 min, and the supernatant was collected. The supernatant was dialyzed twice, each for a for 24 to 36 h periods in 1× phosphate-buffered saline (PBS) at 4°C in the dark using Spectra/Por dialysis membrane tubing (Spectrum Laboratories, Rancho Dominguez, CA) with a molecular weight cutoff at 6,000 to 8,000. A third and final round of dialysis was carried out in DMEM for 24 to 36 h, followed by the addition of Pen-Strep at 20 ml/liter. Virus titers, as determined by viral plaque assay, were approximately 5 × 106 PFU/ml.
Control medium for all mock-infected controls was prepared to closely mimic infected-cell medium as follows. BrdU was added to regular medium, which was then applied to noninfected cells. After 10 to 12 days this medium was harvested, dialyzed, and used to mock infect control cells. Importantly, these mock-infected cells were grown concurrently with and processed identically to their infected counterparts described in the protocol above.
Infections.
A total of 150 μl of BrdU or EdU-labeled SV40 stock was absorbed to CV-1 cells growing in 8-well Lab-Tek chamber slides (Nunc, Rochester, NY) for 1 h at 4°C. After 1 h, the virus was removed, and the cells were washed once with DMEM plus 10% FBS. Fresh warm DMEM plus 10% FBS was then added to the wells, and the chamber slides were incubated at 37°C and 5% CO2. The input multiplicity of infection (MOI) in all experiments was approximately 1 PFU/cell. At various times, the infections were stopped by washing the chamber slides three times with 1× PBS and fixed in 100% methanol at −20°C for 10 min. Mock-infected control samples were inoculated with control medium, prepared as described above.
Immunostaining.
In preparation for immunostaining, fixed samples were washed three times with PBS and once with 0.01% BSA. All samples that were infected with BrdU-labeled SV40 were then treated with 20 KU (Kunitz units) of DNase (Sigma-Aldrich) per well for 20 min at 37°C. After 20 min, the cells were washed three times with 0.01% BSA solution. The samples were then incubated with primary antibodies for 1 h at 37°C and washed three times with 0.01% BSA. The following primary antibodies were utilized in various combinations with each other: mouse anti-BrdU (1:100), rabbit anti-VP2/3 (1:1,000), mouse anti-PDI (1:100), rabbit anti-PDI (1:100), mouse anti-SV40 LT (1:200), and PAb597 anti-VP1 (1:25). They were next incubated with secondary antibodies for 50 min at room temperature and again washed three times with 0.01% BSA. The secondary antibodies FITC-conjugated donkey anti-rabbit IgG(H+L) (1:100), and Cy3-conjugated goat anti-mouse IgG(H+L) (1:200) were used in conjunction with the appropriate primaries. Primary and secondary antibodies were diluted in 0.01% BSA, and the staining procedure was carried out in the dark. After the final wash, the cells were mounted in either Fluoromount-G or mounting medium with DAPI. Staining for EdU-labeled SV40 DNA was carried out according to the manufacturer's protocol in the Click-iT Alexa Fluor high-throughput imaging (HCS) assay kit from Invitrogen.
Microscopy.
Immunofluorescence microscopy was carried out using a Nikon E600 epifluorescence microscope. An ORCA-ER-cooled charge-coupled device camera (Hamamatsu) and OPENLAB software (Improvision) were used for all image acquisition and processing. Image acquisition parameters were as follows: a 100- to 200-ms exposure using the neutral ND4 filter (neutral density filter 4), the filter sets #86013 and #89006 (Chroma), and identical calibration curves were applied to all sets of images (infected and uninfected). For many micrographs, z-sections were acquired at 0.25-μm intervals. Both a phase-contrast and a fluorescence image was captured for each field.
RESULTS
Exposure of the SV40 genome to two independent staining procedures occurs after transport of partially disassembled SV40 particles from the ER into the cytoplasm.
We began by confirming our earlier experimental finding that internal capsid proteins VP2/3 (VP2 and VP3) become accessible to immunostaining within the ER (19). (We use the designation “VP2/3” since the anti-VP2/3 antibody used in our experiments recognizes a common epitope on VP2 and VP3 [19].) In this experiment, and in all that follow here, the ER is identified by immunostaining against the ER marker, protein disulfide isomerase (PDI). No immunostaining of VP2/3 was seen in the 3-h samples. A representative image is shown in Fig. 1A. However, we observed VP2/3 immunostaining by 6 hpi (Fig. 1B). Moreover, at this time point, virtually all of the VP2/3 stain colocalized with PDI, as indicated by the yellow color of the VP2/3 immunostain in the merged image (Fig. 1B, lower right). However, between 6 and 12 hpi, the fraction of cells containing VP2/3 that entirely colocalized with PDI diminished, concomitant with a rise in VP2/3 staining seen in the cytoplasm. A representative 12-h image is shown in Fig. 1C, and the data over these time points are summarized in Fig. 2. Multiple VP2/3-stained entities may be seen within a single cell. At 12 hpi, in a number of cells, some of the VP2/3 stain is associated with PDI, and some is not. This finding most likely reflects the fact that the movement of the disassembly intermediates through the infection pathway is not synchronous in individual cells. Cells containing both ER-associated and cytoplasmic VP2/3 stain were included in each of the columns at their respective time points in our summary (Fig. 2). A VP2/3 antibody control is shown in Fig. S2 in the supplemental material.
Fig 1.
Exposure of minor capsid proteins VP2/3 within the ER. CV-1 cells were infected with SV40, fixed in cold methanol, and then stained for VP2/3 (FITC) and ER marker PDI (Cy3). (A) A 3-h sample. No green VP2/3 stain can be seen, while the PDI stain is seen in red. The nucleus is highlighted by the blue DAPI stain. (B) Sample fixed at 6 h. The top two panels show a punctate VP2/3 stain, seen in green, and the PDI stain, seen in red. The bottom two panels show a 5-fold magnification of the VP2/3 stain and of a merge image. The yellow VP2/3 stain in the 5× merge indicates the colocalization of VP2/3 with the ER marker. (C) Sample fixed at 12 h. Note that the VP2/3 stain in the merge is green. Compare this image to panel B. The scale bars for the three images in panel A, the top two images in panel B, and the three images in panel C correspond to 10 μm.
Fig 2.
Localization of the VP2/3 immunostain over time. Experiments were carried out in triplicate, in which a total of 100 × 3 cells, immunostained for VP2/3, were examined at each time point. The average numbers of cells in which the VP2/3 stain colocalized or did not colocalize with the PDI immunostain at each time are indicated. Note that some cells contained VP2/3 associated with PDI and also VP2/3 not associated with PDI. These cells were included in each of the columns at their respective time points. No VP2/3 immunostain was detected at 3 hpi.
We next sought to determine whether the partial disassembly of SV40 particles in the ER, as demonstrated by exposure of VP2/3 to immunostaining, likewise makes the genomic DNA accessible to immunostaining. To do so, we used BrdU for labeling parental SV40 genomes and anti-BrdU antibodies to detect the exposure of the BrdU-labeled DNA within partially disassembled viral particles. We began by confirming that BrdU could be incorporated into SV40 genomes and that antibody detection of the BrdU-labeled genomes is possible only after the particles have undergone some degree of disassembly, in this instance as induced by exposure to various temperatures ranging from 25 to 100°C. In a dot blot assay, no signal was observed in any of the unlabeled samples after incubation at any temperature or in the BrdU-labeled samples incubated at or below 37°C. However, at incubation temperatures of 45°C and higher, a positive signal was seen in the BrdU-labeled samples. Moreover, the magnitude of the BrdU signal correlated with the increasing temperature of incubation (see Fig. S1 in the supplemental material). Thus, we were able to incorporate BrdU into SV40 genomes. In addition, the native SV40 capsid was impermeable to the anti-BrdU antibodies and, finally, the BrdU-labeled parental viral DNA was accessible to detection with the antibodies only within viral particles that underwent sufficient thermal-induced capsid disassembly. The presence of SV40 particles in the samples was confirmed by stripping the blot and probing for the major capsid protein VP1 (data not shown).
We next assessed the affect that the incorporated BrdU might have on the kinetics of SV40 entry and subsequent infectivity (i.e., viral gene expression) as follows. CV-1 cells were seeded on chamber slides and infected with either BrdU-labeled or unlabeled SV40 stocks. At various times postinfection, the cells were fixed in 100% cold methanol and immunostained for SV40 large T-antigen (LT), an SV40 early gene product. LT-positive nuclei were then counted. Experiments were carried out in triplicate, in which a total of 8,000 × 3 cells were examined at each time point and the number of LT-positive cells were counted (see Fig. S3 in the supplemental material). The slopes of the best-fit lines (not shown) indicated that the time courses of infection of the BrdU-labeled and unlabeled SV40 stocks were statistically equivalent. These experimental findings are in complete agreement with the earlier report of Calothy et al. (3), in which SV40 stocks grown in BrdU (1 μg/ml or less) were found to produce LT. Under these conditions, up to 15% of the thymine in SV40 DNA may be substituted with BrdU (3).
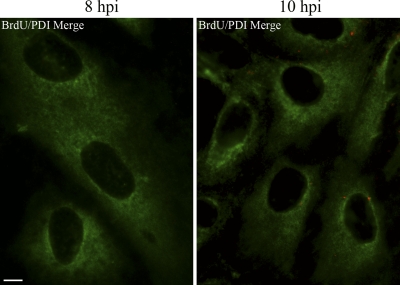
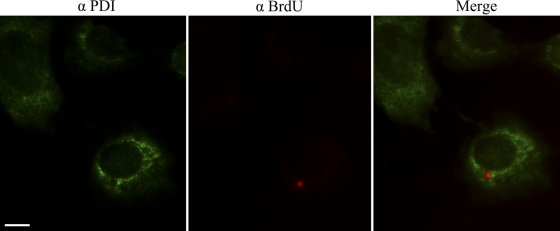
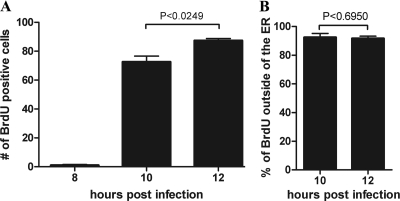
With the above experimental findings at hand, we sought to determine whether the genomes of incoming SV40 particles, like their internal capsid proteins VP2/3, might become accessible within the ER to immunodetection with antibodies. CV-1 cells, infected with BrdU-labeled SV40, were fixed at 8, 10, and 12 h hpi and examined for the accessibility of the BrdU-labeled viral DNA to immunostaining with antibodies. The same cells were also immunostained for the ER marker, PDI. Representative merge images are shown in Fig. 3 (8 and 10 hpi) and Fig. 4 (12 hpi). (A BrdU antibody control is shown in Fig. S2 in the supplemental material.) Experiments were carried out in triplicate, in which a total of 500 × 3 cells were examined at each time point. The means and standard errors of the mean (SEM) of three samples of 500 cells each, in which BrdU was detected by immunostaining at 8, 10, and 12 hpi, were 1.00 ± 0.577, 72.67 ± 3.93, and 87.33 ± 1.45, respectively (Fig. 5A). These experimental results show that viral disassembly intermediates, in which the parental viral DNA is accessible to immunostaining, accumulate between 8 and 10 hpi. At or prior to 8 h the viral particles had not yet undergone sufficient disassembly to expose the DNA to the anti-BrdU antibodies (Fig. 3). Moreover, and importantly, whereas VP2/3 were detected within the ER by immunostaining at 6 hpi (Fig. 1), we did not see any colocalization of the punctate BrdU DNA stain with the ER marker PDI at any time in any of the more than 400 cells in which the BrdU-labeled DNA was detected (Fig. 5B). Colocalization of the BrdU stain with the green PDI stain would have been indicated by yellow in the merge images (Fig. 3 and 4). For comparison, colocalization was indicated above, in the instance of VP2/3 with the PDI stain at 6 hpi (Fig. 1). The set of images from the 12-h BrdU sample provides confirmation that the red BrdU stain is in a region of the cell that is devoid of PDI (Fig. 4). Moreover, multiple BrdU-staining disassembly intermediates were generally seen in each of these cells. Thus, these experimental findings demonstrate that the viral particles become sufficiently disassembled to make the viral genomes accessible to immunostaining, only after the particles are transported out from the ER. Note that in this and all subsequent experiments, Z stacks were examined to ascertain that we were examining central planes through the cells.
Fig 3.
BrdU-labeled SV40 genomes are first detected outside of the ER. CV-1 cells were infected with BrdU-labeled SV40 and fixed in cold 100% methanol at 8 and 10 hpi. Cells were treated with DNase for 10 min and immunostained for BrdU (Cy3) and ER marker PDI (FITC). A representative merge image is shown for each time point. Scale bar, 10 μm.
Fig 4.
Cells from the same experiment as in Fig. 3 were fixed at 12 hpi and likewise were immunostained for BrdU (Cy3) and ER marker PDI (FITC). A representative merge image is shown, as well as separate BrdU and PDI immunostains. Scale bar, 10 μm.
Fig 5.
Time-dependent exposure of BrdU-labeled SV40 genomes occurs outside of the ER. (A) Cells were infected with BrdU-labeled SV40 and fixed and immunostained for BrdU and PDI at 8, 10, and 12 hpi. Experiments were carried out in triplicate, in which a total of 500 × 3 cells were examined at each time point. The mean and SEM of three samples of 500 cells each, in which BrdU was detected by immunostaining at each time point, are indicated. An unpaired two-tail t test was completed, and P values were calculated. Note that a cell was counted to be BrdU positive if it contained at least one BrdU puncta, even though many cells contained multiple BrdU puncta staining. (B) Cells that were counted in panel A were further examined for the location of the BrdU stain within the cell. In both the 10- and the 12-h time points nearly all of the BrdU-labeled SV40 genomes were found to be outside of the ER.
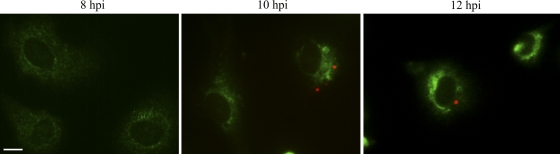
To be certain that the preceding experimental findings were not an artifact of our antibody-based SV40 DNA detection procedure, we used an antibody-independent method to detect exposed SV40 DNA, in which we incorporated EdU into the DNA of our stock SV40 particles during active DNA synthesis. We then infected cells with EdU-labeled virus particles and detected exposed DNA using a copper (Cu+)-catalyzed reaction between the alkyne on EdU and the azide on an Alexa Fluor dye. The small size of the dye-azide compound allows for the detection of the target DNA without the need for DNA denaturation with HCl or DNase. Using the EdU-based method, viral genomes could not be detected until between 10 and 12 hpi (Fig. 6). Importantly, and in agreement with our findings using our antibody-based detection procedure, exposed viral genomes were never detected within the ER but instead were seen in the cytoplasm (Fig. 6).
Fig 6.
EdU-labeled SV40 genomes are first detected outside of the ER. CV-1 cells were infected with EdU-labeled SV40. The cells were then fixed and stained for EdU (Alexa Fluor 594 [red]) and PDI (FITC) at 8, 10, and 12 hpi. Representative merge images are shown for each time point. The initial detection of the EdU-labeled viral genomes is seen as red punctate staining at 10 hpi. The lack of colocalization between the EdU and PDI stains demonstrates that the virus is not in the ER at either 10 or 12 hpi. Scale bar, 10 μm.
Exposure of internal capsid proteins VP2/3 precedes exposure of genomic DNA.
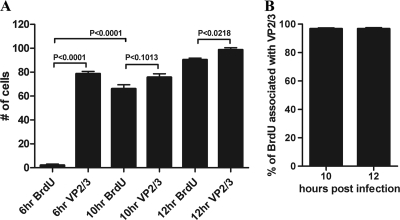
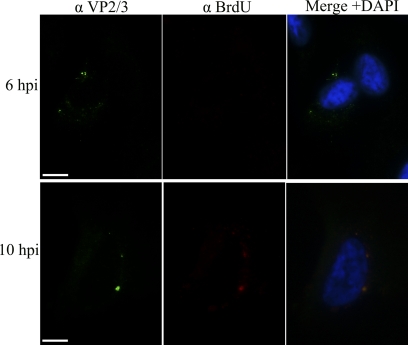
The experimental findings reported above show that SV40 particles undergo sufficient disassembly during transit through the ER to expose internal capsid proteins VP2/3 to immunostaining with antibodies. In contrast, SV40 disassembly in the ER did not appear sufficient to expose the viral DNA to either of our two detection procedures. Instead, BrdU-labeled and EdU-labeled SV40 genomes became accessible to detection only in the cytoplasm. Together, these experimental findings suggest that SV40 disassembly events in the ER expose the internal capsid proteins VP2/3 but that later disassembly in the cytoplasm exposes the genomic DNA. To test this premise further, we compared the time course of exposure of VP2/3 with that of the BrdU-labeled genomic DNA in the same cells. Cells that were infected with BrdU-labeled SV40 were simultaneously immunostained for both VP2/3 and for BrdU-labeled viral genomes. Experiments were carried out in triplicate, in which a total of 500 × 3 cells were examined at 6, 10, and 12 hpi. At 6 hpi the mean number of cells, per 500-cell sample, in which BrdU was detected was 2.00 ± 1.00, while the mean number of cells in which VP2/3 was detected was 78.67 ± 2.02 (Fig. 7A). At 10 hpi we detected 66.00 ± 3.46 cells in which BrdU is exposed to detection by anti-BrdU antibodies. Concurrently, within the same population of cells, we detected 75 ± 2.96 cells that immunostained for VP2/3. Similar statistical results were observed at 12hpi (Fig. 7A). Representative experimental samples are shown in Fig. 8. These experimental findings show that exposure of VP2/3 precedes exposure of the genomic DNA.
Fig 7.
Exposure of VP2/3 precedes exposure of genomic DNA, and genomic DNA is seen in association with VP2/3. (A) Cells infected with BrdU-labeled SV40 were fixed at 6, 10, and 12 hpi and stained for BrdU and VP2/3. Cell counts were carried out in triplicate with a total of n = 500/count. Means ± the standard errors of the means and the results of an unpaired two-tail t test are shown. (B) The same cells as in panel A were examined for the association of BrdU-labeled DNA with VP2/3. Representative micrograph images are shown in Fig. 8.
Fig 8.
Exposure of VP2/3 precedes exposure of the genomic DNA. Representative 6- and 10-h samples from the experiment summarized in Fig. 7 are shown. Samples were stained for BrdU (Cy3) and VP2/3 (FITC), and the nucleus was stained with DAPI (blue). Scale bar, 10 μm.
Together, these findings demonstrate that exposure of the internal capsid proteins and the genomes occur sequentially as distinct events. Disassembly in the ER exposes VP2/3. However, events in the ER do not yet expose the genomic DNA. Instead, further particle disassembly, evident only after the viral disassembly intermediates are transported into the cytoplasm, exposes the viral DNA to our detection methods. Note that the punctate anti-BrdU staining of the SV40 genomes seen in these experiments, as well as the punctate staining of the associated internal VP2/3 capsid proteins, is similar to what is seen in the case of BrdU-labeled papillomavirus genomes and the associated papillomavirus internal L2 protein (11).
Cytoplasmic parental SV40 genomes remain associated with VP2/3 and VP1 after transport out of the ER.
Next, we examined the same cells from which the data in Fig. 7A were obtained to determine whether parental SV40 genomes might remain associated with VP2/3 after being transported to the cytoplasm. Indeed, cytoplasmic, immunostained viral genomes almost always remained in association with VP2/3. At 10 hpi, all of the punctate BrdU staining was seen to be associated with VP2/3 in 192 of 198 cells (Fig. 7B, 8). These data are even more compelling, considering that multiple entities that stain positively for both BrdU and VP2/3 are often seen within a single cell. The association between cytoplasmic BrdU and VP2/3 continued to remain strong even at 12 hpi (data not shown). Since the BrdU-labeled parental viral genomes became susceptible to immunostaining only upon transfer to the cytoplasm, these experimental findings demonstrate that virtually all viral genomes remain in association with at last some VP2/3 during and after their transport to the cytoplasm.
VP1, the major SV40 capsid protein, comprises the outer shell of the capsid. At least until 12 hpi, virtually all of the viral disassembly intermediates that were accessible to immunostaining with anti-VP2/3 were still associated with at least some VP1, including particles seen around the nucleus (Fig. 9).
Fig 9.
VP1 remains in association with VP2/3-containing SV40 disassembly intermediates. Cells were infected with BrdU-labeled SV40, fixed at 12 hpi, and immunostained for VP1 (Cy3) and VP2/3 (FITC). Scale bar, 10 μm.
DISCUSSION
All polyomaviruses examined to date appear to traffic to the nucleus via the ER (12, 16, 24). In the case of SV40, experimental results reported previously (19) and above show that this virus goes through a sequence of discrete disassembly steps while traversing this pathway. First, SV40 capsids undergo partial disassembly in the ER, which exposes internal capsid proteins VP2/3 to immunostaining with antibodies. Then, upon reaching the cytoplasm, the partially disassembled particles undergo further disassembly, which makes the viral DNA accessible to either of two independent detection procedures: one involving immunostaining with anti-BrdU antibodies and the other an EdU-based chemical reaction.
In the following discussion, we interpret our findings with respect to ER-associated processes and factors that are believed to play a role in initiating the uncoating of SV40 and its penetration across the ER membrane. We begin by recounting the unique organization of polyomavirus capsids. Icosahedral polyomavirus capsids are comprised mainly of 360 copies of the major capsid protein VP1, all of which are arranged as pentamers. This is a rather uncommon arrangement of capsid subunits, since the subunits of icosahedral capsids are more usually arranged as pentamers only at the 12 vertices of the icosahedron and as hexamers everywhere else. In order for polyomaviruses to display the characteristic 3- and 5-fold rotational axes of symmetry of an icosahedron (14), despite being comprised only of pentamers, five C-terminal arms of VP1 extend from each pentamer and insert into the neighboring pentamers in three distinct kinds of interactions. Moreover, and atypically, these interactions are stabilized by interchain disulfide bonds, as well as by calcium ions (13, 14). More commonly, viral capsids are held together by noncovalent interactions. The internal face of each polyomavirus pentamer binds one copy of either of the minor capsid proteins, VP2 or VP3.
Bearing in mind the organization of polyomavirus capsids, the oxidoreductase ERp57, which resides in the ER lumen, catalyzes a disulfide isomerization reaction in vitro that disconnects a subset of SV40 pentamers (most likely the 12 vertex pentamers) from the network of VP1 molecules cross-linked by disulfide bonds in the capsid (23). Importantly, while 20% of SV40 VP1 molecules no longer participate in the disulfide-bonded network following exposure to ERp57, the pentamers that they comprise nevertheless remain associated with the virus particle as long as the VP1-bound Ca2+ ions are in place to stabilize the interpentamer associations. In the case of the related mouse polyomavirus, the ER chaperone ERp29 causes local unfolding of the VP1 C termini to expose VP2 (21).
Considering this, it is plausible that ERp57, and possibly other ER chaperones, catalyze reactions in the ER that make VP2/3 accessible to the immunostaining as reported here. Next, the genomic DNA might become accessible to our DNA staining procedures when the SV40 disassembly intermediates enter the reducing and low-calcium environment of the cytoplasm, conditions that would cause the vertex pentamers to disassociate from the particles (23). Interestingly, when SV40 particles devoid of viral DNA (i.e., “empty particles”) were subjected to ERp57, then no remnant capsids were seen, implying that the viral DNA plays a role in maintaining the structural integrity of the disassembly intermediates (23). That result is consistent with our findings that the viral DNA remains associated with capsid proteins after transfer of the disassembly intermediates to the cytoplasm.
Next, we interpret our findings with respect to the means by which SV40 might cross the ER membrane into the cytoplasm. While enveloped viruses commonly cross cellular membranes by membrane fusion, it is generally not well understood how nonenveloped viruses might cross cellular membranes. In the case of SV40, hydrophobic domains on the exposed VP2 and VP3 molecules might perhaps insert into the ER membrane to somehow facilitate transport of the disassembly intermediates out of the ER. The following experimental findings are consistent with this possibility. First, the hydrophobic N terminus of the murine polyomavirus VP2 protein is linked to entry (22). Second, SV40 VP2 and VP3 each efficiently inserts into purified ER membranes (6). Third, SV40 VP2 and VP3 (but not VP1), when expressed in transfected bacterial cells, permeabilize bacterial membranes. Finally, VP3 expression leads to lysis of these bacterial cells (7).
Recent experimental evidence suggests that the ER-associated degradation (ERAD) pathway, which removes misfolded proteins from the ER into the cytoplasm (25), may be involved in SV40 translocation out of the ER. For example, SV40 entry is impaired by proteasomal inhibitors and by the silencing of retrotranslocation proteins Derlin-1 and Sel1L (23). Moreover, the related murine polyomavirus was found to require the ER protein Derlin-2 to initiate infection (15), and expression of dominant-negative Derlin-1 impaired infection by the BK human polyomavirus (10). Perhaps an altered conformation, possibly resulting from the ERp57-mediated isomerization of the interpentamer disulfide bonds, is recognized by an ERAD sensor.
More recently, SV40 infection was shown to depend on four different DNAJ molecular cochaperones and BiP, the Hsp70 partner of DNAJB11 (9). In this regard, the DNAJ proteins bind specifically to Hsp70 ATPases to mediate protein folding and rearrangement in the ER. Interestingly, although BiP formed a complex with SV40 particles in the ER in a DNAJB11-dependent manner, the complex did not appear to play a role in exposing VP2/3. Instead, binding of BiP appeared to be necessary for transport of the particles out of the ER. Three of the cellular proteins involved in this recent study—BiP, DNAJB11, and DNAJB12—are known to be involved in ERAD, which is consistent with the premise that ERAD plays a role in SV40 entry. Although it is not at all clear how the ERAD translocation system might work for particles as large as SV40 disassembly intermediates, it might well depend upon the protein unfolding and rearrangements within the SV40 capsid that occur inside the ER.
The following plausible scheme for SV40 disassembly and trafficking out from the ER, as modified from that proposed by Goodwin et al. (9), takes all of the above findings into account. ERp57, and possibly other ER chaperones, catalyze reactions in the ER that expose hydrophobic peptide segments on VP2/3 (21). These segments are then recognized by the DNAJ-BiP complex, which may promote insertion of VP2/3 into the ER membrane (9). Regardless, the disassembly intermediates are transferred through the membrane by the DNAJ proteins, perhaps acting in association with associated cytosolic Hsc70s. The virus particles might possibly undergo further disassembly during this penetration step through the ER membrane. In any case, viral genomes do not become accessible to our two detection procedures until after the disassembly intermediates emerge in the cytoplasm. The disassembly step that exposes the genomic DNA might be triggered by the reducing and low-calcium environment of the cytoplasm.
Our findings might also be relevant regarding targeting of the cytoplasmic SV40 disassembly intermediates to the nucleus. That is, nuclear localization signals (NLSs) exposed on VP2/3 molecules, which remain associated with the viral genome, might target the disassembly intermediates to nuclear pore complexes. This premise is consistent with reports that microinjecting anti-VP2/3 antibodies into the cytoplasm blocks SV40 infection (17). Also, SV40 virus-like particles, which contain NLS-deficient VP3, enter cells normally but do not transport their DNA into the nucleus (18).
Supplementary Material
ACKNOWLEDGMENT
This study was supported by Public Health Service grant CA100479 from the National Cancer Institute.
Footnotes
Published ahead of print 16 November 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Anderson HA, Chen Y, Norkin LC. 1996. Bound simian virus 40 translocates to caveolin-enriched membrane domains, and its entry is inhibited by drugs that selectively disrupt caveolae. Mol. Biol. Cell 11:1825–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Breau WC, Atwood WJ, Norkin LC. 1992. Class I major histocompatibility proteins are an essential component of the simian virus 40 receptor. J. Virol. 66:2037–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Calothy G, Hirai K, Defendi V. 1973. 5-Bromodeoxyuridine incorporation into simian virus 40 deoxyribonucleic acid. Effects on simian virus 40 replication in monkey cells. Virology 55:329–338 [DOI] [PubMed] [Google Scholar]
- 4. Chen XS, Stehle T, Harrison SC. 1998. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J. 17:3233–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen Y, Norkin LC. 1999. Extracellular simian virus 40 transmits a signal that promotes virus enclosure within caveolae. Exp. Cell Res. 246:83–90 [DOI] [PubMed] [Google Scholar]
- 6. Daniels R, Rusan NM, Wadsworth P, Hebert DN. 2006. SV40 VP2 and VP3 insertion into ER membranes is controlled by the capsid protein VP1: implications for DNA translocation out of the ER. Mol. Cell 24:955–966 [DOI] [PubMed] [Google Scholar]
- 7. Daniels R, et al. 2006. Simian virus 40 late proteins possess lytic properties that render them capable of permeabilizing cellular membranes. J. Virol. 80:6575–6587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eash S, Querbes W, Atwood WJ. 2004. Infection of vero cells by BK virus is dependent on caveolae. J. Virol. 78:11583–11590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goodwin EC, et al. 2011. BiP and multiple DNAJ molecular chaperones in the endoplasmic reticulum are required for efficient simian virus 40 infection. mBio 2(3):e00101-11 doi:10.1128/mBio.00101.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiang M, Abend JR, Tsai B, Imperiale MJ. 2009. Early events during BK virus entry and disassembly. J. Virol. 83:1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kämper N, et al. 2006. A membrane-destabilizing peptide in capsid protein L2 is required for egress of papillomavirus genomes from endosomes. J. Virol. 80:759–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kartenbeck J, Stukenbrok H, Helenius A. 1989. Endocytosis of simian virus 40 into the endoplasmic reticulum. J. Cell Biol. 109:2721–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li PP, et al. 2003. Importance of Vp1 calcium-binding residues in assembly, cell entry, and nuclear entry of simian virus 40. J. Virol. 77:7527–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liddington RC, et al. 1991. Structure of simian virus 40 at 3.8-Å resolution. Nature 354:278–284 [DOI] [PubMed] [Google Scholar]
- 15. Lilley BN, Gilbert JM, Ploegh HL, Benjamin TL. 2006. Murine polyomavirus requires the endoplasmic reticulum protein Derlin-2 to initiate infection. J. Virol. 80:8739–8744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mannová P, Forstová J. 2003. Mouse polyomavirus utilizes recycling endosomes for a traffic pathway independent of COPI vesicle transport. J. Virol. 77:1672–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakanishi A, Clever J, Yamada M, Li PP, Kasamatsu H. 1996. Association with capsid proteins promotes nuclear targeting of simian virus 40 DNA. Proc. Natl. Acad. Sci. U. S. A. 93:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nakanishi A, Shum D, Morioka H, Otsuka E, Kasamatsu H. 2002. Interaction of the Vp3 nuclear localization signal with the importin alpha 2/beta heterodimer directs nuclear entry of infecting simian virus 40. J. Virol. 76:9368–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Norkin LC, Anderson HA, Wolfrom SA, Oppenheim A. 2002. Caveolar endocytosis of simian virus 40 is followed by brefeldin A-sensitive transport to the endoplasmic reticulum, where the virus disassembles. J. Virol. 76:5156–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pelkmans L, Kartenbeck J, Helenius A. 2001. Caveolar endocytosis of simian virus 40 reveals a novel two-step vesicular transport pathway to the ER. Nat. Cell Biol. 3:473–483 [DOI] [PubMed] [Google Scholar]
- 21. Rainey-Barger EK, Magnuson B, Tsai B. 2007. A chaperone-activated nonenveloped virus perforates the physiologically relevant endoplasmic reticulum membrane. J. Virol. 81:12996–133004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sahli R, et al. 1993. Defect in entry and altered pathogenicity of a polyoma virus mutant blocked in VP2 myristylation. Virology 192:142–153 [DOI] [PubMed] [Google Scholar]
- 23. Schelhaas M, et al. 2007. Simian virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131:516–529 [DOI] [PubMed] [Google Scholar]
- 24. Tsai B, et al. 2003. Gangliosides are receptors for murine polyoma virus and SV40. EMBO J. 22:4346–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vembar SS, Brodsky JL. 2008. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell. Biol. 9:944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.