Abstract
The influenza A virus M2 ion channel protein has the longest cytoplasmic tail (CT) among the three viral envelope proteins and is well conserved between different viral strains. It is accessible to the host cellular machinery after fusion with the endosomal membrane and during the trafficking, assembly, and budding processes. We hypothesized that identification of host cellular interactants of M2 CT could help us to better understand the molecular mechanisms regulating the M2-dependent stages of the virus life cycle. Using yeast two-hybrid screening with M2 CT as bait, a novel interaction with the human annexin A6 (AnxA6) protein was identified, and their physical interaction was confirmed by coimmunoprecipitation assay and a colocalization study of virus-infected human cells. We found that small interfering RNA (siRNA)-mediated knockdown of AnxA6 expression significantly increased virus production, while its overexpression could reduce the titer of virus progeny, suggesting a negative regulatory role for AnxA6 during influenza A virus infection. Further characterization revealed that AnxA6 depletion or overexpression had no effect on the early stages of the virus life cycle or on viral RNA replication but impaired the release of progeny virus, as suggested by delayed or defective budding events observed at the plasma membrane of virus-infected cells by transmission electron microscopy. Collectively, this work identifies AnxA6 as a novel cellular regulator that targets and impairs the virus budding and release stages of the influenza A virus life cycle.
INTRODUCTION
Influenza A virus (IAV) is an enveloped virus with a segmented negative-sense RNA genome. The eight RNA segments encode 11 proteins (44). Influenza virus is pleomorphic (23), forming spherical virions that are ∼100 nm in diameter as well as filamentous virions that are ∼100 nm in diameter and over 20 μm in length. The viral lipid envelope, derived by budding from the apical plasma membrane of the host cell, contains two major envelope glycoproteins, the receptor-binding/membrane fusion protein hemagglutinin (HA) and the enzyme neuraminidase (NA). A third minor (∼16 to 20 molecules/virion) integral membrane protein, M2, is a proton-selective ion channel that allows virion interior acidification for efficient uncoating after fusion in endosomes. M2 contains 97 amino acid residues and assembles into a homotetramer. This small type III integral membrane protein contains a single 19-residue transmembrane domain that forms the pore of the ion channel, a short (24-residue) amino-terminal ectodomain, and a long (54-residue) carboxy-terminal cytoplasmic domain (27, 34, 45). This cytoplasmic tail (CT) is highly conserved among viral strains (57).
In the early stages of the replication cycle, after virus internalization by endocytosis, endosomal acidification activates M2 ion channel activity, causing acidification of the virus interior and leading to dissociation of the matrix protein M1 from the viral ribonucleoprotein (vRNP) complex (reviewed in reference 47). M2 is the target of the antiviral drug amantadine, which inhibits M2 ion channel activity, thus preventing M1-vRNP dissociation and virus uncoating (reviewed in reference 46). M2 also functions to equilibrate the pH gradient between the lumen of the trans-Golgi network (TGN) and the cytoplasm to prevent premature low-pH-dependent conformational changes of HA for some viral strains (9). M2 ion channel activity is also required for activation of inflammasomes by perturbing the ion concentration in the Golgi compartment (30).
In addition to ion channel activity, Gannagé et al. reported a role of M2 in autophagy (17). M2 blocks autophagosome maturation, preventing fusion with the lysosome and significantly affecting host cell apoptosis. Recent work has also suggested that M2 CT, in particular the amphipathic helix residues 44 to 62, may play a crucial role in virus assembly and budding (reviewed in references 41 and 51). Indeed, M2 CT interacts with M1 and mediates budding of filamentous virions by stabilizing the virus budding site (6, 31, 38, 39, 49, 50). It is also important for vRNP incorporation and the production of infectious virions (20, 38, 39). Recently, Rossman et al. indicated that the M2 CT amphipathic helix modifies membrane curvature at the neck of the budding virus in a cholesterol-dependent manner, causing membrane scission and completion of the budding process with the release of the progeny virion (50).
Altogether, these studies support the notion that M2 is an essential multifunctional viral protein that plays important roles at many steps of the virus life cycle. However, little is known about host factors that interact with M2. Few host cell interactants have been identified, and the functional relevance of these interactions is not fully determined (21, 55). In this study, we used a yeast two-hybrid (Y2H) system to screen a human placenta cDNA library to identify host proteins that interact with M2 CT. We hypothesized that cellular factors interacting with M2 CT could either help or restrict processes involving M2. The human protein annexin A6 (AnxA6), a Ca2+- and phospholipid-binding protein, was identified as a novel cellular interactant of M2, and their physical interaction was confirmed in virus-infected human cells. By modulating the expression of AnxA6, we found that AnxA6 negatively regulates IAV infection by impairing virus budding without affecting the early steps of the virus life cycle and virus replication.
MATERIALS AND METHODS
Yeast two-hybrid screening.
Bait cloning and Y2H screening with influenza A virus M2 CT (from strain A/Goose/Guangdong/1/96; GenBank accession no. 81975894) as bait were performed by Hybrigenics, S.A., Paris, France. Briefly, M2 CT was subcloned into the pB27 vector, enabling its fusion with the LexA binding domain. The bait construct pM2/CT was transformed into the L40ΔGAL4 yeast strain (16) and then mated with the Y187 yeast strain transformed by a random-primed human placenta cDNA library containing 10 million independent fragments. After selection on medium lacking leucine, tryptophan, and histidine, the 273 positive clones were picked. The corresponding prey fragments were subjected to PCR and sequencing. Sequences were then filtered, divided into contigs, and compared to the latest release of the GenBank database by using BLASTN. A predicted biological score was attributed to assess the reliability of the interaction, as described earlier (15).
Cells and viruses.
Human embryonic kidney (293T), human alveolar basal epithelial (A549), human vulval squamous epithelial (A431), and Madin-Darby canine kidney (MDCK) cells were all purchased from ATCC and grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA).
Influenza A virus strains A/WSN/33 (H1N1), A/HK/1/68 (H3N2), and A/HK/54/98 (H1N1) were propagated in MDCK cells with serum-free medium supplemented with 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Thermo Scientific), and viral titers were determined by plaque assay on MDCK cells. Briefly, virus stocks were serially diluted in phosphate-buffered saline (PBS) (Invitrogen, Carlsbad, CA) and adsorbed onto confluent MDCK cells for 1 h at 37°C. Later, the inoculum was removed and cells were washed twice with PBS and covered with an agar overlay (1% agarose, 1 μg/ml TPCK-treated trypsin in DMEM). After 3 days of incubation, the cells were stained with crystal violet and plaques were counted to obtain the virus concentration as PFU per ml.
Plasmids and antibodies.
Plasmid pCMV-myc-AnxA6, corresponding to the DNA sequence of human AnxA6 (GenBank accession no. NM_001155.3) with a C-terminal myc tag, was purchased from OriGene Technologies. Plasmid pCMV-AnxA6-GFP was generated by inserting the AnxA6 open reading frame (ORF) into the pCMV-GFP vector at the KpnI and NotI restriction sites. For production of an A431 stable cell line expressing AnxA6, the AnxA6 cDNA containing a myc tag was amplified from pCMV-myc-AnxA6 using the forward primer AGTCGAACTAGTGCCGCCATGGCCAAACCAGC and the reverse primer GATGCTGGGCCCTTACAGATCCTCTTCAGAGATGAGTTTC and subsequently inserted into pLenti-6.3/V5-Dest vector (Invitrogen) at the SpeI and ApaI restriction sites. All constructs were sequenced at the Genome Research Centre of the University of Hong Kong.
Primary antibodies used in this study were mouse anti-M2 monoclonal antibody (MAb) (clone 14C2; Affinity BioReagent Inc.), mouse anti-M1 MAb (clone GA2B; Abcam, Cambridge, MA), mouse anti-NP MAb (clone AA5H; Abcam), mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) MAb (clone 6C5; Abcam), mouse anti-myc MAb (clone 9E10; Sigma, St. Louis, MO), and mouse anti-AnxA6 MAb (clone 73; BD Transduction Laboratories).
Coimmunoprecipitation and Western blotting.
HEK 293T cells grown in 6-cm dishes were transfected with pCMV-myc-AnxA6 plasmid for 24 h and then infected with influenza A/WSN/33 virus at a multiplicity of infection (MOI) of 0.1. At 24 h postinfection (p.i.), cells were washed and resuspended in 0.5 ml of immunoprecipitation buffer (150 mM NaCl, 15 mM EDTA, 1% Triton X-100, 0.25% sodium deoxycholate, and protease inhibitor cocktail [Roche Diagnostics GmbH]). After centrifugation, the supernatant was incubated with anti-myc, anti-M2, or normal mouse immunoglobulin G (IgG) for 2 h. The immune complexes were recovered by adsorption to protein G-Sepharose beads (Amersham Biosciences, Uppsala, Sweden) overnight at 4°C. After five washes in immunoprecipitation buffer, the immunoprecipitates were analyzed by Western blot analysis.
For Western blotting, protein samples were solubilized in LDS sample buffer (Invitrogen) and 10 mM dithiothreitol (DTT), boiled at 95°C for 8 min, and loaded into a NuPAGE Novex 4 to 12% Bis-Tris minigel for separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Separated proteins were then transferred to a Hybond-P polyvinylidene difluoride (PVDF) membrane (Amersham Biosciences), and the membrane was blocked overnight at 4°C in 5% skim milk prepared in PBS-T (PBS containing 0.1% Tween 20). The membrane was incubated with appropriately diluted primary antibodies for 1 h, washed, and then incubated with a horseradish peroxidase-conjugated secondary antibody (1:5,000; Jackson ImmunoResearch). Blots were visualized using an enhanced chemiluminescence (ECL) system (GE Healthcare). The protein size was estimated using Novex Sharp prestained protein standards (Invitrogen).
siRNA treatment and virus infection.
Gene-specific small interfering RNAs (siRNAs) for AnxA6 (M-011210) were provided as an siGENOME SMARTpool, which is a pool of four siRNAs targeting various sites in a single gene. AnxA6_17 siRNA (si_AnxA6 [CGGGCAAGUUUGAACGGUU]) and the nontargeting siRNA 3 (si_Control; D-001210) were provided as individual siRNA duplexes. All siRNAs and transfection reagents (DharmaFECT 1 and T-2001) were purchased from Dharmacon (Dharmacon Research Inc., Lafayette, CO). Low-passage A549 cells were transfected with siRNA at a concentration of 100 nM for 60 h according to the manufacturer's instructions, and the knockdown efficiency was checked by immunoblot analysis. To study the effect of AnxA6 knockdown on the replication of influenza virus, three strains (A/WSN/33 [H1N1], A/HK/1/68 [H3N2], and A/HK/54/98 [H1N1]) were used to infect the siRNA-treated A549 cells at an MOI of 0.01. Supernatants were collected at 24 and 48 h p.i., and the virus titers were determined by plaque assay on MDCK cells as described above.
Establishment of A431 stable cell lines expressing AnxA6 and virus infection.
The ViraPower lentiviral expression system (Invitrogen) was used to generate an A431-based stable cell line expressing recombinant human AnxA6 (A431-AnxA6). The pLenti-AnxA6 construct was generated as described above. Briefly, subconfluent monolayers of 293T cells grown in a 10-cm dish were transfected with pLenti-AnxA6 or pLenti-eGFP transfer vector (5 μg), pGag-pol (5 μg) packaging plasmid (5 μg), and pVSV-G plasmid (5 μg) using a CalPhos transfection kit (Clontech). At 48 h posttransfection, the cell culture medium containing pseudotyped lentiviral particles was harvested and centrifuged to remove cell debris. The viral supernatant was used directly for infection of A431 cells seeded into 6-well plates and grown to 70% confluence before infection. Two days later, cell culture medium was replaced with culture medium supplemented with 5 μg/ml blasticidin for selection. The medium was changed every 2 days, and the surviving clones were propagated and screened for AnxA6 expression by Western blotting, flow cytometry, and immunofluorescence assays.
Immunofluorescence assay.
To study the colocalization of AnxA6 and M2 on virus-infected cells, A549 or A431 cells grown on coverslips were transiently transfected with pCMV-AnxA6-GFP plasmid and then infected with influenza A/WSN/33 virus at an MOI of 5. A low-temperature preincubation method was used to allow synchronized infection. At 14 h p.i., cells were fixed with 4% paraformaldehyde (PFA) for 10 min and permeabilized in PBS containing 0.1% Triton X-100 for 10 min. The viral M2 protein was stained with mouse anti-M2 14C2 MAb, followed by incubation with Alexa Fluor 555-conjugated goat anti-mouse IgG (Invitrogen). Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole), and images were captured with a Zeiss LSM 700 confocal fluorescence microscope at the Faculty Imaging Core Facility of the University of Hong Kong.
To study the effects of AnxA6 on the early stages of influenza virus infection, virus infectivity was determined on siRNA-treated (si_AnxA6 or si_Control) A549 cells or A431 cells and the A431-AnxA6 stable cell line. Briefly, cells grown on glass coverslips in a 24-well plate were infected with influenza A/WSN/33 virus at an MOI of 5. At 3 h p.i., cells were fixed and permeabilized in PBS containing 0.1% Triton X-100 for 10 min. Indirect immunofluorescence was then performed using a mouse anti-NP MAb as a marker of infection, followed by incubation with Alexa Fluor 555-conjugated goat anti-mouse IgG (Invitrogen). Nuclei were stained with DAPI to count the total number of cells. Images were acquired using a Zeiss Axio Observer wide-field inverted fluorescence microscope. The percentage of infected cells was then calculated using the cell scoring application module of Metamorph image analysis software (Molecular Devices).
To investigate the effect of AnxA6 depletion on viral protein trafficking, the subcellular localization of NP and M2 was studied. Briefly, siRNA-treated A549 cells were infected with influenza A/WSN/33 virus at an MOI of 5 and fixed at 4, 6, and 8 h p.i. Viral NP was immunostained as a marker for export of the vRNP complex, and nuclei were stained with DAPI. Images of randomly chosen microscopic fields of duplicates were acquired (>200 cells) using a Zeiss Axio Observer wide-field inverted fluorescence microscope. The percentage of cells with nuclear NP only was determined using Metamorph image analysis software (Molecular Devices).
Quantitative RT-PCR assay for vRNA and mRNA.
siRNA-treated A549 cells were infected with influenza A/WSN/33 virus at an MOI of 3, and the total RNA was extracted at 4 h p.i. by using an RNeasy kit (Qiagen). One microgram of total RNA was reverse transcribed using SuperScript II reverse transcriptase (Invitrogen). For the detection of viral genomic RNA (vRNA), a 10 μM concentration of a vRNA-specific primer complementary to the 3′ end of vRNA was used in the reverse transcription (RT) reaction mix (26), whereas for mRNA detection, 500 ng of oligo(dT) primer was used. For quantification of M gene vRNA and mRNA, a SYBR green-based real-time PCR method (Roche) was used, and β-actin mRNA was quantified to normalize the total RNA concentration between different samples, as described previously (29). The primers for M gene detection were M-F (5′-AAGACCAATCCTGTCACCTCTGA-3′) and M-R (5′-CAAAGCGTCTACGCTGAGTCC-3′), and those for β-actin detection were B-F (5′-CCCAAGGCCAACCGCGAGAAGAT-3′) and B-R (5′-GTCCCGGCCAGCCAGGTCCAG-3′). The PCR experiments were performed using a LightCycler system (Roche). A reaction mix of 20 μl was composed of 1 μl of each gene-specific primer at 10 μM, 10 μl of SYBR green master mix, 5 μl of 10-fold-diluted cDNA, and 3 μl of distilled water. The amplification program was as follows: 95°C for 5 min followed by 45 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 10 s. The specificity of the assay was confirmed by melting curve analysis at the end of the amplification program.
Luciferase assay for viral polymerase complex activity.
siRNA-treated 293T cells were cotransfected with pPolI-Luc-RT (36) together with plasmids encoding PB2, PB1, PA and NP of influenza A/WSN/33 virus. In addition, a green fluorescent protein (GFP) expression plasmid was also cotransfected, and GFP signals were used for data normalization. At 48 h posttransfection, cells were lysed and then incubated with Steady-Glo assay reagent (Promega) for 5 min, and the luminescence was measured using a luminometer (Victor3; PerkinElmer). Means and standard deviations were determined for triplicates of two independent experiments.
Flow cytometry.
A431 cells and the A431-AnxA6 stable cell line were infected with influenza A/WSN/33 virus at an MOI of 3. Cells were fixed at 8 and 10 h p.i., and the cell surface-expressed viral M2 protein was stained using mouse anti-M2 14C2 MAb and Alexa Fluor 488-conjugated goat anti-mouse IgG (Invitrogen), without permeabilization treatment. After washing, the cell suspensions were subjected to flow cytometry on an LSRII flow cytometer (BD Biosciences), and more than 20,000 singlet living cells were collected. The postacquisition data analysis was performed using FlowJo software (TreeStar).
Electron microscopy.
Thin-section transmission electron microscopy (TEM) was performed as previously described (1). A431 cells and the A431-AnxA6 stable cell line were infected with influenza A/WSN/33 virus at an MOI of 5. At 10 h p.i., cells were washed with PBS and prefixed with 2.5% glutaraldehyde in cacodylate buffer (0.1 M sodium cacodylate-HCl buffer, pH 7.4) overnight at 4°C. Fixed cells were then scraped off and pelleted by low-speed centrifugation. After being washed in the same buffer, cells were postfixed with 1% osmium tetroxide for 30 min at room temperature, dehydrated in increasing concentrations of ethanol, and embedded in epoxy resin. Ultrathin sections were then stained with 2% aqueous uranyl acetate and Reynold's lead citrate. Samples were imaged on a Philips EM208s electron microscope at the Electron Microscopy Unit of the University of Hong Kong.
RESULTS
M2 CT interacts with human AnxA6 in yeast.
The IAV M2 ion channel protein is a highly conserved transmembrane protein and possesses the longest CT among the three viral envelope proteins. This 54-residue CT is accessible to the cellular machinery after fusion with the endosomal membrane and during production, maturation, and trafficking of viral envelope proteins and virus assembly and budding (25, 31, 33, 51). We hypothesized that cellular factors interacting with this CT could either help or restrict processes involving M2. To identify these factors, we performed a genomic Y2H screen, using IAV M2 CT as bait (Fig. 1A). A random-primed cDNA library from human placenta was screened (63.89 million interactions were tested), leading to the identification of 273 positive clones corresponding to 47 cellular factors. One of the most prominent results of the screen was the identification of the interaction between M2 CT and human AnxA6, with a total of 15 positive clones (5%) corresponding to this protein (GenBank accession number NM_001155.3) (Table 1). This interaction was classified with the highest confidence score (predicted biological score of A). AnxA6 was not found in parallel screens performed with other influenza virus baits (data not shown). Among the 15 AnxA6 cDNA clones, clones 270 and 196 constituted the smallest and largest cDNA fragments, respectively. Sequence alignment of the 15 AnxA6 cDNA clones revealed a minimal interacting domain located between repeats 2 and 3 of the first core domain (amino acid residues 44 to 147) (Fig. 1B). These data indicate that AnxA6 interacts specifically with the IAV M2 CT in yeast.
Fig 1.
Identification of AnxA6 as a cellular interactant of M2 CT. (A) Diagram of structural motifs in the M2 protein and sequence of the CT used as bait for Y2H screening. ED, ectodomain; TM, transmembrane domain. (B) Schematic representation of full-length AnxA6 and of clones 196 and 270. (a) The eight repeats of AnxA6 are shown with gray numbered boxes. Among the 15 AnxA6-positive clones isolated, clone 196 (b) and clone 270 (c) contained the largest and smallest AnxA6 cDNA fragments, respectively. Numbers indicate amino acid positions.
Table 1.
Yeast two-hybrid screening for AnxA6-M2 CT interaction
| Characteristic | Value or description |
|---|---|
| Bait | M2 cytoplasmic tail (aa 44–97) |
| Library | Random-primed human placenta cDNA |
| No. of interactions tested | 63.89 million |
| Prey | AnxA6 |
| No. of positive clones | 15 |
| Confidence scorea | A |
| Minimal interacting domain | aa 44-147 |
Predicted biological score (A to E), a statistical confidence score assigned to each interaction by Hybrigenics.
M2 binds to AnxA6 in influenza virus-infected human cells.
To confirm the interaction between M2 CT and AnxA6 during the IAV life cycle in human cells, we performed coimmunoprecipitation experiments with HEK 293T cells transiently transfected with a plasmid encoding a myc-tagged AnxA6 protein (myc-AnxA6) and infected with influenza A/WSN/33 virus. Expression of the recombinant myc-AnxA6 and viral M2 proteins was detected with anti-myc and anti-M2 MAbs, respectively (Fig. 2A, upper panel). Cell lysates were then immunoprecipitated with anti-myc MAb and subsequently immunoblotted with anti-M2 MAb. As shown in the middle panel of Fig. 2A, coimmunoprecipitation of M2 with AnxA6 was detected, confirming their physical interaction in virus-infected cells. The specificity of the M2-AnxA6 interaction was further demonstrated by detection of myc-AnxA6 protein in reciprocal coimmunoprecipitation assays using anti-M2 MAb and by the absence of coimmunoprecipitation of myc-AnxA6 in cell lysate from noninfected cells (Fig. 2A, lower panel). These results clearly demonstrate that the IAV M2 protein physically binds to recombinant AnxA6 in virus-infected cells.
Fig 2.
AnxA6 interacts with M2 in virus-infected human cells. (A) Reciprocal coimmunoprecipitation assay of myc-AnxA6 and M2 in 293T cells. 293T cells were transfected with a plasmid encoding a myc-tagged AnxA6 protein and were infected 24 h later with A/WSN/33 virus at an MOI of 0.1. Cells were lysed at 24 h p.i., and a coimmunoprecipitation assay was performed using either anti-myc (middle panel) or anti-M2 (lower panel) MAb. Immunoprecipitated proteins were detected by Western blotting using either anti-M2 or anti-myc MAb. The input samples were 10-fold-diluted cell lysates (upper panel). (B) AnxA6 colocalizes with M2 protein close to the plasma membranes of infected A549 cells. A549 cells were transfected with AnxA6-GFP plasmid and then infected with influenza A/WSN/33 virus at an MOI of 5 as indicated. Cells were fixed at 14 h p.i., stained with mouse anti-M2 MAb followed by Alexa Fluor 555-conjugated goat anti-mouse IgG, and analyzed by confocal microscopy. Bars, 10 μm.
To investigate the subcellular localization where the M2-AnxA6 interaction occurs in infected cells, we performed colocalization studies with A549 cells transiently transfected with a plasmid encoding AnxA6 fused to GFP (AnxA6-GFP) (12) and infected with influenza virus strain WSN at an MOI of 5 for 14 h. In noninfected but transfected control cells, AnxA6 was evenly distributed in the cytosol (Fig. 2B, left panels), as reported previously by others (12, 14). For the infected but nontransfected control cells, the M2 protein was distributed in intracellular compartments close to the perinuclear region, suggestive of its localization at the endoplasmic reticulum and Golgi apparatus and at the plasma membrane (Fig. 2B, middle panels). For transfected and infected double-positive cells, there was no change in the M2 staining pattern compared to that of nontransfected infected control cells. Interestingly, confocal imaging revealed a partial colocalization of M2 and AnxA6 in distinct punctum-like areas at or close to the plasma membrane of virus-infected cells (Fig. 2B, right panels, arrows).
Altogether, these results confirm the Y2H result that the IAV M2 protein physically interacts with AnxA6 in virus-infected human cells and suggest that the interaction occurs close to or at the plasma membrane.
AnxA6 negatively modulates influenza virus infection.
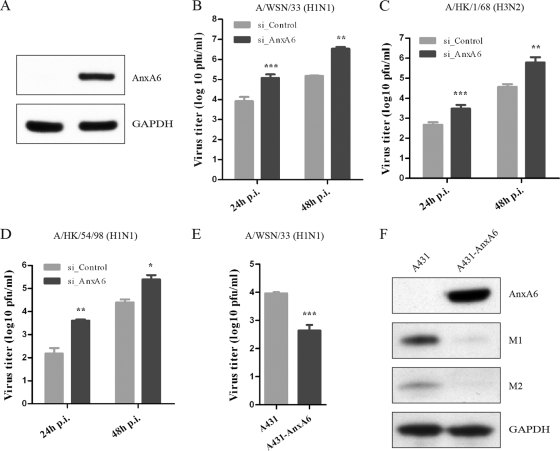
To define the role and impact of M2-AnxA6 interaction on the viral replication cycle, we first studied the effect of AnxA6 gene knockdown on the replication of IAV in A549 human lung epithelial cells. Immunoblot analyses confirmed that a pool of AnxA6-specific siRNA duplexes (data not shown), as well as one individual siRNA duplex (si_AnxA6), efficiently and reproducibly reduced cellular AnxA6 expression levels, but nonspecific control siRNA (si_Control) did not (Fig. 3A). Using this protocol, we investigated whether silencing of AnxA6 expression had any impact on the production of infectious influenza virus particles. Twenty-four hours after infection with influenza virus strain WSN at an MOI of 0.01, we found that silencing of AnxA6 expression led to a significant increase (∼10-fold) of progeny virus titers in cell culture supernatants compared with those of si_Control-treated cells (Fig. 3B). In order to test whether this inhibitory role is also applicable to other IAV strains, two clinical isolates were tested. Similar enhancements of virus progeny titers were also observed for influenza A/HK/1/68 (H3N2) (Fig. 3C) and A/HK/54/1998 (H1N1) (Fig. 3D) viruses at both 24 and 48 h p.i. These results demonstrate that silencing of AnxA6 expression increases the production of infectious influenza A virus progeny, suggesting that AnxA6 negatively modulates IAV infection.
Fig 3.
AnxA6 negatively modulates influenza virus infection. (A) siRNA-mediated gene knockdown efficiency analysis. Cell lysates of siRNA-treated cells were collected at 60 h posttransfection, and expression levels of the AnxA6 and GAPDH proteins were determined by Western blotting. (B, C, and D) AnxA6 depletion in A549 cells increases progeny virus titer. A549 cells treated with the indicated individual siRNAs were infected with influenza A/WSN/33 (H1N1) virus (B), A/HK/1/68 (H3N2) virus (C), or A/HK/54/98 (H1N1) virus (D) at an MOI of 0.01. Cell culture supernatants were collected at 24 and 48 h p.i., and virus titers were determined by plaque assay on MDCK cells. Data are shown as means ± standard deviations (SD) for triplicates from four independent experiments (B) and duplicates from three independent experiments (C and D). **, P < 0.01; ***, P < 0.0001 by unpaired Student's t test. (E) Overexpression of AnxA6 in A431 cells decreases progeny virus titer. A431 cells were used to establish a stable cell line overexpressing AnxA6 that was infected with influenza A/WSN/33 virus at an MOI of 0.01. Cell culture supernatants were collected at 24 h p.i., and virus titers were determined by plaque assay on MDCK cells. Data are shown as means ± SD for measurements from quadruplicates in a representative experiment. ***, P < 0.0001 by unpaired Student's t test. (F) Overexpression of AnxA6 in A431 cells impairs virus propagation. The same experiment as that in panel E was conducted with cell lysates collected at 24 h p.i. Expression levels of viral (M1 and M2) and cellular (AnxA6 and GAPDH) proteins were determined by Western blotting.
To corroborate these findings, we determined the impact of AnxA6 overexpression on virus replication. We used the A431 human vulval squamous epithelial cell line, which naturally lacks endogenous AnxA6 (54), and established an A431 cell line stably expressing AnxA6 (A431-AnxA6), and we infected these cells with influenza virus WSN at an MOI of 0.01. At 24 h p.i., the progeny virus titer from A431 cells reached 9 × 103 PFU/ml. In contrast, a viral titer of 5 × 102 PFU/ml was measured for the A431-AnxA6 stable cell line (Fig. 3E). Although the virus replicates less efficiently in A431 cells than in A549 cells, a significant decrease of progeny virus titers was observed with the A431-AnxA6 stable cell line (∼18-fold). In addition, the viral M1 and M2 protein levels were determined by immunoblot analysis at 24 h p.i. As shown in Fig. 3F, immunoblot analyses confirmed the absence of endogenous AnxA6 and the overexpression of recombinant AnxA6 in normal A431 and A431-AnxA6 cells, respectively (upper panel), and showed a clear reduction of viral proteins detected in the A431-AnxA6 stable cell line (middle panels). These results demonstrate that AnxA6 overexpression decreases the titer of released infectious influenza virus particles, and they further support the negative regulatory role of AnxA6 in virus infection.
Collectively, these results demonstrate that modulation of AnxA6 expression (silencing or overexpression) impacts the production of infectious progeny viruses, which confirms that AnxA6 negatively regulates IAV infection and its propagation in human cell lines.
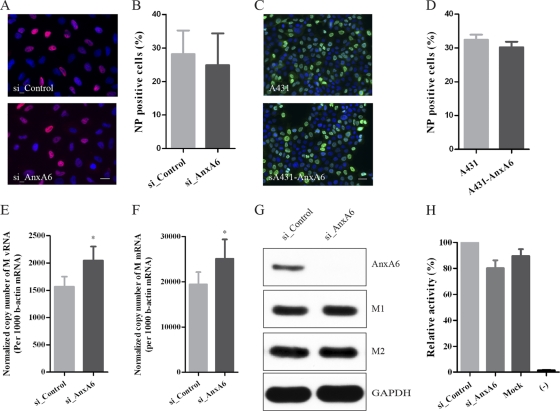
Modulation of AnxA6 expression does not impair the early stages of the viral life cycle or viral gene expression.
To address which step(s) of the virus life cycle is regulated by the modulation of AnxA6 expression, we performed a comprehensive and systematic dissection of the viral life cycle. We first wondered whether silencing or overexpression of AnxA6 might impair the early stages of the influenza virus life cycle, including viral entry. To address this point, we determined the virus infectivity in A549 cells pretreated with si_AnxA6 or si_Control and in A431 cells or the A431-AnxA6 stable cell line after infection with influenza virus WSN at MOIs of 1 and 5, respectively. Cells were fixed at 4 h p.i. and assayed for infection by immunostaining for NP, and the percentage of infected cells was determined. Similar fluorescence signals (Fig. 4A) and comparable percentages of NP-positive cells (Fig. 4B) were observed for cells treated with si_AnxA6 or si_Control. Similar results were obtained for A431 cells and the A431-AnxA6 stable cell line, and no significant difference between percentages of infected cells was observed (Fig. 4C and D). These results suggest that modulation of AnxA6 expression does not impair influenza virus entry or viral gene expression. We further confirmed this result by investigating the effects of AnxA6 silencing on HA-mediated viral entry, binding, and internalization, NP nuclear import after analysis of transduction with HA-pseudotyped lentiviral particles (HApp) (42), and NP subcellular localization at early time points of virus infection (30, 60, 120, and 180 min p.i.) (13). No significant difference was observed in transduction, measured as HApp-expressed luciferase activity, or in NP subcellular localization, showing that virus entry and NP nuclear import were normal in A549 cells pretreated with si_AnxA6 (see Fig. S1 in the supplemental material).
Fig 4.
AnxA6 does not affect the early stages of the viral life cycle or viral gene expression. (A and B) Silencing of AnxA6 expression does not affect the early stages of the viral life cycle. (A) A549 cells treated with the indicated siRNAs were infected with influenza A/WSN/33 virus at an MOI of 5. At 3 h p.i., cells were fixed and immunostained for NP as a marker of infection. Nuclei were stained using DAPI. Bar, 20 μm. (B) Percentages of infected (NP-positive) cells (n, >100,000) plotted against the siRNA treatments used. (C and D) AnxA6 overexpression does not affect the early stages of the viral life cycle. The same experiment as that in panels A and B was performed with the A431-AnxA6 stable cell line and wild-type A431 cells (n, >10,000). (E and F) Silencing of AnxA6 expression does not affect viral RNA replication and transcription. A549 cells treated with the indicated siRNAs were infected with influenza A/WSN/33 virus at an MOI of 3, and total RNA was then extracted at 4 h p.i. M gene vRNA and mRNA levels were determined by quantitative RT-PCR. Data are shown as means ± SD for measurements from triplicates and are representative of 2 independent experiments. (G) Silencing of AnxA6 expression does not affect viral M gene expression. A549 cells treated with the indicated siRNAs were infected with influenza A/WSN/33 virus at an MOI of 3. Cell lysates were collected at 4 h p.i., and expression levels of viral (M1 and M2) and cellular (AnxA6 and GAPDH) proteins were determined by Western blotting. (H) Silencing of AnxA6 expression does not affect viral polymerase complex activity. 293T cells treated with the indicated siRNAs were transfected with plasmids encoding the polymerase complex components and NP derived from influenza A/WSN/33 virus, along with a reporter plasmid containing the noncoding sequence from the M segment as well as the luciferase gene driven by the human polymerase I promoter. Luciferase activity was determined at 48 h posttransfection, and relative activities were compared. Data are shown as means ± SD for measurements from triplicates and are representative of 2 independent experiments.
To further investigate a potential effect of modulation of AnxA6 expression on vRNA replication or mRNA transcription, the copy number of the viral M gene was measured by quantitative RT-PCR at 4 h p.i. on siRNA-treated A549 cells infected with influenza virus WSN at an MOI of 3. Slight but statistically significant increases in the copy numbers of M gene vRNA (Fig. 4E) and mRNA (Fig. 4F) were detected for infected si_AnxA6-treated cells (∼1.3-fold). Nevertheless, we could not observe any change in protein level for the M1 and M2 proteins at the same time point by immunoblot analysis (Fig. 4G), indicating that modulation of AnxA6 expression does not affect viral gene expression. In addition, the potential effect of silencing of AnxA6 expression on viral polymerase activity was also investigated using an influenza virus minireplicon system. siRNA-pretreated 293T cells were cotransfected with plasmids encoding the influenza virus WSN polymerase complex proteins (PB1, PB2, PA, and NP), along with a reporter plasmid containing noncoding sequences from the influenza virus M segment as well as the firefly luciferase gene driven by the human RNA polymerase I promoter (36, 60). Consistently, AnxA6 depletion had no significant effect on the luciferase reporter enzymatic activity (∼1.25-fold) (Fig. 4H), suggesting that modulation of AnxA6 expression does not impair viral polymerase activity.
Taken together, these results indicate that the impact of modulation of AnxA6 expression on production of infectious progeny viruses does not occur during the early stages of the virus life cycle or during viral RNA synthesis and gene expression.
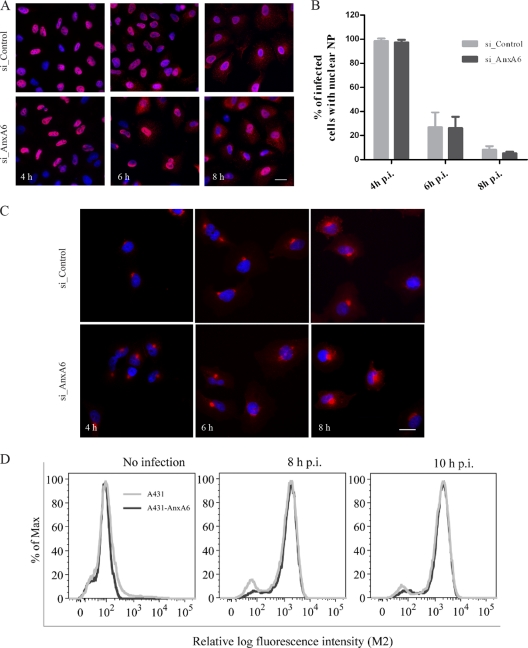
Modulation of AnxA6 expression does not impair vRNP export or M2 protein expression and trafficking.
To address whether silencing of AnxA6 expression could affect vRNP export to the cytoplasm, NP subcellular localization was analyzed during the time course of infection of siRNA-treated A549 cells (Fig. 5A). NP subcellular localization was scored according to whether the NP staining was predominantly nuclear or cytoplasmic (Fig. 5B) (43). At 4 h p.i., for both si_AnxA6- and si_Control-treated cells, NP proteins were found to be mainly nuclear, as expected for an intermediate time point in the virus life cycle (Fig. 5A, left panels, and B). For both types of siRNA-treated cells, the percentage of cells exhibiting nuclear NP decreased with time as NP was progressively exported to the cytoplasm (Fig. 5A, middle and right panels, and B). No significant difference in NP subcellular localization was measured between si_AnxA6- and si_Control-treated A549 cells. This observation indicates that silencing of AnxA6 expression has no effect on vRNP export to the cytoplasm.
Fig 5.
AnxA6 does not affect vRNP export or M2 protein expression and trafficking. (A and B) NP subcellular localization in AnxA6-depleted A549 cells. A549 cells treated with the indicated siRNAs were infected with influenza A/WSN/33 virus at an MOI of 5. Cells were fixed at 4, 6, and 8 h p.i. for immunostaining of NP as a marker for vRNP complex export, and nuclei were stained using DAPI. Bar, 20 μm. Cytoplasmic and nuclear NP localization was quantified using Metamorph software, and the plotted histogram represents the percentage of infected cells with nuclear NP only. Data are shown as means ± SD for measurements from at least 200 cells in duplicates of a representative experiment. (C) M2 subcellular localization in AnxA6-depleted A549 cells. A549 cells treated with the indicated siRNAs were infected with influenza A/WSN/33 virus at an MOI of 5. Cells were fixed at 4, 6, and 8 h p.i. for immunostaining of M2 protein. Bar, 20 μm. (D) AnxA6 overexpression does not affect M2 cell surface expression. A431 cells and the A431-AnxA6 stable cell line were infected with influenza A/WSN/33 virus at an MOI of 3. Cells were fixed at 8 and 10 h p.i., left nonpermeabilized, and labeled with mouse anti-M2 MAb followed by Alexa Fluor 488-conjugated goat anti-mouse IgG for M2 cell surface analysis by flow cytometry. The graph shows the amount of cell surface expression and fluorescence intensity of the M2 viral antigen.
Subcellular localization of viral M2 protein was also analyzed during the time course of infection of siRNA-treated A549 cells (Fig. 5C). No difference in M2 subcellular localization was observed between si_AnxA6- and si_Control-treated A549 cells. We also wondered whether AnxA6 overexpression could affect cell surface expression of M2. We quantified cell surface expression of M2 antigen on nonpermeabilized infected A431 cells and the A431-AnxA6 stable cell line by using flow cytometry. As shown in Fig. 5D, no significant change in M2 cell surface expression was observed at 8 or 10 h p.i. These results indicate that AnxA6 depletion has no effect on M2 cell surface expression.
Taken together, these data indicate that the regulation of production of infectious progeny viruses by AnxA6 does not occur during NP nuclear export or M2 cell surface expression, and they suggest that a very late stage of the virus life cycle is affected.
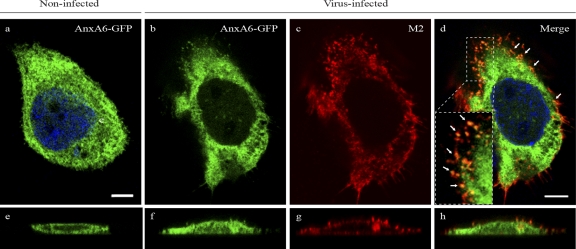
AnxA6 modulates influenza A virus replication by disrupting virus budding processes.
To determine whether late stages of the virus life cycle are regulated by the modulation of AnxA6 expression, we reexamined the colocalization of M2 and AnxA6 at 14 h p.i. A431 cells transiently transfected with a plasmid encoding a GFP-fused AnxA6 protein (AnxA6-GFP) (12) were infected with influenza virus strain WSN at an MOI of 5. Compared to A549 cells, A431 cells naturally lack AnxA6 expression, which allowed us to exclude any interaction between M2 and endogenous AnxA6. The results obtained were similar to those observed for A549 cells (Fig. 2B). In noninfected but transfected controls, AnxA6 was evenly distributed in the whole-cell cytosol (Fig. 6a), as reported previously by others (12). In infected but nontransfected control cells, M2 was localized mainly at the cell plasma membrane and also in some punctate areas (data not shown). For transfected and infected double-positive cells, the M2 staining pattern was similar to that for infected but nontransfected control cells (Fig. 6c). However, the distribution of AnxA6 was less diffuse in the cytosol and exhibited many enriched punctate areas compared with control cells without infection, in which AnxA6 colocalized with M2 (Fig. 6d, arrows and inset). In the z-plane reconstruction, the confocal imaging revealed that M2 and AnxA6 colocalized at the plasma membrane (Fig. 6h). This observation indicates that both proteins most likely interact at the plasma membrane, where viral assembly and virion budding occur.
Fig 6.
AnxA6 colocalizes with M2 at the plasma membrane of virus-infected human cells. A431 cells were transfected with a plasmid encoding the AnxA6-GFP fusion protein and then infected 24 h later with influenza A/WSN/33 virus at an MOI of 5. After fixation at 14 h p.i., cells were stained with a mouse anti-M2 MAb followed by Alexa Fluor 555-conjugated goat anti-mouse IgG. Single optical sections (upper panels) and representative z-plane reconstructions (lower panels) are shown. The inset in panel d shows a magnified (×3.7) region at the plasma membrane. Arrows indicate colocalization of AnxA6 and M2 at the plasma membrane. Bar, 10 μm.
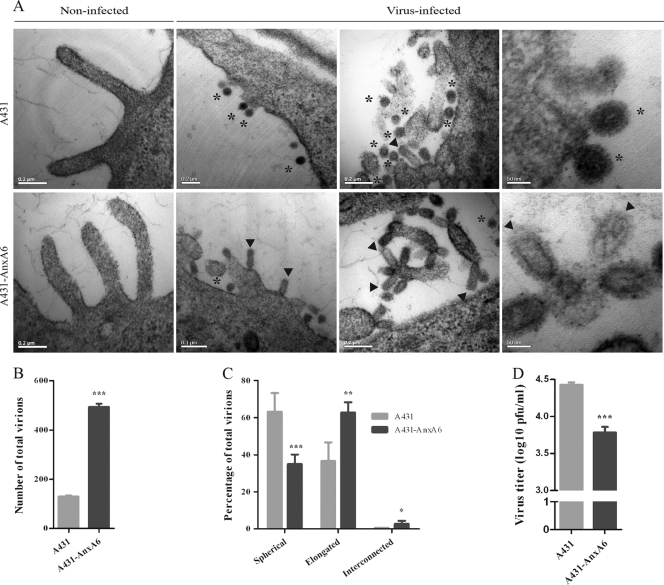
Using thin-section TEM, we examined virus budding and morphology on the surfaces of A431 cells and the A431-AnxA6 stable cell line after infection with influenza virus strain WSN at an MOI of 5 for 10 h. For A431 cells, consistent with previous observations of infection with influenza virus WSN (10), a majority of spherical particles that were 60 to 80 nm in diameter and completely released from the plasma membrane was observed (Fig. 7A, upper panels, asterisks). Some spherical budding virions connected to the cell surface by a neck were also detected, occasionally retaining an elongated morphology (Fig. 7A, upper panels, arrowhead). In contrast, in the A431-AnxA6 stable cell line, virions were more abundant and displayed a higher degree of pleomorphism. Most virions were viral particles exhibiting an elongated shape rather than a spherical one (Fig. 7A, lower panels, arrowheads and asterisks, respectively). In some rare cases, interconnected virions were observed, resembling budding-defective influenza virus particles found in cells depleted of Rab11 (3) or overexpressing viperin (61) or seen when M2 protein is truncated (38). For both A431 and A431-AnxA6 noninfected controls, only smooth plasma membranes with some filopodial protrusions of 100 nm in diameter and less than 1 μm in length were observed (Fig. 7A, left panels). To confirm these observations, the total number of virions was determined for both cell types, and virions were categorized into three different types according to morphology: spherical, elongated, and interconnected virions. Interestingly, 4 times more total virions were counted in the A431-AnxA6 stable cell line than in A431 cells (Fig. 7B), and 60.7% of the virions were elongated, while 36.6% were spherical ones (Fig. 7C). For A431 cells, a reverse distribution was observed, with 36.1% elongated viral particles and 63.5% spherical ones. Interconnected virions were marginal and were observed mostly in the A431-AnxA6 stable cell line (2.7% compared to 0.4%). Using the same single-cycle replication conditions, we determined the PFU titers of progeny virus from normal A431 and A431-AnxA6 cells (Fig. 7D). A significant drop in virus titer was observed when AnxA6 was overexpressed. Conversely, virus titers were increased after siRNA-mediated silencing of AnxA6 expression in A549 cells at 10, 12, and 14 h p.i. under single-cycle replication conditions (data not shown). These results suggest that AnxA6 overexpression impairs or delays viral budding. Taking these data in conjunction with the larger number of virions detected on the A431-AnxA6 stable cell line, it appears that the absence of AnxA6 results in more efficient budding and release of infectious viral particles.
Fig 7.
AnxA6 overexpression impairs influenza virus budding. (A) Analysis of budding virions from AnxA6-overexpressing cells by TEM. A431 cells (upper panels) and the A431-AnxA6 stable cell line (lower panels) were infected with influenza A/WSN/33 virus at an MOI of 5. At 10 h p.i., cells were fixed and processed for thin sectioning and TEM. Asterisks indicate spherical virions, and arrowheads indicate elongated virions. Noninfected controls are shown on the left. (B) Semiquantitation of total numbers of virions. Virions observed in infected cells (n = 13 cells with >100 virions) of each cell type were manually counted blinded by five different investigators, and the total numbers of virions were compared. ***, P < 0.0001 by unpaired Student's t test. (C) Semiquantitation of virions according to morphology. During counting, virions were categorized into three types according to their morphology and size, as indicated. The numbers of virions for each type were determined, and their percentages were subjected to unpaired Student's t test and plotted as means ± SD (***, P < 0.0001; **, P < 0.01; *, P < 0.05). (D) Progeny virus titration of released virions. The same experiment as that in panel A was performed with cell culture supernatants collected at 9 h p.i. Virus titers were determined by plaque assay on MDCK cells. Data are shown as means ± SD for triplicates in a representative experiment. ***, P < 0.0001 by unpaired Student's t test.
Taken together, these results demonstrate that AnxA6 overexpression results in defects in virus budding, leading to slower budding and release of infectious viral particles.
DISCUSSION
Morphogenesis and virus budding of influenza virions comprise a complex, multistep process that occurs in lipid raft domains on the apical membrane of infected cells (reviewed in references 41 and 51). This process requires involvement of both host and viral components. Lipid rafts and actin microfilaments are host components that play important roles in virus assembly and budding. Unlike other enveloped viruses, influenza virus does not utilize the host endosomal sorting complex required for transport (ESCRT) machinery to mediate membrane scission and virus release, suggesting that influenza virus buds independently of the host ESCRT pathway (reviewed in references 4, 5, and 62). Among the viral components, it has been suggested that M2 provides this membrane scission mechanism, causing budding completion and virus release. Rossman et al. proposed an influenza virus budding model (51) in which M2 interacting with M1 is first recruited to a cholesterol-rich environment stabilizing the budding site (49). M2 is then localized to a lower-cholesterol environment at the neck of the budding virion, where it induces a negative membrane curvature, causing membrane scission and release of the progeny virus (50). Nevertheless, involvement of unknown host factors in morphogenesis and virus budding cannot be ruled out.
In the present study, we hypothesized that cellular factors interacting with the 54-residue M2 CT could either help or restrict processes involving M2. Few host factors interacting with M2 have been identified, and the functional relevance of these interactions is not fully characterized (21, 55). In order to identify and characterize human cellular regulators of influenza virus infection, we used a genomic Y2H approach with M2 CT as bait. Previous studies have shown that such an approach was successful for the IAV PB1 (28), M1 (37), NS1 (35), and NS2 (7) proteins. We identified a novel interaction between M2 CT and human AnxA6, which most likely occurs at the plasma membrane, the virus assembly and budding site. We showed that AnxA6 negatively modulates influenza virus infection by affecting virus budding, as indicated by a drop in progeny virus PFU titer in supernatants and an increased number of virions at the surface of A431-AnxA6 cells, which exhibit an elongated shape rather than a spherical one.
AnxA6 is a member of the annexin family, a conserved multigene family of Ca2+-dependent membrane-binding proteins, which provides a link between Ca2+ signaling and membrane functions (18). It has been demonstrated that AnxA6 interacts with the μ1 and μ2 subunits of the AP1-clathrin complex (11) of the clathrin-mediated endocytosis process, which is exploited by IAV to enter host cells (32). In the present study, when AnxA6 was depleted we measured no difference in infectivity rate (Fig. 4B) or transduction with HApp (see Fig. S1 in the supplemental material), suggesting that AnxA6 does not affect viral entry. Previous studies indicated that AnxA6 has a tumor suppressor activity by facilitating the Ca2+-dependent targeting of p120GAP (GTPase-activating protein) at the cell plasma membrane, which leads to inhibition of Ras/Raf-1 activity (59). Interestingly, it has been shown that inhibition of the Raf signaling pathway results in nuclear retention of vRNPs and impairs production of infectious influenza virus virions (48). In our study, when AnxA6 was depleted we observed no difference in NP subcellular localization over time, suggesting that NP nuclear export was normal (Fig. 5B). AnxA6 plays important roles in several biological processes involving host components which are also essential for virus assembly and budding, such as cholesterol, a critical component of lipid rafts, and cortical actin (reviewed in reference 14). Each step of virus assembly and budding (i.e., assembly of viral components, budding initiation, growth, and completion) requires different interactions with host components, some of which may facilitate some step(s) of viral budding but may also interfere with other steps (41).
At a low intracellular Ca2+ ([Ca2+]i) concentration, AnxA6 is distributed diffusely throughout the cytosol (14). However, when [Ca2+]i increases, stimulated AnxA6 is targeted to distinct subcellular membrane locations, such as the plasma membrane, endosomes, or secretory vesicles. Thus, cytosolic AnxA6 is probably translocated to distinct subcellular membrane locations upon virus infection, due to the dysregulation of Ca2+ homeostasis observed after IAV infection (24, 58). If AnxA6 is overexpressed and targeted at the plasma membrane, it has been shown that it interacts with, recruits, and causes the rearrangement of F-actin, resulting in stabilization of the cortical cytoskeleton (40). In the present study, we have demonstrated that M2 physically interacts with AnxA6 in infected cells and colocalizes with AnxA6-GFP at the plasma membrane of infected cells (Fig. 2A and B, panel i, and 6d and h), where more budding or budded virions with an elongated shape are observed when AnxA6 is overexpressed (Fig. 7A, B, and C). Rossman et al. showed that M2 localizes to the neck of budding virions at the virus budding site, using immunogold labeling and electron microscopy (50). At the point of membrane scission, M2 accumulates where its CT is accessible to AnxA6. A tempting hypothesis would be that AnxA6 serves as a linker to stabilized cortical F-actin. This linking may affect the M2-mediated late stage of virus budding and explain the virus budding completion defect or delay observed by TEM after AnxA6 overexpression in A431 cells. Indeed, cortical actin filaments may interfere with the membrane scission step, as actin disruption has been shown to help in bud release in HeLa (22) and MDCK (53) cells. AnxA6-mediated changes observed in the cortical actin cytoskeleton may also impact other actin-dependent stages of virus assembly and budding. Recently, it was suggested that an intact actin network is necessary for clustering of M2 with HA at the virus assembly site (56), since cortical actin may drive the formation of protein clusters and/or organize the maintenance of lipid raft domains (19). Moreover, Bruce et al. have shown that release of influenza virus virions is attenuated when expression of Rab11 is silenced (3). This small GTP-binding protein is involved in the trafficking of proteins to the apical membrane in polarized cells and in actin remodeling. It has further been shown that silencing of Rab11 expression reduces M2 cell surface expression, indicating a defect in proper M2 transport to the plasma membrane along the actin network rather than a direct role for Rab11 in membrane scission (50). In the present study, when AnxA6 was overexpressed we observed no changes in M2 cell surface expression, suggesting that AnxA6 instead affects virus budding (Fig. 5D).
Interestingly, AnxA6 can also indirectly regulate cholesterol homeostasis. Cubells et al. have shown that AnxA6 overexpression impairs cholesterol export from late endosomes, contributing to an imbalance in cholesterol homeostasis and to decreased pools of cholesterol in other compartments, such as the Golgi apparatus and the plasma membrane (12). In addition, direct visualization of the membrane lipid structure indicated that AnxA6 overexpression induces a significant decrease of condensed membrane domains (C. Enrich, personal communication). Cholesterol depletion and architectural changes at the plasma membrane induced by high levels of AnxA6 may have important biological consequences on virus assembly and budding. Indeed, virus assembly occurs at lipid rafts in the apical plasma membrane, where HA is supposed to organize the budozone (52). We could therefore hypothesize that AnxA6 overexpression would impair the coalescence of HA-containing lipid rafts forming the budozone that is required for budding initiation. Interestingly, the interferon-inducible restriction factor viperin has been shown to exert its effects at the late stages of the influenza virus life cycle, preventing the release of viral particles by disrupting lipid rafts (61). At the surface of viperin-expressing cells, 43% of the viral particles were either budding virions with an abnormally elongated and large neck or a “daisy chain” structure in which two or more viral particles appeared to be interconnected. Moreover, in the influenza virus budding model proposed by Rossman et al., M2 is sequentially located to cholesterol-rich and then lower-cholesterol regions to allow budding initiation/growth and completion steps, respectively (51). Barman and Nayak have shown that cholesterol depletion at the plasma membrane of infected cells by use of methyl-β-cyclodextrin (MβCD) for a short time induces more rapid virus budding completion and release (2). Indeed, MβCD treatment may favor the association of M2 with lower-cholesterol regions, triggering membrane scission. In the present study, when AnxA6 was overexpressed we could observe a drop in progeny virus titer and more virions at the plasma membrane exhibiting a rather elongated shape than a spherical one (Fig. 7). Interconnected virions were also detected, but they were only marginally represented. These results do not support the notion that reduction of cholesterol pools at the plasma membrane upon AnxA6 overexpression may facilitate virus budding and release but rather suggest that it may impair or delay this process. This discrepancy with MβCD-mediated cholesterol depletion might be explained by the differences between the mechanisms of action of a specific drug for a short time and a multifunctional host cellular factor that is constitutively overexpressed and interacts transiently with its partners. Indeed, AnxA6-mediated reduction of cholesterol pools at the plasma membrane may favor the association of M2 with lower-cholesterol regions at the point of membrane scission, where AnxA6 could serve as a linker between M2 and stabilized cortical F-actin as discussed previously, resulting in a membrane scission defect.
In the present study, we have demonstrated that AnxA6 interacts with M2 CT and negatively modulates influenza virus infection by affecting viral budding. We propose that AnxA6 acts as a host cellular negative regulator of influenza virus infection, by interacting with M2, which is able to mediate membrane scission, or by modulating cellular components involved in viral budding, or both. Using a recombinant mutant WSN influenza virus lacking M2 by mutation of the splicing signal of the M gene (8), no significant difference in virus titers was observed when AnxA6 was silenced or overexpressed, suggesting that regulation of influenza virus production by AnxA6 is dependent on M2 (data not shown). The detailed molecular mechanism underlying this AnxA6-mediated negative modulation of influenza virus budding awaits further investigation. Work is ongoing to determine the functional correlation between M2-AnxA6 interaction and the defective viral budding phenotype and the contributions of AnxA6-mediated plasma membrane changes through cholesterol homeostasis modulation and cortical actin remodeling.
Supplementary Material
ACKNOWLEDGMENTS
We thank Carlos Enrich (Departamento de Biologia Cellular, Facultad de Medicina, Universidad de Barcelona, Spain) for expert advice and helpful discussions and Anthony Tsolaki (Centre for Infection, Immunity and Disease Mechanisms, School of Health Sciences and Social Care, Brunel University, United Kingdom) for a critical reading of the manuscript. We acknowledge the Core Imaging Facility and Electron Microscopy Unit of the Faculty of Medicine of the University of Hong Kong.
This work was supported by the Research Fund for the Control of Infectious Disease (project 09080892) of the Hong Kong Government, the Area of Excellence Scheme of the University Grants Committee (grant AoE/M-12/-06 of the Hong Kong Special Administrative Region, China), the French Ministry of Health, the RESPARI Pasteur Network, and the Li Ka Shing Foundation. H.M. and Y.Z. are Ph.D. and M.Phil. students supported by The University of Hong Kong. M.M. is an M.Phil. student supported by the University of Montpellier II, France.
Footnotes
Published ahead of print 23 November 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Barman S, Adhikary L, Kawaoka Y, Nayak DP. 2003. Influenza A virus hemagglutinin containing basolateral localization signal does not alter the apical budding of a recombinant influenza A virus in polarized MDCK cells. Virology 305:138–152 [DOI] [PubMed] [Google Scholar]
- 2. Barman S, Nayak DP. 2007. Lipid raft disruption by cholesterol depletion enhances influenza A virus budding from MDCK cells. J. Virol. 81:12169–12178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bruce EA, Digard P, Stuart AD. 2010. The Rab11 pathway is required for influenza A virus budding and filament formation. J. Virol. 84:5848–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carlton JG, Martin-Serrano J. 2009. The ESCRT machinery: new functions in viral and cellular biology. Biochem. Soc. Trans. 37:195–199 [DOI] [PubMed] [Google Scholar]
- 5. Chen BJ, Lamb RA. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen BJ, Leser GP, Jackson D, Lamb RA. 2008. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 82:10059–10070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen J, Huang S, Chen Z. 2010. Human cellular protein nucleoporin hNup98 interacts with influenza A virus NS2/nuclear export protein and overexpression of its GLFG repeat domain can inhibit virus propagation. J. Gen. Virol. 91:2474–2484 [DOI] [PubMed] [Google Scholar]
- 8. Cheung TK, et al. 2005. Generation of recombinant influenza A virus without M2 ion-channel protein by introduction of a point mutation at the 5′ end of the viral intron. J. Gen. Virol. 86:1447–1454 [DOI] [PubMed] [Google Scholar]
- 9. Ciampor F, Cmarko D, Cmarkova J, Zavodska E. 1995. Influenza virus M2 protein and haemagglutinin conformation changes during intracellular transport. Acta Virol. 39:171–181 [PubMed] [Google Scholar]
- 10. Compans RW, Dimmock NJ. 1969. An electron microscopic study of single-cycle infection of chick embryo fibroblasts by influenza virus. Virology 39:499–515 [DOI] [PubMed] [Google Scholar]
- 11. Creutz CE, Snyder SL. 2005. Interactions of annexins with the mu subunits of the clathrin assembly proteins. Biochemistry 44:13795–13806 [DOI] [PubMed] [Google Scholar]
- 12. Cubells L, et al. 2007. Annexin A6-induced alterations in cholesterol transport and caveolin export from the Golgi complex. Traffic 8:1568–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eierhoff T, Hrincius ER, Rescher U, Ludwig S, Ehrhardt C. 2010. The epidermal growth factor receptor (EGFR) promotes uptake of influenza A viruses (IAV) into host cells. PLoS Pathog. 6:e1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Enrich C, et al. 2011. Annexin A6-linking Ca(2+) signaling with cholesterol transport. Biochim. Biophys. Acta 1813:935–947 [DOI] [PubMed] [Google Scholar]
- 15. Formstecher E, et al. 2005. Protein interaction mapping: a Drosophila case study. Genome Res. 15:376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fromont-Racine M, Rain JC, Legrain P. 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16:277–282 [DOI] [PubMed] [Google Scholar]
- 17. Gannagé M, et al. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6:367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gerke V, Creutz CE, Moss SE. 2005. Annexins: linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell. Biol. 6:449–461 [DOI] [PubMed] [Google Scholar]
- 19. Goswami D, et al. 2008. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell 135:1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grantham ML, Stewart SM, Lalime EN, Pekosz A. 2010. Tyrosines in the influenza A virus M2 protein cytoplasmic tail are critical for production of infectious virus particles. J. Virol. 84:8765–8776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guan Z, et al. 2010. Interaction of Hsp40 with influenza virus M2 protein: implications for PKR signaling pathway. Protein Cell 1:944–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gujuluva CN, Kundu A, Murti KG, Nayak DP. 1994. Abortive replication of influenza virus A/WSN/33 in HeLa229 cells: defective viral entry and budding processes. Virology 204:491–505 [DOI] [PubMed] [Google Scholar]
- 23. Harris A, et al. 2006. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. U. S. A. 103:19123–19127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hartshorn KL, et al. 1988. Effects of influenza A virus on human neutrophil calcium metabolism. J. Immunol. 141:1295–1301 [PubMed] [Google Scholar]
- 25. Henkel JR, Weisz OA. 1998. Influenza virus M2 protein slows traffic along the secretory pathway. pH perturbation of acidified compartments affects early Golgi transport steps. J. Biol. Chem. 273:6518–6524 [DOI] [PubMed] [Google Scholar]
- 26. Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275–2289 [DOI] [PubMed] [Google Scholar]
- 27. Holsinger LJ, Lamb RA. 1991. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology 183:32–43 [DOI] [PubMed] [Google Scholar]
- 28. Honda A. 2008. Role of host protein Ebp1 in influenza virus growth: intracellular localization of Ebp1 in virus-infected and uninfected cells. J. Biotechnol. 133:208–212 [DOI] [PubMed] [Google Scholar]
- 29. Hui KP, et al. 2009. Induction of proinflammatory cytokines in primary human macrophages by influenza A virus (H5N1) is selectively regulated by IFN regulatory factor 3 and p38 MAPK. J. Immunol. 182:1088–1098 [DOI] [PubMed] [Google Scholar]
- 30. Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. 2009. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J. Exp. Med. 206:79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iwatsuki-Horimoto K, et al. 2006. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J. Virol. 80:5233–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lakadamyali M, Rust MJ, Zhuang X. 2004. Endocytosis of influenza viruses. Microbes Infect. 6:929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lamb RA, Choppin PW. 1981. Identification of a second protein (M2) encoded by RNA segment 7 of influenza virus. Virology 112:729–737 [DOI] [PubMed] [Google Scholar]
- 34. Lamb RA, Zebedee SL, Richardson CD. 1985. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 40:627–633 [DOI] [PubMed] [Google Scholar]
- 35. Lee JH, et al. 2010. Direct interaction of cellular hnRNP-F and NS1 of influenza A virus accelerates viral replication by modulation of viral transcriptional activity and host gene expression. Virology 397:89–99 [DOI] [PubMed] [Google Scholar]
- 36. Li OT, et al. 2009. Full factorial analysis of mammalian and avian influenza polymerase subunits suggests a role of an efficient polymerase for virus adaptation. PLoS One 4:e5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu X, et al. 2009. Cyclophilin A interacts with influenza A virus M1 protein and impairs the early stage of the viral replication. Cell. Microbiol. 11:730–741 [DOI] [PubMed] [Google Scholar]
- 38. McCown MF, Pekosz A. 2006. Distinct domains of the influenza A virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J. Virol. 80:8178–8189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McCown MF, Pekosz A. 2005. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J. Virol. 79:3595–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Monastyrskaya K, et al. 2009. Plasma membrane-associated annexin A6 reduces Ca2+ entry by stabilizing the cortical actin cytoskeleton. J. Biol. Chem. 284:17227–17242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nayak DP, Balogun RA, Yamada H, Zhou ZH, Barman S. 2009. Influenza virus morphogenesis and budding. Virus Res. 143:147–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nefkens I, et al. 2007. Hemagglutinin pseudotyped lentiviral particles: characterization of a new method for avian H5N1 influenza sero-diagnosis. J. Clin. Virol. 39:27–33 [DOI] [PubMed] [Google Scholar]
- 43. Noton SL, et al. 2009. Studies of an influenza A virus temperature-sensitive mutant identify a late role for NP in the formation of infectious virions. J. Virol. 83:562–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Palese P, Shaw ML. 2007. Orthomyxoviridae: the viruses and their replication, p 1647–1689 In Knipe DM, et al. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 45. Pinto LH, Holsinger LJ, Lamb RA. 1992. Influenza virus M2 protein has ion channel activity. Cell 69:517–528 [DOI] [PubMed] [Google Scholar]
- 46. Pinto LH, Lamb RA. 2007. Controlling influenza virus replication by inhibiting its proton channel. Mol. Biosyst. 3:18–23 [DOI] [PubMed] [Google Scholar]
- 47. Pinto LH, Lamb RA. 2006. The M2 proton channels of influenza A and B viruses. J. Biol. Chem. 281:8997–9000 [DOI] [PubMed] [Google Scholar]
- 48. Pleschka S, et al. 2001. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat. Cell Biol. 3:301–305 [DOI] [PubMed] [Google Scholar]
- 49. Rossman JS, et al. 2010. Influenza virus M2 ion channel protein is necessary for filamentous virion formation. J. Virol. 84:5078–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rossman JS, Jing X, Leser GP, Lamb RA. 2010. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 142:902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rossman JS, Lamb RA. 2011. Influenza virus assembly and budding. Virology 411:229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scheiffele P, Roth MG, Simons K. 1997. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 16:5501–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Simpson-Holley M, et al. 2002. A functional link between the actin cytoskeleton and lipid rafts during budding of filamentous influenza virions. Virology 301:212–225 [DOI] [PubMed] [Google Scholar]
- 54. Smythe E, Smith PD, Jacob SM, Theobald J, Moss SE. 1994. Endocytosis occurs independently of annexin VI in human A431 cells. J. Cell Biol. 124:301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sun L, Hemgard GV, Susanto SA, Wirth M. 2010. Caveolin-1 influences human influenza A virus (H1N1) multiplication in cell culture. Virol. J. 7:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thaa B, Herrmann A, Veit M. 2010. Intrinsic cytoskeleton-dependent clustering of influenza virus M2 protein with hemagglutinin assessed by FLIM-FRET. J. Virol. 84:12445–12449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Thomas JM, Stevens MP, Percy N, Barclay WS. 1998. Phosphorylation of the M2 protein of influenza A virus is not essential for virus viability. Virology 252:54–64 [DOI] [PubMed] [Google Scholar]
- 58. Ueda M, et al. 2010. Highly pathogenic H5N1 avian influenza virus induces extracellular Ca2+ influx, leading to apoptosis in avian cells. J. Virol. 84:3068–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vila de Muga S, et al. 2009. Annexin A6 inhibits Ras signalling in breast cancer cells. Oncogene 28:363–377 [DOI] [PubMed] [Google Scholar]
- 60. Wang P, et al. 2009. Nuclear factor 90 negatively regulates influenza virus replication by interacting with viral nucleoprotein. J. Virol. 83:7850–7861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang X, Hinson ER, Cresswell P. 2007. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2:96–105 [DOI] [PubMed] [Google Scholar]
- 62. Welsch S, Muller B, Krausslich HG. 2007. More than one door—budding of enveloped viruses through cellular membranes. FEBS Lett. 581:2089–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.