Abstract
Respiratory syncytial virus (RSV) is one of the major causes of respiratory infections in children, and it is the main pathogen causing bronchiolitis in infants. The binding and entry mechanism by which RSV infects respiratory epithelial cells has not yet been determined. In this study, the earliest stages of RSV infection in normal human bronchial epithelial cells were probed by tracking virions with fluorescent lipophilic dyes in their membranes. Virions colocalized with cholesterol-containing plasma membrane microdomains, identified by their ability to bind cholera toxin subunit B. Consistent with an important role for cholesterol in RSV infection, cholesterol depletion profoundly inhibited RSV infection, while cholesterol repletion reversed this inhibition. Merger of the outer leaflets of the viral envelope and the cell membrane appeared to be triggered at these sites. Using small-molecule inhibitors, RSV infection was found to be sensitive to Pak1 inhibition, suggesting the requirement of a subsequent step of cytoskeletal reorganization that could involve plasma membrane rearrangements or endocytosis. It appears that RSV entry depends on its ability to dock to cholesterol-rich microdomains (lipid rafts) in the plasma membrane where hemifusion events begin, assisted by a Pak1-dependent process.
INTRODUCTION
Respiratory syncytial virus (RSV) is one of the most prevalent respiratory pathogens targeting all age groups. RSV has been found to cause 2.4% of community-acquired pneumonias (20). However, infants (<18 months) and the elderly suffer from the most severe RSV disease compromising the lower respiratory tract (i.e., bronchiolitis and pneumonia) (75). By 2 years of age, 90% of children show serological evidence of having been infected by RSV (2, 75). Complete immunity is not developed, allowing new infections throughout life. Although cleverly designed candidate vaccines for RSV have been developed, none is currently approved. Passive immunoprophylaxis has rendered tremendous benefits for children at risk for suffering severe RSV disease. Unfortunately, the cost of this treatment limits its use as a prophylactic in the general ward in developing countries (23, 37). The development of new antivirals requires a thorough understanding of the earliest molecular events of RSV infection, particularly attachment and fusion, that deliver the virus genome into the target cell.
Iduronic acid-enriched heparan sulfate proteoglycans (HSPGs) present on HEp-2 cell membranes have been reported to mediate virus attachment (28, 30). However, it appears that this receptor is not the one used by the virus in attaching to human airway epithelial cells, since several reports have concluded that HSPGs are not localized on the ciliated apical side of fully differentiated bronchial epithelial cells cultured at the air-liquid interface or tracheal tissue sections (29, 44, 90, 91). In addition, the RSV envelope protein F, which triggers envelope fusion with the target cell membrane, may also independently attach virions to cells, though such attachment seemed less dependent on HSPGs than G protein-mediated attachment to HEp-2 cells (82). The F and G proteins have been reported to form a complex on the virion envelope (50). The structure of a soluble F protein, the transmembrane and cytosolic tail domains of which were both replaced with a hexahistidine tag, appeared spherical by electron microscopic analysis and was not aggregated, consistent with a native pretriggered trimer; but after exposure to low-molarity buffer, it acquired a “hatpin” shape that aggregated as rosettes characteristic of the posttriggered form (8). During the maturation process of the F protein, a furin-like protease cleaves the F0 precursor at two sites releasing a short peptide of 27 amino acids and resulting in the formation of two subunits stabilized by two disulfide bridges (F2-F1) (15, 25, 79). This cleavage is necessary to make the F protein fusogenic. Although the crystal structure for the prefusion conformation of RSV F protein has not yet been solved, the prefusion structure of the parainfluenza virus type 5 (PIV5) F protein has been (89) and is likely to be a representative of the paramyxovirus F structure. Each subunit of the trimeric PIV5 prefusion structure contributes to the globular head, which is attached to a stalk formed by the C-terminal heptad repeat (HRB) region. The fusion peptides of each subunit at the N termini of the heptad repeat A (HRA) regions are sequestered between the adjacent subunits (89).
The stimulus that triggers the RSV F protein is not known, but the refolding of the F protein probably follows what is known for other class I fusion proteins. The apical, spring-loaded portion of the F1 protein reforms to generate a very long HRA α-helix with the fusion peptide at its N terminus that reaches out to target the cell membrane. The fusion peptide inserts into the target membrane, and the F protein folds back on itself, bringing the C-terminal HRB into an antiparallel assembly with the N-terminal HRA that is adjacent to the fusion peptide (11, 54, 76), forming a trimeric hairpin or 6-helix bundle while bringing the two membranes together. Based on other class I fusion proteins, this conformational change is required for pore stabilization and enlargement (11, 57, 58).
Currently, it is unclear where RSV fuses its envelope with the target cell membrane to deliver its ribonucleoprotein complex to the cytoplasm. Previous studies suggested that RSV fusion occurs at the plasma membrane based on the fact that fusion is independent of endosomal acidification (33, 63, 69, 78). However, Kolokoltsov et al. (41) reported that RSV could deliver its genome after fusing with early endosomes in the clathrin-mediated pathway. These findings were obtained in HeLa cells by inhibiting virus entry with small interfering RNAs (siRNAs) against genes important for clathrin-mediated endocytosis and dominant negative versions of clathrin-associated proteins such as Eps-15 (41).
We adopted the approach of Sakai et al. (73) to identify membrane microdomains used for virus docking to primary normal human bronchial epithelial (NHBE) cells and to track virus fusion with these cells in real time. The tracking approach is based on the physics of light diffraction, which allows nanoparticles such as fluorescently labeled viruses to be visualized in living cells. Similar strategies have been applied to track influenza virus, dengue virus, hepatitis C virus (HCV), HIV, and poliovirus entry (4, 5, 12, 40, 61, 72, 73, 87).
Our data show that docking takes place on cholesterol-rich microdomains that are similar to lipid rafts and that RSV hemifusion events begin in the plasma membrane. We also found that cholesterol in the cell membrane is essential for RSV to successfully infect bronchial epithelial cells and that RSV infection is sensitive to Pak1 inhibition, suggesting that complete fusion may require plasma membrane rearrangement processes and/or endocytosis, which are dependent on actin polymerization and cytoskeletal reorganization.
MATERIALS AND METHODS
NHBE cells culture.
NHBE cells at passage 1 were provided by Clonetics (Cambrex Bio Science, Walkersville, MD), which had obtained the cells from healthy volunteers after informed consent, and were grown following the provided guidelines. Briefly, the cells were grown in BEGM, which consists of basal medium (BEBM) supplemented with bovine pituitary extract, insulin, hydrocortisone, human epithelial growth factor, epinephrine, transferrin, retinoic acid, and triiodothyronine, for two passages before being collected and stored at −135°C in a liquid nitrogen storage cabinet. For experiments, cells at passages 4 and 5 were used and grown in BEGM. On the day of experimentation, cells were washed twice with phosphate-buffered saline (PBS) before BEBM containing either RSV or the various drugs was added.
Production and purification of respiratory syncytial virus.
For these experiments, we used RSV-A2 and recombinant strains of RSV that expressed the green fluorescent protein (GFP; rgRSV) and contained different combinations of the three RSV envelope proteins (rgRSV-GF, lacking SH; rgRSV-SF, lacking G; and rgRSV-F, lacking SH and G). All of them were partially purified by pelleting through a glycerol layer following the general recommendations of Carpenter et al. (6). HEp-2 cells were seeded into T-175 flasks in Opti-MEM medium (Invitrogen Corporation, Carlsbad, CA) at a density of 8.75 × 106 cells in order to reach 50% to 60% confluence the next day. On the day of infection, each flask was washed once with Ca2+-and Mg2+-free Dulbecco's 1× PBS (Mediatech Inc., Herndon, VA). The inoculum was prepared by resuspending 0.1 to 0.2 PFU/cell of virus in Opti-MEM (Invitrogen Corporation, Carlsbad, CA) containing 2% fetal bovine serum (FBS) (Mediatech Inc., Herndon, VA). Inoculation was allowed to take place at 37°C, 5% CO2, and humid atmosphere for 2 h, with gentle rocking every 15 min after which the inoculum was replaced with Opti-MEM containing 2% FBS (40 ml per flask). The flasks were returned to the CO2 incubator, and the infection was allowed to progress until cytopathological effects were evident in 70% to 80% of the cell monolayer (about 2 to 2.5 days after infection). The cell monolayer was then scraped into the culture medium, transferred to a 50-ml conical tube, vortexed at half speed for 10 s, and centrifuged at 3,200 rpm for 10 min at 4°C to remove the cells. The supernatant was transferred to a new conical tube and mixed with 0.1 volume (approximately 4.2 ml) of sterile 1 M MgSO4. RSV was pelleted through a glycerol layer consisting of 30% glycerol in 0.1 M MgSO4 and 50 mM HEPES, pH 7.5 (filtered through a 0.22-μm membrane filter) by centrifuging at 24,000 × g in an SW28 rotor for 3 h at 4°C. The supernatant was suctioned off from the top, avoiding the viral pellet, which was gently rinsed with serum-free Opti-MEM without disturbing it and resuspended in 750 μl of precooled (4°C) buffer containing 0.22-μm-filtered 50 mM HEPES, pH 7.5, 0.1 M MgSO4, and 150 mM NaCl. The RSV suspension was aliquoted and stored at −135°C in liquid nitrogen (vapor phase). For these experiments, the RSV titer was determined on NHBE cell cultures by a fluorescence-based flow cytometry assay. The titer was estimated at those dilutions in which the multiplicity of infection (MOI) was such as to produce an infection rate of 0.2 to 24%. This range was found to define the linear zone of infection where one infected cell corresponds to a single infectious event fitting a Poisson distribution (27, 45, 47). The formula used to estimate the titer was as follows: (% of infected cells × total number of cells × dilution factor)/(100 × volume of infectious aliquot). The titer we obtained correlated with the fluorescence-based titration assay described by Techaarpornkul et al. (81).
Fluorescent labeling.
Purified RSV (100 mg in 1 ml) was simultaneously labeled with DiOC18 and R18 (Invitrogen) following the general recommendations of Sakai et al. (73). DiOC18 and R18 at 10 nM and 20 nM, respectively, five times the suggested concentrations, were used in order to enhance the signal-to-noise ratio. After the dyes were added to the virus suspension, it was mixed vigorously and then gently shaken for 1 h at room temperature away from light. Finally, unincorporated dyes present in the reaction mixture were removed by gel filtration using G-50 MacroSpin columns (Harvard Apparatus).
To determine if the labeling procedure impaired the virus infectivity, we subjected a recombinant version of RSV encoding GFP (rgRSV) (30) to the same procedure. Then, we used the dually labeled rgRSV to infect NHBE cells at an equivalent titer of 0.1 MOI. After 16 h, the number of green cells was evaluated by fluorescence microscopy and compared to the expected number of green cells based on the MOI that was inoculated. Although DiOC18 emits in the green channel, there is no confusion with the GFP fluorescence since DiOC18-labeled virus is punctiform while the GFP protein is found throughout the cytoplasm.
Real-time tracking assays.
NHBE cells were seeded on glass-bottom culture dishes (Fluorodish, World Precision Instruments) and grown to 80% confluence. On the day of the experiment, cell cultures were washed with ice-cold phosphate-buffered saline (PBS) and then incubated with dually labeled virus (seen as red; see Results) at 4°C for 30 min to synchronize virus attachment. The virus was added at the equivalent of 3 PFU/cell based on the titer determined prior to labeling. During the final 10 min of the attachment stage, either Alexa-647-labeled cholera toxin subunit B (CTB; 50 ng/ml; Invitrogen, seen as blue) or Cell Mask Deep Red plasma membrane marker (5 μg/ml; Invitrogen, seen as blue) was added to the cell cultures. The cell culture dish was placed on a microincubator (PDMI-2; Harvard Apparatus) and warmed to 37°C while simultaneously being imaged using an Olympus FV1000 laser scanning confocal upright microscope with an Olympus LUMPLFL 60× 0.9 numerical aperture (NA) water-immersion lens. The cells were scanned with a 488-nm laser and two fluorescence emissions, 500 to 530 nm and 555 to 618 nm, were simultaneously detected. A 635-nm laser was used to excite markers in the 655- to 755-nm range.
Probing dynamin-dependent or -independent pathways.
The highest concentrations of the respective inhibitors at which there was no cell toxicity were chosen for these experiments. Dynasore, which inhibits dynamin-dependent endocytic processes, was used at 80 μM, while IPA-3, which targets PAK1 and consequently prevents the actin polymerization and cytoskeletal reorganization that results in the formation of lamellipodia, ruffling, and/or endocytosis/macropinocytosis, was used at 10 μM. The concentrations used were effective in preventing transferrin and dextran uptake, respectively. The cell cultures were treated with inhibitor prior to infection (1 h in the case of dynasore or 30 min in the case of IPA3). As a control, cell cultures were incubated with dimethyl sulfoxide (DMSO) (1:1,000). The virus inoculum (<0.3 PFU/cell) containing inhibitor or DMSO was then added to the cell cultures, and incubation was continued for 2 h. Afterwards, the infectious inoculum was removed, the cell culture was washed with Hanks balanced salt solution (HBSS) buffer to remove the unbound virions and placed on ice. Cold trypsin-EDTA (0.9 mg/ml) was added to remove bound but unfused virions. After 10 min, the trypsin-EDTA was inactivated with Opti-MEM–10% FBS. Cells were washed with HBSS, fresh BEGM without inhibitor was added to each well, the plate was incubated at 37°C, and the infection was allowed to proceed for 16 h. The fluorescence associated with infected cells was detected by flow cytometry.
Copatching assays.
NHBE cells were seeded on 8-well chamber slides (Lab-Tek II; NuncBrand) at an appropriate cell density to reach 70 to 80% confluence. On the day of the experimental assay, NHBE cell cultures were washed with ice-cold PBS followed by washing with HEPES-buffered saline solution (HEPES-BSS). Each set of cell cultures was exposed to rgRSV-SGF, -GF, -SF, or -F (MOI of 20 PFU/cell). To synchronize virus attachment, cell cultures were kept at 4°C for 45 min then switched to 37°C for 30 min. After washing, both Alexa-555-labeled cholera toxin subunit B (20 μg/ml in PBS with 0.1% bovine serum albumin [BSA]; Invitrogen, seen as red) and Alexa-488 (Zenon)-labeled anti-RSV-F envelope protein antibody (Millipore, seen as green) were added to the respective wells and incubated at 12°C for 30 min. The cells were then washed with ice-cold PBS and fixed with 4% paraformaldehyde (PFA; Electron Microscopy Sciences) in PBS for 20 min on ice followed by 40 min at room temperature. The cells were washed and prepared for confocal microscopy by adding Prolong-Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen, seen as blue).
Cholesterol dependency of RSV entry (confocal assay).
NHBE cells were seeded on 8-well chamber slides (Lab-Tek II; NuncBrand) at an appropriate cell density to reach 70 to 80% confluence. On the day of the experiment, NHBE cell cultures were washed with PBS before being incubated as (i) “treatment” (methyl-beta-cyclodextrin [MBCD; 2.5 mM] and lovastatin [Lov; 2.5 μg/ml]) for 30 min; (2) “cholesterol replenishment” (control for MBCD; same as “treatment” followed by MBCD-balanced water-soluble cholesterol [400 μg/ml; Sigma-Aldrich]) for 45 min; or (3) “basal conditions” (cell cultures fed with basal medium). After the respective treatments, each set of cell cultures was washed with ice-cold PBS followed by washing with HEPES-BSS. Control cultures were fed with basal medium and both “treatment” and “cholesterol replenishment” cultures were fed with basal medium and Lov (2.5 μg/ml). Each set of cell cultures was exposed to RSV (MOI of 20 PFU/cell). In order to synchronize virus attachment and fusion, the cell cultures were kept at 4°C for 1 h and then switched to 37°C for 1 h. After the inoculation, the cells were washed with ice-cold PBS and fixed with 4% paraformaldehyde (PFA; Electron Microscopy Sciences) in PBS for 20 min on ice followed by 40 min at room temperature. Cells were washed with PBS and then incubated with Alexa-488–wheat germ agglutinin (WGA; 5 μg/ml in 1× PBS; Invitrogen, seen as green) for 10 min at room temperature. After washing three times, the cells were incubated with PBS buffer containing 3% goat serum, 1% glycine, and 0.1% saponin for 20 min at room temperature in order to block, quench, and permeabilize the cells. Viral ribonucleoproteins in the cytoplasm were identified by incubating the cells with Alexa 555-(Zenon)-labeled anti-RSV-N antibody (MAB858-3; Millipore, seen as red) in the blocking buffer mentioned above for 1 h at 20°C. Cells were prepared for confocal microscopy by adding Prolong-Gold antifade reagent with DAPI (Invitrogen).
Cholesterol dependency of RSV entry (flow cytometry assay).
NHBE cells were seeded on 6-well plates (Costar-Corning) at an appropriate cell density to reach 70 to 80% confluence. On the day of the experiment, NHBE cell cultures were washed with PBS and subjected to (i) mock treatment, (ii) cholesterol depletion by incubation with 1.25 to 5 mM MBCD (as indicated) and 2.5 μg/ml Lov for 30 min (culture 2), or (iii) cholesterol depletion by incubation with 2.5 mM MBCD and 2.5 μg/ml Lov followed by reversal by treatment with MBCD-balanced water-soluble cholesterol (400 μg/ml; Sigma-Aldrich) for 45 min (culture 3). After the respective treatments, each set of cell cultures was washed with PBS followed by washing with HEPES-BSS. For maintenance conditions, control cultures were fed with basal medium, and cultures 2 and 3 were fed with basal medium and Lov (2.5 μg/ml). Each set of cell cultures was next exposed to wild-type (wt) RSV expressing GFP (rgRSV-SGF) or the mutants rgRSV-GF, -SF, or -F (1 PFU/cell) for 2 h. After washing with PBS and HEPES-BSS, each set of cell culture was fed with the respective maintenance medium for 16 h. Afterwards, the cells were washed with PBS and fixed, and the percentage of cells expressing GFP in each cell culture was determined by flow cytometry (BD Canto II analyzer).
RESULTS
Real-time tracking during the early stages of RSV infection.
As an initial approach to studying the mechanism of RSV attachment and entry, we performed real-time tracking of the movement of RSV particles in living normal human bronchial epithelial cells by using confocal microscopy. In order to facilitate the tracking and to determine the time point when fusion has probably occurred, we used the dual-wavelength imaging technique developed by Sakai et al. (73). By simultaneously loading the lipophilic fluorochromes octadecylrhodamine (R18, seen as red) and 3,3′-dioctadecylcarbocyanine (DiOC18, seen as green) into the viral envelope, this approach takes advantage of both the self-quenching of lipophilic fluorochromes loaded at high density in the membrane (32) and the fluorescence resonance energy transference (FRET) from DiOC18 to R18 (73). The concentration of the dyes in the viral membrane is sufficiently high so that its fluorescence is largely quenched but still allows single-labeled virions to be detected. During the observation window, DiOC18 was excited at 488 nm but the fluorescence emissions were simultaneously recorded at both wavelengths 500 to 530 nm (DiOC18, seen as green) and 555 to 618 nm (R18, seen as red). The fluorescence emission of virions before fusion is detected only at wavelengths 555 to 618 nm, and consequently they are seen as red spots due to the energy transfer from DiOC18 to R18. The fusion event was registered as a shift in the fluorescence color to orange-yellow or green depending on the amount of R18 relative to DiOC18 remaining in the virion as the lipophilic tracers diffuse into the cell membrane.
This approach also allowed us to identify whether fusion had occurred at the plasma membrane or at the endosomal level, since the disappearance of the fluorescence signal indicates the dilution of the dyes into a large membrane. However, R18 and DiOC18 are lipophilic dyes, and since both of them may be rapidly transferred to the plasma membranes in hemifusion events (defined as the mixing of the outer membrane leaflets) or during pore formation and opening, we operationally define the fluorescence color shift as the beginning of the fusion event. If fusion occurs in the plasma membrane, the color shift will be followed by a loss of fluorescent signal due to dilution in the vast plasma membrane. However, if the color shift occurs in an endosome, the color shift will occur inside the cell without loss of the fluorescence signal because of the limited size of the endosomal membrane (73).
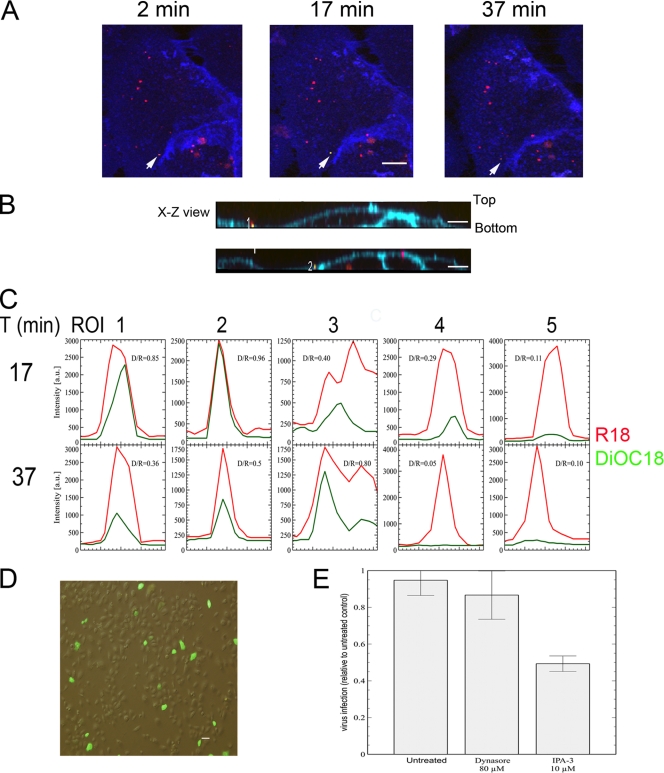
To experimentally detect the fluorescence color shift indicative of fusion, NHBE cells were grown on 35-mm glass-bottom fluorodishes to 80% confluence. On the day of the experiment, the Sephadex G50-purified dually labeled virions (seen as red) were allowed to attach to the cells at 4°C for 30 min at an MOI of 3 PFU/cell (based on titer prior to labeling) to synchronize virus fusion to cell membranes. During the last 10 min of the attachment stage, CellMask Deep Red plasma membrane marker (Invitrogen, seen as blue) was added to the cell cultures to label the plasma membrane and facilitate the identification of virus association with the plasma membrane. The culture dish was placed into a microincubator (PDMI-2; Harvard Apparatus) on the confocal microscope and maintained at 37°C during the 1-h observation period. Figure 1A shows that any movement of a virion was confined to a very restricted area on the cell membrane. In the image, the arrow indicates a representative example of a virion undergoing fusion to the cell membrane. At 2 min after switching the temperature to 37°C, the virion appeared as a red particle, indicative of its nonfused stage. At 17 min, the fluorescence color had shifted to yellow. By 37 min, the DiOC18 fluorescence had significantly declined, but the red signal was still present. On the other hand, a fraction of the virions did not fuse, remaining as red spots during the entire observation period. This could be due to the presence of inactivated virions or some cell debris contaminants; the ratio of nonfused virions to fused virions was estimated to be 4:1. In an x-z view (Fig. 1B), we did not detect endosomes present deep inside the cells with fluorescence color shift to yellow or orange. Contrary to what was found for RSV, when dually labeled vesicular stomatitis virus (a virus known to fuse in acidic endosomes) was followed in real time, there was a clear progression from red to green in several endosomes, as is shown in Fig. S1 in the supplemental material, which agreed with a study tracking dually labeled influenza virus (73). In Fig. 1C is shown the changes of the fluorescence intensities associated to R18 and DiOC18 in 3 regions of interest (ROI 1 to 3) taken as representatives of those 25 that showed fusion in three different experiments. For comparison, the changes in fluorescence intensity of both ROI 4 and 5 containing virions that did not fuse to the cell membrane are shown. In those virions where a clear fluorescence switch happened (ROI 1 to 3), the ratio of the fluorescence intensity between DiOC18 and R18 was higher than 0.80. In most of the virions, after reaching a peak, the green DiOC18 fluorescence declined to a steady level, which indicates that RSV hemifusion starts at the cell surface. However, the continuing presence of the red signal suggests that all these spots actually represented clusters of virions. In each cluster, those virions that did not fuse with cell membrane remained as red particles. Van der Schaar et al. (87) have suggested that the presence of high fluorescence intensity (higher than 500 arbitrary units) in DiD-labeled dengue virus particles represents clusters of multiple particles, which were either aggregated virions in the preparation or are formed during inoculation, and that confocal fluorescence microscopy considered each cluster of virions as a single particle. However, this does not preclude the identification of clusters or regions in which any virion of the cluster has hemifused, as shown by the appearance and disappearance of the green signal.
Fig 1.
RSV hemifusion/fusion begins at the plasma membrane. (A) Time dependence color shift of dually labeled viruses (visualized as red prior to fusion). NHBE cells grown on glass-bottom culture dishes were incubated with dually labeled virus at 4°C for 30 min to allow and synchronize virus attachment. The amount of input virus represented 3 PFU per cell based on the titer of the preparation measured prior to labeling. Cell Mask Deep Red plasma membrane marker (blue) was added to the cell cultures to label the contours of the plasma membrane. Cell cultures were placed on a microincubator (PDMI-2; Harvard Apparatus), and the temperature raised to 37°C for the entire observation period. The time at 37°C is indicated in minutes. Green and red fluorescences were simultaneously detected with two detectors (500 to 530 nm and 555 to 618 nm) using an Olympus FV1000 laser scanning confocal upright microscope with Olympus LUMPLFL 60× 0.9 NA water-immersion lens. To detect the labeled viruses as red spots, the red channel detector was tuned with greater sensitivity than the green detector in all experiments. All images have been merged. An arrow indicates the time dependence of the color shift of a dually labeled virion from red (nonfused) through yellow to the disappearance of the DiOC18 fluorescence signals (hemifusion/fusion). Bar = 10 μm. (B) Representative computer reconstructed x-z views as seen from the side of NHBE cells incubated with dually labeled virions as in A. Cell Mask Deep Red plasma membrane marker (blue) highlights the plasma membrane (PM). “Bottom” indicates the region of the cell plasma membrane attached to the glass enclosing the cytoplasm. The numbers indicate virus particles in fusion (yellow). There is no such yellow signal inside the cells. On the other hand, blue- and red-stained spots are visible in the cytoplasm, compatible with endosomes. (C) Changes of the fluorescence intensities associated with R18 and DiOC18 in 5 clusters of virions (ROI 1 to 5). ROI 1 to 3 indicate virions representative of those that underwent fusion events, while ROI 4 and 5 show two groups of unfused virions. Virions shown in ROI 1 and 2 are representative of the behavior observed for most of those virions that underwent fusion. After reaching a peak, the fluorescence associated with DiOC18 declined to a steady level, suggesting a process in which hemifusion begins at the plasma membrane. (D) Evaluation of the infectious capability of the dually labeled virions. NHBE cell cultures grown on 6-well plates were infected with dually labeled rgRSV (0.1 MOI, based on the titer of the preparation prior to labeling). Infected cells were identified by the expression of green fluorescence protein. (E) Role of Pak1 during RSV entry. NHBE cells were preincubated with dynasore or IPA-3 for 1 h or 30 min, respectively. Cells were then exposed to the infectious inoculum (rgRSV, <0.3 PFU/cell) in the continued presence of the respective drugs for 2 h. Subsequently, cells were washed to remove unbound virions and incubated with cold trypsin-EDTA to remove bound but unfused virions. After trypsin neutralization and washing, fresh medium was added and cells were placed at 37°C for 16 h. Infected cells were detected by flow cytometry and results are expressed as the percentage of infected cells in inhibitor-treated cells relative to that in untreated cells (exposed to DMSO). The data represents the average ± standard deviation (SD) of three independent experiments, each performed in triplicate.
We evaluated whether reversible inhibitors targeting either dynamin-dependent or -independent pathways prevent RSV from entering NHBE cells. These inhibitors were used at the highest concentration not associated with toxicity to NHBE cells. Dynasore, which blocks all the dynamin-dependent endocytic processes (38, 52) such as clathrin-, caveola-, flotillin-, interleukin-2 receptor β (IL-2Rβ)-, and circular dorsal ruffle-mediated endocytosis, was used at 80 μM. IPA-3, which targets PAK1-dependent activity (16, 59, 88), was used at 10 μM. The presence of the inhibitors in the cell cultures was restricted to what is considered the entry period of RSV. Ice-cold trypsin was used to remove bound but still unfused virions in the plasma membrane that could potentially enter once the inhibitor was taken away. Dynasore treatment of NHBE cells did not prevent RSV infection, while IPA-3 significantly reduced the number of RSV-infected cells to about half of that seen for untreated cell cultures (Fig. 1E).
We also evaluated whether loading virions with the fluorochromes impaired the infectious capabilities of RSV. For this assay, an aliquot corresponding to an MOI of 0.1 PFU/cell of rgRSV, based on titer prior to labeling was used to infect NHBE cells. After 16 h, the infection was evaluated by fluorescence microscopy. As shown in Fig. 1C, approximately 8% of the cells contained virus-produced GFP, indicating that the infectivity of RSV subjected to the dual labeling procedure was close to the expected value and hence that most of the virions were still infectious after labeling.
RSV association with lipid rafts.
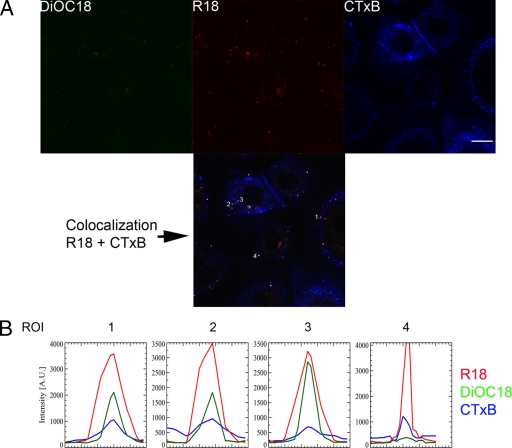
By taking advantage of the real-time imaging approach, we evaluated whether RSV docked on lipid raft microdomains in the plasma membrane of normal human bronchial epithelial cells. Lipid rafts are defined as small (10- to 200-nm) cholesterol- and sphingolipid-enriched domains that compartmentalize cellular processes. Small rafts can sometimes be stabilized to form larger platforms through protein-protein and protein-lipid interactions (66). Since RSV was found to be confined to a very restricted area on the plasma membrane (Fig. 1A) and such behavior might indicate attachment to the membrane microdomains (14), we probed the position of GM1-rich microdomains relative to the sites of RSV binding. Cholera toxin subunit B (Alexa 647-CTB, seen as blue) can be used to label the lipid rafts in living NHBE cells because its receptor, ganglioside GM1, is located in lipid rafts. CTB has also been used to identify distinct endocytic pathways (10, 31, 39, 83). This tracer was added along with the dually labeled virions (red) during virus attachment at 4°C. Cells were under continuous imaging from the time the culture dish was placed into the microincubator at 37°C. The setup of the experiment allowed us to determine the adsorption and fusion stages of the virus as a continuum and their relationship with lipid raft microdomains present in the plasma membrane. As shown in Fig. 2A, taken at 3 min after the beginning of incubation at 37°C, most of the virions (red) and all those that were in the hemifusion stage (shown as green dots in the corresponding DiOC18 panel) were associated with lipid rafts (blue). The colocalization (white dots) is clearly seen in the colocalization panel obtained in the analysis by using the Olympus Fluoroview software. The overlap index between the red and blue pixels was found to be 0.74 with a colocalization index of 0.0007, indicative of a high colocalization between both fluorophores. This is seen more clearly in Fig. 2B, where the profile of each of the fluorescence signals (R18, DiOC18, and CTB) per virion (ROI 1 to 4) is shown. There is an almost perfect alignment of the fluorescence peaks, indicating colocalization of RSV with the lipid rafts.
Fig 2.
RSV docks at lipid raft microdomains. (A) Colocalization of dually labeled RSV (red) with Alexa-647-labeled cholera toxin subunit B (CTxB, blue). NHBE cells grown on glass-bottom culture dishes were incubated with dually labeled virus at 4°C for 30 min at an input MOI of 3 PFU/cell (based on the titer prior to labeling) before adding Alexa-647-labeled cholera toxin subunit B for 10 min. RSV docking and fusion were followed in real time, beginning as soon as the culture dish was placed at the microincubator at 37°C. The image shows the fluorescence associated with DiOC18 (green), R18 (red), CTB (blue), and a merged picture of both R18 and CTB channels, indicating those pixels where colocalization (white) was present as analyzed by Olympus Fluoroview software. Some examples of colocalization are indicated with numbers. Bar = 10 μm. (B) Fluorescence intensities (in arbitrary units) for R18, DiOC18, and Alexa-647-CTxB are shown for virions 1 to 4. There is an almost perfect correlation for all the respective fluorescence signals.
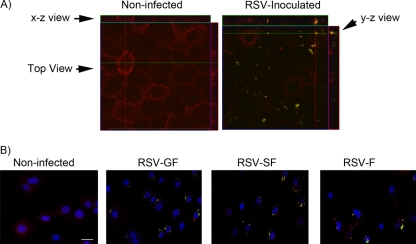
As an additional test of lipid raft microdomains serving as docking sites for RSV, we performed antibody-mediated copatching experiments to determine whether virion patches caused by anti-RSV envelope protein antibodies are clustered at the same spots as lipid rafts that are identified by CTB (31, 48). Unlabeled RSV was allowed to adsorb at 4°C for 30 min, after which the temperature was shifted to 37°C for 30 min. After discarding the infectious medium and washing with cold PBS, Alexa 555-CTB (red) and Alexa 488-anti-RSV F antibody (green) were added. The antibody facilitates the segregation of the membrane regions that have been bound by the virus, which consequently form patches. Cells were incubated an additional 30 min at 12°C to allow patch formation, followed by fixation. In Fig. 3A, lipid rafts (red) are homogeneously distributed in noninfected cells. However, in those cells inoculated with RSV (labeled with green-tagged antibody), virions that colocalized with the lipid rafts are visualized as yellow. Note that the antibody affected the distribution of the CTB signal, causing a shift from a diffuse pattern to the formation of patches (Fig. 3A and B). This likely reflects antibody-mediated aggregation of membrane microdomains containing bound virions.
Fig 3.
Copatching analysis of NHBE cells infected with RSV. (A) NHBE cells seeded on 8-well chamber slides were exposed to rgRSV-SGF (recombinant wild-type virus) at a titer of 20 MOI for 30 min at 4°C followed by 30 min at 37°C. The cells were washed and incubated with a mixture of Alexa-555-labeled CTxB and Alexa-488 (Zenon)-labeled anti-RSV F protein antibody at 12°C for 30 min: these are red (CTxB) and green (anti-F antibody) separately and yellow when colocalized. Cells were fixed, stained with DAPI to identify nuclei (blue), and visualized by confocal microscopy. The stained images corresponding to a Z-section were merged to show the colocalization (yellow) of anti-RSV antibody and the GM1 ligand CTxB. Computer reconstructions of both x-z and y-z views as seen from the side facilitate the identification of the copatches. (B) Evaluation of copatch formation by recombinant RSV engineered to express different sets of its three envelope proteins (GFΔSH, lacking SH; SHFΔG, lacking G; or FΔGSH, lacking G and SH) as described for A. Note that F glycoprotein alone was sufficient for RSV to bind to the lipid rafts. Bar = 10 μm. Note that, in A and B, the antibody facilitates the segregation of the membrane regions that have been bound by the virus, which consequently changes from a diffuse distribution to form patches.
Using the same approach with red-tagged CTB and green-tagged anti-RSV-F antibody, we determined which RSV envelope protein or combination thereof favored the formation of copatches with lipid rafts. Using RSV strains engineered to express different combinations of the three envelope proteins (rgRSV-GF, -SF, or -F), we found that each one of them copatched with the lipid rafts (Fig. 3B, yellow), which suggests that the RSV F protein, the only viral glycoprotein present in all of these viruses, is sufficient for RSV binding to cholesterol-rich microdomains.
Cholesterol-rich microdomains are required for successful RSV entry.
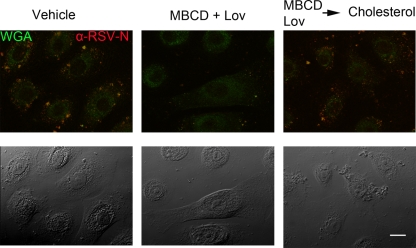
Since cholesterol is essential for the formation of lipid rafts, we depleted the cholesterol content of NHBE cultures by treatment with methyl-β-cyclodextrin (MBCD) to sequester cholesterol and the drug lovastatin (Lov) to inhibit its de novo synthesis. Cultures exposed to the vehicle in which these compounds were dissolved were used as the control. To confirm that the observed effect was due exclusively to cholesterol, water-soluble cholesterol was added back to a set of cultures previously treated with MBCD and Lov. In this way, we controlled for potential confounding effects of Lov such as its impact on the isoprenylate modification of intracellular proteins including RhoA, and any side effects of MBCD. The effect of these treatments was evaluated by confocal microscopy (Fig. 4) and flow cytometry (Fig. 5). For confocal microscopy, the ribonucleoprotein complexes were stained using anti-RSV-N antibodies tagged with Alexa-555 (red). The plasma membrane dye wheat germ agglutinin tagged with Alexa-488 (green) was used to identify cells and serve as a counterstain. As is shown in Fig. 4, cholesterol depletion by MBCD and Lov resulted in a significant reduction in the number of ribonucleoprotein complexes in the NHBE cells. These were visualized as red spots, indicating their presence in the cytoplasm, and yellow spots when the virions were either at the plasma membrane or in the endosomes due to merging with the background green counterstain. These results demonstrated a reduction in the number of infected cells, which was reversed when cholesterol was replenished.
Fig 4.
Cholesterol depletion blocks RSV entry. NHBE cells seeded on 8-well chamber slides were mock treated (left panel), subjected to cholesterol depletion by MBCD (2.5 mM) and Lov (2.5 μg/ml) for 30 min (middle), or subjected to cholesterol depletion that was reversed by the subsequent addition of MBCD-balanced water-soluble cholesterol (400 μg/ml; Sigma-Aldrich) for 45 min (right). After washing and adding the maintenance medium, cells were exposed to unlabeled RSV (MOI of 20 PFU/cell) at 4°C for 1 h, followed by 37°C for 1 h. After fixation, the cell membranes were stained with Alexa 488-WGA (identified with the green color) as a counterstain to define cell boundaries. Cells were then permeabilized with saponin, and the presence of RSV ribonucleoproteins in the cytoplasm was probed by Alexa 555-(Zenon)-labeled anti-RSV-N (identified with the red color). The stained images were merged after visualization by confocal microscopy (top). Bright-field images are shown on the bottom. Cholesterol depletion by MBCD significantly reduced the number of viral ribonucleoproteins in the cytoplasm, and this effect was reversed by cholesterol replenishment. Bar = 10 μm.
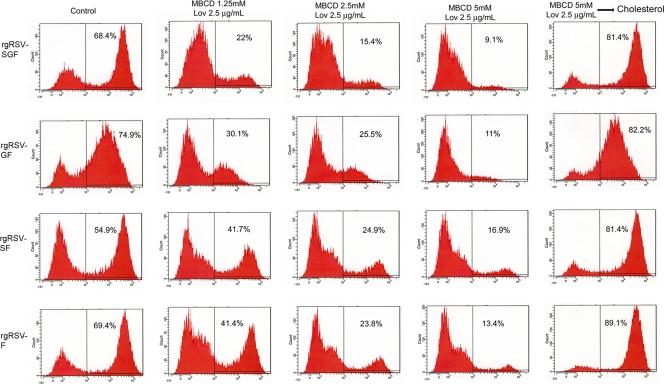
Fig 5.
Evaluation by flow cytometry of the cholesterol requirement for RSV infection. NHBE cells grown on 6-well plates were mock treated (first column); subjected to cholesterol depletion by incubation for 30 min with 1.25 mM MBCD and 2.5 μg/ml Lov (second column), 2.5 mM MBCD and 2.5 μg/ml Lov (third column), or 5 mM MBCD and 2.5 μg/ml Lov (fourth column); or subjected to cholesterol depletion by incubation for 30 min with 5 mM MBCD and 2.5 μg/ml Lov followed by reversal by incubation for 45 min with 400 μg/ml MBCD-balanced water-soluble cholesterol (fifth column). Cells were then washed and infected at an MOI of 1 PFU/cell with rgRSV-SGF or with rgRSV-GFΔSH, -SHFΔG, or -FΔGSH in the presence of lovastatin (2.5 μg/ml). After 16 h, the cells were fixed and then gated for GFP expression. Cholesterol depletion by MBCD significantly reduced the number of infected cells; this effect was reversed by cholesterol replenishment, and the presence of F protein as the sole viral surface protein was sufficient for infection.
These observations were confirmed and extended using flow cytometry. After treating the cultures as described above, the cells were infected with recombinant RSV containing different combinations of envelope proteins. Whereas the previous experiment measured the intracellular presence of viral N protein shortly after infection, the present experiment measured the expression of GFP. This is dependent on robust viral gene expression and thus was confirmed to be monitoring the pathway leading to productive infection. As is shown in Fig. 5, cholesterol depletion of NHBE cell membranes reduced the number of infected cells by each of the evaluated recombinant viruses, although MBCD at low concentration had a larger inhibitory effect on the infectivity of the two viruses containing the G protein than on the two viruses lacking the G protein. Replenishment of cholesterol increased infectivity to a level greater than the control in each case.
DISCUSSION
The present work indicates the importance of GM1-rich microdomains (lipid rafts) and the requirement of cholesterol for RSV entry into normal human bronchial epithelial cells, which are considered to be one of the primary targets in natural infection. In addition, RSV hemifusion events appear to begin in the plasma membrane of NHBE cells.
The mixing of the outer membrane leaflets between the virus and the cell membrane seems to be triggered at the plasma membrane. This is supported by the fact that after the FRET release (indicated by the appearance of the green signal), there was a reduction in DiOC18-associated fluorescence intensity (Fig. 1A to C). This does not mean that DiOC18 is the only fluorophore that moves to the cell membrane. In order for the FRET release to happen, it is necessary that both R18 and DiOC18 fluorophores move away from the viral envelope into an area of larger volume such as the plasma membrane or an endosome. Regarding the loss of fluorescence, endosomal trafficking could not be the cause, since such traffic is expected to generate green endosomes due also to the FRET release. Moreover, based on what was previously reported by Sakai et al. (73) in the context of tracking dually labeled influenza virus compared to what we found by tracking dually labeled VSV, there is a progression to the emergence of green endosomes in these viruses that are known to fuse in endosomes.
Our initial exploration using small-molecule inhibitors suggested that RSV infection may be facilitated by Pak1-driven rearrangements of the plasma membrane. Kolokoltsov et al. (41) previously identified Pak1 as participating in RSV infection of HeLa cells. Pak1 activation is associated with actin phosphorylation and cytoskeletal reorganization (18). However, Pak1 activation also accompanies endocytosis and macropinocytosis (17, 86). The intracellular trafficking of an endosome/macropinosome was not unambiguously identified by live-cell imaging in those experiments shown in Fig. 1. In a seminal paper describing the entry of human immunodeficiency virus by endocytosis, Miyauchi et al. (61) labeled virions with both a lipophilic dye (DiD) and a diffusible content marker (NC-GFP). They found that viral markers were released (disappeared) sequentially in a subset of regular fusion events, often exhibiting a considerable delay between lipid and content transfer. They suggested that such events indicated a two-step fusion mechanism, in which the lipid transfer happens at the plasma membrane while the content is delivered from an endosome (61). It is important to emphasize here that we have labeled only the virus envelope by using lipophilic dyes; we did not label the diffusible content marker, so we could not identify the site at which the content transfer occurred. However, in our case, the decline in DiOC18 after FRET release at the cell surface and the prevention of RSV infection by PAK1 inhibition suggest a two-step fusion phenotype for RSV, similar to the one described by Miyauchi et al. (61). Moreover, they noted that the temporal separation of lipid and content transfer events suggests that the two-step fusion proceeds through a remarkably long-lived hemifusion intermediate.
On the other hand, RSV has been shown to inhibit the amiloride-sensitive sodium channels (ENaC) within minutes of contacting the bronchial epithelium, and such effect is reproduced by recombinant viral F protein (43). Although amiloride and its analog 5-(N-ethyl-N-isopropyl)-amiloride (EIPA) are considered classical inhibitors of macropinocytosis, Karjalainen et al. (35) have shown, using ruthenium red-based electron microscopy and fluorescence-based confocal microscopy, that EIPA does not block the internalization and formation of tubulovesicular structures inside the cell. However, EIPA does prevent the traffic of such structures from the peripheral cytoplasm. Consequently, it would be interesting to explore in a further study if RSV triggers the formation of intracellular tubulovesicular structures via Pak1 activation but prevents the trafficking of such structures from the peripheral cytoplasm via its amiloride-like effects. If this were the case, RSV would complete full fusion close to the cell plasma membrane, but possibly in an early endosome.
A requirement for clathrin-mediated endocytosis during RSV entry of HeLa cells was suggested by Kolokoltsov et al. (41) based on the observation that RSV infection of HeLa cells was inhibited by knockdown of clathrin pathway-associated proteins. However, the fact that dynamin inhibition, using dynasore, did not prevent RSV entry implied that RSV may not be using clathrin-mediated endocytosis in NHBE cells. Dynamin is a large GTPase, with a remarkable ability to self-assemble into complex polymers that deform the membranes through a constriction process dependent on GTP hydrolysis. In association with other effector proteins, dynamin serves as a pinchase, releasing vesicles from donor membrane compartments (42). Dynamin has been found to be a key player in clathrin-, caveola-, IL-2Rβ-, flotillin-, and circular dorsal ruffle-mediated endocytosis pathways as well as in phagocytosis (19). Consequently, our finding that dynasore failed to prevent RSV infection suggests that RSV did not use dynamin-dependent endocytic mechanisms to enter NHBE cells. It is possible that HeLa cells, being an immortalized cell line, could behave differently from primary bronchial epithelial cells regarding the use of clathrin-mediated endocytosis for virus entry.
Although dynasore has been proven to be a very specific inhibitor of dynamin (1, 38, 52), it will be important in future studies to determine if RSV localizes to clathrin-coated pits. On the other hand, the knockdown of those clathrin pathway-associated proteins identified in the siRNA screening by Kolokoltsov et al. (41) may interfere with RSV infection through mechanisms different from clathrin-mediated endocytosis, such that inhibition of the clathrin pathway cannot distinguish between clathrin-mediated endocytosis and vesicular traffic from the trans-Golgi network (71, 77). Kolokoltsov et al. (41) reported that the expression of a dominant negative version of Eps-15 inhibited viral infection of HeLa cells without affecting viral attachment and suggested that fusion, the only remaining step in entry, must be the target for this inhibition. However, this conclusion does not take into account the possibility that the F protein also has a receptor required for infection (13, 24, 53, 74, 82) that may be sensitive to blockade of Eps-15 and, therefore, clathrin-mediated transport from the trans-Golgi network (9, 41). Overall, our data indicated that RSV hemifusion events in NHBE cells begin at the plasma membrane. It remains possible that the virus releases its contents from endosomes following the “two-step” scenario observed for HIV (61), which begins in the plasma membrane but is completed in an endosome, or which it takes place very close to the cell membrane, as observed for poliovirus (4).
Although it is possible that host cell proteins present in the RSV envelope could attach the virions to lipid rafts, it is tempting to speculate that the F protein mediates the viral attachment to such microdomains. Since cholesterol and sphingomyelin are the major constituents of the lipid rafts, it is interesting that the ectodomain of F protein has two cholesterol recognition amino acid consensus sites (CRACs)—[V/L-(X1-5)-Y-(X1-5)-K/R, where X1-5 represents 1 to 5 of any amino acid] at the regions from 414 to 419 (VSCYGK) and from 452 to 461 (VGNTLYYVNK) (22, 64). These regions flank antigenic site IV, which is recognized by the neutralizing monoclonal antibodies 101F and MAb19 (3, 49, 56, 84). It could be that initially, or in any of the intermediary stages of the fusion process, the predicted CRACs bind to cholesterol present in the lipid rafts and that such a process could be sterically interrupted by a monoclonal antibody (56).
The earliest stages of RSV entry were very sensitive to cholesterol depletion. Cholesterol may be a requirement for the entry of many viruses as demonstrated for Sindbis virus and Semliki Forest virus (36, 51, 65, 85). However, flavivirus penetration seems to be impaired by cholesterol (46). We found that NHBE cultures depleted of cholesterol and subjected to cholesterol replenishment showed a clear increase in the percentage of RSV-infected cells to levels higher than those observed for control cultures. This observation is surprising, since HIV-1 and human herpesvirus 6, which both require cholesterol for successful entry, only partially recovered their infectivity with cholesterol replenishment (7, 80).
Although clathrin-mediated endocytosis also depends on the presence of cholesterol to facilitate the proper curvature at the cell membrane for endocytosis to ensue (70), the sum of the observations reported here suggests that cholesterol is required for the formation of particular microdomains that enable RSV binding and fusion on the cell membrane of bronchial epithelial cells.
Zhang et al. (91) reported that RSV preferentially targets the ciliated cells of the airway epithelium and that infection occurs exclusively via the apical surface. Interestingly, the lipid composition of the ciliary membrane is enriched in cholesterol-rich microdomains, consistent with lipid rafts having high liquid order, as determined by laurdan two-photon microscopy (21). A classic study using electron microscopy and taking advantage of the ability of filipin to bind cholesterol found that filipin-sterol complexes appeared densely and uniformly distributed over most of the ciliary membrane (62). In addition, both GM1 and GM3 gangliosides are found in the primary cilia (34). Overall, the abundance of cholesterol-rich microdomains in the cilia may partially explain the preference of RSV to infect ciliated bronchial epithelial cells.
We have presented evidence that cholesterol-rich microdomains are of critical importance during the initial stages of attachment and fusion of RSV. The prophylactic and therapeutic implications of this finding might be exploited with cholesterol-tagged peptide inhibitors such as those developed by Porotto et al. (67, 68). The cholesterol moiety was found to target inhibitory peptides to the membrane site where fusion occurs, thereby increasing their antiviral potency to inhibit paramyxovirus (human parainfluenza virus type 3, Hendra virus, and Nipah virus) infection both in vitro and in an animal model (67, 68).
Lipid rafts have also been reported to be the virion assembly site for RSV budding (55). Targeting cholesterol-rich microdomains on the cell membrane might therefore impair both the production of infectious virions and their ability to spread. Gower and Graham (26) reported that statins showed antiviral activity against RSV in vivo. The pharmacological potential of cholesterol-lowering drugs to curb virus infection has also been proposed and evaluated in hepatitis C virus with promising results in which statins significantly improved the rapid antiviral response caused by pegylated interferon in an open-labeled randomized controlled study (60).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID; R01-AI075523) to H.S.-J.-V. and S.S.M., by the VA Career Scientist Award and the VA Merit Review Award to S.S.M., and by two grants from the Colombian Administrative Department for Science, Technology and Innovation (COLCIENCIAS; code 121540820542, contract no. 312-2007; and code 121549326215, contract no. 651-2009). Neither sponsor had any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. Peter L. Collins was supported by the NIAID Intramural Program. The authors declare no conflict of interest related to this work.
Confocal microscopy was performed in the Lisa Muma Weitz Laboratory for Advanced Microscopy & Cell Imaging at the Health Sciences Center at the University of South Florida (Tampa, FL) under the direction of Byeong Cha. Flow cytometry was performed in the Fred Wright Flow Cytometry Core Facility at the Health Sciences Center at the University of South Florida (Tampa, FL) under the direction of Charlie Szekeres. Flow cytometry was also performed in the Fundación Grupo Estudio Barranquilla (Laboratorios Rey-Fals) under the direction of Orlando Fals and with the collaboration of Gema Miranda (B.Sc.) and Elvira Manrique (B.Sc.) (Barranquilla, Colombia). The editing contributions and insightful questions of Subhra Mohapatra, Gary Hellermann, and John David Rodríguez are all greatly appreciated. We also thank the anonymous reviewers for their valuable comments and suggestions.
Footnotes
Published ahead of print 16 November 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Abban CY, Bradbury NA, Meneses PI. 2008. HPV16 and BPV1 infection can be blocked by the dynamin inhibitor dynasore. Am. J. Ther. 15: 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anonymous. 2000. Respiratory syncytial virus activity–United States, 1999–2000 season. MMWR Morb. Mortal. Wkly. Rep. 49: 1091–1093 [PubMed] [Google Scholar]
- 3. Arbiza J, et al. 1992. Characterization of two antigenic sites recognized by neutralizing monoclonal antibodies directed against the fusion glycoprotein of human respiratory syncytial virus. J. Gen. Virol. 73: 2225–2234 [DOI] [PubMed] [Google Scholar]
- 4. Brandenburg B, et al. 2007. Imaging poliovirus entry in live cells. PLoS Biol. 5: e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brandenburg B, Zhuang X. 2007. Virus trafficking - learning from single-virus tracking. Nat. Rev. Microbiol. 5: 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carpenter LR, Moy JN, Roebuck KA. 2002. Respiratory syncytial virus and TNF alpha induction of chemokine gene expression involves differential activation of Rel A and NF-kappa B1. BMC Infect. Dis. 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carter GC, et al. 2009. HIV entry in macrophages is dependent on intact lipid rafts. Virology 386: 192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaiwatpongsakorn S, Epand RF, Collins PL, Epand RM, Peeples ME. 2011. Soluble respiratory syncytial virus fusion protein in the fully cleaved, pretriggered state is triggered by exposure to low-molarity buffer. J. Virol. 85: 3968–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chi S, Cao H, Chen J, McNiven MA. 2008. Eps15 mediates vesicle trafficking from the trans-Golgi network via an interaction with the clathrin adaptor AP-1. Mol. Biol. Cell 19: 3564–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chinnapen DJ, Chinnapen H, Saslowsky D, Lencer WI. 2007. Rafting with cholera toxin: endocytosis and trafficking from plasma membrane to ER. FEMS Microbiol. Lett. 266: 129–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cianci C, et al. 2004. Targeting a binding pocket within the trimer-of-hairpins: small-molecule inhibition of viral fusion. Proc. Natl. Acad. Sci. U. S. A. 101: 15046–15051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coller KE, et al. 2009. RNA interference and single particle tracking analysis of hepatitis C virus endocytosis. PLoS Pathog. 5: e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crim RL, Audet SA, Feldman SA, Mostowski HS, Beeler JA. 2007. Identification of linear heparin-binding peptides derived from human respiratory syncytial virus fusion glycoprotein that inhibit infectivity. J. Virol. 81: 261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Damm EM, Pelkmans L. 2006. Systems biology of virus entry in mammalian cells. Cell Microbiol. 8: 1219–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Day ND, et al. 2006. Contribution of cysteine residues in the extracellular domain of the F protein of human respiratory syncytial virus to its function. Virol. J. 3: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deacon SW, et al. 2008. An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chem. Biol. 15: 322–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Vries E, et al. 2011. Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 7: e1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dharmawardhane S, Brownson D, Lennartz M, Bokoch GM. 1999. Localization of p21-activated kinase 1 (PAK1) to pseudopodia, membrane ruffles, and phagocytic cups in activated human neutrophils. J. Leukoc. Biol. 66: 521–527 [DOI] [PubMed] [Google Scholar]
- 19. Doherty GJ, McMahon HT. 2009. Mechanisms of endocytosis. Annu. Rev. Biochem. 78: 857–902 [DOI] [PubMed] [Google Scholar]
- 20. Dowell SF, et al. 1996. Respiratory syncytial virus is an important cause of community-acquired lower respiratory infection among hospitalized adults. J. Infect. Dis. 174: 456–462 [DOI] [PubMed] [Google Scholar]
- 21. Emmer BT, Maric D, Engman DM. 2010. Molecular mechanisms of protein and lipid targeting to ciliary membranes. J. Cell Sci. 123: 529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Epand RM. 2008. Proteins and cholesterol-rich domains. Biochim. Biophys. Acta 1778: 1576–1582 [DOI] [PubMed] [Google Scholar]
- 23. Falsey AR, et al. 1995. Respiratory syncytial virus and influenza A infections in the hospitalized elderly. J. Infect. Dis. 172: 389–394 [DOI] [PubMed] [Google Scholar]
- 24. Feldman SA, Audet S, Beeler JA. 2000. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J. Virol. 74: 6442–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gonzalez-Reyes L, et al. 2001. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 98: 9859–9864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gower TL, Graham BS. 2001. Antiviral activity of lovastatin against respiratory syncytial virus in vivo and in vitro. Antimicrob. Agents Chemother. 45: 1231–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gueret V, Negrete-Virgen JA, Lyddiatt A, Al-Rubeai M. 2002. Rapid titration of adenoviral infectivity by flow cytometry in batch culture of infected HEK293 cells. Cytotechnology 38: 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hallak LK, Collins PL, Knudson W, Peeples ME. 2000. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology 271: 264–275 [DOI] [PubMed] [Google Scholar]
- 29. Hallak LK, Kwilas SA, Peeples ME. 2007. Interaction between respiratory syncytial virus and glycosaminoglycans, including heparan sulfate. Methods Mol. Biol. 379: 15–34 [DOI] [PubMed] [Google Scholar]
- 30. Hallak LK, Spillmann D, Collins PL, Peeples ME. 2000. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J. Virol. 74: 10508–10513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harder T, Scheiffele P, Verkade P, Simons K. 1998. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 141: 929–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoekstra D, Klappe K. 1986. Use of a fluorescence assay to monitor the kinetics of fusion between erythrocyte ghosts, as induced by Sendai virus. Biosci. Rep. 6: 953–960 [DOI] [PubMed] [Google Scholar]
- 33. Huang K, Incognito L, Cheng X, Ulbrandt ND, Wu H. 2010. Respiratory syncytial virus-neutralizing monoclonal antibodies motavizumab and palivizumab inhibit fusion. J. Virol. 84: 8132–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Janich P, Corbeil D. 2007. GM1 and GM3 gangliosides highlight distinct lipid microdomains within the apical domain of epithelial cells. FEBS Lett. 581: 1783–1787 [DOI] [PubMed] [Google Scholar]
- 35. Karjalainen M, et al. 2008. A Raft-derived, Pak1-regulated entry participates in alpha2beta1 integrin-dependent sorting to caveosomes. Mol. Biol. Cell 19: 2857–2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kielian MC, Helenius A. 1984. Role of cholesterol in fusion of Semliki Forest virus with membranes. J. Virol. 52: 281–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kimpen JL. 2002. Prevention and treatment of respiratory syncytial virus bronchiolitis and postbronchiolitic wheezing. Respir. Res. 3: S40–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kirchhausen T, Macia E, Pelish HE. 2008. Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzymol. 438: 77–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kirkham M, et al. 2005. Ultrastructural identification of uncoated caveolin-independent early endocytic vehicles. J. Cell Biol. 168: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koch P, et al. 2009. Visualizing fusion of pseudotyped HIV-1 particles in real time by live cell microscopy. Retrovirology 6: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kolokoltsov AA, et al. 2007. Small interfering RNA profiling reveals key role of clathrin-mediated endocytosis and early endosome formation for infection by respiratory syncytial virus. J. Virol. 81: 7786–7800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kruchten AE, McNiven MA. 2006. Dynamin as a mover and pincher during cell migration and invasion. J. Cell Sci. 119: 1683–1690 [DOI] [PubMed] [Google Scholar]
- 43. Kunzelmann K, Sun J, Meanger J, King NJ, Cook DI. 2007. Inhibition of airway Na+ transport by respiratory syncytial virus. J. Virol. 81: 3714–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kwilas S, et al. 2009. Respiratory syncytial virus grown in Vero cells contains a truncated attachment protein that alters its infectivity and dependence on glycosaminoglycans. J. Virol. 83: 10710–10718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lambeth CR, White LJ, Johnston RE, de Silva AM. 2005. Flow cytometry-based assay for titrating dengue virus. J. Clin. Microbiol. 43: 3267–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee CJ, Lin HR, Liao CL, Lin YL. 2008. Cholesterol effectively blocks entry of flavivirus. J. Virol. 82: 6470–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Z, Ling L, Liu X, Laus R, Delcayre A. 2010. A flow cytometry-based immuno-titration assay for rapid and accurate titer determination of modified vaccinia Ankara virus vectors. J. Virol. Methods 169: 87–94 [DOI] [PubMed] [Google Scholar]
- 48. Lingwood D, Kaiser HJ, Levental I, Simons K. 2009. Lipid rafts as functional heterogeneity in cell membranes. Biochem. Soc. Trans. 37: 955–960 [DOI] [PubMed] [Google Scholar]
- 49. Lopez JA, et al. 1998. Antigenic structure of human respiratory syncytial virus fusion glycoprotein. J. Virol. 72: 6922–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Low KW, Tan T, Ng K, Tan BH, Sugrue RJ. 2008. The RSV F and G glycoproteins interact to form a complex on the surface of infected cells. Biochem. Biophys. Res. Commun. 366: 308–313 [DOI] [PubMed] [Google Scholar]
- 51. Lu YE, Cassese T, Kielian M. 1999. The cholesterol requirement for Sindbis virus entry and exit and characterization of a spike protein region involved in cholesterol dependence. J. Virol. 73: 4272–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Macia E, et al. 2006. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 10: 839–850 [DOI] [PubMed] [Google Scholar]
- 53. Malhotra R, et al. 2003. Isolation and characterisation of potential respiratory syncytial virus receptor(s) on epithelial cells. Microbes Infect. 5: 123–133 [DOI] [PubMed] [Google Scholar]
- 54. Matthews JM, Young TF, Tucker SP, Mackay JP. 2000. The core of the respiratory syncytial virus fusion protein is a trimeric coiled coil. J. Virol. 74: 5911–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McCurdy LH, Graham BS. 2003. Role of plasma membrane lipid microdomains in respiratory syncytial virus filament formation. J. Virol. 77: 1747–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. McLellan JS, et al. 2010. Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J. Virol. 84: 12236–12244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Melikyan GB. 2008. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology 5: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Melikyan GB, Barnard RJ, Abrahamyan LG, Mothes W, Young JA. 2005. Imaging individual retroviral fusion events: from hemifusion to pore formation and growth. Proc. Natl. Acad. Sci. U. S. A. 102: 8728–8733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mercer J, Helenius A. 2009. Virus entry by macropinocytosis. Nat. Cell Biol. 11: 510–520 [DOI] [PubMed] [Google Scholar]
- 60. Milazzo L, et al. 2010. Fluvastatin as an adjuvant to pegylated interferon and ribavirin in HIV/hepatitis C virus genotype 1 co-infected patients: an open-label randomized controlled study. J. Antimicrob. Chemother. 65: 735–740 [DOI] [PubMed] [Google Scholar]
- 61. Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. 2009. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137: 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Montesano R. 1979. Inhomogeneous distribution of filipin-sterol complexes in the ciliary membrane of rat tracheal epithelium. Am. J. Anat. 156: 139–145 [DOI] [PubMed] [Google Scholar]
- 63. Ohki S, Liu JZ, Schaller J, Welliver RC. 2003. The compound DATEM inhibits respiratory syncytial virus fusion activity with epithelial cells. Antiviral Res. 58: 115–124 [DOI] [PubMed] [Google Scholar]
- 64. Palmer M. 2004. Cholesterol and the activity of bacterial toxins. FEMS Microbiol. Lett. 238: 281–289 [DOI] [PubMed] [Google Scholar]
- 65. Phalen T, Kielian M. 1991. Cholesterol is required for infection by Semliki Forest virus. J. Cell Biol. 112: 615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pike LJ. 2006. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 47: 1597–1598 [DOI] [PubMed] [Google Scholar]
- 67. Porotto M, et al. 2010. Inhibition of Nipah virus infection in vivo: targeting an early stage of paramyxovirus fusion activation during viral entry. PLoS Pathog. 6: e1001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Porotto M, et al. 2010. Viral entry inhibitors targeted to the membrane site of action. J. Virol. 84: 6760–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Razinkov V, Huntley C, Ellestad G, Krishnamurthy G. 2002. RSV entry inhibitors block F-protein mediated fusion with model membranes. Antiviral Res. 55: 189–200 [DOI] [PubMed] [Google Scholar]
- 70. Rodal SK, et al. 1999. Extraction of cholesterol with methyl-beta-cyclodextrin perturbs formation of clathrin-coated endocytic vesicles. Mol. Biol. Cell 10: 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Royle SJ, Bright NA, Lagnado L. 2005. Clathrin is required for the function of the mitotic spindle. Nature 434: 1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rust MJ, Lakadamyali M, Zhang F, Zhuang X. 2004. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat. Struct. Mol. Biol. 11: 567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sakai T, et al. 2006. Dual wavelength imaging allows analysis of membrane fusion of influenza virus inside cells. J. Virol. 80: 2013–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shields B, Mills J, Ghildyal R, Gooley P, Meanger J. 2003. Multiple heparin binding domains of respiratory syncytial virus G mediate binding to mammalian cells. Arch. Virol. 148: 1987–2003 [DOI] [PubMed] [Google Scholar]
- 75. Simoes EA. 1999. Respiratory syncytial virus infection. Lancet 354: 847–852 [DOI] [PubMed] [Google Scholar]
- 76. Smith BJ, Lawrence MC, Colman PM. 2002. Modelling the structure of the fusion protein from human respiratory syncytial virus. Protein Eng. 15: 365–371 [DOI] [PubMed] [Google Scholar]
- 77. Sorkin A, von Zastrow M. 2009. Endocytosis and signalling: intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 10: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Srinivasakumar N, Ogra PL, Flanagan TD. 1991. Characteristics of fusion of respiratory syncytial virus with HEp-2 cells as measured by R18 fluorescence dequenching assay. J. Virol. 65: 4063–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sugrue RJ, Brown C, Brown G, Aitken J, Rixon HWM. 2001. Furin cleavage of the respiratory syncytial virus fusion protein is not a requirement for its transport to the surface of virus-infected cells. J. Gen. Virol. 82: 1375–1386 [DOI] [PubMed] [Google Scholar]
- 80. Tang H, Kawabata A, Takemoto M, Yamanishi K, Mori Y. 2008. Human herpesvirus-6 infection induces the reorganization of membrane microdomains in target cells, which are required for virus entry. Virology 378: 265–271 [DOI] [PubMed] [Google Scholar]
- 81. Techaarpornkul S, Barretto N, Peeples ME. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 75: 6825–6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Techaarpornkul S, Collins PL, Peeples ME. 2002. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology 294: 296–304 [DOI] [PubMed] [Google Scholar]
- 83. Torgersen ML, Skretting G, van Deurs B, Sandvig K. 2001. Internalization of cholera toxin by different endocytic mechanisms. J. Cell Sci. 114: 3737–3747 [DOI] [PubMed] [Google Scholar]
- 84. Ulbrandt ND, et al. 2008. Identification of antibody neutralization epitopes on the fusion protein of human metapneumovirus. J. Gen. Virol. 89: 3113–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Umashankar M, et al. 2008. Differential cholesterol binding by class II fusion proteins determines membrane fusion properties. J. Virol. 82: 9245–9253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Van den Broeke C, Radu M, Chernoff J, Favoreel HW. 2010. An emerging role for p21-activated kinases (Paks) in viral infections. Trends Cell Biol. 20: 160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. van der Schaar HM, et al. 2007. Characterization of the early events in dengue virus cell entry by biochemical assays and single-virus tracking. J. Virol. 81: 12019–12028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Viaud J, Peterson JR. 2009. An allosteric kinase inhibitor binds the p21-activated kinase autoregulatory domain covalently. Mol. Cancer Ther. 8: 2559–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439: 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhang L, et al. 2005. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 79: 1113–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang L, Peeples ME, Boucher RC, Collins PL, Pickles RJ. 2002. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J. Virol. 76: 5654–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.