Abstract
Genotyping of 21 varicella-zoster virus (VZV) strains using a scattered single nucleotide polymorphism (SNP) method revealed ambiguous SNPs and two nontypeable isolates. For a further genetic characterization, the genomes of all strains were sequenced using the 454 technology. Almost-complete genome sequences were assembled, and most remaining gaps were closed with Sanger sequencing. Phylogenetic analysis of 42 genomes revealed five established and two novel VZV genotypes, provisionally termed VIII and IX. Genotypes VIII and IX are distinct from the previously reported provisional genotypes VI and VII as judged from the SNP pattern. The alignments showed evidence of ancient recombination events in the phylogeny of clade 4 and recent recombinations within single strains: 3/2005 (clade 1), 11 and 405/2007 (clade 3), 8 and DR (clade 4), CA123 and 413/2000 (clade 5), and strains of the novel genotypes VIII and IX. Bayesian tree inference of the thymidine kinase and the polymerase genes of the VZV clades and other varicelloviruses revealed that VZV radiation began some 110,000 years ago, which correlates with the out-of-Africa dispersal of modern humans. The split of ancestral clades 2/4 and 1/3/5/VIII/IX shows the greatest node height.
INTRODUCTION
Varicella-zoster virus (VZV) is a member of the genus Varicellovirus within the subfamily Alphaherpesvirinae, family Herpesviridae (8). The alphaherpesviruses are characterized by their short reproduction cycle, fast spreading, efficient destruction of infected cells, and persistence in sensory ganglia. VZV replication is limited exclusively to cells of human and simian origins. The VZV genome consists of double-stranded DNA with a size of 125 kb and comprises 73 genes, 70 of which are unique and 3 of which are duplicated (9). The virus genome includes two main coding regions, unique long (UL) and unique short (US), and flanking inverted repeats, termed terminal and internal repeats long (TRL, IRL) and short (TRS, IRS). In addition to these repeats, there are five genomic regions with tandem reiterations, designated R1 to R5.
Primary infection with VZV causes varicella (chickenpox), a condition characterized by fever and exanthema with small blisters. Virus uptake occurs via the mucous membranes of the respiratory tract, with primary replication seen in the regional lymph nodes. During a short phase of primary cell-associated viremia, the virus infects peripheral blood mononuclear cells. In a secondary viremia, the virus spreads to cutaneous epithelial cells, where it induces rash. Usually, the virus is spread by excretion of aerosolized virus particles from the respiratory tract and highly infectious vesicle fluid from rash (2). After primary infection, VZV establishes a lifelong latency in trigeminal and dorsal root ganglia. Endogenous viral reactivation, e.g., after decline of VZV-specific cell-mediated immunity, may lead to viral replication and inflammation in the ganglion. Then, progeny virus migrates along the axons of neurons and is released in the skin, where it causes herpes zoster (shingles). Zoster is characterized by a unilateral vesicular rash within a single cutaneous dermatome. In most cases, the thoracic region is involved.
Genetic characterization of VZV DNA is achieved by analysis of restriction fragment length polymorphism (RFLP), single nucleotide polymorphisms (SNPs), and full-genome sequencing. Early RFLP analyses revealed both interstrain variations among wild-type isolates and differences between wild- and vaccine-type viruses. The most commonly used RFLP markers of VZV included the polymorphism of open reading frames (ORFs) 38 (PstI), 54 (BglI), and 62 (SmaI) (19, 23, 45). These markers allowed the majority of wild-type strains in North America and Europe to be typed as PstI+ BglI−, whereas African and Asian strains were BglI+ and Japanese Oka-like wild-type strains were PstI+/PstI− BglI+ SmaI−; the Oka vaccine strains were PstI− BglI+ SmaI+ (20, 40, 47). Genotyping was improved when DNA sequencing was employed to screen for SNPs in different ORFs of the VZV genome. The scattered SNP method proposed by Barrett-Muir and coworkers (4) reported SNPs present in ORFs 1, 21, 50, and 54 to distinguish four main viral clades (termed A, B, C, and J). Two similar approaches by Faga et al. and Wagenaar et al. analyzed the sequences of the VZV IE62 gene and the glycoprotein gH, gI, gL, gB, and gE genes (12, 62). Sequences clustered in four major clades, designated A, B, C, and D. Finally, Loparev and coworkers (24, 25) combined ORF 22-based genotyping with the analysis of either ORF 21 or ORF 50 and described five confirmed clades, E1, E2, J, M1, and M2, and the two provisional clades M3 and M4. Furthermore, different nucleotide positions in ORFs 51 to 58 (49, 50, 52) were used to classify VZV wild-type strains into the five clades corresponding to clades A, B, C, D (12, 62), and M1 (25). Another study investigated full-genome sequences and presented a phylogenetic analysis of VZV and a description of an origin-of-replication-based genotyping scheme (38). These authors analyzed 18 VZV strains, 11 sequences of which were novel, and observed four major clades, similar to the genotyping scheme of Wagenaar et al. (62). At present, full-genome sequencing is not practical for the genotyping of most clinical specimens, since most VZV strains cannot be propagated in cell cultures and the amount of viral DNA is limited. Recently, a new universal nomenclature which aims to unify the different approaches has been introduced. This genotyping scheme separates five genotype clades, termed 1 to 5, and two provisional genotypes (VI and VII) (5). In this context, “genotype” refers to particular alleles at specified loci; a “clade” signifies a single branch with at least two leaves on the phylogenetic tree.
In the present study, we analyzed the genomes of 19 German VZV field isolates and 2 Oka-derived strains. These strains represent the known clades 1, 2, 3, and 5 and two novel genotypes. SNP analysis indicates that both novel genotypes are distinct from the provisional genotypes VI and VII.
MATERIALS AND METHODS
VZV strains and cell culture.
Nineteen VZV strains were isolated from clinical samples obtained from varicella or zoster patients; two strains were derivatives of the vOka and pOka strains received from the Department of Virology, Charité, Berlin, Germany. The patient isolates were obtained after 5 to 10 passages. Virus isolation and propagation were conducted in human embryonic lung fibroblast (HELF) cell cultures from vesicle fluid or skin biopsy specimens (46). Before sequencing, both the vOka and pOka strains were propagated for six passages after receipt. The viruses are described in Table 1, which indicates the genotype as well as the virus origin, sampling date, and age, gender, and immune status of the patients. Cells were maintained in Eagle's minimum essential medium (EMEM) supplemented with 25 mM HEPES, 5% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate.
Table 1.
VZV strains
| Clade | Strain designationa | Internal lab designation | Sampling place | Sampling date | Strain Origin | Patient information (gender, age [yr], immune statusc) | GenBank accession. no. |
|---|---|---|---|---|---|---|---|
| 1 | VZVi/Erfurt.GER/12.00/Z[1] | 243/2000 | Erfurt | 23 Mar. 2000 | Zoster | m, 31 | JN704690 |
| 1 | VZVi/Jena.GER/28.04/Z[1] | 1256/2004 | Jena | 8 July 2004 | Zoster | f, 87 | JN704691 |
| 1 | VZVi/Jena.GER/14.05/V[1] | 551/2005 | Jena | 5 April 2005 | Varicella | m, 2 | JN704692 |
| 1 | VZVi/Weimar.GER/16.05/V[1] | 667/2005 | Weimar | 22 April 2005 | Varicella | f, 2 | JN704693 |
| 1 | VZVi/Weimar.GER/45.07/V[1] | 1883/2007 | Weimar | 7 Nov. 2007 | Varicella | f, 4 | JN704694 |
| 1 | VZVi/Jena.GER/06.08/Z[1] | 432/2008 | Jena | 7 Feb. 2008 | Zoster | m, 57 | JN704695 |
| 1 | VZVi/Berlin.GER/14.08/V[1] | 925/2008 | Berlin | 1 Apr. 2008 | Varicella | f, 5 | JN704696 |
| 2 | Not applicable | 1002/2008 | 9 Apr. 2008b | vOka (Varilrix) | JN704697 | ||
| 2 | Not applicable | 1003/2008 | 9 Apr. 20082 | pOka | JN704698 | ||
| 3 | VZVi/Jena.GER/47.03/V[3] | 2308/2003 | Jena | 22 Nov. 2003 | Varicella | f, 5, immunocompr. | JN704699 |
| 3 | VZVi/Erfurt.GER/01.05/V[3] | 3/2005 | Erfurt | 3 Jan. 2005 | Varicella | m, 3 | JN704700 |
| 3 | VZVi/Jena.GER/02.07/Z[3] | 52/2007 | Jena | 8 Jan. 2007 | Zoster | m, 18, immunocompr. | JN704701 |
| 3 | VZVi/Heidelberg.GER/10.07/V[3] | 405/2007 | Heidelberg | 8 Mar. 2007 | Varicella | m, 26 | JN704702 |
| 3 | VZVi/Munich.GER/30.07/Z[3] | 1219/2007 | Munich | 26 July 2007 | Zoster | f, 22, immunocompr. | JN704703 |
| 5 | VZVi/Erfurt.GER/20.00/V[5] | 413/2000 | Erfurt | 18 May 2000 | Varicella | m, 2 | JN704704 |
| 5 | VZVi/Erfurt.GER/19.04/V[5] | 875/2004 | Erfurt | 7 May 2004 | Varicella | m, 2 | JN704705 |
| 5 | VZVi/Weimar.GER/04.05/V[5] | 134/2005 | Weimar | 24 Jan. 2005 | Varicella | m, 4 | JN704706 |
| 5 | VZVi/Erfurt.GER/11.07/V[5] | 446/2007 | Erfurt | 15 Mar. 2007 | Varicella | m, 1 | JN704707 |
| 5 | VZVi/Erfurt.GER/44.07/V[5] | 1805/2007 | Erfurt | 30 Oct. 2007 | Varicella | f, 8 | JN704708 |
| VIII | VZVi/Jena.GER/37.05/V[VIII] | 1483/2005 | Jena | 17 Sept. 2005 | Varicella | m, 5 | JN704709 |
| IX | VZVi/Jena.GER/06.08(2)/V[IX] | 457/2008 | Jena | 8 Feb. 2008 | Varicella | f, 5 | JN704710 |
Genotyping.
All strains were typed using a modified SNP method based on genotyping schemes described by Barrett-Muir et al. (4) and Loparev and coworkers (24, 25). Open reading frames (ORFs) 1, 21, 22, 50, 54, and 60 were amplified and sequenced as described previously (44, 48).
DNA preparation and genome sequencing.
Prior to DNA preparation, VZV virions were sedimented by ultracentrifugation (125,000 × g, 2 h, 4°C) from the supernatants of virus-infected HELF and the lysates of virus-infected cells after freezing and thawing three times. DNA was prepared from sedimented virus using the QIAamp blood kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
Multiplex sequencing was performed on a genome sequencer, GS FLX (Roche Diagnostics). For bar coding of VZV DNA fragments, about 10 μg of each virus strain DNA was separately fragmented for 1 min at 3 × 105 Pa N2 by nebulizers (part of the GS DNA library preparation kit; Roche Diagnostics). The nebulized DNAs were purified by using MinElute columns (Qiagen, Hilden, Germany) and eluted in 20 μl Tris-EDTA (TE) buffer each. Blunt-end repair, ligation of the heptamer bar coding adaptors to the fragments, and all other steps prior to preparation of the 454 sequencing library were carried out as described by Meyer et al. (30) using 200 to 500 ng of each nebulized DNA as starting material (Quant-IT PicoGreen ds DNA assay; Invitrogen).
The appropriate oligonucleotides (desalted) were purchased at Metabion (Martinsried, Germany) and consist of the bar code (3′ end) and its reverse complement (5′ end), separated by the SrfI recognition site, 5′-GCCCGGGC-3′. After careful quantification of the bar-coded fragments by the PicoGreen assay, ∼100 ng of each DNA was pooled, and the pool was allowed to proceed to the dephosphorylation step, followed by SrfI digestion and small fragment removal (30). After small-fragment removal, the SrfI-digested fragment pool (in 20 μl elution buffer) was used to prepare the 454 sequencing library, employing the GS Titanium library preparation kit (Roche Diagnostics). Single-stranded 454 sequencing libraries were quantified by a quantitative PCR (qPCR) assay (31) and processed by emulsion PCR, following the manufacturer's instructions. Sequencing was done on two lanes of a 70 by 75 GS Titanium picotiter plate of a Roche GS FLX sequencer. For bar code identification and separation of sequences, the reads were sorted according to their bar-coding adaptors using pts-untag software (Uwe Stenzel; http://bioinf.eva.mpg.de/pts/) in combination with the 454 command-line tools included in the GS FLX software package and in-house Perl scripts. Only adapter sequences with a full match were accepted, whereas the recognition of the SrfI site had to be flexible due to sequencing errors and homopolymers.
The sequence data were assembled using VZV strain Dumas (GenBank accession no. NC_001348) as a reference. For the closing of the remaining gaps, the respective DNA fragments were amplified with specific primer pairs. PCR products were gel purified and sequenced with the cycle sequencing method (54, 56) using the CEQ DTCS Quick Start kit and the CEQ8000 genetic analysis system (Beckman Coulter, Krefeld, Germany). The procedure followed the previously described protocol (48). Sequences of the PCR products were obtained from both DNA strands.
Genetic analysis of sequences.
Two types of VZV data sets were analyzed. One data set (A) comprised 42 complete genomes of VZV. This data set included several pOka-derived clade 2 strains. The second data set (B) comprised sequences of 37 independent VZV isolates without pOka-derived strains. VZV sequences retrieved from GenBank are those for vOka (accession no. AB097932), pOka (AB097933), HJ0 (AJ871403), MSP (AY548170), BC (AY548171), VarilRix (DQ008354), VariVax (DQ008355), DR (DQ452050), CA123 (DQ457052), SD (DQ479953), Kel (DQ479954), strain 11 (DQ479955), strain 22 (DQ479956), 03-500 (DQ479957), strain 36 (DQ479958), strain 49 (DQ479959), strain 8 (DQ479960), strain 32, passage 5 (DQ479961), NH29_3 (DQ674250), SVETA (EU154348), and Dumas (NC_001348).
For phylogenetic analyses, sequence information determined in this study and sequence data retrieved from GenBank were used. Alignments and phylogenetic analyses were conducted with the MEGA5 software program (59). For maximum-likelihood (ML) tree inference, gaps or missing data were completely deleted, the “nearest-neighbor-interchange” (NNI) option was selected, and the initial tree was made automatically. Bootstrap analyses were performed with 1,000 replications. The optimal substitution model was selected on the basis of the Bayesian information criterion (BIC) and the corrected Akaike information criterion (AICc) using a model test implemented in MEGA5. If gamma-distributed rates among sites were proposed, five discrete gamma categories were selected.
Divergence times within the varicelloviruses were inferred using Bayesian Metropolis-coupled Markov chain (MCMC) analysis and a relaxed clock (uncorrelated log normal) as implemented in the BEAST 1.4.7 software program (11). Again, the optimal substitution model was determined with MEGA5, and the data set was analyzed using the general time-reversible substitution model assuming gamma distribution and invariable sites (GTR+G+I). A chain length of 20 million generations was analyzed. Log parameters were sampled every 1,000 generations; burn-in was set to 25%. A maximum-credibility tree was constructed using the software program FigTree 1.1.2 (11). Posterior probabilities of nodes and node heights were displayed.
The phylogenetic network of the VZV sequences was computed with the Splitstree4 software program (17). Recombination of VZV was analyzed using the Recombination Detection Program (RDP3), v.3.44, which implements seven distinct methods, i.e., the original RDP method, Bootscan, Geneconv, MaxChi, Chimaera, SiScan, and 3Seq (16). For these analyses, sets A and B were analyzed using default settings.
Bootstrap analyses were performed with the software program SimPlot v3.5.1 (22). The following parameters were chosen throughout: neighbor-joining method, Kimura 2-parameter model, transition/transversion (T/t) ratio of 2. Complete VZV genomes (set A) were investigated using composites of four aligned sequences. Due to the large size of VZV genomes, 100 replications were performed in bootstrap analyses; the window size was set to 3,000 bp, which was moved in steps of 1,000 bp along the genome. Variable sites of the VZV genome (set B) were analyzed using a window size of 60 nucleotides (nt) and a step size of 10 nt; bootstrapping was done with 1,000 replications.
The evolutionary divergence between VZV clades was estimated as the number of differences and the number of differences per site obtained in pairwise comparisons. A bootstrapping procedure (1,000 replications) was used for variance estimation. The evolutionary divergence was calculated using MEGA5 (59).
Nucleotide sequence accession numbers.
The sequences were deposited in GenBank; accession numbers are presented in Table 1.
RESULTS
Genotyping of VZV field strains.
For a preliminary characterization of the VZV isolates, all strains were genotyped as a first step. Table 2 demonstrates that 11 SNPs of six genes (ORFs 1, 21, 22, 50, 54, and 60) are sufficient to distinguish seven genotypes. However, it was observed that both clade 4 genomes exhibited a G residue at SNP 38019 (ORF 22), whereas the proposal of Breuer et al. (5) postulates a characteristic A residue (Table 2). The data indicate further variations at positions 23294 (strain 3/2005), 24533 (strains 875/2004, 134/2005, and 1805/2007), and 113243 (strains 11 and 405/2007). Analysis of variable sites adjacent to these SNPs revealed additional deviant nucleotides, suggesting recombination events (data not shown).
Table 2.
Selected SNPs
| GenBank accession no. and/or strain | Clade | Nucleotide for indicated SNP position at ORFa: |
|||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
5 |
6 |
12 |
16 | 17 | 21 |
22 |
35 | 37 |
50 | 54 | 55 | 56 | 60 | 62 |
66 | 113243 | ||||||||||||||||||||
| 508* | 685 | 790* | 4402 | 4644 | 5827 | 6850 | 17834 | 18082 | 23294 | 24533 | 24578 | 33725* | 37902* | 38019* | 38055* | 38081* | 38177* | 39394 | 64703 | 66112 | 66288 | 66646 | 66688 | 66880 | 68101 | 68172 | 68254 | 87841* | 95241* | 98437 | 98825 | 101464* | 105429 | 107026 | |||
| NC_001348, Dumas | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| 243/2000 | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| 432/2008 | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| 551/2005 | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| 667/2005 | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| 925/2008 | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| 1256/2004 | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| DQ479961, strain 32, passage 5 | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| DQ479953, SD | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| AY548171, BC | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| AY548170, MSP | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| EU154348, SVETA | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| DQ674250, NH29_3 | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| DQ479959, strain 49 | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| DQ479958, strain 36 | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| DQ479954, Kel | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| 1883/2007 | 1 | C | G | T | G | C | C | G | C | C | A | A | A | T | A | G | T | A | G | G | G | T | G | G | A | A | T | G | C | C | T | T | T | C | A | A | A |
| AB097933, pOka | 2 | C | A | C | G | C | C | G | T | T | G | A | G | C | G | G | C | C | A | A | G | T | G | T | A | A | T | A | C | T | C | C | T | C | A | A | A |
| AB097932, vOka | 2 | C | A | C | G | C | C | G | T | T | G | A | G | C | G | G | C | C | A | A | G | T | G | T | A | A | T | A | C | T | C | C | T | C | A | A | A |
| DQ008354, VarilRix | 2 | C | A | C | G | C | C | G | T | T | G | A | G | C | G | G | C | C | A | A | G | T | G | T | A | A | T | A | C | T | C | C | T | C | A | A | A |
| DQ008355, VariVax | 2 | C | A | C | G | C | C | G | T | T | G | A | G | C | G | G | C | C | A | A | G | T | G | T | A | A | T | A | C | T | C | C | T | C | A | A | A |
| 1002/2008 | 2 | C | A | C | G | C | C | G | T | T | G | A | G | C | G | G | C | C | A | A | G | T | G | T | A | A | T | A | C | T | C | C | T | C | A | A | A |
| 1003/2008 | 2 | C | A | C | G | C | C | G | T | T | G | A | G | C | G | G | C | C | A | A | G | T | G | T | A | A | T | A | C | T | C | C | T | C | A | A | A |
| DQ479955, strain 11 | 3 | C | G | T | A | C | A | G | C | C | G | A | G | C | A | G | T | A | G | A | G | G | A | G | A | A | C | A | T | T | T | T | T | C | A | G | A |
| DQ479956, strain 22 | 3 | C | G | T | A | C | A | G | C | C | G | A | G | C | A | G | T | A | G | A | G | G | A | G | A | A | C | A | T | T | T | T | T | C | A | G | C |
| AJ871403, HJ0 | 3 | C | G | T | A | C | A | G | C | C | G | A | G | C | A | G | T | A | G | A | G | G | A | G | A | A | C | A | T | T | T | T | T | C | A | G | C |
| DQ479957, 03–500 | 3 | C | G | T | A | C | A | G | C | C | G | A | G | C | A | G | T | A | G | A | G | G | A | G | A | A | C | A | T | T | T | T | T | C | A | G | C |
| 2308/2003 | 3 | C | G | T | G | C | A | G | C | C | G | A | G | C | A | G | T | A | G | A | G | G | G | G | A | A | C | A | T | T | T | T | T | C | A | A | C |
| 52/2007 | 3 | C | G | T | G | C | A | G | C | C | G | A | G | C | A | G | T | A | G | A | G | G | A | G | A | A | C | A | T | T | T | T | T | C | A | A | C |
| 3/2005 | 3a | C | G | T | G | T | A | G | C | C | A | A | G | C | A | G | T | A | G | A | G | G | G | G | A | G | C | A | T | T | T | T | T | C | G | A | C |
| 405/2007 | 3a | C | G | T | G | T | A | G | C | C | G | A | G | C | A | G | T | A | G | A | G | G | G | G | A | G | C | A | T | T | T | T | T | C | G | A | A |
| 1219/2007 | 3a | C | G | T | G | T | A | G | C | C | G | A | G | C | A | G | T | A | G | A | G | G | G | G | A | G | C | A | T | T | T | T | T | C | G | A | C |
| DQ479960, strain 8 | 4 | C | A | C | G | C | C | A | T | C | A | A | A | C | A | G | C | C | A | G | A | T | G | G | A | A | T | A | C | T | C | C | C | C | A | A | A |
| DQ452050, DR | 4 | C | A | C | G | C | C | A | T | C | A | A | A | C | A | G | C | C | A | G | A | T | G | G | A | A | T | A | C | T | C | C | C | C | A | A | A |
| DQ457052, CA123 | 5 | C | G | C | G | C | C | A | C | C | G | C | G | C | A | G | T | C | G | A | A | T | G | G | A | A | T | A | C | T | C | T | C | A | A | A | C |
| 134/2005 | 5 | C | G | C | G | C | C | A | C | C | G | A | G | C | A | G | T | C | G | A | A | T | G | G | A | A | T | A | C | T | C | T | C | A | A | A | C |
| 413/2000 | 5 | C | G | C | G | C | C | A | C | C | G | C | G | C | A | G | T | C | G | A | A | T | G | G | A | A | T | A | C | T | C | T | C | A | A | A | C |
| 446/2007 | 5 | C | G | C | G | C | C | A | C | C | G | C | G | C | A | G | T | C | G | A | A | T | G | G | A | A | T | A | C | T | C | T | C | A | A | A | C |
| 875/2004 | 5 | C | G | C | G | C | C | A | C | C | G | A | G | C | A | G | T | C | G | A | A | T | G | G | A | A | T | A | C | T | C | T | C | A | A | A | C |
| 1805/2007 | 5 | C | G | C | G | C | C | A | C | C | G | A | G | C | A | G | T | C | G | A | A | T | G | G | A | A | T | A | C | T | C | T | C | A | A | A | C |
| 1483/2005 | VIII | C | A | C | G | C | A | G | T | C | G | A | G | C | A | G | T | C | A | A | G | G | G | G | A | A | T | A | T | T | T | C | T | C | A | A | A |
| 457/2008 | IX | C | G | C | G | C | A | G | C | C | A | A | A | C | A | G | C | A | G | G | G | G | G | G | A | A | T | A | C | T | C | C | C | C | A | A | C |
Sites marked with “∗” were used for genotyping in this study.
Genotyping revealed that the present set of VZV field isolates includes seven strains that belong to clade 1, five strains of clade 3, and five strains of clade 5 (Table 1). Three of the five clade 3 strains have additional SNPs characteristic of subclade 3a (formerly E2a), which was previously proposed (51). Clades 2 and 4 were not detected. Two strains, however, 1483/2005 and 457/2008, exhibited unique features that did not fit any of the known genotypes shown in Table 2. Strain 1483/2005 deviates from the present scheme since it combines features of clades 2 (SNPs of ORFs 1 and 21) and VI (SNPs of ORFs 22, 50, 54, and 60), whereas isolate 457/2008 has a characteristic SNP pattern in ORF 22. Since our virus collection does not comprise clade 2 field isolates, one vOka-derived strain (1002/2008) and one pOka-derived strain (1003/2008) were also included to demonstrate the robustness of our genotyping scheme. Despite the differences of certain SNPs, genotyping suggested two novel VZV lineages, termed VIII and IX.
Sequencing of VZV genomes.
The VZV genomes were sequenced by the 454 technology. Two runs were performed on a Roche GS FLX genome sequencer. In a first run, DNA of VZV strain 52/2007 was sequenced on six lanes of a picotiter plate using the GS FLX chemistry to establish the procedure and to assess the applicability of this next-generation-sequencing (NGS) technique to the VZV genome, which may bear difficulties due to several repeats of variable lengths. This yielded 40,443 high-quality reads, which were mapped against the sequence of the VZV reference strain Dumas (GenBank accession no. NC_001348). Some 3,700 mappable reads were obtained and assembled into 20 contigs, with a total length of 115,698 bp (corresponding to 92.6% of the genome). After manual inspection and editing, 98.4% of the VZV genome was covered and 14 gaps with a total length of 1,707 bp remained to be determined with conventional methods.
In the second run, DNA libraries of 20 VZV isolates were prepared, bar coded, and sequenced in a pool on a half-picotiter plate. This run yielded 937,038 high-quality reads (314 Mb; average read length, 335 bp). The results are summarized in Table 3. The mean number of reads per library was 45,893, of which on average 4,441 reads (9.9%) were mappable to the VZV strain Dumas. The number of contigs per virus averaged 7.3, and the mean coverage was 117,240 bp of the VZV genome (corresponding to 99.7% of the UL, US, and IR). The sequence depths ranged from 5.3- to 37.1-fold (mean, 16.3-fold). The remaining gaps per strain differed considerably in number (3 to 22; average, 6.3) and cumulative length (16 to 2,146 bp; average, 286 bp). The average length of all gaps was 33 bp (Table 3). Gaps were most frequently observed in the repeat regions R1 (ORF 11), R2 (ORF 14), and R3 (ORF 22), the internal repeat IRS, and nonrepeat regions of ORF 22, ORF 29, and ORF 31. Most gaps were closed with conventional sequencing methods.
Table 3.
Sequencing of a pool of 20 bar-coded VZV genome libraries
| VZV strain | Total no. of reads | No. of mapped reads | % mapped reads | No. of contigs | Total contig length (bp) | % coverage | Sequence depth (fold) | No. of gaps | Total gap length (bp) | Avg gap length (bp) |
|---|---|---|---|---|---|---|---|---|---|---|
| 243/2000 | 48,752 | 4,332 | 8.9 | 7 | 117,420 | 99.9 | 15.7 | 6 | 134 | 22 |
| 413/2000 | 46,829 | 5,077 | 10.8 | 5 | 117,475 | 99.9 | 18.5 | 4 | 36 | 9 |
| 2308/2003 | 49,586 | 7,952 | 16.0 | 6 | 117,491 | 99.9 | 28.7 | 5 | 63 | 13 |
| 875/2004 | 58,477 | 9,514 | 16.3 | 5 | 117,471 | 99.9 | 34.4 | 4 | 39 | 10 |
| 1256/2004 | 51,853 | 2,517 | 4.9 | 9 | 117,199 | 99.7 | 9.5 | 8 | 324 | 41 |
| 3/2005 | 36,160 | 2,470 | 6.8 | 6 | 117,360 | 99.8 | 9.3 | 5 | 181 | 36 |
| 134/2005 | 39,313 | 8,465 | 21.5 | 5 | 117,469 | 99.9 | 30.3 | 4 | 16 | 4 |
| 551/2005 | 39,720 | 1,923 | 4.8 | 11 | 116,569 | 99.2 | 7.3 | 10 | 917 | 92 |
| 667/2005 | 42,461 | 1,912 | 4.5 | 8 | 117,184 | 99.7 | 7.2 | 7 | 354 | 51 |
| 1483/2005 | 55,072 | 3,399 | 6.2 | 6 | 117,211 | 99.7 | 12.8 | 5 | 290 | 58 |
| 405/2007 | 38,006 | 4,383 | 11.5 | 7 | 117,340 | 99.8 | 15.9 | 6 | 229 | 38 |
| 446/2007 | 38,081 | 10,203 | 26.8 | 4 | 117,482 | 99.9 | 37.1 | 3 | 22 | 7 |
| 1219/2007 | 57,213 | 2,686 | 4.7 | 7 | 117,163 | 99.7 | 11.0 | 6 | 391 | 65 |
| 1805/2007 | 49,646 | 3,708 | 7.5 | 7 | 117,367 | 99.8 | 13.7 | 6 | 151 | 25 |
| 1883/2007 | 51,247 | 3,350 | 6.5 | 6 | 117,474 | 99.9 | 12.4 | 5 | 63 | 13 |
| 432/2008 | 49,857 | 4,096 | 8.2 | 4 | 117,485 | 99.9 | 15.0 | 3 | 61 | 20 |
| 457/2008 | 43,393 | 1,387 | 3.2 | 23 | 115,394 | 98.2 | 5.3 | 22 | 2,146 | 98 |
| 925/2008 | 45,186 | 2,881 | 6.4 | 7 | 117,441 | 99.9 | 10.6 | 6 | 87 | 15 |
| 1002/2008 | 47,557 | 4,761 | 10.0 | 7 | 117,431 | 99.9 | 17.3 | 6 | 97 | 16 |
| 1003/2008 | 29,454 | 3,797 | 12.9 | 6 | 117,382 | 99.8 | 14.0 | 5 | 128 | 26 |
| Mean | 45,893 | 4,440.65 | 9.9 | 7.3 | 117,240 | 99.7 | 16.3 | 6.3 | 286.45 | 33 |
Phylogenetic analysis of VZV.
Twenty-one VZV genome sequences determined in the present study were aligned to 21 VZV genomes obtained from GenBank. Two complete genomes of highly passaged variants of strain 32 (accession no. DQ479962 and DQ479963) were excluded, since these sequences were derived from a single strain (38). For further analyses, gaps and reiterated regions of ORFs 11, 14, 22, and the IRS/TRS (R1 to R5) were deleted, resulting in an alignment with 123,321 positions. This data set A revealed 942 variable sites and, after removal of the singletons, 420 parsimony informative (Pi) sites. Pairwise estimation of divergence revealed values of between 2 and 305 differences among any two sequences and a mean number of differences of 150.6 (standard error [SE], 4.4). The average p-distance (proportion of nucleotide sites at which two compared sequences are different) was 1.1215 × 10−3 (SE, 3.2 × 10−5). Since all clade 2 strains were derived from the pOka strain, those sequences were also deleted, yielding a set of 37 sequences (123,370 positions) with 906 variable sites and 344 parsimony informative sites (data set B).
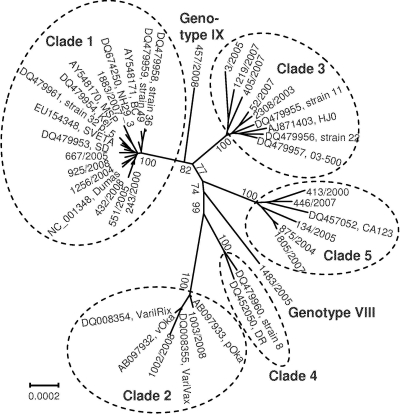
The phylogenetic analysis of data set A was conducted with two statistical methods, the maximum-likelihood (ML) and neighbor-joining (NJ) methods; both approaches yielded virtually identical results (data not shown). The ML bootstrap consensus tree shown in Fig. 1 reveals five clades that have been described previously (5) and two novel distinct branches, which were provisionally termed genotypes VIII (VZV 1483/2005) and IX (VZV 457/2008). In this analysis, sequences of the provisional genotypes VI and VII were not included since no complete sequences of these strains are available. Branch lengths of all clades indicate a considerable genetic divergence. Another ML tree with data set B (345 parsimony informative sites) revealed an essentially identical tree topology (data not shown). However, due to the deletion of the high-passage clade 2 strains and the removal of the singletons, branch lengths of clades 2, VIII, and IX appear to be short. In both trees, clades 1, 3, and IX on the one hand and clades 2 and 4 on the other hand cluster together.
Fig 1.
Phylogenetic analysis of VZV. The maximum-likelihood tree of 42 aligned complete VZV genomes shows five established clusters (clades 1 to 5) and two novel genotypes. Strain designations, GenBank accession numbers, and clade designations are presented. The bar indicates substitutions per site. Numbers at major nodes indicate bootstrap values obtained after 1,000 replications.
Genetic distances (number of base differences per site from averaging over all sequence pairs) within and between the seven clades were calculated to substantiate the finding of two novel VZV genotypes (Table 4). For the novel lineages VIII and IX, the mean number of substitutions per site obtained in intertypic pairwise sequence comparisons ranges from 9.82 × 10−4 to 1.722 × 10−3, which is comparable to the values of the established clades 1 to 5, i.e., 1.907 × 10−3 to 1.81 × 10−3 but 2- to 5-fold higher than the genetic distances characteristic of intratypic comparisons (2.87 × 10−4 to 5.79 × 10−4). Figure 1 and Table 4 show a low intratypic sequence heterogeneity of clades 1 and 2 and a higher heterogeneity of clades 3, 4, and 5. This is reflected by longer branch lengths and roughly doubled genetic distances for the latter ones (Table 4). Both the phylogenetic tree and the genetic distances support the establishment of novel clades, even though the analysis of additional strains may be required to confirm their existence.
Table 4.
Genetic distances
| Clade | Distance (SE) within or between clades |
||||||
|---|---|---|---|---|---|---|---|
| Clade 1 | Clade 2 | Clade 3 | Clade 4 | Clade 5 | Clade VIII | Clade IX | |
| Clade 1 | 2.870 × 10−4 (1.7 × 10−5) | ||||||
| Clade 2 | 1.706 × 10−3 (8.1 × 10−5) | 2.90 × 10−4 (2.3 × 10−5) | |||||
| Clade 3 | 1.198 × 10−3 (5.2 × 10−5) | 1.191 × 10−3 (8 × 10−5) | 5.790 × 10−4 (2.6 × 10−5) | ||||
| Clade 4 | 1.330 × 10−3 (9.5 × 10−5) | 1.358 × 10−3 (6.6 × 10−5) | 1.793 × 10−3 (7.1 × 10−5) | 4.920 × 10−4 (5.6 × 10−5) | |||
| Clade 5 | 1.494 × 10−3 (3 × 10−5) | 1.794 × 10−3 (7.8 × 10−5) | 1.348 × 10−3 (4.9 × 10−5) | 1.445 × 10−3 (6.2 × 10−5) | 5.640 × 10−4 (4.3 × 10−5) | ||
| Clade VIII | 1.508 × 10−3 (6 × 10−5) | 1.684 × 10−3 (6.2 × 10−5) | 1.410 × 10−3 (9.6 × 10−5) | 1.407 × 10−3 (5.5 × 10−5) | 1.538 × 10−3 (5.4 × 10−5) | NAa | |
| Clade IX | 9.820 × 10−4 (4.9 × 10−5) | 1.722 × 10−3 (8 × 10−5) | 1.242 × 10−3 (7.2 × 10−5) | 1.317 × 10−3 (5.9 × 10−5) | 1.431 × 10−3 (6.7 × 10−5) | 1.486 × 10−3 (6.3 × 10−5) | NA |
NA, not applicable.
Recombination of VZV.
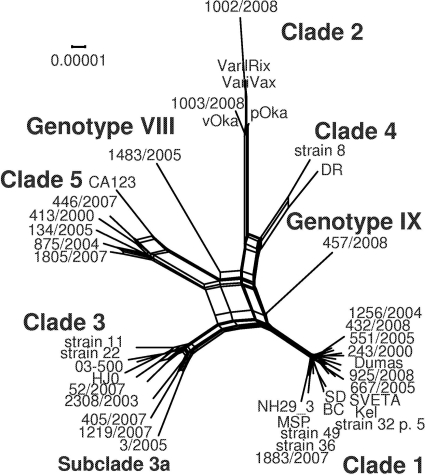
Previous studies have suggested recombination as one driver of VZV evolution (10, 27, 36, 37, 38). Since recombination events may lead to conflicting tree topologies in phylogenetic studies, a splits analysis using the SplitsTree4 software program was performed. The resultant phylogenetic network, based on 42 complete VZV genomes, is shown in Fig. 2. Numerous parallel edges (splits) between the sequence clusters indicate possible recombination events in the evolution of the clades.
Fig 2.
Splits analysis of 42 complete VZV genomes. The edges (splits) of the phylogenetic network reveal possible recombination events. The network resembles those presented by Norberg (36) and Schmidt-Chanasit and Sauerbrei (53).
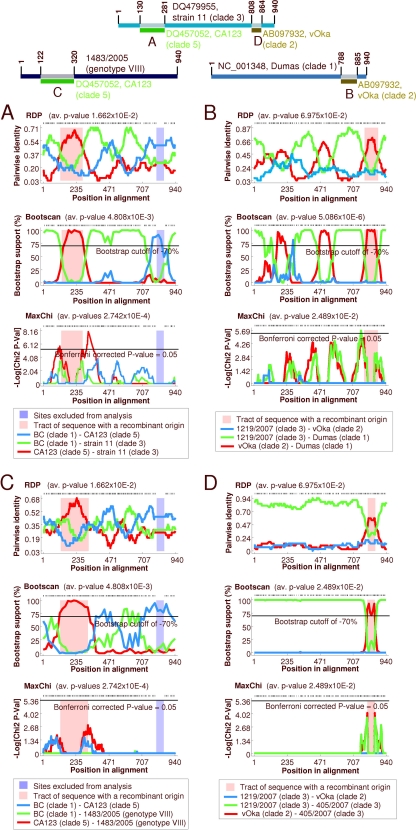
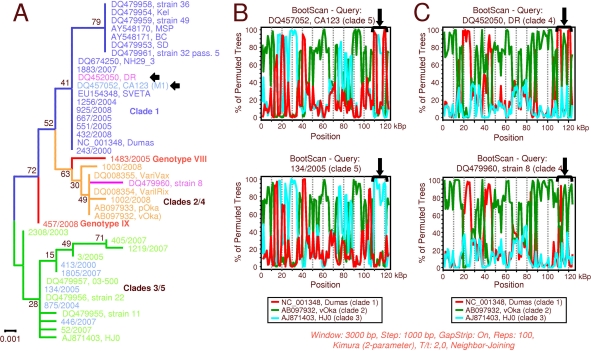
For a further characterization, recombination detection analyses were performed using data set A (942 variable sites). The recombination detection program RDP3 proposed several recombination events. Among these events are the following: (i) the replacement of the clade 3 R1-R3 region by clade 5 sequences (Fig. 3A), (ii) the transposition of the clade 2 US (donor) with the respective gene region of a clade 1 ancestor (acceptor) (Fig. 3B), and (iii) the exchange of ORFs 25 to 35 of genotype VIII with clade 5 sequences (Fig. 3C). In addition, two of nine clade 3 sequences (strains 11 and 405/2007) exhibit exchange of the US region by clade 2 US (Fig. 3D).
Fig 3.
Recombination in VZV as analyzed with RDP3. Analysis was performed with data set A, comprising the 942 variable sites of 42 aligned VZV genomes. The upper panel shows acceptor genomes and the proposed exchanged regions. The results of three of seven recombination detection methods are presented in panels A to D. (A) Proposed replacement of the R1-R3 region of 9 clade 3 strains with clade 5 sequences. (B) Proposed exchange of the US region of 17 investigated clade 1 genomes by clade 2 sequences. (C) Proposed exchange of ORFs 25 to 35 of genotype VIII by clade 5 sequences. (D) Proposed exchange of the US region of strains 11 and 405/2000.
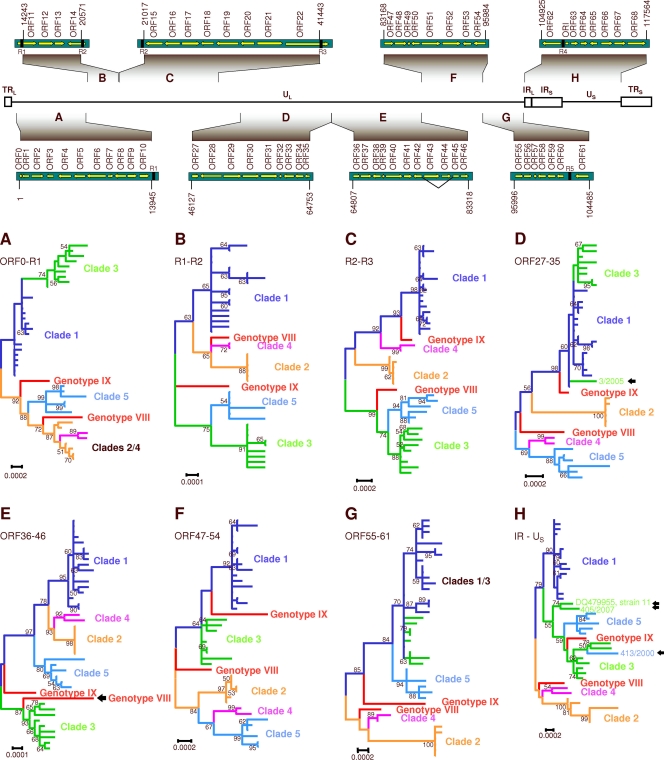
For independent support of the recombination events proposed by RDP3, phylogenetic analyses of 8 distinct subgenomic regions of the VZV genome were performed; these regions were as follows: (i) nucleotides 1 to 13945 (numbering related to the Dumas strain; NC_001348), including HHVgp01 and ORFs 1 to 10, (ii) nucleotides 14243 to 20571 (R1-R2; ORFs 11 to 14), (iii) nucleotides 21017 to 41443 (R2-R3; ORFs 15 to 22), (iv) nucleotides 46127 to 64753 (ORFs 27 to 35), (v) nucleotides 64807 to 83318 (ORFs 36 to 46), (vi) nucleotides 83168 to 95984 (ORFs 47 to 54), (vii) nucleotides 95996 to 104485 (ORFs 55 to 61), and (viii) nucleotides 104925 to 117564 (IR-US, including R4, the origin of replication, and ORFs 62 to 68). Phylogenetic trees of these 8 regions were inferred using the ML method and optimal substitution models and rates among sites. An overview of the trees is presented in Fig. 4. In these phylogenetic trees, recombination events are indicated by divergent clustering of a given strain in relation to the other strains of its clade. The following isolates exhibit aberrant clustering of their IR-US region: strains CA123 and 413/2000 (both clade 5) and the clade 3 strains 11 and 405/2007 (Fig. 4H). The recombinant region of the respective strains was mapped when analyzing the origin of replication (nucleotides 109908 to 110580) and the US region (nucleotides 112332 to 117679) in two additional computations. The phylogenetic tree of the ori region and the related bootscan analyses are presented in Fig. 5. In this tree, CA123 clusters together with the clade 1 strains (Fig. 5A); the corresponding bootscan plot confirms a significant similarity of the ori regions with those of clade 1 (Fig. 5B). Two parsimony informative (Pi) sites of each ori region characteristic of clade 1 are detectable, and 8 Pi sites which were shared with clade 3 disappear (data not shown). Surprisingly, strain DR also clusters with clade 1 (Fig. 5A), which is not evident from the IR-US tree (Fig. 4H). This observation is also supported by the bootstrap analysis: two peaks appear in the plot, representing the ori regions of IRS and TRS (Fig. 5C). Here, DR and Dumas share two Pi sites within the ori region (data not shown).
Fig 4.
Phylogenetic analysis of subgenomic regions. The upper panel shows schematic organization of the VZV genome, showing unique (UL and US) and repeated terminal and internal (TR and IR) sequences. Nucleotide positions and genes of the analyzed fragments are presented. The diagrams are not drawn to scale. Lower panels: overview of maximum-likelihood trees computed with MEGA5. Each established clade is indicated in a different color; proposed clades are indicated in red. Different clustering of certain strains along the genome suggests recombination events (indicated with arrows). (A) Fragment TR-R1. (B) Fragment R1-R2. (C) Fragment R2-R3 . (D) Fragment ORF27-35 . (E) Fragment ORF36-46. (F) Fragment ORF47-54. (G) Fragment ORF55-61. (H) IR-US. Scale bars indicate substitutions per site. Numbers at major nodes indicate bootstrap values obtained after 1,000 replications.
Fig 5.
Phylogenetic analysis and bootstrap analyses of the VZV ori region (nucleotides 109908 to 110580). (A) Maximum-likelihood tree computed using MEGA5. Numbers at nodes indicate bootstrap values obtained after 1,000 replications. Scale bar indicates substitutions per site. (B) Bootstrap analyses using strains CA123 (recombinant, upper panel) and 134/2005 (lower panel) as query sequences. The bracket indicates the region of interest. (C) Bootstrap analyses using strain DR (recombinant, upper panel) and strain 8 (lower panel) as query sequences. The bracket indicates the region of interest.
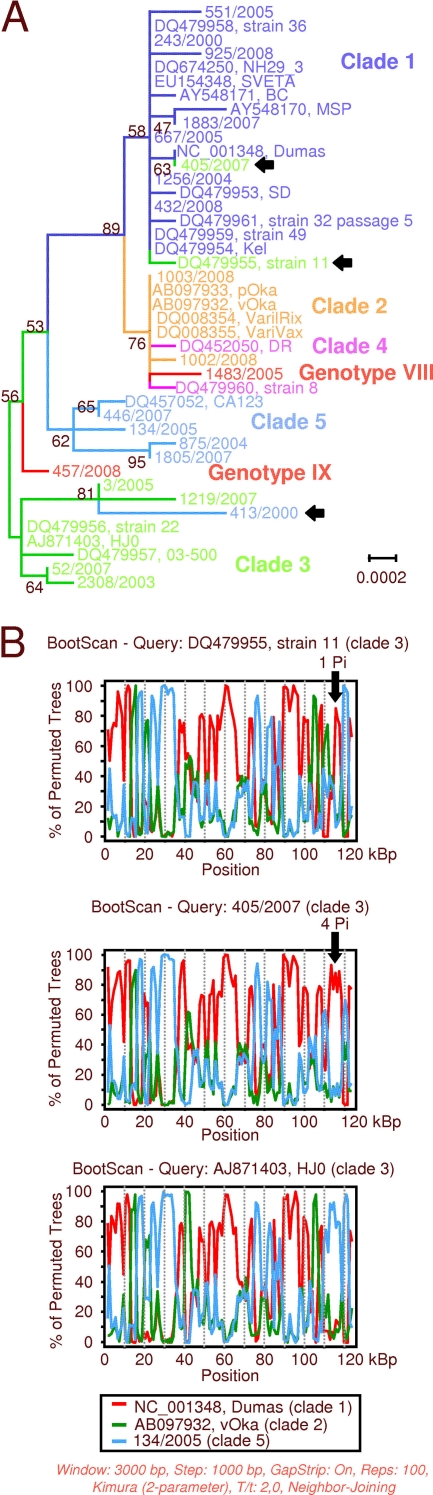
Analysis of the US region reveals three recombinant strains. Strains 11 and 405/2007 (both clade 3) but none of the other clade 3 strains share a striking similarity to clade 1 (Fig. 6A; see also 4H). The corresponding bootscan plots show different exchanged regions of strains 11 and 405/2007 (Fig. 6B). This finding is supported by an analysis of Pi sites, which reveals that the respective genome region of the Dumas strain shares one Pi site with strain 11 but four with strain 405/2007. The third strain with a recombinant US region is 413/2000 (clade 5), which clusters with clade 3 (Fig. 6A and 4H). Due to the general similarity of this genome region among clade 3 and clade 5 strains, the bootscan analysis is a tool unsuited for demonstrating this finding graphically. However, analysis of the Pi sites reveals 9 and 10 additional sites when 413/2000 is compared to 3/2005 and 1219/2007, respectively; comparison to other clade 3 strains reveals two additional Pi sites (data not shown).
Fig 6.
Phylogenetic analysis and bootstrap analyses of the VZV US region (nucleotides 112332 to 117679). (A) Maximum-likelihood tree computed with MEGA5. Numbers at nodes indicate bootstrap value obtained after 1,000 replications. Scale bar indicates substitutions per site. Arrows indicate proposed recombinant strains. (B) Bootstrap analyses using strains 11 (recombinant, upper panel), strain 405/2007 (recombinant, middle panel), and strain HJ0 (lower panel) for query sequences. The arrows indicate the region of interest. The numbers of parsimony informative (Pi) sites supporting the bootstrap scan are given.
Beside single strains with unexpected sequence homologies to other clades, the phylogenetic analyses indicate recombination events in early stages of phylogeny: ORFs 14 to 17 (nucleotides 19431 to 25516), ORFs 18 to 21 (nucleotides 25573 to 33875), and ORFs 22 to 26 (nucleotides 34083 to 46263) were analyzed, since a previous study suggested recombination among clades 1 and 2 (38). As shown in Fig. 7, the previous results could be verified: the phylogenetic trees as well as the bootstrap analysis support recombination of a clade 1 ancestor (donor) with a clade 2 ancestor (acceptor) to yield clade 4 (recombinant). In addition, ORFs 27 to 35 and 47 to 54 of clade 4 show a high similarity to clade 5 (Fig. 4D and F). Accordingly, the bootstrap analysis with SimPlot identifies genotypes 1, 2, and 5 as parental viruses in the evolution of clade 4 (Fig. 7D and E). Since clade 4 is linked to the novel genotype IX, it is likely that genotype IX might have received a considerable portion of its genome from clade 4.
Fig 7.
Recombination events in early stages of VZV phylogeny. (A) Phylogenetic tree of the gene region presenting ORFs 14 to 17. (B) Phylogenetic tree of the gene region presenting ORFs 18 to 21. (C) Phylogenetic tree of the gene region presenting ORFs 22 to 26. All trees are maximum-likelihood trees computed with MEGA5. Scale bars indicate substitutions per site. Clustering of clades 4 and IX with clade 1 (A and C) or clade 2 (B) indicates recombination. Likewise, clustering of genotype VIII with clades 3/5 (A and B) or clade 2 (C) indicates further recombination events. (D) Bootstrap scanning using clade 4 (query sequence) and clades 1, 2, and 3 confirms the results of the phylogenetic trees. Data set B (905 variable sites) was used for this analysis. (E) Bootstrap scanning using clade 4 (query sequence) and clades 2, 3, and 5 indicates further recombination events in the history of clade 4. Data set B (906 variable sites) was used for this analysis.
In general, phylogenetic analyses of different genome fragments and bootstrap analyses indicate numerous inconsistencies that are a strong hint of previous recombination events in the evolution of these clades.
Estimation of divergence times within the Alphaherpesvirinae.
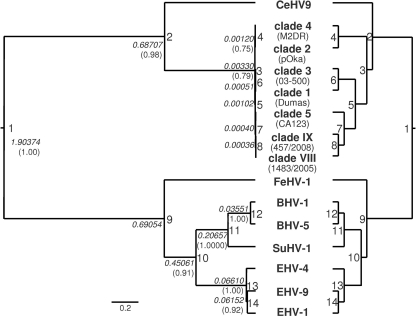
Since herpesviruses are thought to have evolved with their hosts (28, 29), the divergence times of seven VZV genotypes and other members of the genus Varicellovirus were estimated using the BEAST 1.4.7 program. For this estimation, the polymerase gene (ORF 28) and the thymidine kinase gene (TK) (ORF 36) of one representative strain of each VZV clade (Dumas [accession no. NC_001348], pOka [AB097932], 05-300 [DQ479957], DR [DQ452050], 446/2007, 1483/2005, and 457/2008) were included, as well as the respective genes of cercopithecine herpesvirus 9 (CeHV-9) (AF275348), bovine herpesviruses 1 and 5 (BHV-1 and -5) (NC_001847 and NC_005261), feline herpesvirus 1 (FeHV-1) (FJ478159), suid herpesvirus 1 (SuHV-1) (NC_006151), and equine herpesviruses 1, 4, and 9 (EHV-1, -4, and -9) (AY464052, AF030027, and NC_011644). The phylogenetic tree (Fig. 8) inferred using Bayesian statistics revealed two branches, one comprising the herpesviruses of primates (human and vervet) and the other including sequences of the herpesviruses infecting species of Carnivora (cat), Artiodactyla (cow and pig), and Perissodactyla (horse). Branching of FeHV, EHV, and BHV/SuHV reflects some published molecular trees of Mammalia (e.g., see references 21, 26, 34, and 61). For a calibration of the phylogenetic tree, the split between CeHV-9 and the VZV clades was dated 23 million years before the present, which is in line with the estimated time of separation of their hosts, the Old World monkeys (Cercopithecidae) and the Hominoidea (15, 18). Divergence times were calculated from the node heights (Table 5). According to the external calibration, VZV radiation started some 110,000 years before the present and was completed 12,000 years ago. On the basis of the evolutionary divergence (p-distance) of the aligned TK/pol sequences of VZV and CeHV-9 and the assumed split of Old World monkeys and the Hominoidea 23 million years ago, the evolutionary rate was determined to equal 3.9 × 10−9 substitutions/site/year.
Fig 8.
Evolution of varicelloviruses. The polymerase and thymidine kinase genes of seven VZV clades and eight animal varicelloviruses were aligned, and a phylogenetic tree was inferred with BEAST. The left tree is drawn to scale, with branch lengths indicating the node heights. The scale bar indicates substitutions per site. The right tree shows the tree topology only. Node numbers (1 to 14) are presented to identify certain splits (compare Table 5). Node heights (in italics) and posterior probabilities greater than 0.7500 (in parentheses) are also presented.
Table 5.
Divergence times of varicelloviruses as calculated from node heights
| Node no. | Node hta | Divergence time (Myrb) | Corresponding event in mammalian/human evolution |
|---|---|---|---|
| 1 | 1.90374 | 63.729 | Split of Laurasiatheria and Euarchontoglires |
| 2 | 0.68707 | 23c | Split of Cercopithecidae and Hominoidea |
| 3 | 0.00330 | 0.110 | Human dispersal out of Africa |
| 4 | 0.00120 | 0.040 | |
| 5 | 0.00102 | 0.034 | |
| 6 | 0.00051 | 0.017 | |
| 7 | 0.00040 | 0.013 | |
| 8 | 0.00036 | 0.012 | |
| 9 | 0.69054 | 23.116 | Split of Carnivora and Perissodactyla/Cetartiodactyla |
| 10 | 0.45061 | 15.084 | Split of Perissodactyla and Cetartiodactyla |
| 11 | 0.20657 | 6.915 | Split of Suidae and Bovidae |
| 12 | 0.03551 | 1.189 | |
| 13 | 0.06610 | 2.213 | |
| 14 | 0.06152 | 2.059 |
Node height.
Myr, million years.
This date was used for calibration.
DISCUSSION
Genotyping of VZV field isolates.
Recently, Breuer and coworkers proposed a minimum complement of 27 SNPs of 15 genes for the genotyping of VZVs (5). This novel genotyping scheme combines four previous studies describing VZV genotypes based on the following: (i) five glycoprotein genes and the IE62 gene (61 polymorphisms) (12), (ii) the scattered SNP method (92 polymorphism in 37 ORFs) (4, 33), (iii) the ORF 22 (24), and (iv) whole-genome sequencing (37, 38). It reliably identifies the five established VZV genotypes and two novel clades, provisionally termed VI and VII. In the present study, 19 VZV field isolates from Germany and two Oka-derived strains were typed using a simplified genotyping scheme based on the minimum-complement SNPs. It involves only 11 SNPs of 6 genes but also allows distinguishing the 5 established clades and the proposed genotypes VI and VII (Table 2). Two of the German strains, 1483/2005 and 457/2008, did not match the SNP patterns of either of those genotypes. Assuming that they represent novel genotypes, these strains together with 19 others were subjected to complete genome sequencing.
A number of single nucleotide variants were observed. Some of them (SNPs 23294, 24533, and 113243) are most likely effects of recombination. For SNP 24533 (strains 134/2005, 875/2004, and 1805/2007), neighboring sequences also differ. For SNP 113243 (strains 11 and 405/2007), the recombination could be detected with RDP3, SimPlot, and phylogenetic analyses (Fig. 3, 4, and 6). In contrast, both complete genomes of clade 4 exhibit a G residue rather than an adenine at SNP 38019. This A/G polymorphism was described previously (24). Since neighboring sites were not mutated, a naturally occurring intratypic polymorphism could be one explanation. Therefore, inclusion of such nucleotide positions in the minimum complement of SNPs appears to be of little significance. Genotyping of VZV by SNP analysis may be hampered if such sites are detected. Distinction between recombination and natural polymorphisms requires additional sequence information which might not be available in some cases.
Next-generation sequencing of VZV field isolates.
In order to elucidate the molecular cause of the genotyping inconsistencies, the complete genomes of the present set of field isolates were determined. In addition, identification of two nontypeable strains in the SNP analysis demanded complete sequence analysis for further characterization. Finally, investigation of VZV recombination requires comprehensive sequence data. Previous VZV sequence analyses involved strategies either based on construction of DNA libraries after restriction enzyme digestion or shotgun cloning followed by sequencing of numerous plasmid clones (38) or based on the amplification of large PCR fragments and subsequent fragment sequencing by primer walking (37). In the present study, the 454 pyrosequencing technology was used since this method generates rather long reads (mean length, 335 nt) that may span most if not all of the reiterated elements of the R1 to R5 regions within the VZV genome. The test run yielded 40,443 reads, whereas multiplex sequencing yielded on average 45,893 reads per library (+13%). Finally, 3,700 reads (9.1%) were mapped to VZV in the test run, and on average, 4,441 reads (9.7%) were mapped in the multiplex sequencing run (Table 3). This allowed almost complete UL, US, and IR regions to be assembled, corresponding to 99.7% of the respective genomes. The mean sequence depth was 16.3-fold, which exceeds that of most sequences generated by conventional methods. With the exception of the test run and strain 457/2008, the numbers of gaps and gap lengths were rather low (mean number of gaps,: 6.3; mean gap length, 33 nt). Most of the remaining gaps were closed using conventional sequencing. Alignment and translation of the respective genome sequences reveals that some sequences at the reiteration regions R1 to R3 may still be incomplete. Nevertheless, it was observed that the subunit composition of the repeat elements is not clade specific but varies from strain to strain (data not shown).
Phylogenetic analysis and genetic characterization of VZV clades.
The phylogenetic analyses using the ML and NJ methods confirmed the genotyping results without exception and yielded two main results. First, five clusters representing the established genotypes can be demonstrated in the unrooted ML tree of Fig. 1. Second, in accordance with the genotyping results, two strains (1483/2005 and 457/2008) formed distinct branches. The peculiar SNP patterns (Table 2), a distinct branching (Fig. 1), and pronounced genetic distances (Table 4) support the proposal of two novel genotypes, provisionally named VIII and IX. Since the phylogenetic trees of Fig. 1 were calculated from complete genome alignments, the tree topology may be confounded by recombination events in the evolution of VZV. The novel genotypes have to be considered provisional, since sequence data of genotypes VI and VII are not available and a final evaluation requires complete sequence data. Although the SNP patterns of genotypes VI and VII are distinct from those of genotypes VIII and IX (5), it cannot be excluded yet that they are phylogenetically related.
The genetic analysis of data sets A and B revealed a diversity (p-distance) ranging from 2.87 × 10−4 to 5.79 × 10−4 in pairwise intratypic comparisons and values ranging from 9.82 × 10−4 to 1.794 × 10−3 in pairwise intertypic comparisons. These results exceed a previous estimation of 6.3 × 10−4, which may not be representative since it was determined on approximately 19% of the VZV genome (33).
Recombination in VZV.
Recombination events are important for the maintenance of internal and terminal repeats and are thought to contribute to VZV evolution (27). Previously, several authors described and discussed recombination and the role of these events in the phylogeny of VZV, albeit with mutually contradictory conclusions (24, 27, 37, 38). For example, Norberg and coworkers (37) investigated sequences of clades 1, 2, 3, and 4 and proposed that clade 1 and 2 strains recombined at least twice to yield the clade 4 and 5 strains. Similar observations were described by Peters et al. (38), who compared clade 1, 2, 4, and 5 sequences. They suggested that two fragments of clade 1, ORFs 14 to 17 and ORFs 22 to 26, replaced the corresponding regions of clade 2 to yield clade 4. Another study came to the conclusion that clade 1 derived from a recombinant of clades 3 and 4 (27). In fact, interpretation of recombination results is difficult without knowledge of a reliable VZV phylogeny. We have investigated data sets that comprised up to 42 complete VZV genomes. The bootstrap analyses indicate that virtually all circulating VZV strains represent mosaic genomes, but in the retrospective analysis it is hardly possible to decide which clade was the donor and which clade was the acceptor in the recombination events. Several strains of established clusters are probably recombinants, since certain genome regions do not cluster with other strains of their clade (strain 8, strain 11, DR, CA123, 413/2000, 3/2005, and 405/2007). Also, strains 1483/2005 and 457/2008, though single representatives of genotypes VIII and IX, have mosaic genomes and therefore are most likely recombinants.
The opinion on the relevance of recombination as a means of VZV evolution is presently changing. Whereas Muir et al. (33) assumed “that recombination had little effect on the evolution of the genotypes,” other authors provided evidence that some of the circulating genotypes arose by recombination (27, 37, 38). Based on the small number of complete VZV genomes, a low recombination rate is generally considered. However, this view may change if more VZV sequences are available. Appraisal of VZV recombination leads to the question of whether it occurs only during primary infection. The rationale of this scenario is the perception that seropositive hosts are basically resistant to a secondary infection by another VZV strain. If this scenario is adopted, only a relatively short time frame from the beginning of the primary infection to the establishment of an immune response would allow infection by a different VZV genotype, which would be a prerequisite for a recombination event. Primary infection of a patient with two different clades of VZV was recently described (41). An alternative scenario could be recombination after reinfection with another VZV genotype despite the presence of detectable serum VZV antibodies at the time of exposure. This would certainly increase the probability of a given virus coming in contact with a VZV strain of another clade, but it is unclear how the secondary virus evades the host immune response. Analysis of the VZV MSP variant revealed a mutation of glycoprotein E (gE) that resulted in the loss of an immunodominant B-cell epitope (43). Such a mutation may explain the failure of protection from infection with such strains. Although our isolates had no such mutations, exogenic reinfection may occur in immunocompetent and immunocompromised individuals, and recombinant virus could be released during subclinical virus replication or a shingles episode. Both serologic evidence and confirmed cases of reinfection with VZV have been described (3, 13, 40; see also the review of Quinlivan and Breuer [39]). Another open question is the mechanism of recombination. The mosaic genomes of VZV exhibit exchanged regions that vary from several hundred to several thousand nucleotides. It appears that recombination preferably occurs in a site-specific mechanism at the reiteration regions, as previously observed for herpes simplex virus (7).
Estimation of divergence times between the VZV clades.
Knowledge of VZV diversification is an important issue, since a reliable estimation of the root of the VZV tree sets a benchmark for the evaluation of recombination in VZV evolution. According to Fig. 8, the ancestors of clades 2/4 and 1/3/5/VIII/IX separated some 110,000 years ago. Then, further evolution by accumulation of nucleotide substitutions and recombination led to the contemporary clades and genotypes. Due to the lifelong persistence of VZV in the human host and the lack of sufficient sequence data representing virus isolates of several human generations, a precise measurement of evolutionary rates is not possible at present. However, there is one accepted method to roughly estimate the evolutionary rate of a virus and hence determine the divergence dates of a phylogenetic tree. This approach requires one or several reliable outgroup sequences and at least one calibration date determined with molecular and/or fossil data. Applying this approach, the split of the ancestors of CeHV-9 and VZV was assumed to have taken place 23 million years ago. The rationale for this assumption is supported by several molecular studies (14, 18). Based on this assumption and presuming a relaxed molecular clock, node heights of the phylogenetic tree allowed us to calculate the divergence times. VZV radiation began some 110,000 years ago and ended 12,000 years before the present (Table 5). Interestingly, the beginning of VZV diversification roughly coincides with the dispersal of modern humans out of Africa some 100,000 years ago (6, 60). This event is a hallmark in human history and could explain the development of different VZV clades (15, 62). Assuming this model of VZV evolution, geographical strain variation was observed and Asian and European/North American clades were described (33, 62). Similar observations were made by Quinlivan et al. (40). A recent meta-analysis outlined a worldwide distribution of VZV, with clade 2 being most prevalent in Asia and clades 1 and 3 in Europe, America, and Australia. Clade 5 shows a worldwide distribution but is the only genotype which has been detected in Africa so far (53). Whereas our study estimated the divergence times of the VZV clades in accordance with human history, a previous study (33) proposed considerably shorter divergence times (from 19,000 to 3,000 years before the present), which is inconsistent with the out-of-Africa hypothesis. Moreover, such short divergence times lack other plausible explanations for the emergence of VZV clades, e.g., fossil and molecular data (substitution rates). Our estimation of 3.9 × 10−9 substitutions/site/year for the TK/pol genes is similar to the evolutionary rates determined by McGeoch and Cook (28) for the highly conserved gB gene of alphaherpesviruses (2.7 × 10−9 substitutions/site/year) but is 26 times lower than the substitution rate of 10−7 assumed by Muir et al. (33). The latter value appears to be an overestimation. However, the rate of change of the whole alphaherpesvirus genome was esteemed to be an order of magnitude higher than that of the highly conserved gB gene (28), which allows rate heterogeneity of certain genes among the virus species.
The phylogenetic tree of the varicellovirus sequences (Fig. 8) correlates with the morphological tree of mammals (e.g., see reference 55), since it places those viruses that infect cows and pigs (order Artiodactyla) and horses (order Perissodactyla) close together. Also, several molecular studies support the morphological tree (21, 26, 34, 61). Whether those orders are actually monophyletic or evolved convergently is still an open question, since other authors observe the branching of Artiodactyla prior to the separation of Carnivora and Perissodactyla (e.g., see references 1, 18, 32, 35, 42, and 58; reviewed by Springer and coworkers [57]). Despite variation of parameters in the phylogenetic analyses of varicelloviruses, no tree of the present study reflected a close relationship between FeHV-1 and the EHVs. In their seminal study, McGeoch and Cook analyzed additional genes but achieved only a “poorly defined” position for FHV-1, which did not “approximate the host tree's topology” (28). Similar observations were made with other sets of 15 sequences and up to 7 genes (ORFs 28, 29, 30, 31, 36, 40, and 42/45; data not shown). In general, the topology of phylogenetic trees strongly depends on the selection of the investigated genes. This should be interpreted as a hint regarding a significant heterogeneity of the evolutionary rates within and between the different virus species. Therefore, it may be questionable whether herpesvirus phylogeny can contribute to resolving questions of mammalian evolution and vice versa, although coevolution (cospeciation) of herpesviruses and their hosts is generally accepted. Assuming the split of the ancestors of CeHV-9 and VZV 23 million years ago, the resulting diversification of the pol and TK genes of VZV clades started some 110,000 years before the present. However, the divergence times of the nonprimate varicelloviruses as calculated from the node heights of the pol and TK genes are inconsistent with the fossil data of their hosts (Table 5). A lack of additional valuable calibration points, a too-small database, and evolutionary rate heterogeneity between the investigated virus genes could be explanations. This finding urges the need to complete the genomes of further varicelloviruses.
ACKNOWLEDGMENTS
This work was supported by grants from the Deutsche Forschungsgemeinschaft awarded to A.S. (SA 964/3-2).
The excellent technical assistance of Martina Müller, Monika Alexi, and Cornelia Luge is acknowledged.
Footnotes
Published ahead of print 30 November 2011
REFERENCES
- 1. Arnason U, Gullberg A, Janke A. 1998. Molecular timing of primate divergences as estimated by two nonprimate calibration points. J. Mol. Evol. 48:718–727 [DOI] [PubMed] [Google Scholar]
- 2. Arvin AM, Gershon AA. 1996. Live attenuated varicella vaccine. Annu. Rev. Microbiol. 50:59–100 [DOI] [PubMed] [Google Scholar]
- 3. Arvin AM, Koropchak CM, Wittek AE. 1983. Immunologic evidence of reinfection with varicella-zoster virus. J. Infect. Dis. 148:200–205 [DOI] [PubMed] [Google Scholar]
- 4. Barrett-Muir W, et al. 2003. Genetic variation of varicella-zoster virus: evidence for geographical separation of strains. J. Med. Virol. 70(Suppl. 1):S42–S47 [DOI] [PubMed] [Google Scholar]
- 5. Breuer J, Grose C, Norberg P, Tipples G, Schmid DS. 2010. A proposal for a common nomenclature for viral clades that form the species varicella-zoster virus: summary of VZV Nomenclature Meeting 2008, Barts and the London School of Medicine and Dentistry, 24–25 July 2008. J. Gen. Virol. 91:821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cavalli-Sforza LL. 1998. The DNA revolution in population genetics. Trends Genet. 14:60–65 [DOI] [PubMed] [Google Scholar]
- 7. Chou J, Roizman B. 1985. Isomerization of herpes simplex virus 1 genome: identification of the +-acting and recombination sites within the domain of the a sequence. Cell 41:803–811 [DOI] [PubMed] [Google Scholar]
- 8. Davison A, et al. 2005. Family Herpesviridae, p 193–212 In Fauquet C, Mayo M, Maniloff J, Desselberger E, Ball L. (ed), Virus taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA [Google Scholar]
- 9. Davison AJ. 2000. Molecular evolution of alphaherpesviruses, p 25–50 In Arvin AM, Gershon AA. (ed), Varicella-zoster virus. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 10. Dohner DE, Adams SG, Gelb LD. 1988. Recombination in tissue culture between varicella-zoster virus strains. J. Med. Virol. 24:329–341 [DOI] [PubMed] [Google Scholar]
- 11. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Faga B, Maury W, Bruckner DA, Grose C. 2001. Identification and mapping of single nucleotide polymorphisms in the varicella-zoster virus genome. Virology 280:1–6 [DOI] [PubMed] [Google Scholar]
- 13. Gershon AA, Steinberg SP, Gelb L. 1984. Clinical reinfection with varicella-zoster virus. J. Infect. Dis. 149:137–142 [DOI] [PubMed] [Google Scholar]
- 14. Glazko GV, Nei M. 2003. Estimation of divergence times for major lineages of primate species. Mol. Biol. Evol. 20:424–434 [DOI] [PubMed] [Google Scholar]
- 15. Grose C. 1999. Varicella-zoster virus: less immutable than once thought. Pediatrics 103:1027–1028 [DOI] [PubMed] [Google Scholar]
- 16. Heath L, van der Walt E, Varsani A, Martin DP. 2006. Recombination patterns in aphthoviruses mirror those found in other picornaviruses. J. Virol. 80:11827–11832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267 [DOI] [PubMed] [Google Scholar]
- 18. Kumar S, Hedges SB. 1998. A molecular timescale for vertebrate evolution. Nature 392:917–920 [DOI] [PubMed] [Google Scholar]
- 19. LaRussa P, et al. 1992. Restriction fragment length polymorphism of polymerase chain reaction products from vaccine and wild-type varicella-zoster virus isolates. J. Virol. 66:1016–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. LaRussa PS, Gershon AA. 2001. Biologic and geographic differences between vaccine and clinical varicella-zoster virus isolates. Arch. Virol. Suppl. 2001:41–48 [DOI] [PubMed] [Google Scholar]
- 21. Liu FGR, et al. 2001. Molecular and morphological supertrees for eutherian (placental) mammals. Science 291:1786–1789 [DOI] [PubMed] [Google Scholar]
- 22. Lole KS, et al. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loparev VN, Argaw T, Krause PR, Takayama M, Schmid DS. 2000. Improved identification and differentiation of varicella-zoster virus (VZV) wild-type strains and an attenuated varicella vaccine strain using a VZV open reading frame 62-based PCR. J. Clin. Microbiol. 38:3156–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loparev VN, et al. 2004. Global identification of three major genotypes of varicella-zoster virus: longitudinal clustering and strategies for genotyping. J. Virol. 78:8349–8358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Loparev VN, et al. 2007. Identification of five major and two minor genotypes of varicella-zoster virus strains: a practical two-amplicon approach used to genotype clinical isolates in Australia and New Zealand. J. Virol. 81:12758–12765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Madsen O, et al. 2001. Parallel adaptive radiations in two major clades of placental mammals. Nature 409:610–614 [DOI] [PubMed] [Google Scholar]
- 27. McGeoch DJ. 2009. Lineages of varicella-zoster virus. J. Gen. Virol. 90:963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McGeoch DJ, Cook S. 1994. Molecular phylogeny of the alphaherpesvirinae subfamily and a proposed evolutionary timescale. J. Mol. Biol. 238:9–22 [DOI] [PubMed] [Google Scholar]
- 29. McGeoch DJ, Davison AJ, Dolan A, Gatherer D, Sevilla-Reyes EE. 2008. Molecular evolution of the Herpesvirales. In Domingo E, Parrish CR, Holland JJ. (ed), Origin and evolution of viruses, 2nd ed. Academic Press, London, United Kingdom [Google Scholar]
- 30. Meyer M, Stenzel U, Hofreiter M. 2008. Parallel tagged sequencing on the 454 platform. Nat. Protoc. 3:267–278 [DOI] [PubMed] [Google Scholar]
- 31. Meyer M, et al. 2008. From micrograms to picograms: quantitative PCR reduces the material demands of high-throughput sequencing. Nucleic Acids Res. 36:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Montgelard C, Catzeflis FM, Douzery E. 1997. Phylogenetic relationships of artiodactyls and cetaceans as deduced from the comparison of cytochrome b and 12S rRNA mitochondrial sequences. Mol. Biol. Evol. 14:550–559 [DOI] [PubMed] [Google Scholar]
- 33. Muir WB, Nichols R, Breuer J. 2002. Phylogenetic analysis of varicella-zoster virus: evidence of intercontinental spread of genotypes and recombination. J. Virol. 76:1971–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murphy WJ, et al. 2001. Molecular phylogenetics and the origins of placental mammals. Nature 409:614–618 [DOI] [PubMed] [Google Scholar]
- 35. Murphy WJ, et al. 2001. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294:2348–2351 [DOI] [PubMed] [Google Scholar]
- 36. Norberg P. 2010. Divergence and genotyping of human alphaherpesviruses: an overview. Infect. Genet. Evol. 10:14–25 [DOI] [PubMed] [Google Scholar]
- 37. Norberg P, et al. 2006. Complete-genome phylogenetic approach to varicella-zoster virus evolution: genetic divergence and evidence for recombination. J. Virol. 80:9569–9576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peters GA, et al. 2006. A full-genome phylogenetic analysis of varicella-zoster virus reveals a novel origin of replication-based genotyping scheme and evidence of recombination between major circulating clades. J. Virol. 80:9850–9860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Quinlivan M, Breuer J. 2006. Molecular studies of varicella zoster virus. Rev. Med. Virol. 16:225–250 [DOI] [PubMed] [Google Scholar]
- 40. Quinlivan M, et al. 2002. The molecular epidemiology of varicella-zoster virus: evidence for geographic segregation. J. Infect. Dis. 186:888–894 [DOI] [PubMed] [Google Scholar]
- 41. Qinlivan M, Sengupta N, Breuer J. 2009. A case of varicella caused by co-infection with two different genotypes of varicella-zoster virus. J. Clin. Virol. 44:66–69 [DOI] [PubMed] [Google Scholar]
- 42. Reyes A, et al. 2004. Congruent mammalian trees from mitochondrial and nuclear genes using Bayesian methods. Mol. Biol. Evol. 21:397–403 [DOI] [PubMed] [Google Scholar]
- 43. Santos RA, et al. 2000. Varicella-zoster virus gE escape mutant VZV-MSP exhibits an accelerated cell-to-cell spread phenotype in both infected cell cultures and SCID-hu mice. Virology 275:306–317 [DOI] [PubMed] [Google Scholar]
- 44. Sauerbrei A, et al. 2011. Monitoring prevalence of varicella-zoster virus clades in Germany. Med. Microbiol. Immunol. 200:99–107 [DOI] [PubMed] [Google Scholar]
- 45. Sauerbrei A, et al. 2003. Characterisation of varicella-zoster virus strains in Germany and differentiation from the Oka vaccine strain. J. Med. Virol. 71:313–319 [DOI] [PubMed] [Google Scholar]
- 46. Sauerbrei A, Philipps A, Zell R, Wutzler P. 2007. Genotyping of varicella-zoster virus strains after serial passages in cell culture. J. Virol. Methods 145:80–83 [DOI] [PubMed] [Google Scholar]
- 47. Sauerbrei A, Wutzler P. 2007. Different genotype pattern of varicella-zoster virus obtained from patients with varicella and zoster in Germany. J. Med. Virol. 79:1025–1031 [DOI] [PubMed] [Google Scholar]
- 48. Sauerbrei A, Zell R, Philipps A, Wutzler P. 2008. Genotypes of varicella-zoster virus wild-type strains in Germany. J. Med. Virol. 80:1123–1130 [DOI] [PubMed] [Google Scholar]
- 49. Schmidt-Chanasit J, et al. 2007. Novel genotyping approach for varicella-zoster virus strains from Germany. J. Clin. Microbiol. 45:3540–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schmidt-Chanasit J, et al. 2008. Novel varicella-zoster virus glycoprotein E gene mutations associated with genotypes A and D. J. Clin. Microbiol. 46:325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schmdit-Chanasit J, et al. 2009. Novel approach to differentiate subclades of varicella-zoster virus genotypes E1 and E2 in Germany. Virus Res. 145:347–349 [DOI] [PubMed] [Google Scholar]
- 52. Schmidt-Chanasit J, et al. 2008. Molecular analysis of varicella-zoster virus strains circulating in Tanzania demonstrating the presence of genotype M1. J. Clin. Microbiol. 46:3530–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schmidt-Chanasit J, Sauerbrei A. 2011. Evolution and world-wide distribution of varicella-zoster virus clades. Infect. Genet. Evol. 11:1–10 [DOI] [PubMed] [Google Scholar]
- 54. Sears LE, et al. 1992. Circumvent thermal cycle sequencing and alternative manual and automated DNA sequencing protocols using the highly thermostable VentR (exo-) DNA polymerase. Biotechniques 13:626–633 [PubMed] [Google Scholar]
- 55. Shoshani J, McKenna MC. 1998. Higher taxonomic relationships among extant mammals based on morphology, with selected comparisons of results from molecular data. Mol. Phylogenet. Evol. 9:572–584 [DOI] [PubMed] [Google Scholar]
- 56. Slatko BE. 1996. Thermal cycle dideoxy DNA sequencing. Methods Mol. Biol. 58:413–423 [DOI] [PubMed] [Google Scholar]
- 57. Springer MS, Stanhope MJ, Madsen O, de Jong WW. 2004. Molecules consolidate the placental mammal tree. Trends Ecol. Evol. 19:430–438 [DOI] [PubMed] [Google Scholar]
- 58. Springer MS, Murphy WJ, Eizirik E, O'Brien SJ. 2003. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc. Natl. Acad. Sci. U. S. A. 100:1056–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 60. Templeton AR. 1997. Out of Africa? What do genes tell us? Curr. Opin. Genet. Dev. 7:841–847 [DOI] [PubMed] [Google Scholar]
- 61. Waddell PJ, Kishino H, Ota R. 2001. A phylogenetic foundation for comparative mammalian genomics. Genome Inform. 12:141–154 [PubMed] [Google Scholar]
- 62. Wagenaar TR, Chow VT, Buranathai C, Thawatsupha P, Grose C. 2003. The out of Africa model of varicella-zoster virus evolution: single nucleotide polymorphisms and private alleles distinguish Asian clades from European/North American clades. Vaccine 21:1072–1081 [DOI] [PubMed] [Google Scholar]