Abstract
Herpes simplex virus type 1 (HSV-1) infection induces profound nucleolar modifications at the functional and organizational levels, including nucleolar invasion by several viral proteins. One of these proteins is US11, which exhibits several different functions and displays both cytoplasmic localization and clear nucleolar localization very similar to that of the major multifunctional nucleolar protein nucleolin. To determine whether US11 interacts with nucleolin, we purified US11 protein partners by coimmunoprecipitations using a tagged protein, Flag-US11. From extracts of cells expressing Flag-US11 protein, we copurified a protein of about 100 kDa that was further identified as nucleolin. In vitro studies have demonstrated that nucleolin interacts with US11 and that the C-terminal domain of US11, which is required for US11 nucleolar accumulation, is sufficient for interaction with nucleolin. This association was confirmed in HSV-1-infected cells. We found an increase in the nucleolar accumulation of US11 in nucleolin-depleted cells, thereby revealing that nucleolin could play a role in US11 nucleocytoplasmic trafficking through one-way directional transport out of the nucleolus. Since nucleolin is required for HSV-1 nuclear egress, the interaction of US11 with nucleolin may participate in the outcome of infection.
INTRODUCTION
The genome of herpes simplex virus type 1 (HSV-1) contains more than 80 genes that have been grouped into two different categories related to the function of their products. In vivo, most are “essential genes” that code for proteins that are essential for viral gene expression, DNA synthesis, and the assembly of viral particles; the remaining are “nonessential genes” that encode products that allow viral production under various cellular conditions (29). Because all of these nonessential genes are dispensable for virus growth in cell culture, the precise functions of most of them remain to be elucidated.
The HSV-1 US11 gene was initially described as one of these nonessential genes (5, 21, 22). US11 is an abundant 21-kDa late viral protein that is packaged within the viral tegument and delivered into newly infected cells prior to the expression of viral genes. Although US11 is dispensable for infection of cell cultures, it may play a role in the replication of HSV-1 in the adrenal gland, an organ important for viral penetration into the spinal cord and the brain (18, 26), and in cells subjected to thermal stress (5, 13). Also, US11 is involved in the antiviral response (20) and displays antiapoptotic activity, notably against heat-induced apoptosis, which appears to be located at the level of mitochondria or upstream signaling (19).
Since the first identification of US11 as a DNA-binding protein (9), US11 has been demonstrated to exhibit several different functions. Strong evidence shows that US11 is an RNA-binding protein that binds RNA in a sequence- and conformation-specific fashion and also displays a role in posttranscriptional regulation of gene expression (30, 31). Moreover, by its ability to bind certain mRNAs, we have demonstrated that US11 displays striking functional similarities to HIV-1 Rev and human T-cell leukemia/lymphoma virus type I (HTLV-I) Rex proteins. US11 can substitute for Rev and Rex and intervenes posttranscriptionally in the life cycle of these retroviruses by transactivating expression of the genes that encode HIV-1 and HTLV-I envelope (env) glycoproteins (10).
In addition to RNAs, US11 interacts with cellular proteins. Until now, the only proteins known to interact with US11 were human ubiquitous kinesin heavy chain (14), PAT1, which is a homolog of kinesin light chain (2), PKR, a double-stranded RNA (dsRNA)-dependent protein kinase (27), PACT, which is a dsRNA-independent protein activator of PKR (20, 27), and 2′-5′-oligoadenylate synthetase (33).
During HSV-1 infection, the incoming US11 protein is delivered early into the cells. Soon after infection, US11 is found in the cytoplasm, either as a heterogeneous oligomer or associated with the ribosomes or both. Later during infection, US11 accumulates in nucleoli but is also found in ribonucleoprotein fibrils and in clusters of interchromatin granules (12, 25, 28).
The nucleolus consists of fibrillar centers (FC), dense fibrillar components (DFC), and granular components (GC). rRNA genes are localized in the FC, pre-rRNA resides in the DFC, and the late processing steps of ribosome biogenesis occur in the GC. When US11 concentrates in nucleoli, it is abundant in the DFC and GC but absent from the FC (3), an intranucleolar distribution that is similar to that of nucleolin.
Indeed, nucleolin, one of the most abundant nucleolar components, is usually found in the DFC and GC of nucleoli. Its interaction with ribosomal proteins and with specific pre-rRNA sequences and its implication in pre-rRNA maturation suggest that nucleolin could be an important ribosome assembly factor (4). Several of our observations suggest that nucleolin is also a major factor in promoting cell proliferation (39, 40).
During HSV-1 infection, a fraction of nucleolin is depleted from the nucleolus and is found in the viral replication compartments (6, 23, 24). Nucleolin is one of the few cellular proteins that are required for HSV-1 infection (6); recent data indicate that nucleolin is needed for efficient nuclear egress of HSV-1 nucleocapsids (32).
Since US11 and nucleolin share a similar localization and because we have shown that HSV-1 infection is prevented in nucleolin-depleted cells, we designed a strategy to analyze whether these two proteins interact. Using a coimmunoprecipitation approach coupled with Western blot and mass spectrometry analysis, we show that nucleolin and US11 are present in the same protein complexes in cells expressing recombinant US11 proteins and, more importantly, in HSV-1-infected cells. In vitro interaction experiments and far-Western blot analysis revealed that US11 interacts with nucleolin and that the polyproline type II helix-containing domain of US11 is responsible for this interaction. The findings that this domain is also responsible for US11 nucleolar accumulation and that nucleolin is a nucleocytoplasmic shuttling protein suggest that the interaction between US11 and nucleolin could regulate the intracellular trafficking of US11 or the molecular processes in which these proteins are involved during infection. Indeed, we show that in nucleolin-depleted cells, US11 accumulates in nucleoli, indicating that nucleolin contributes to US11 nucleolar export and strongly supporting the notion that the interaction between nucleolin and US11 plays a role in the outcome of infection.
MATERIALS AND METHODS
Plasmids.
The pHTLV-env, pcREX, and pcTax plasmids that express HTLV env, Rex, and Tax, respectively, the pCMV-US11 plasmid that expresses US11, and the plasmids that code for the recombinant proteins GST-US11, GST-US11 (Δ1–40), GST-US11-m5, GST-US11-m6, and GST-US11-m15 derived from pGEX-2T that have been described previously (7, 10, 34) were used in this study. Briefly, GST-US11 contains the 149 amino acids (aa) of full-length US11 from the KOS strain, whereas GST-US11 (Δ1–40) contains aa 41 to 149 of the US11 protein, GST-US11-m5 contains aa 36 to 149, GST-US11-m6 contains aa 62 to 149, and GST-US11-m15 contains aa 1 to 40. pSG5-Flag-US11 vector expressing flag-US11 (full-length US11 fused to the Flag sequence) was obtained by inserting the HindIII/BglII fragment of plasmid pGEX-2T-US11 (7) into the pSG5-Flag vector.
Cell culture, transfection, infection, and functional assay.
HeLa cells were grown in Eagle's minimal essential medium (E-MEM) containing 100 U/ml penicillin and 100 μg/ml streptomycin with or without 5% heat-inactivated fetal calf serum (FCS) at 37°C under 5% CO2. Cells were transfected for 48 h using the calcium phosphate precipitation procedure (17). When indicated, HeLa cells were infected just before confluence with the HSV-1 17+ wild-type (wt) strain at 10 PFU per cell. After 1 h of adsorption of the virus at 33°C under 5% CO2, the medium was removed, and the cells were washed and then incubated in E-MEM at 37°C until harvesting at the indicated times postinfection. US11 can substitute for Rex in transactivating expression of the retroviral genes encoding HTLV-I env glycoproteins (10).
Nucleolin silencing by siRNA.
A mixture of functional small interfering RNAs (siRNAs) (Eurogentec) specific for human nucleolin siRNA 4 (UUCUUUGACAGGCUCUUCCUU) and siRNA 2 (UCCAAGGUAACUUUAUUUCUU) was used as previously described (6, 40). siRNAs were reconstituted at a concentration of 100 μM and stored at −20°C. The HL5a1 clones of HeLa cells that stably expressed US11 (13, 37) were transfected in a 6-well dish using siRNA at a 2 nM final concentration. siRNAs were diluted in 200 μl of Opti-MEM (Invitrogen) and loaded in a well. Eighty microliters of INTERFERin (Polyplus) diluted 1:10 in RNase-free water was added. After 10 min of incubation, 2 ml of media containing 3 ×105 cells was added.
Immunofluorescence and fluorescence quantification.
Cells were either infected for 8 h with the HSV-1 17+ wt strain or transfected for 48 h with the indicated expression vectors. Then, cells were washed with phosphate-buffered saline (PBS), fixed, and submitted to immunohistochemistry as described previously (6). For nucleolin siRNA treatment, cells were plated at 4 × 104 cells/well in 24-well dishes onto glass coverslips 2 days after nucleolin siRNA transfection. Four days after siRNA transfection, cells were fixed and treated as previously described (40). Rabbit polyclonal antibodies for US11 (12) or nucleolin (15) and mouse monoclonal anti-nucleolin (4E2 Assay Designs; Immunogen) (human nucleolin; 1/1,000 dilution) or M2 anti-Flag antibodies were the primary antibodies used. Secondary antibodies were coupled to Alexa dyes (A647 or A488, Invitrogen; A546 or A555, Interchim). For Fig. 1, appropriate confocal emission filters were used for double-labeling experiments. Images were merged by computer. To control for cross-reactivity, samples were stained with one primary antibody and appropriate secondary antibodies. No overlap between the channels was observed for any of the samples at the settings used. For the bottom panels of Fig. 1B (see also Fig. 6), 12-bit images were acquired with the same exposure times using a Cool Snap HQ charge-coupled-device (CCD) camera mounted on a Zeiss Axio-Imager equipped with a 63× oil-immersion objective lens (numerical aperture [NA] = 1.4) and fluorescence filters suited to visualization of DAPI (4′-6-diamidino-2-phenylindole), A488, and A647. For each field of view, Z-stacks of about 10 images with a pixel size of 102 nm were obtained by setting the Z-step at 200 nm. For each analyzed cell, the optical section in which the nucleoli were the most in focus was chosen for quantification. We ensured that the choice of the section did not significantly affect the US11 nucleocytoplasmic ratio (US11 ratio variations < 0.1 among 5 successive optical sections). Quantification of US11 intensities was performed on the raw selected 12-bit section using ImageJ freeware. Outlines of the cells were hand drawn on the green (US11) channel section and used to obtain the mean integrated intensity, referred to as the US11 cellular level (see Fig. 6F). The US11 nucleocytoplasmic ratio was calculated for each cell by dividing the mean nucleolar intensity by the mean cytoplasmic intensity obtained on the green channel section as follows. For each cell, outlines of every nucleolus were hand drawn, and the corresponding integrated intensities were summed and divided by the sum of their areas to obtain the mean nucleolar intensity. The mean cytoplasmic intensity was calculated by subtracting the US11 integrated intensity of the nucleus from that of the cytoplasm divided by the difference of their areas. The outline of the nucleus was hand drawn on the blue (DAPI) channel section and was copied onto the green channel to determine the mean US11 nuclear integrated intensity. This analysis was performed on at least 16 individual cells and observed in 5 (control untransfected [C], mock-transfected [MT], and scramble siRNA-transfected [SC] conditions) or 10 (nucleolin siRNA-transfected [NCL] condition) different fields of view. The corresponding means and standard deviations are plotted for each set of conditions (see Fig. 6E and F).
Fig 1.
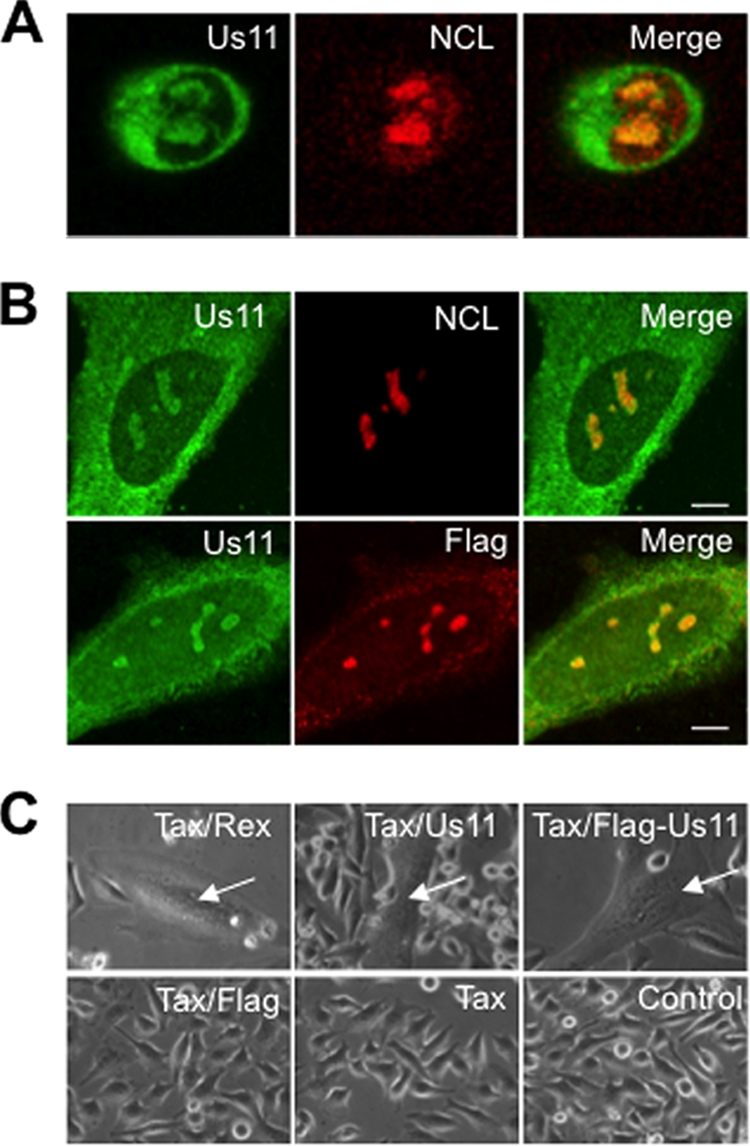
Intracellular localization and functional analysis of US11 protein. (A) Intracellular localization of US11 and nucleolin (NCL) in HeLa cells infected for 8 h with HSV-1 (wt 17+ strain, 10 PFU/cell) was visualized by indirect immunofluorescence using anti-US11 polyclonal and anti-nucleolin monoclonal antibodies. A merged image is presented. (B) Subcellular localization of wt US11 and Flag-US11 proteins in HeLa cells transfected with vectors expressing either untagged US11 (upper panels) or Flag-US11 fusion protein (lower panels). At 48 h after transfection, cells were fixed and the localization of the proteins was assayed with a polyclonal anti-US11 antibody and a monoclonal anti-nucleolin antibody (upper panels) or with a monoclonal anti-Flag antibody and a polyclonal anti-US11 antibody (lower panels). Merged images are presented. Scale bars represent 5 μm. (C) Transactivation of HTLV-I envelope glycoprotein expression by Rex, wt US11, and Flag-US11 proteins. HeLa cells were transfected with vectors expressing HTLV-env and Tax plus vectors expressing HTLV-Rex (upper left panel) or US11 (upper middle panel) or Flag-US11 fusion protein (upper right) or Flag only (lower left panel). Transactivation of the expression of HTLV-I env by either Rex or wt US11 led to the formation of syncytia (arrows in left and middle upper panels) as described in detail previously (10). The same transactivating property was displayed by Flag-US11, as visualized by syncytium formation (upper right panel). As control experiments, HeLa cells were either left untransfected (lower right panel) or transfected with vectors expressing HTLV-env and Tax plus vectors expressing either Flag only (lower left panel) or no additional vector (lower middle panel). As expected, in the three lower panels, only individual cells are visible and no syncytium was detected.
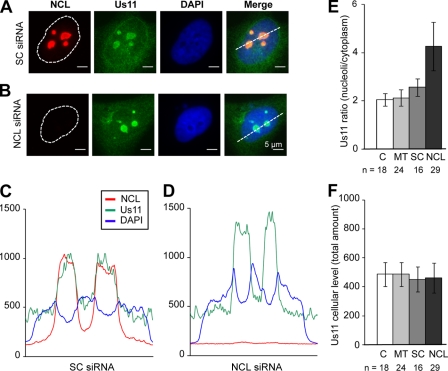
Fig 6.
Nucleolin depletion leads to US11 accumulation in nucleoli. (A and B) Immunofluorescence in a HeLa cell line stably expressing untagged US11 (clone HL5a1) transfected with scramble siRNA (SC) (A) or siRNA against nucleolin (B). Nucleolin (NCL) is shown in red and US11 in green, and DNA is stained with DAPI (blue). Scale bars represent 5 μm. (C and D) Fluorescence intensity profiles (gray level values) obtained on 16-bit images along the white lines indicated in panels A and B, respectively. Profiles correspond to nucleolin (red), US11 (green), and DAPI (blue) fluorescence intensities. (E) Quantification of the nucleolar/cytoplasmic ratio of US11. The ratio of nucleolar to cytoplasmic US11 fluorescence intensity was calculated for control untransfected cells (C; n = 18), mock-transfected cells (MT; n = 24), cells transfected with scramble siRNA (SC; n = 16), and cells transfected with nucleolin siRNA (NCL; n = 29). Five to 10 fields of view were analyzed for each set of conditions. Error bars correspond to standard deviations. Mann-Whitney and Wilcoxon statistical tests showed that the US11 ratio in nucleolin-depleted cells was significantly higher than in all the control cell populations (P < 2 × 10−7). (F) US11 cellular intensity in control untransfected cells (C), mock-transfected cells (MT), cells transfected with scramble siRNA (SC), and cells transfected with nucleolin siRNA (NCL) (camera gray level).
Immunoprecipitation and 1-DE of proteins and protein identification by mass spectrometry and Western blot analysis.
Cells were harvested 48 h posttransfection or 8 and 16 h after infection with the HSV-1 17+ wt strain. To immunoprecipitate cellular multiprotein complexes that contained US11, several cell lysis procedures were evaluated, including the use of different detergents such as NP-40, Triton X-100, SDS (sodium dodecyl sulfate), DOC (deoxycholic acid), and CHAPS {3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate}. The experimental conditions using an anti-Flag antibody were optimized to efficiently and reproducibly copurify Flag-US11 and proteins that specifically bind to Flag-US11 and not to Flag alone (data not shown). The optimal lysis buffer retained contained 0.1% Triton X-100 and 0.1% CHAPS. Cells were washed with PBS and then lysed at 4°C for 30 min in 1 ml of radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.2], 0.1% Triton X-100, 0.1% CHAPS) that contained protease inhibitor and was either supplemented with NaCl from 50 to 500 mM or not supplemented. Cell extracts were centrifuged at 75,000 rpm (Beckman TLA-100.3 rotor) for 30 min at 4°C. Supernatants were incubated overnight with the M2 anti-Flag antibody or a rabbit polyclonal anti-nucleolin antibody (P. Bouvet, unpublished data). Immunocomplexes were immobilized for 30 min at 4°C on protein-A Sepharose beads. After extensive washing with RIPA buffer supplemented with the appropriate amount of NaCl, bound proteins were eluted by addition of Laemmli buffer and were then separated by monodimensional gel electrophoresis (1-DE) (11) and stained with either Coomassie blue R250 Biosafe or silver nitrate. Western blot analyses were performed as described previously (6). For mass spectrometry analyses, proteins present at around 100 kDa in Coomassie blue-stained gel slices were submitted to in-gel digestion as described previously (35). The resulting peptides were analyzed by nanoliquid chromatography coupled to tandem mass spectrometry as described in reference 8, and proteins were identified using Phenyx software (Genebio).
Protein purification and protein-protein interaction assays.
Recombinant GST-US11 proteins and glutathione S-transferase (GST) protein were produced in Escherichia coli DH5a and purified from bacterial lysates by affinity chromatography on glutathione-agarose beads as previously described (34). Nucleolin-p50 (also called R1234G in reference 36) was produced in E. coli and purified as previously described (36). Nucleolin purified from CHO cells was biotinylated as previously described (15). Biotinylated nucleolin-US11 complexes were recovered with magnetic streptavidin beads (Dynal). Beads were first extensively washed with binding buffer (BB; 15 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 150 mM NaCl, 0.1% Tween 20, 0.1% bovine serum albumin [BSA]). One hundred nanograms of biotinylated nucleolin was incubated with approximately 200 ng of GST or GST-US11 protein in 100 μl of BB for 60 min at 4°C. The beads were then extensively washed with BB, and proteins bound to streptavidin beads were eluted in Laemmli electrophoresis buffer and then separated by 1-DE.
Far-Western blot analysis.
Far-Western blot analysis of the interaction of nucleolin with US11 deletion mutants was performed as previously described (4, 15). After 1-DE separation, proteins were transferred to nitrocellulose membranes, and immobilized proteins were stained with Ponceau dye–5% trichloroacetic acid (TCA). Membranes were washed twice with PBS and then in 10 mM Tris-HCl (pH 7.5)–150 mM KCl–0.1% Tween 20. Membranes were incubated for 90 min at room temperature (RT) in buffer A (15 mM Tris-HCl [pH 7.5], 150 mM KCl, 2 mM MgCl2, 1 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride [PMSF], 0.1% Tween 20, 0.1 mg/ml BSA). Nucleolin was added at a final concentration of 2.5 μg/ml for 2 h at RT. Membranes were washed four times in buffer A and then subjected to Western blot analysis.
RESULTS
Intracellular localization and functional analysis of the Flag-US11 protein.
For the purification of the native multiprotein complexes containing US11, we generated an expression vector allowing the expression of Flag-US11 protein. To validate this strategy, we first verified whether Flag-US11 and wild-type (wt) US11 proteins have the same intracellular distribution. HeLa cells were either infected with HSV-1 or transfected with expression vectors coding for US11 or Flag-US11. The results shown in Fig. 1A confirm that in HSV-1-infected cells, US11 was present in the cytoplasm and accumulated in the nucleoli, where it colocalized with nucleolin, a nucleolar marker. We then analyzed the intracellular distribution of wt US11 and Flag-US11 (Fig. 1B). HeLa cells were transfected with the pCMV-US11 vector to allow expression of wt US11 (Fig. 1B, upper panels) or with the pSG5-Flag-US11 vector to allow expression of Flag-US11 (Fig. 1B, lower panels). This analysis showed that the two proteins displayed very similar intracellular distributions (Fig. 1B, upper and lower panels). They were concentrated within the nucleoli and distributed throughout the cytoplasm. Nucleolin localization was unchanged in cells expressing untagged US11 (Fig. 1B, upper panels). Therefore, the intracellular distribution of Flag-US11 protein is very similar to that of wt US11 protein present in HSV-1-infected cells.
We then determined whether Flag-US11 protein retained the known function of the wt US11 protein. We have previously demonstrated that US11 can substitute for Rex in transactivating expression of the genes encoding HTLV-I env glycoproteins (10). We therefore determined whether Flag-US11, like US11 and Rex, exhibited the ability to transactivate expression of an mRNA coding for HTLV-I env (10). HeLa cells were cotransfected with expression vectors coding for the envelope protein and the Tax transcriptional activator of HTLV-I combined with the expression vector coding for Rex or wt US11 or Flag-US11. Control experiments included the cotransfection of HeLa cells with the env vector only, as well as with the env and tax vectors. After transfection, expression of functional envelope glycoproteins was evaluated by examining the formation of multinucleated cells (Fig. 1C). As expected, expression of Rex or wt US11 together with env and tax led to the formation of syncytia (indicated by arrows in Fig. 1C). No such multinucleated cells were observed in the negative controls (Fig. 1C, lower panels) (10). Syncytia were also observed in cells expressing Flag-US11 protein (Fig. 1C, arrow in right upper panel). Altogether, these data demonstrate that Flag-US11 displays an intracellular localization and a function similar to wt US11.
Nucleolin is associated with Flag-US11 protein in transfected cells.
Immunoprecipitation experiments with the Flag tag were optimized to allow the coimmunoprecipitation of US11-associated proteins. Because the ionic strength of the buffer can modify the electrostatic protein-protein interactions between Flag-US11 and its binding partners, we have evaluated various NaCl concentrations for purification of the US11-containing complexes. The proteins that coimmunoprecipitated with Flag-US11 in buffers containing NaCl concentrations ranging from 0 to 500 mM are shown in Fig. 2A. Numerous proteins coimmunoprecipitated with Flag-US11 when no NaCl was added to the extraction buffer. Most of these proteins were also detected after immunoprecipitation with Flag alone, although in smaller amounts (Fig. 2A, lane 1). Increasing the NaCl concentration from 0 to 150 mM led to an enrichment of proteins that bind specifically to Flag-US11 (Fig. 2A, lanes 1 to 6). Concentrations higher than 150 mM induced a strong reduction in the number of proteins coimmunoprecipitated. Therefore, the concentration of 150 mM NaCl was selected for the affinity proteomic analyses.
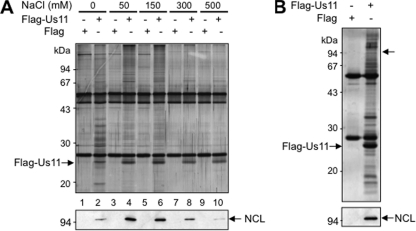
Fig 2.
Nucleolin is associated with Flag-US11 fusion protein in transfected cells: purification of the Flag-US11-containing complexes. (A) Optimization of the coimmunoprecipitation conditions of the Flag-US11 binding partners. HeLa cells were transfected with pSG5-Flag (lanes 1, 3, 5, 7, and 9) or with pSG5-Flag-US11 (lanes 2, 4, 6, 8, and 10) expression vectors. (Upper panel) Effect of NaCl concentration on the isolation of protein complexes containing Flag-US11 protein in a coimmunoprecipitation experiment using an anti-Flag antibody. Protein complexes were purified with an optimized buffer (see Materials and Methods) in the presence of an anti-Flag antibody and of various NaCl concentrations as indicated on the figure, and they were then analyzed by 1-DE and silver staining. The positions of the Flag-US11 protein and of molecular mass markers are indicated on the left. (Lower panel) The presence of nucleolin (NCL) in the corresponding purified complexes was checked by Western blot analysis using a polyclonal anti-nucleolin antibody. (B) Identification of nucleolin as a Flag-US11-interacting protein. Protein complexes (purified in the presence of 150 mM NaCl as described for panel A) from cells transfected with a plasmid expressing either Flag or Flag-US11 were separated by 1-DE and stained with Coomassie blue (upper panel). The portion of the gel corresponding to proteins of about 100 kDa in size (arrow on the right) was cut out, and the corresponding nucleolin protein was identified by mass spectrometry and bioinformatics analyses (see Table 1). The identity of this protein in the purified complexes was confirmed by Western blot analysis using a specific anti-nucleolin antibody (lower panel). The position of nucleolin (NCL) is indicated by an arrow on the right.
To determine whether nucleolin was a US11 binding partner, the presence of nucleolin in these purified protein complexes was evaluated by Western blot analysis with an anti-nucleolin antibody (Fig. 2A, lower panel). Results clearly showed that nucleolin was present in the pool of proteins coimmunoprecipitated with the anti-Flag antibody in cells expressing Flag-US11 (Fig. 2A, lanes 2, 4, 6, 8, and 10) and not detected in cells expressing Flag only (Fig. 2A, lanes 1, 3, 5, 7, and 9). The amount of nucleolin was proportional to that of Flag-US11 and was maximal at 50 and 150 mM NaCl.
We then used mass spectrometry to confirm the presence of nucleolin in the US11 binding partners. Copurified proteins were separated by 1-DE and stained with Biosafe Coomassie blue (Fig. 2B). Several gel fragments of molecular masses of approximately 100 kDa were cut out, and the proteins contained in each fragment were subjected to in-gel digestion with trypsin. Peptides were then analyzed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) and quadrupole TOF (Q-TOF) mass spectrometry. The sequences of the 5 peptides (Table 1) obtained in the portion of the gel indicated by an arrow in Fig. 2B (upper panel) unambiguously demonstrate the presence of nucleolin. Western blot analysis with the anti-nucleolin antibody (Fig. 2B, lower panel) validates the presence of nucleolin in complexes purified from cells expressing Flag-US11. Altogether, these results demonstrate that US11 protein and nucleolin are binding partners.
Table 1.
Nucleolin is present in Flag-US11-containing complexes purified from transfected cellsa
| Name | DB entry | MW | pI | Peptide no. | Coverage (%) | Sequence of peptide | Score |
|---|---|---|---|---|---|---|---|
| Nucleolin | NUCL_HUMAN | 76,614 | 4.6 | 5 | 7 | R/SISLYYTGEK/G | 10.49 |
| K/GFGFVDFNSEEDAK/A | 8.3 | ||||||
| K/EVFEDAAEIR/L | 7.67 | ||||||
| K/GIAYIEFK/T | 6.85 | ||||||
| R/LELQGPR/G | 6.31 |
Proteins coimmunoprecipitated with an anti-Flag antibody from cells expressing Flag-US11 protein were separated by 1-DE, and the proteins present in the portion of the gel corresponding to the proteins, the molecular weights (MW) of which were approximately 100 (the apparent nucleolin MW), were identified by mass spectrometry (MS) analysis. Only data from peptides identified as corresponding to nucleolin are presented. DB (database) entry, database entry name of nucleolin in SWISS-PROT, TrEMBL, or GenBank databases. MW and pI (isoelectric point) values corresponding to the theoretical MW and pI values were calculated using Compute pI/Mw software (www.expasy.org). Coverage, percentage of coverage of peptides in the nucleolin sequence.
Nucleolin interacts with US11 in vitro.
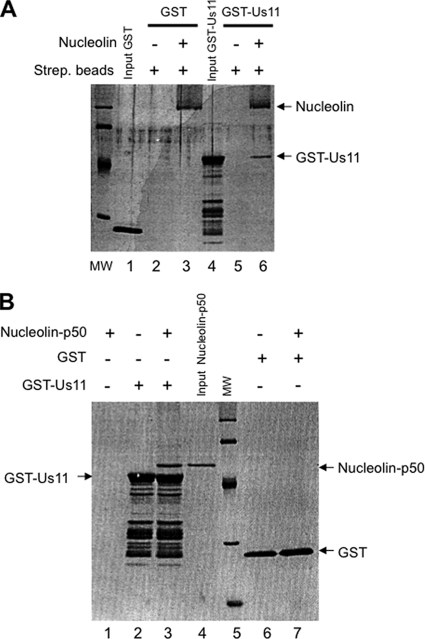
The experiments described above show that nucleolin and US11 were coimmunoprecipitated. To determine whether these two proteins interact with each other, we performed in vitro interaction experiments using purified nucleolin and purified GST-US11. Full-length nucleolin was purified and biotinylated as previously described (15). GST-US11 was expressed and purified from E. coli (34). GST-US11 or GST was incubated with streptavidin beads that were either loaded or not loaded with biotinylated nucleolin. The results in Fig. 3A clearly show that GST-US11 is specifically bound to nucleolin (lane 6) and not to streptavidin beads alone (lane 5). This finding was specific to US11, since GST alone did not interact at all with nucleolin (lane 3). To strengthen this result, and in particular to exclude the possibility that the interaction was mediated by a contaminating protein, we performed an additional pulldown experiment with nucleolin-p50, a recombinant nucleolin protein produced in E. coli, and with GST-US11 (Fig. 3B). GST-US11 and GST alone were bound to glutathione Sepharose beads and incubated with purified nucleolin-p50 protein (lanes 3 and 7). Only GST-US11 was able to retain nucleolin-p50 on the beads (lane 3), indicating that this interaction was mediated by US11. Altogether, these experiments demonstrated an association between nucleolin and US11. To further determine which domain(s) of US11 is required for the interaction with nucleolin, several mutants of US11 protein (Fig. 4A) were produced in E. coli and used in a far-Western blot analysis (Fig. 4C). Each GST-US11 protein mutant was loaded on a denaturing gel and transferred to a nitrocellulose membrane (Fig. 4B). Transferred proteins were then renatured and incubated with purified nucleolin protein. After extensive washing, the presence of nucleolin bound to US11 proteins was revealed with an anti-nucleolin antibody (Fig. 4C). As a control experiment (Fig. 4D), a Western blot analysis performed with the anti-nucleolin antibody on a membrane similar to that of Fig. 4C, but which had not been incubated with the purified hamster nucleolin, allowed us to conclude that the signal visualized in panel C was indeed due to the interaction of nucleolin with the US11 protein and not to a cross-reactivity of nucleolin antibody with these proteins. This far-Western blot analysis indicated that full-length US11 (lane 2) and US11 mutants Δ1–40, m5, and m6 (lanes 3 to 5) efficiently bind nucleolin whereas US11 mutant m15 (lane 6) and GST alone (lanes 1and 7) do not bind nucleolin. This indicates that the C-terminal domain of US11, which is required for the nucleolar localization of US11, contains residues also required for direct interaction with nucleolin (7, 43).
Fig 3.
Nucleolin interacts in vitro with full-length US11. (A) Nucleolin purified from CHO cells was biotinylated as previously described (15) and used for interaction studies with E. coli-expressed GST (lanes 1 to 3) or GST-US11 (lanes 4 to 6) proteins. Streptavidin beads (Strept. beads) were incubated with GST (lanes 2 and 3) or GST-US11 (lanes 5 and 6) in the absence (lanes 2 and 5) or presence (lanes 3 and 6) of biotinylated nucleolin. After extensive washing, streptavidin beads were incubated in Laemmli loading buffer, and recovered proteins were separated by 1-DE and then visualized by silver staining. The positions of nucleolin and GST-US11 are indicated. (B) Nucleolin-p50 protein was produced in E. coli and purified as previously described (36). Glutathione Sepharose beads were incubated with binding buffer (lane 1), GST-US11 (lanes 2 and 3), or GST protein (lanes 6 and 7). Nucleolin-p50 protein was then added to the beads (lanes 1, 3, and 7). After extensive washing, beads were incubated with Laemmli buffer and proteins were separated by 1-DE and then visualized by silver staining.
Fig 4.
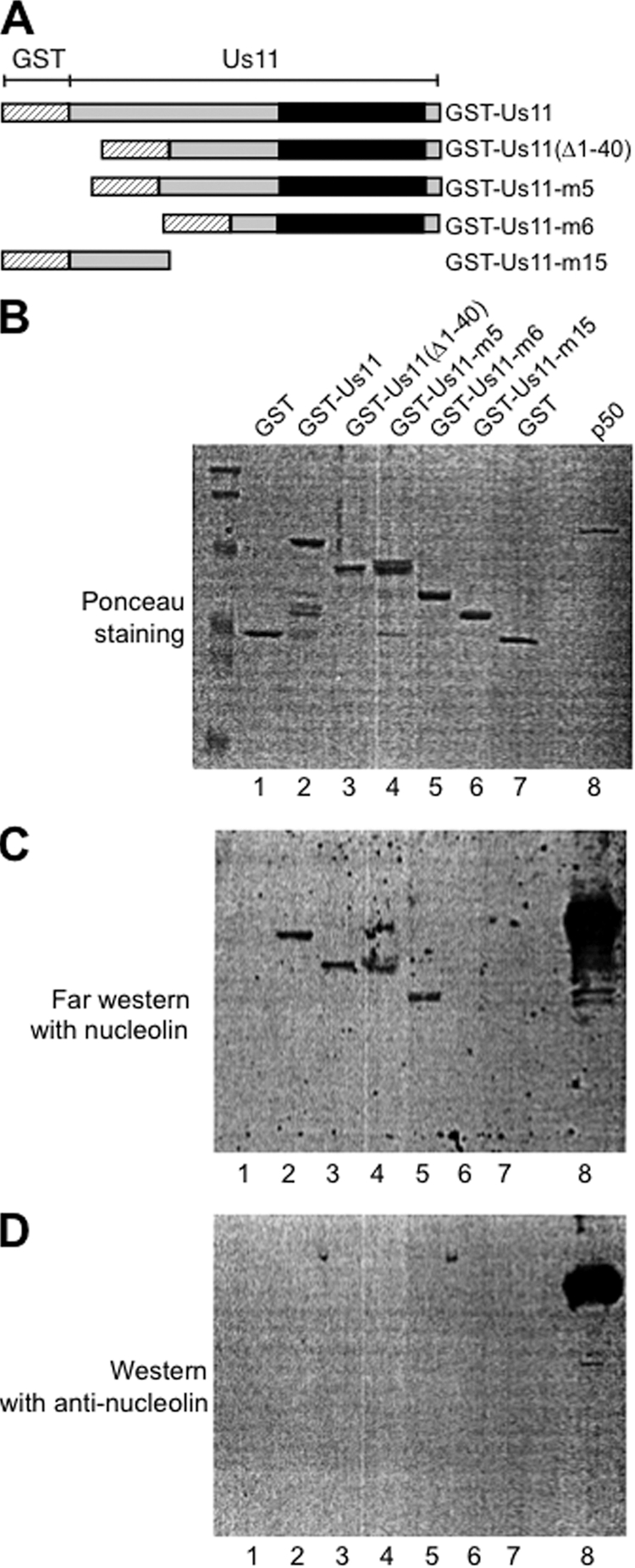
Interaction of nucleolin with US11 mutants. (A) Schematic representation of the different GST-US11 mutants used in this study. (B) The different proteins (1 μg) were separated by 1-DE, transferred on a nitrocellulose membrane, and then stained with Ponceau dye. The nucleolin-p50 protein (lane 8) corresponding to the C-terminal domain of nucleolin was used as a positive control for the Western blot analysis with anti-nucleolin antibody. (C) The membrane was incubated with the purified nucleolin protein as described in Materials and Methods, and after extensive washing, it was subjected to Western blot analysis with a polyclonal anti-nucleolin antibody. Nucleolin detection was then revealed using enhanced chemiluminescence (ECL). (D) As a control, a membrane similar to the one shown in panel B was subjected directly to a Western blot analysis with the anti-nucleolin antibody. Only the p50 nucleolin protein could be detected, showing that the nucleolin antibody does not cross-react with any of the GST-US11 proteins.
Nucleolin is associated with native wild-type US11 protein in HSV-1-infected cells.
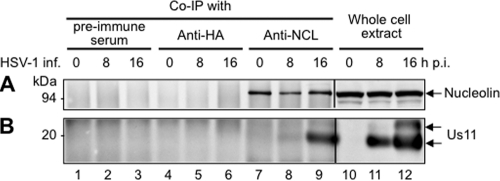
To determine whether this association occurs with the native proteins in HSV-1-infected cells, we performed reverse coimmunoprecipitation experiments using an anti-nucleolin antibody on protein extracts prepared from cells infected for 8 h and 16 h with wt HSV-1. Extracts from mock-infected cells were used as the negative controls. Results in Fig. 5 demonstrate that nucleolin was specifically immunoprecipitated by the rabbit polyclonal anti-nucleolin antibody (Fig. 5A, lanes 7 to 9) and not by the rabbit polyclonal anti-hemagglutinin (anti-HA) antibody (Fig. 5A, lanes 4 to 6) or by rabbit preimmune sera (Fig. 5A, lanes 1 to 3), which were used as negative controls. In addition, US11 was present only in complexes containing nucleolin that were purified from HSV-1-infected cells and not in those from mock-infected cells (Fig. 5B, lanes 7 to 9 and lanes 10 to 12). Furthermore, US11 was detected in larger amounts in complexes purified from cells infected for 16 h (Fig. 5B, lane 9) compared to cells infected for 8 h (Fig. 5B, lane 8). These results further validate the specific association between nucleolin and US11 in HSV-1-infected cells.
Fig 5.
US11 is present in nucleolin-containing complexes purified from extracts of HSV-1-infected (HSV-1 inf.) cells. HeLa cells were mock infected at 0 h (lanes 1, 4, 7, and 10) or infected with HSV-1 for 8 h (lanes 2, 5, 8, and 11) and 16 h (lanes 3, 6, 9, and 12) at 10 PFU/cell. Coimmunoprecipitations (Co-IP) (lanes 1 to 9) were performed with the corresponding total cell extract proteins and used a polyclonal anti-nucleolin (anti-NCL; lanes 7 to 9) or an anti-HA (lanes 4 to 6) antibody or a preimmune serum (lanes 1 to 3), as indicated at the top of the figure. The presence of nucleolin (A) and of US11 (B) in the purified complexes was checked by Western blot analysis using anti-nucleolin- and anti-US11-specific antibodies, respectively. The positions of nucleolin and of US11 are indicated. Total cell extract proteins not submitted to the coimmunoprecipitation were used as positive controls (lanes 10 to 12).
Nucleolin depletion leads to US11 accumulation in nucleoli.
In order to analyze the functional implications of nucleolin and US11 association, the cellular localization of US11 in nucleolin-depleted cells was investigated (Fig. 6). To this end, we performed siRNA nucleolin silencing in a HeLa cell line stably expressing wt US11 (13, 37). As previously described for HSV-1-infected cells (Fig. 1), US11 localized to nucleoli and to the cytoplasm in this stable cell line (Fig. 6A and C). In nucleolin-depleted cells, US11 was still present in nucleoli (Fig. 6B), thereby ruling out the possibility that nucleolar localization of US11 is nucleolin dependent. Fluorescence quantification of US11 intensities in a total of 87 cells (Fig. 6E) revealed that the US11 ratio between nucleolar and cytoplasmic compartments is 2-fold higher in nucleolin-depleted cells than in any type of control cells (Fig. 6E). This significantly higher level of nucleolar US11 was not a consequence of overexpression, since the US11 level remained constant after nucleolin silencing (Fig. 6F). Therefore, our results point to a redistribution of US11 toward the nucleolar compartment in nucleolin-depleted cells, thereby indicating that the absence of nucleolin leads to US11 accumulation within nucleoli to the detriment of the US11 cytoplasmic fraction. In summary, our results reveal that nucleolin plays a role in US11 export from nucleoli to the cytoplasm.
DISCUSSION
The function of US11, an abundant HSV-1 protein, in viral infection is still not well understood. Previous studies have demonstrated that US11 is present in different cell compartments during the time course of HSV-1 infection, and in particular it seems to accumulate within nucleoli (3, 12, 25, 28). The function of US11 during HSV-1 infection probably requires the interaction with cellular proteins, but so far, only a very limited number of proteins have been shown to interact with US11 (2, 14, 20, 27, 33).
In order to better understand US11 function and trafficking within the cell, we have identified and characterized a new nucleolar US11 protein partner. Taking advantage of a Flag-US11-tagged protein that behaves as a wild-type US11 protein produced during the infection (Fig. 1), we determined by proteomic analysis that nucleolin, an abundant nucleolar protein, interacts with US11 (Fig. 2).
What could be the role of the interaction of nucleolin with US11? In addition to showing a strong nucleolar accumulation, US11 is also found associated with cytoplasmic ribosomes (12, 31). We and others have shown that upon HSV-1 infection, rRNA synthesis is altered (1, 41), whereas ribosomal protein synthesis is maintained until later in the infection (16, 38). We have also shown that nucleolin interacts with certain ribosomal proteins and might be involved in their assembly within ribosomal subunits (4). Therefore, the interaction between nucleolin and US11 could be involved in the association of US11 with the ribosomal subunits.
Another possibility is that interaction of nucleolin with US11 participates in the intracellular trafficking of US11. US11 shuttling between the nucleus and the cytoplasm in HSV-1-infected cells or in transiently transfected cells (7) has recently been confirmed in living cells (43). This nucleocytoplasmic shuttling requires the C-terminal extremity of US11 (7). This C-terminal domain of US11 is composed of the 20 XPR tripeptide repetitions that are organized into a polyproline type II helix structure that is commonly involved in protein-protein interaction (42). In this study, using several in vitro protein interaction assays (Fig. 3 and 4), we showed that this C-terminal domain is indeed involved in the interaction with nucleolin. A previous study using saturated mutagenesis and computer modeling of this C-terminal domain of US11 that interacts with nucleolin has shown that this domain carries the capacity of US11 to be concentrated within nucleoli and also to be exported to the cytoplasm (7, 34).
Interestingly, the silencing of nucleolin prevents HSV-1 infection (6), and the expression of nucleolin is required for nuclear egress of HSV-1 nucleocapsids (32), demonstrating that nucleolin is an important protein for HSV-1 infection and that the interaction with US11 could play an important role.
We demonstrate that the silencing of nucleolin affects the trafficking of US11 between the nucleolus and the cytoplasm (Fig. 6). Indeed, we observed a significant increase in the accumulation of US11 within nucleoli when nucleolin expression was inhibited (more than 4-fold higher in the nucleoli than in the cytoplasm in silenced cells compared to 2-fold higher in control cells; Fig. 6), suggesting that nucleolin participates in US11 nucleocytoplasmic transport by promoting US11 nucleolar export.
The role of nucleolin in US11 nucleocytoplasmic trafficking seems to be a one-way directional transport out of the nucleoli, since US11 is still present in the nucleoli of nucleolin-silenced cells.
This hypothesis is also supported by the fact that US11 exhibits a noncanonical nuclear export signal (7) and is based on recent data that show that US11 nucleocytoplasmic export does not require the CRM1 conventional export factor (43). We show here that the level of cytoplasmic localization of US11 depends on the presence of nucleolin. These results, together with the findings that nucleolin has been recently shown to be required for efficient nuclear egress of HSV-1 nucleocapsids (32) and that US11 is a structural protein that is cotransported with capsids (14), suggest that nucleolin, through its interaction with US11, could participate in the transport of the viral particles.
ACKNOWLEDGMENTS
We acknowledge the contribution of the PLATIM platform of SFR BioSciences Gerland-Lyon Sud (UMS3444/US8). We are grateful to Marie Chapoton for her technical assistance.
This work was supported by CNRS, INSERM, Université Claude Bernard Lyon-1, and Ecole Normale Supérieure de Lyon. It was funded by grants from the Région Rhône-Alpes (thématiques prioritaires and Cluster 10 and MIRA 2007 and 2010), the Institut National contre le Cancer (RIBOCAN), the Agence Nationale de la Recherche (ANR-07-BLAN-0062-01), and the Association pour la recherche sur le Cancer (ECL2010R01122).
A.C., A.G., J.-J.D., and Y.C. are members of INSERM.
Footnotes
Published ahead of print 30 November 2011
REFERENCES
- 1. Belin S, et al. 2010. Uncoupling ribosome biogenesis regulation from RNA polymerase I activity during herpes simplex virus type 1 infection. RNA 16:131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benboudjema L, Mulvey M, Gao Y, Pimplikar SW, Mohr I. 2003. Association of the herpes simplex virus type 1 Us11 gene product with the cellular kinesin light-chain-related protein PAT1 results in the redistribution of both polypeptides. J. Virol. 77:9192–9203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Besse S, et al. 1996. In situ hybridization and immuno-electron microscope analyses of the Us11 gene of herpes simplex virus type 1 during transient expression. Chromosoma 104:434–444 [DOI] [PubMed] [Google Scholar]
- 4. Bouvet P, Diaz JJ, Kindbeiter K, Madjar JJ, Amalric F. 1998. Nucleolin interacts with several ribosomal proteins through its RGG domain. J. Biol. Chem. 273:19025–19029 [DOI] [PubMed] [Google Scholar]
- 5. Brown SM, Harland J. 1987. Three mutants of herpes simplex virus type 2: one lacking the genes US10, US11 and US12 and two in which Rs has been extended by 6 kb to 0.91 map units with loss of Us sequences between 0.94 and the Us/TRs junction. J. Gen. Virol. 68:1–18 [DOI] [PubMed] [Google Scholar]
- 6. Callé A, et al. 2008. Nucleolin is required for an efficient herpes simplex virus type 1 infection. J. Virol. 82:4762–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Catez F, et al. 2002. Unique motif for nucleolar retention and nuclear export regulated by phosphorylation. Mol. Cell. Biol. 22:1126–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Couté Y, et al. 2010. Early activation of the fatty acid metabolism pathway by chronic high glucose exposure in rat insulin secretory beta-cells. Proteomics 10:59–71 [DOI] [PubMed] [Google Scholar]
- 9. Dalziel RG, Marsden HS. 1984. Identification of two herpes simplex virus type 1-induced proteins (21K and 22K) which interact specifically with the a sequence of herpes simplex virus DNA. J. Gen. Virol. 65:1467–1475 [DOI] [PubMed] [Google Scholar]
- 10. Diaz JJ, et al. 1996. Post-transcriptional transactivation of human retroviral envelope glycoprotein expression by herpes simplex virus Us11 protein. Nature 379:273–277 [DOI] [PubMed] [Google Scholar]
- 11. Diaz JJ, Giraud S, Greco A. 2002. Alteration of ribosomal protein maps in herpes simplex virus type 1 infection. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 771:237–249 [DOI] [PubMed] [Google Scholar]
- 12. Diaz JJ, et al. 1993. The herpes simplex virus type 1 Us11 gene product is a phosphorylated protein found to be non-specifically associated with both ribosomal subunits. J. Gen. Virol. 74:397–406 [DOI] [PubMed] [Google Scholar]
- 13. Diaz-Latoud C, et al. 1997. Herpes simplex virus Us11 protein enhances recovery of protein synthesis and survival in heat shock treated HeLa cells. Cell Stress Chaperones 2:119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Diefenbach RJ, et al. 2002. Herpes simplex virus tegument protein US11 interacts with conventional kinesin heavy chain. J. Virol. 76:3282–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ghisolfi-Nieto L, Joseph G, Puvion-Dutilleul F, Amalric F, Bouvet P. 1996. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J. Mol. Biol. 260:34–53 [DOI] [PubMed] [Google Scholar]
- 16. Greco A, Laurent AM, Madjar JJ. 1997. Repression of ß-actin synthesis and persistence of ribosomal protein synthesis after infection of HeLa cells by herpes simplex virus type 1 are under translational control. Mol. Gen. Genet. 256:320–327 [DOI] [PubMed] [Google Scholar]
- 17. Greco A, et al. 1994. The DNA sequence coding for the 5′ untranslated region of herpes simplex type 1 ICP22 mRNA mediates a high level of gene expression. J. Gen. Virol. 75:1693–1702 [DOI] [PubMed] [Google Scholar]
- 18. Hill TJ, Yirrell DL, Blyth WA. 1986. Infection of the adrenal gland as a route to the central nervous system after viraemia with herpes simplex virus in the mouse. J. Gen. Virol. 67:309–320 [DOI] [PubMed] [Google Scholar]
- 19. Javouhey E, Gibert B, Arrigo AP, Diaz JJ, Diaz-Latoud C. 2008. Protection against heat and staurosporine mediated apoptosis by the HSV-1 US11 protein. Virology 376:31–41 [DOI] [PubMed] [Google Scholar]
- 20. Khoo D, Perez C, Mohr I. 2002. Characterization of RNA determinants recognized by the arginine- and proline-rich region of Us11, a herpes simplex virus type 1-encoded double-stranded RNA binding protein that prevents PKR activation. J. Virol. 76:11971–11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Longnecker R, Roizman B. 1987. Clustering of genes dispensable for growth in culture in the S component of the HSV-1 genome. Science 236:573–576 [DOI] [PubMed] [Google Scholar]
- 22. Longnecker R, Roizman B. 1986. Generation of an inverting herpes simplex virus 1 mutant lacking the L-S junction a sequences, an origin of DNA synthesis, and several genes including those specifying glycoprotein E and the alpha 47 gene. J. Virol. 58:583–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. López-Iglesias C, Puvion-Dutilleul F, Cebrian J, Christensen ME. 1988. Herpes simplex virus type 1-induced modifications in the distribution of nucleolar B-36 protein. Eur. J. Cell Biol. 46:259–269 [PubMed] [Google Scholar]
- 24. Lymberopoulos MH, Pearson A. 2007. Involvement of UL24 in herpes-simplex-virus-1-induced dispersal of nucleolin. Virology 363:397–409 [DOI] [PubMed] [Google Scholar]
- 25. MacLean CA, Rixon FJ, Marsden HS. 1987. The products of gene Us11 of herpes simplex virus type 1 are DNA-binding and localize to the nucleoli of infected cells. J. Gen. Virol. 68:1921–1937 [DOI] [PubMed] [Google Scholar]
- 26. Nishiyama Y, Kurachi R, Daikoku T, Umene K. 1993. The US 9, 10, 11, and 12 genes of herpes simplex virus type 1 are of no importance for its neurovirulence and latency in mice. Virology 194:419–423 [DOI] [PubMed] [Google Scholar]
- 27. Peters GA, Khoo D, Mohr I, Sen GC. 2002. Inhibition of PACT-mediated activation of PKR by the herpes simplex virus type 1 Us11 protein. J. Virol. 76:11054–11064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Puvion-Dutilleul F. 1987. Localization of viral-specific 21 kDa protein in nucleoli of herpes simplex infected cells. Eur. J. Cell Biol. 43:487–498 [PubMed] [Google Scholar]
- 29. Roizman B, Sears AE. 1993. Herpes simplex viruses and their replication, p 11–68 In Roizman B, Whitley RJ, Lopez C. (ed), The human herpes viruses, 3rd ed Raven Press, New York, NY [Google Scholar]
- 30. Roller RJ, Roizman B. 1991. Herpes simplex virus 1 RNA-binding protein US11 negatively regulates the accumulation of a truncated viral mRNA. J. Virol. 65:5873–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roller RJ, Roizman B. 1992. The herpes simplex virus 1 RNA binding protein Us11 is a virion component and associates with ribosomal 60S subunits. J. Virol. 66:3624–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sagou K, Uema M, Kawaguchi Y. 2010. Nucleolin is required for efficient nuclear egress of herpes simplex virus 1 nucleocapsids. J. Virol. 84:2110–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sànchez R, Mohr I. 2007. Inhibition of cellular 2′-5′ oligoadenylate synthetase by the herpes simplex virus type 1 Us11 protein. J. Virol. 81:3455–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schaerer-Uthurralt N, Erard M, Kindbeiter K, Madjar JJ, Diaz JJ. 1998. Distinct domains in herpes simplex virus type 1 US11 protein mediate post-transcriptional transactivation of human T-lymphotropic virus type I envelope glycoprotein gene expression and specific binding to the Rex responsive element. J. Gen. Virol. 79:1593–1602 [DOI] [PubMed] [Google Scholar]
- 35. Scherl A, et al. 2002. Functional proteomic analysis of the human nucleolus. Mol. Biol. Cell 13:4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Serin G, et al. 1997. Two RNA-binding domains determine the RNA-binding specificity of nucleolin. J. Biol. Chem. 272:13109–13116 [DOI] [PubMed] [Google Scholar]
- 37. Simonin D, Diaz JJ, Kindbeiter K, Pernas P, Madjar JJ. 1995. Phosphorylation of herpes simplex virus type 1 Us11 protein is independent of viral genome expression. Electrophoresis 16:1317–1322 [DOI] [PubMed] [Google Scholar]
- 38. Simonin D, Diaz JJ, Masse T, Madjar JJ. 1997. Persistence of ribosomal protein synthesis after infection of HeLa cells by herpes simplex virus type 1. J. Gen. Virol. 78:435–443 [DOI] [PubMed] [Google Scholar]
- 39. Storck S, Thiry M, Bouvet P. 2009. Conditional knockout of nucleolin in DT40 cells reveals the functional redundancy of its RNA-binding domains. Biol. Cell 101:153–167 [DOI] [PubMed] [Google Scholar]
- 40. Ugrinova I, et al. 2007. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol. Biol. 8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wagner EK, Roizman B. 1969. Ribonucleic acid synthesis in cells infected with herpes simplex virus. I. Patterns of ribonucleic acid synthesis in productively infected cells. J. Virol. 4:36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Williamson MP. 1994. The structure and function of proline-rich regions in proteins. Biochem. J. 297:249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xing J, Wu F, Pan W, Zheng C. 2010. Molecular anatomy of subcellular localization of HSV-1 tegument protein US11 in living cells. Virus Res. 153:71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]