Abstract
Immature retroviral particles are assembled by self-association of the structural polyprotein precursor Gag. During maturation the Gag polyprotein is proteolytically cleaved, yielding mature structural proteins, matrix (MA), capsid (CA), and nucleocapsid (NC), that reassemble into a mature viral particle. Proteolytic cleavage causes the N terminus of CA to fold back to form a β-hairpin, anchored by an internal salt bridge between the N-terminal proline and the inner aspartate. Using an in vitro assembly system of capsid-nucleocapsid protein (CANC), we studied the formation of virus-like particles (VLP) of a gammaretrovirus, the xenotropic murine leukemia virus (MLV)-related virus (XMRV). We show here that, unlike other retroviruses, XMRV CA and CANC do not assemble tubular particles characteristic of mature assembly. The prevention of β-hairpin formation by the deletion of either the N-terminal proline or 10 initial amino acids enabled the assembly of ΔProCANC or Δ10CANC into immature-like spherical particles. Detailed three-dimensional (3D) structural analysis of these particles revealed that below a disordered N-terminal CA layer, the C terminus of CA assembles a typical immature lattice, which is linked by rod-like densities with the RNP.
INTRODUCTION
The assembly of fully infectious retroviruses comprises two different phases: immature- and mature-particle formation. Initially, the virus is formed in immature form by self-association of the major structural polyprotein Gag into spherical particles either at the plasma membrane (alpharetroviruses, gammaretroviruses, or lentiviruses) or at a distinct site within the infected cell (betaretroviruses). The particles are organized by lattices of Gag and Gag-derived polyproteins that associate with the plasma membrane that becomes the outer layer of the immature particle released during budding. Upon release from the host cell, the Gag precursor is proteolytically processed into N-terminal membrane binding matrix protein (MA), two-domain capsid protein (CA), and RNA-binding nucleocapsid protein (NC). This cleavage leads to a dramatic morphological change and reorganization of the virus particle from its immature form into a mature, infectious virion.
Gag itself is sufficient for immature-particle assembly. The interactions critical for immature-particle assembly are mediated by CA-CA and NC-RNA interfaces. CA consists of two subdomains, the CA–N-terminal domain (NTD) and the CA–C-terminal domain (CTD), connected by a short sequence. Despite a low level of sequence homology, all available retroviral CA-NTD structures show conserved structural motifs consisting of six or seven α-helices and an N-terminal β-hairpin anchored by a salt bridge between the N-terminal proline and an aspartate in helix 3 (10, 20, 29, 30, 42–44). CA-CTD contains a dimerization domain and is essential for the assembly of Gag. In the alpharetroviruses (e.g., Rous sarcoma virus [RSV]) and lentiviruses (e.g., HIV-1), a short spacer peptide (SP) (12 or 14 amino acids, respectively) separates CA from NC. This SP domain plays a critical role in assembly, as mutations or deletions in the SP influence immature-spherical-particle assembly (1, 28, 33, 37). Both HIV-1 and RSV SP sequences were also suggested to function as molecular switches, because the deletion of SP1 of HIV-1 Gag led to the formation of tubular instead of spherical particles (23). Similarly, mutations within the SP sequence of RSV Gag change spherical-particle formation to tubular-particle formation (28).
The appearances of all immature retroviral particles observed by transmission electron microscopy (TEM) are similar; they are roughly spherical, with an inner electron-dense ring. Based on cryo-electron microscopy (cEM) studies, the Gag polyprotein is arranged radially and is associated with the plasma membrane through its MA domain, with the NC domain oriented toward the center of the particle (65, 67). Three-dimensional (3D) reconstructions of immature HIV-1 and RSV particles obtained by cryo-electron tomography have shown that the immature Gag lattice adopts a hexagonal arrangement (5, 66). To form a closed sphere from a hexagonal lattice, the incorporation of pentamers or irregular defects is required. Pentamers have not been observed in the immature Gag lattice; instead, the Gag hexameric lattice closes through the incorporation of heterogeneously shaped defects (6, 35). Upon maturation, NC with bound RNA condenses in the center of the particle, surrounded by a core shell assembled from released CA. The shapes of the mature cores differ according to the retrovirus genus: in gamma-, delta-, and alpharetroviruses, they are spherical; in betaretroviruses, cylindrical; and in lentiviruses, conical. These assemblies are composed of a curved hexagonal lattice closed by incorporation of CA pentamers. The final shapes of the retroviral cores are then determined by the locations of the pentamers. The core of the most studied retrovirus, HIV-1, is a fullerene-like cone composed of a hexameric lattice with 12 pentamers, 5 at the narrow end of the cone and 7 at the broad end of the cone (18, 36, 47). Although the CA domain drives both mature- and immature-particle assembly, the lattices of immature particles differ from those of the mature particles (5–7, 66). The spacing of the immature HIV-1 Gag lattice is smaller than that in the mature particle, and the positions of the CA-NTD and CA-CTD also differ in the immature and mature lattices (6).
Gammaretroviruses, like other type C retroviruses, assemble their immature particles at the plasma membrane. Similarly to other retroviruses, their genome contains three open reading frames: gag, pol, and env. One member of the gammaretroviral family, the xenotropic murine leukemia virus-related virus (XMRV), was recently discovered in association with human prostate tumors (58) and chronic fatigue syndrome (CFS) (38). However, the association has not been confirmed in other studies (15–17, 25, 26, 51–53, 56, 60), thus the relevance of XMRV to human disease remains unclear. Recent publications have provided strong evidence that XMRV does not cause CFS (32, 45, 49, 54). Its genome is most similar to that of exogenous murine leukemia virus (MLV) DG-75, isolated from a human B-lymphoblastoid cell line (48), with which it shares 94% sequence identity (58). The XMRV Gag polyprotein consists of 536 amino acids and is separated from the Pro-Pol sequence by a UAG stop codon. Based on the sequence similarity with the closely related MLV, it is expected that XMRV Gag also comprises four structural proteins: MA, p12, CA, and NC. The MLV capsid protein, like all retroviral CAs, contains an N-terminal β-hairpin that is stabilized by Pro1-Asp54 interaction (43, 44). This hairpin is formed only upon maturation, and prevention of its maturation markedly reduces viral infectivity (61, 62, 64).
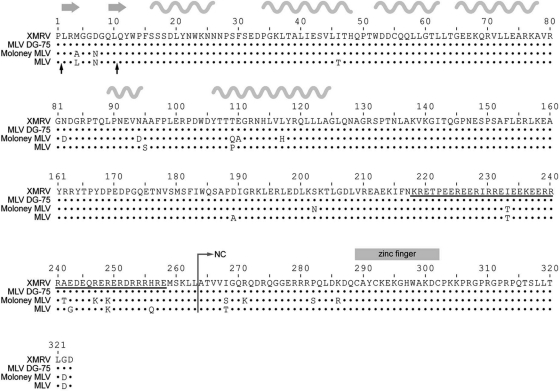
Similarly to betaretroviruses, gammaretroviruses lack a spacer peptide between CA and NC. However, gammaretroviruses possess a unique feature, that is, the presence of a 41-amino-acid stretch rich in both positively and negatively charged residues at the C terminus of CA (Fig. 1, underlined sequence). This region, due to its possible α-helical structure, was named the “charged assembly helix” and is important for the assembly and production of infectious MLV (9, 63).
Fig 1.
Amino acid alignment of XMRV and MLV DG-75, Moloney MLV, and MLV (strain BM5) CANC proteins. The dots represent identical amino acid residues, the black arrows under the alignment show the start sites of XMRV ΔProCANC and Δ10CANC truncated proteins, and the underlined sequence shows the charged residues at the C termini of MLV and XMRV CA. The positions of β-strands and α-helices from the solved MLV CA-NTD structure (pdb 3bp9) are shown by the gray arrows and wavy lines, respectively, above the alignment.
Studies on in vitro-assembled virus-like particles (VLPs) have been proven to be a valuable tool for gaining basic information on retroviral assembly. A number of studies have mimicked the assembly of both immature virus particles and mature cores in vitro using purified recombinant Gag-derived capsid-nucleocapsid (CANC) proteins (8, 21, 22, 31, 41, 57, 68). Although the in vitro assembly of retroviral genera, including the lentivirus HIV-1 (8, 12, 21, 22), the betaretrovirus Mason-Pfizer monkey virus (M-PMV) (31, 57), and the alpharetrovirus RSV (8, 40, 41, 68), has been intensively studied, there is a lack of information on the in vitro assembly of gammaretroviruses. In vitro assembly was achieved by using His-tagged MLV CA molecules anchored to Ni2+-chelating lipid nanotubes as an assembly substrate (24). Recently, a comparison of MLV and HIV-1 monomeric Gag molecules revealed their different behaviors in solution. In contrast to HIV Gag, MLV Gag has a much weaker propensity for interprotein interactions and has a rigid and extended structure, likely caused by a proline-rich region between MA and p12 (11). In this work, Datta et al. also demonstrated that MLV Gag could be assembled in vitro into fragile VLPs with an irregular appearance.
Here, we present the assembly of XMRV CANC protein into immature virus-like spherical VLPs by using an in vitro expression/assembly system. We prepared three N-terminal variants of XMRV CANC and compared their ability to assemble. We found that formation of the N-terminal β-hairpin must be prevented to obtain immature-like particles from XMRV CANC. Using electron microscopy and cryo-electron tomography, we have characterized the structure of assembled immature XMRV VLPs and compared it with that of other retroviruses.
MATERIALS AND METHODS
Cloning.
All expression vectors were prepared by standard cloning techniques, propagated in Escherichia coli DH5α, and verified by sequencing. The fragment encoding XMRV CANC was obtained by PCR (using primers 5′CAT ATG CCA CTC CGC ATG GGG and 3′CTC GAG CTA GTC ACC TAA GGT CAG G) of cDNA from human prostate carcinoma 22Rv1 cells infected with XMRV (kindly provided by M. Hajduch). The PCR fragment was digested with NdeI and XhoI and ligated into an expression vector, pET-22b, under the T7 promoter. The CANC-derived constructs were prepared analogously, with the 5′ primers CAT ATG CAG TAC TGG CCG TTT TCC TC for Δ10CANC and CAT ATG CTC CGC ATG GGG GGA G for ΔProCANC.
Bacterial expression.
Luria-Bertani medium containing ampicillin (100 μg/ml) was inoculated with E. coli BL21(DE3) cells carrying the appropriate construct. At the optical density at 590 nm of ∼0.8, expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.4 mM. The cells were harvested 4 h postinduction.
Protein purification.
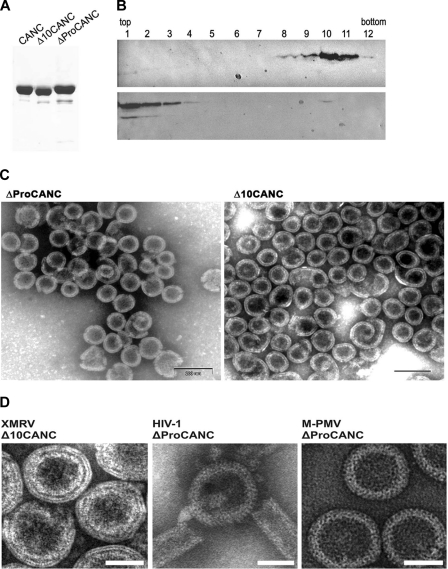
Bacterial cells were resuspended in lysis buffer A (50 mM Tri-HCl, 150 mM NaCl, 1 mM EDTA, pH 8.0), disrupted with lysozyme, sonicated, and incubated with sodium deoxycholate. After centrifugation at 10,000 × g, proteins were solubilized from pellets by the addition of 0.5% Triton X-100 and 1 to 1.5 M NaCl in lysis buffer A. The XMRV protein was precipitated from the supernatant by ammonium sulfate (final concentration, 25% [wt/vol]). After low-speed centrifugation, the pellet was resuspended in buffer E (20 mM Tris-HCl, pH 8.0, 0.1 M NaCl, 50 μM ZnCl2, 10 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]), dialyzed overnight against the same buffer at 4°C, and loaded on the top of a DEAE cellulose column. The flowthrough and washing fractions were pooled and loaded on the top of a phosphocellulose column. The bound protein was eluted by an NaCl gradient from 0.1 M to 1 M NaCl in buffer E. The fractions containing the desired proteins were dialyzed overnight against a storage buffer (buffer E with 0.5 M NaCl), concentrated, and loaded on the top of a Sephadex G-100 column. The purified proteins were concentrated (CANC to 5 mg/ml, ΔProCANC to 2 mg/ml, and Δ10CANC to 4 mg/ml) and stored at −70°C. The purity of each protein was analyzed by SDS-PAGE (see Fig. 3), and the protein identities were confirmed by N-terminal sequencing.
Fig 3.
(A) SDS-polyacrylamide analysis of purified XMRV proteins (Coomassie blue-stained gel). (B) Western blot analysis of in vitro-assembled XMRV particles in sucrose gradient fractions in the presence (upper blot) or in the absence (lower blot) of oligonucleotides. Δ10CANC protein (60 μg) was mixed with 6 μg of oligonucleotide 40-mer, dialyzed overnight against the assembly buffer, and centrifuged to equilibrium through a linear 20 to 65% sucrose gradient. The fractions were analyzed by Western blotting with rabbit anti-XMRV CANC antibodies. The sucrose densities in fractions 8 to 12 were as follows: 8, 1.19 g/ml; 9, 1.21 g/ml; 10, 1.22 g/ml; 11, 1.23 g/ml; 12, 1.25 g/ml. (C) TEM images of negatively stained in vitro-assembled material from XMRV ΔProCANC (left) and XMRV Δ10CANC (right). (D) TEM images of negatively stained VLPs assembled in vitro from XMRV Δ10CANC, HIV-1 ΔProCANC, and M-PMV ΔProCANC. Scale bars,e 50 nm.
In vitro assembly.
An aliquot of 60 μg of purified protein was mixed with 6 μg of DNA (oligonucleotide 30-mer, 40-mer, λDNA, or phage M13 single-stranded DNA [ssDNA]) or RNA (MS2) in a total volume of 100 μl of the storage buffer (20 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 50 μM ZnCl2, 10 mM DTT, 1 mM PMSF). The mixture was dialyzed from 2 h to overnight against the assembly buffer (50 mM Tris buffer, pH 8.0, containing 100 mM NaCl and 1 μM ZnCl2) at 4°C. One hundred microliters of the assembled material (in the presence or absence of oligonucleotide) was loaded on the top of a 20 to 65% (wt/wt) sucrose gradient and centrifuged at 35,000 rpm in a Beckman SW41Ti for 16 h at 4°C. Individual fractions were analyzed by Western blot analysis using rabbit anti-XMRV CANC antibodies. The sucrose density in the fractions was determined using a refractometer.
Electron microscopy.
Particles formed during the assembly reaction were negatively stained with 4% sodium silicotungstate (pH 7.4) on carbon-coated grids and studied by transmission electron microscopy (JEOL JEM-1200EX) with a microscope operated at 60 kV. For thin-section electron microscopic analysis, the bacterial pellets of the induced samples were fixed in 2.5% glutaraldehyde, postfixed in 1% osmium tetroxide, dehydrated by applying an ethanol series, and embedded in Agar 100 epoxy resin. Ultrathin sections (70 nm) were counterstained with saturated uranyl acetate and lead citrate.
Cryo-electron microscopy.
In vitro-assembled particles were deposited on C-flat holey carbon grids and vitrified by plunge freezing in liquid ethane. Tilt series were collected on an FEI Tecnai F30 “Polara” transmission electron microscope with a Gatan GIF 2002 postcolumn energy filter and a 2,000 by 2,000 Multiscan charge-coupled device (CCD) camera. Data collection was performed at 300 kV with an electron dose of approximately 30 e−/Å2. The nominal defocus was 2.0 μm, with a magnification of ×34,000, resulting in a pixel size at the specimen level of 4 Å.
Cryo-electron tomography.
In vitro-assembled particles were mixed with 10-nm gold beads, deposited on C-flat holey carbon grids, and vitrified by plunge freezing in liquid ethane. Tilt series were collected on an FEI Tecnai F30 “Polara” transmission electron microscope with a Gatan GIF 2002 postcolumn energy filter and a 2,000 by 2,000 Multiscan CCD camera. Data collection was performed at 300 kV using the SerialEM software package, and tomograms were reconstructed using the IMOD software package (34). Tilt series were collected between −60° and +60° with an angular increment of 3° and a total electron dose of approximately 70 e/Å2. The defocus range was between 3.0 and 4.0 μm, with a magnification of ×34,000, resulting in a pixel size at the specimen level of 4 Å.
2D Fourier analysis.
Seventy individual particles were boxed out using an EMAN boxer (39) and padded to a final size of 1,024 by 1,024 pixels. A soft spherical mask was applied with a radius of 22 nm for the rotationally averaged power spectrum or 15 nm for the rotationally aligned power spectrum. (The larger mask includes more data, giving the stronger peaks necessary for the alignment. The narrower mask minimizes the smearing of the peak due to particle curvature.) Calculation of power spectra and their rotational average and rotational alignment were performed in IMAGIC (59).
RAF analysis.
Radius-angle-frequency (RAF) plots were generated as described previously (13). Briefly, subtomograms of 384 nm3 were extracted from tomograms of 15 VLPs. The extraction was done along the surface of a sphere centered in the center of each VLP and with a radius equal to the mean radius at CA-CTD level. The subtomograms underwent an orthographic radial projection. The focus of the projection was determined as the geometric center of the particle from which the subtomogram was extracted. The resulting volumes corresponded to flattened subtomograms. The volumes were then padded to 1,024 by 1,024 pixels in xy, and 2D power spectra were calculated at each radius. The rotation-autocorrelation function with a rotation range between 0° and 180° was calculated for each power spectrum. For each subtomogram, this generates a 3D plot with radius on one axis and the other two axes representing those of the rotational autocorrelation function of the power spectrum, namely, angle and frequency. The presence of a peak at a particular point in radius, frequency, and angle indicates that at that radius in the virus, the 2D power spectrum of the protein layer has peaks at that frequency which are arranged rotationally, symmetrically repeating at that angle.
Subtomogram averaging.
The extracted subtomograms were iteratively aligned and averaged using a six-dimensional search as described previously (6). The initial reference used for the alignment was the average of the subtomograms in the extraction position. Sixfold symmetry was applied to the average at each iteration. The threshold for the subtomograms that had to be averaged was set to the mean cross-correlation value of all subtomograms to the reference.
RESULTS
Bacterial and in vitro assembly.
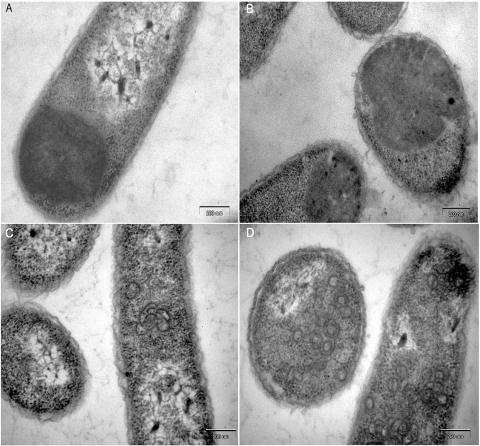
To obtain the XMRV expression vector, the CA and CANC regions were amplified using cDNA of human prostate carcinoma 22Rv1 cells infected with XMRV. DNA sequencing confirmed 100% identity of the CANC with the CANC of XMRV clone VP62 (Fig. 1). To study the ability of XMRV CA and CANC to assemble into VLPs, we used both bacterial and in vitro assembly systems (50, 57). TEM of thin-sectioned E. coli BL21(DE3) expressing XMRV CA and CANC showed amorphous inclusion bodies instead of assembled mature-core-like structures (Fig. 2A and B). This finding was surprising, because several studies have demonstrated that retroviral CANCs of different species (HIV-1 and RSV), with Pro1 at their N termini, assemble into tubular, mature-core-like structures (8, 21, 22, 41).
Fig 2.
Electron micrographs of thin sections of E. coli cells expressing XMRV CA (A), CANC (B), ΔProCANC (C), and Δ10CANC (D).
It is known that the prevention of β-hairpin formation (by either the deletion or substitution of Pro1 or by N-terminal extension of CA) can shift the assembly mode to formation of immature-virus-like structures (21, 46, 50, 57, 61). We therefore decided to truncate the XMRV CANC construct by the deletion of the first proline residue (ΔProCANC) with the aim of assembling immature-like structures. TEM analysis of the cells expressing ΔProCANC protein indicated its tendency to assemble into numerous curved structures (Fig. 2C), together with a small number of approximately spherical particles. In order to delete a substantial portion of the N-terminal β-hairpin, we next generated a further N-terminal truncation of CANC, locating the start of the XMRV CA protein 10 amino acids downstream from Pro1, at Gln11 (Fig. 1). TEM of the thin sections of E. coli expressing the resulting Δ10CANC construct showed the presence of roughly spherical particles (Fig. 2D).
To further analyze the ability of XMRV CANC-derived proteins to assemble in vitro, all studied proteins were purified using procedures similar to those described earlier for M-PMV and HIV-1 CANC (8, 41, 57) (Fig. 3A). An in vitro assembly reaction was performed by overnight dialysis of a mixture of purified XMRV CANC-derived proteins with DNA (oligonucleotide 30-mer, 40-mer, λ DNA, and phage M13 ssDNA) or RNA (MS2). To investigate the homogeneity of the assembled particles, the material was centrifuged through a linear 20 to 65% sucrose gradient, and individual fractions were analyzed by Western blotting using rabbit anti-XMRV CANC antibody. In the presence of the oligonucleotide (40-mer), the majority of the assembled particles were found in the fractions with sucrose densities of 1.19 to 1.23 g/ml, corresponding to the properly assembled spherical particles (Fig. 3B, top). In the absence of the oligonucleotides, the bulk of unassembled material remained at the top of the sucrose gradient (Fig. 3B, bottom). The gradient profiles were similar for both the ΔProCANC and Δ10CANC proteins. The samples of the in vitro-assembled particles were negatively stained with 4% sodium silicotungstate (pH 7.4) and studied by TEM. In contrast to the XMRV CANC protein, where only free or aggregated proteins were observed, the ΔProCANC and Δ10CANC proteins assembled into spherical particles (Fig. 3C). Particles of XMRV ΔProCANC and Δ10CANC were not detected if the in vitro assembly was carried out in the absence of nucleic acid or at an NaCl concentration higher than 0.5 M (data not shown), suggesting that assembly must be initiated by the addition of nucleic acids under suitable conditions. Similar assembly requirements have been observed for the in vitro-assembled particles of RSV CANC (8), HIV-1 CANC (8, 21, 22), and M-PMV ΔProCANC (57).
TEM images of negatively stained in vitro-assembled XMRV Δ10CANC particles were then compared to those of HIV-1 and M-PMV ΔProCANC (Fig. 3D). The major difference between the particles was the appearance of particle protein layers. While the HIV-1 and M-PMV ΔProCANC particles appeared as a single-layer protein shell with visible rod-like projections (Fig. 3D), the XMRV Δ10CANC particle's shell appeared as three smoother protein layers separated by distinct rings of lower electron density (Fig. 3D).
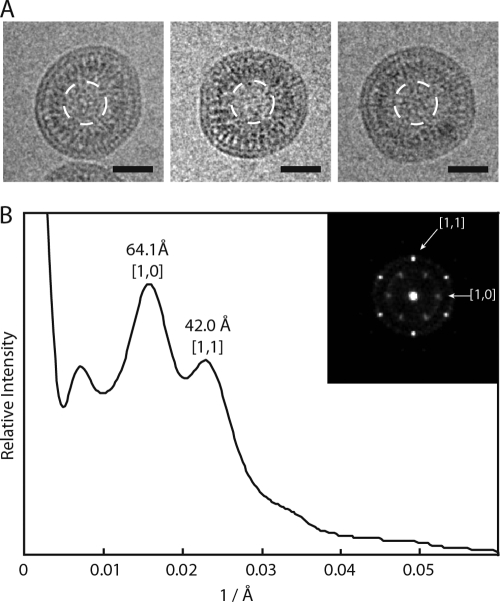
To study the fine structure of the in vitro-assembled XMRV particles in more detail and to avoid artifacts associated with negative staining, we carried out cEM of particles made of Δ10CANC protein. The sample was prepared by the dialysis of Δ10CANC against pH 8.0 buffer in the presence of lambda double-stranded DNA (dsDNA). The CEM images showed three density layers, with the outer two layers close together (Fig. 4A). The mean size of the particles was 82 nm ± 11 nm.
Fig 4.
Cryo-electron microscopy of XMRV Δ10CANC particles. (A) Cryo-electron micrographs of particles assembled in vitro from XMRV Δ10CANC. Scale bars, 50 nm. The white dashed circles show the region used for Fourier analysis. (B) Rotationally averaged power spectra of the particle centers, showing the positions of the [1,0] and [1,1] reflections from a hexameric lattice. The inset displays the sum of the rotational aligned power spectra, indicating the presence of a hexagonal lattice.
We compared our images of the in vitro-assembled XMRV particles (Fig. 4A) with published images of PR− MLV (Fig. 3A and 5 in Yeager et al. [67]). The numbers of layers and the appearances of the layers are extremely similar. For an optimal comparison, we applied the same image analysis procedure that was used for PR− MLV. We calculated the rotationally averaged power spectra of the particle centers, which showed peaks corresponding to repeating elements in the protein lattice (Fig. 4B). Two peaks were seen, centered at 64 ± 9 Å and 42 ± 3 Å. If these are indexed as [1,0] and [1,1] reflections of a hexagonal lattice, they describe a unit spacing of 74 Å or 73 Å. The equivalent analysis for PR− MLV is shown in Fig. 6 in Yeager et al. (67). The pattern of peaks is the same. For PR− MLV, the peaks are centered at 67 ± 11 Å and 45 ± 10 Å, describing a unit cell spacing of 77 Å and 78 Å. We next carried out a rotational alignment and averaging of the power spectra, as described by Briggs et al. (5), revealing that a hexagonal pattern is present in the XMRV particles (Fig. 4B) and confirming the assignment of the [1,0] and [1,1] reflections. Similar patterns were described for individual particles of PR− MLV (Fig. 3 in Yeager et al. [67]).
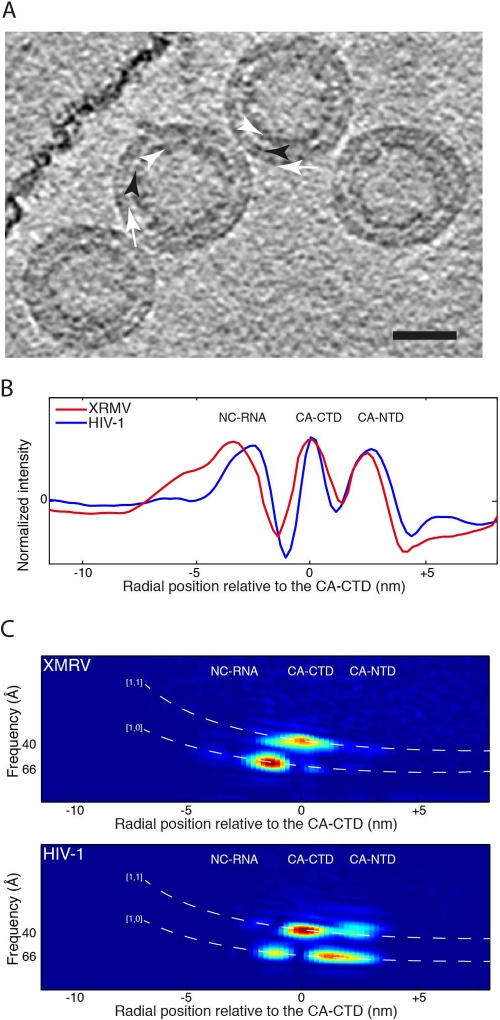
Fig 5.
Cryo-electron tomography of XMRV Δ10CANC particles. (A) Computational slice 0.8 nm thick through a cryo-electron tomogram of particles assembled in vitro from XMRV Δ10CANC. The white arrows point to the CA-NTD, the black arrowhead points to the CA-CTD, and the white arrowheads point to the NC region. The scale bar is 50 nm. (B) Comparison between the radial density profiles of XMRV Δ10CANC (red) and HIV-1 (blue). The two profiles have been aligned on the CA-CTD, and the x axis shows the radial distance from the CA-CTD region. The peaks corresponding to NC and to the NTD and CTD of CA are marked. (C) The two images show slices through the RAF plot at a 60° angle for XMRV Δ10CANC (top) and HIV-1 (bottom). The x axes are aligned to the x axis of the radial density plot in panel B. The expected positions of the [1,0] and [1,1] reflections for a hexameric lattice with a spacing of 7.4 nm at the CA-CTD are indicated by the white dashed lines. Strong peaks indicating a hexagonal order are seen for both XMRV Δ10CANC and HIV-1 in the CA-CTD and downstream regions. Only HIV shows strong peaks in the CA-NTD region. The radial positions of the RNP and CA domains are indicated on the plots.
Fig 6.
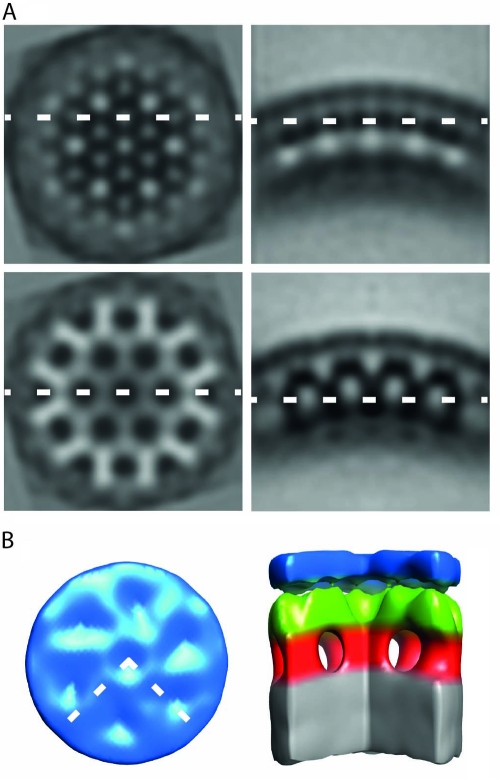
Structure of in vitro-assembled particles of XMRV Δ10CANC. (A) Orthoslices through the average structure. On the right are radial sections, while tangential sections are on the left. The dashed lines show the relative position of the orthoslices between the panels. Density is black. (B) Surface rendering of the average structure. The color scheme follows an approximate radial division of the structure according to the different domains of Gag as previously assigned (12). Gray is the NC-nucleic acid layer, red is the region between NC and the CA-CTD, green is the CA-CTD region, and blue is the CA-NTD. The dashed lines in the top view represent the section cut out in the side view. The CA-NTD region does not show an ordered hexameric lattice.
To further understand the structural organization of the XMRV particles, we collected cryo-electron tomograms (Fig. 5A) and directly compared the radial density profile of XMRV Δ10CANC with those of HIV-1 (using data sets from de Marco et al. [12]). The radial density profile of XMRV Δ10CANC is similar to that of HIV-1 (Fig. 5B). By comparison with HIV-1, the two outer density layers correspond to the NTD and CTD of CA and the inner layer to NC with bound nucleic acid.
In order to identify which layers show 6-fold symmetry, we used the 3D information present in the cryo-electron tomograms and performed RAF analysis (13) (Fig. 5C). Peaks in the RAF plot indicate the presence of symmetrically ordered density at a particular radius and with a particular spacing. We identified two clear peaks: one corresponds to the [1,0] reflection of the expected hexameric lattice in the region between NC and the CA-CTD, the other to the [1,1] reflection of the hexameric lattice in the CA-CTD region. In vitro-assembled HIV particles show the same two peaks, as well as two further peaks in the region between CA-CTD and CA-NTD and in the CA-NTD region. These peaks are missing in XMRV, indicating that, unlike in HIV, the CA-NTD region of the XMRV particles does not show any detectable hexameric lattice.
Knowing that the region between the RNP and the CA-CTD adopts a hexagonal lattice, we performed subtomogram averaging in order to get a 3D representation of the structure (Fig. 6A). Consistent with the RAF analysis, outermost in the structure is a poorly featured layer that corresponds to the CA-NTD (blue in Fig. 6B). In contrast, the CA-CTD domain (green in Fig. 6B) forms an arrangement similar to that seen in other retroviral immature lattices (12). As in HIV and RSV, the CA-CTD of XMRV appears to be connected to the RNP (gray in Fig. 6B) via rod-like structures (red in Fig. 6B).
DISCUSSION
The release of the N and C termini of CA upon maturation leads to local conformational transitions that influence the overall structure of the protein and thus creates novel protein-protein interfaces (for a review, see reference 19). The model for subunit arrangement in immature particles implies that the hexagonal lattice is held together by intra- and interhexamer interactions within SP and CA-CTD (66). The CA-NTD is not absolutely required for immature-particle assembly (2, 4). Cleavage of Gag by viral protease allows the formation of a β-hairpin at the N terminus of CA-NTD and the separation of CA from the putative SP1 helical bundle. This induces structural rearrangement of CA and the formation of mature hexameric CA-NTD rings that hold together the mature lattice. CA-CTD interactions also mediate interhexamer interactions in the mature lattice.
Structural and biochemical data suggest that the ordered arrays of CA are regulated by the presence (β+) or absence (β−) of the β-hairpin at the N terminus and the presence (SP+) or absence (SP−) of the SP sequence at the CA C terminus, thus determining the particle morphology (23, 28). By this convention, within the immature virus, CA is in the form β−/SP+, and within the mature virus, it is in the form β+/SP−. In vitro, the HIV-1 ΔMACANC and RSV ΔPRGag proteins, where β-hairpin formation is prevented by downstream N-terminal sequences of CA but SP is present (β−/SP+), assembled into immature spherical particles (21, 27). In vitro and in vivo assembly studies on HIV CANC and RSV CANC show that the presence of both N- and C-terminal determinants (β+/SP+) of CA results in mature-like tubular (and, in the case of HIV-1, also conical) particle assembly (8, 18, 22). The HIV-1 ΔMACAΔSP1NC and RSV ΔPRGag proteins, which do not form the β-hairpin and in which SP is mutated or deleted (β−/SP−), assemble into tubular (and, in the case of HIV-1, also conical) particles (23, 28). The mature CA proteins (β+/SP−) of HIV and RSV assemble in vitro into tubular structures; however, the conditions under which these structures are formed (∼1 M NaCl) suggest different types of CA-CA interactions (14, 22, 30).
M-PMV, which lacks the SP sequence between CA and NC, appears to behave differently in vitro. In the absence of the β-hairpin and in the presence of downstream NC, M-PMV forms spherical particles. In the presence of the β-hairpin, M-PMV cannot be assembled in vitro. Here, we show that XMRV behaves in the same way: it is unable to form mature-like structures in vitro from CA or CANC but can form immature-like ΔProCANC and Δ10CANC particles. In contrast to HIV-1 and RSV, XMRV and M-PMV do not have SP separating the CA and NC domains. This begs the question whether some part of XMRV CA or NC might substitute for the SP-like domain. Based on recent data showing that MLV, in contrast to other retroviruses, does not have strict structural requirements for the N-terminal portion of NC (55), it is likely that the SP-like domain occurs within the CA CTD. Here, we show that the region between CA and NC forms a rod-like structure similar to that previously described for HIV and RSV and consistent with the presence of a structured region between the two folded domains. A possible candidate is the “charged assembly helix” at the C terminus of MLV CA (9). This region was suggested to form an amphipathic α-helix, and mutations within the region showed its necessity for MLV assembly (9). The SP-like domain in M-PMV was localized at the CANC junction (3); however, its precise length is currently being studied. It is possible that such sequences at the C terminus of CA in XMRV and M-PMV are what prevents the in vitro assembly of mature virus-like assemblies.
Given the high degree of sequence similarity to MLV Gag, the arrangement in the immature-like in vitro-assembled XMRV particles should be comparable to that in immature MLV particles, as described by Yeager et al. (67). Indeed, we found that the two particles have similar appearances and adopt a hexameric lattice with the same spacing. The CA and NC domains formed noticeable layers in both CEM and TEM images of the in vitro-assembled XMRV Δ10CANC, which correspond well to the features in the immature MLV particles shown by Yeager et al. (67).
3D structural analysis of the particles revealed that they form an ordered hexameric lattice with structural features mostly similar to those previously described for HIV, M-PMV, and RSV particles. Unexpectedly, in contrast to the other viruses, we found that the outermost layer in the XMRV Δ10CANC particles, corresponding to the CA-NTD region, does not form an ordered hexameric lattice. Two formal explanations can be considered: the CA-NTD may be laterally disordered in the in vitro-assembled XMRV particles, or gammaretroviruses may adopt a structural arrangement different from that of other retroviruses, in which, for example, part of CA-NTD may fill the typical holes in the CA-NTD layer, giving a smooth appearance. Given the high degree of secondary-structure similarity between the CA-NTD of MLV and those of other retroviruses, we consider the second possibility highly unlikely. It is sensible to conclude that the in vitro-assembled XMRV Δ10CANC particles faithfully mimic the structure of the immature XMRV Gag lattice in the CA-CTD and regions immediately downstream but show some lateral disorder in the CA-NTD region. We hypothesize that the difference in the order of CA-NTD (as shown in the RAF plots and in the 3D structure [Fig. 5C and 6B, respectively]) could explain the different appearances of the TEM images of negatively stained XMRV particles compared to HIV-1 and M-PMV (Fig. 3D). Although the cEM radial densities of XMRV and HIV-1 are very similar (Fig. 5B), showing the same radial positions of the CA-NTD, CA-CTD, and RNP domains, the cryo-electron tomography clearly shows that the degree of order in the XMRV CA-NTD (as shown in the RAF plots and in the 3D structure [Fig. 5C and 6B, respectively]) is very different.
Many previous studies indicated the importance of in vitro assembly studies of different retroviruses to obtain valuable information on the principles of retroviral assembly. A system for the in vitro assembly of MLV was described for CA protein (24) recruited to lipid nanotubes as an assembly template. Another in vitro assembly was reported recently for MLV Gag (11). However, in this system, the MLV Gag VLPs were fragile and of “irregular appearance.” The protocol presented here for the efficient in vitro assembly of virus-like Gag-derived particles in solution provides the basis for future studies of detailed protein packing and arrangement in gammaretroviral particles. Importantly, although the region downstream of XMRV CA does not contain a spacer peptide and has an unusual run of charged residues (9), we found that it assembles into a typical immature retroviral lattice with rod-like densities linking CA to the RNP. Further investigation of this system could therefore help us to better understand the critical role of the region in retrovirus assembly.
ACKNOWLEDGMENTS
We thank Marian Hajduch for kindly providing us with the cDNA of 22Rv1 cells and Romana Cubínková for excellent technical assistance. This work was technically supported by EMBL IT Services.
The work was supported by the Grant Agency of the Czech Republic (grant 204/09/1388) and by research projects 1M0508, 1M0520, Z40550506, and MSM 6046137305 from the Czech Ministry of Education and NIH grant CA 27834. J.A.G.B. was supported by a grant from the Deutsche Forschungsgemeinschaft within SPP 1175.
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Accola MA, Hoglund S, Gottlinger HG. 1998. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72: 2072–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Accola MA, Strack B, Gottlinger HG. 2000. Efficient particle production by minimal gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74: 5395–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bohmova K, et al. 2010. Effect of dimerizing domains and basic residues on in vitro and in vivo assembly of Mason-Pfizer monkey virus and human immunodeficiency virus. J. Virol. 84: 1977–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borsetti A, Ohagen A, Gottlinger HG. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 72: 9313–9317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Briggs JA, Johnson MC, Simon MN, Fuller SD, Vogt VM. 2006. Cryo-electron microscopy reveals conserved and divergent features of gag packing in immature particles of Rous sarcoma virus and human immunodeficiency virus. J. Mol. Biol. 355: 157–168 [DOI] [PubMed] [Google Scholar]
- 6. Briggs JA, et al. 2009. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. U. S. A. 106: 11090–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Briggs JA, et al. 2004. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11: 672–675 [DOI] [PubMed] [Google Scholar]
- 8. Campbell S, Vogt VM. 1995. Self-assembly in vitro of purified Ca-Nc proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69: 6487–6497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheslock SR, et al. 2003. Charged assembly helix motif in murine leukemia virus capsid: an important region for virus assembly and particle size determination. J. Virol. 77: 7058–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cornilescu CC, Bouamr F, Yao X, Carter C, Tjandra N. 2001. Structural analysis of the N-terminal domain of the human T-cell leukemia virus capsid protein. J. Mol. Biol. 306: 783–797 [DOI] [PubMed] [Google Scholar]
- 11. Datta SA, et al. 2011. Solution properties of murine leukemia virus Gag protein: differences from HIV-1 Gag. J. Virol. 85: 12733–12741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Marco A, et al. 2010. Conserved and variable features of Gag structure and arrangement in immature retrovirus particles. J. Virol. 84: 11729–11736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Marco A, et al. 2010. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog. 6: e1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ehrlich LS, Agresta BE, Carter CA. 1992. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 66: 4874–4883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Erlwein O, et al. 2011. Investigation into the presence of and serological response to XMRV in CFS patients. PLoS One 6: e17592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fischer N, et al. 2008. Prevalence of human gammaretrovirus XMRV in sporadic prostate cancer. J. Clin. Virol. 43: 277–283 [DOI] [PubMed] [Google Scholar]
- 17. Furuta RA, et al. 2011. No association of xenotropic murine leukemia virus-related virus with prostate cancer or chronic fatigue syndrome in Japan. Retrovirology 8: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283: 80–83 [DOI] [PubMed] [Google Scholar]
- 19. Ganser-Pornillos BK, Yeager M, Sundquist WI. 2008. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18: 203–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gitti RK, et al. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273: 231–235 [DOI] [PubMed] [Google Scholar]
- 21. Gross I, Hohenberg H, Huckhagel C, Krausslich HG. 1998. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J. Virol. 72: 4798–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gross I, Hohenberg H, Krausslich HG. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249: 592–600 [DOI] [PubMed] [Google Scholar]
- 23. Gross I, et al. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19: 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hilditch L, et al. 2011. Ordered assembly of murine leukemia virus capsid protein on lipid nanotubes directs specific binding by the restriction factor, Fv1. Proc. Natl. Acad. Sci. U. S. A. 108: 5771–5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hohn O, et al. 2009. Lack of evidence for xenotropic murine leukemia virus-related virus(XMRV) in German prostate cancer patients. Retrovirology 6: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hong P, Li J, Li Y. 2010. Failure to detect xenotropic murine leukaemia virus-related virus in Chinese patients with chronic fatigue syndrome. Virol. J. 7: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson MC, Scobie HM, Vogt VM. 2001. PR domain of rous sarcoma virus Gag causes an assembly/budding defect in insect cells. J. Virol. 75: 4407–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keller PW, Johnson MC, Vogt VM. 2008. Mutations in the spacer peptide and adjoining sequences in Rous sarcoma virus Gag lead to tubular budding. J. Virol. 82: 6788–6797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khorasanizadeh S, Campos-Olivas R, Summers MF. 1999. Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J. Mol. Biol. 291: 491–505 [DOI] [PubMed] [Google Scholar]
- 30. Kingston RL, et al. 2000. Structure and self-association of the Rous sarcoma virus capsid protein. Structure 8: 617–628 [DOI] [PubMed] [Google Scholar]
- 31. Klikova M, Rhee SS, Hunter E, Ruml T. 1995. Efficient in-vivo and in-vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J. Virol. 69: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Knox K, et al. 2011. No evidence of murine-like gammaretroviruses in CFS patients previously identified as XMRV-infected. Science 333: 94–97 [DOI] [PubMed] [Google Scholar]
- 33. Krausslich HG, Facke M, Heuser AM, Konvalinka J, Zentgraf H. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69: 3407–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116: 71–76 [DOI] [PubMed] [Google Scholar]
- 35. Kuznetsov YG, Ulbrich P, Haubova S, Ruml T, McPherson A. 2007. Atomic force microscopy investigation of Mason-Pfizer monkey virus and human immunodeficiency virus type 1 reassembled particles. Virology 360: 434–446 [DOI] [PubMed] [Google Scholar]
- 36. Li S, Hill CP, Sundquist WI, Finch JT. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407: 409–413 [DOI] [PubMed] [Google Scholar]
- 37. Liang C, Hu J, Whitney JB, Kleiman L, Wainberg MA. 2003. A structurally disordered region at the C terminus of capsid plays essential roles in multimerization and membrane binding of the Gag protein of human immunodeficiency virus type 1. J. Virol. 77: 1772–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lombardi VC, et al. 2009. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science 326: 585–589 [DOI] [PubMed] [Google Scholar]
- 39. Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128: 82–97 [DOI] [PubMed] [Google Scholar]
- 40. Ma YM, Vogt VM. 2002. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J. Virol. 76: 5452–5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ma YM, Vogt VM. 2004. Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro. J. Virol. 78: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Macek P, et al. 2009. NMR structure of the N-terminal domain of capsid protein from the Mason-Pfizer monkey virus. J. Mol. Biol. 392: 100–114 [DOI] [PubMed] [Google Scholar]
- 43. Mortuza GB, et al. 2008. Structure of B-MLV capsid amino-terminal domain reveals key features of viral tropism, gag assembly and core formation. J. Mol. Biol. 376: 1493–1508 [DOI] [PubMed] [Google Scholar]
- 44. Mortuza GB, et al. 2004. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature 431: 481–485 [DOI] [PubMed] [Google Scholar]
- 45. Paprotka T, et al. 2011. Recombinant origin of the retrovirus XMRV. Science 333: 97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Phillips JM, Murray PS, Murray D, Vogt VM. 2008. A molecular switch required for retrovirus assembly participates in the hexagonal immature lattice. EMBO J. 27: 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pornillos O, Ganser-Pornillos BK, Yeager M. 2011. Atomic-level modelling of the HIV capsid. Nature 469: 424–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Raisch KP, et al. 2003. Molecular cloning, complete sequence, and biological characterization of a xenotropic murine leukemia virus constitutively released from the human B-lymphoblastoid cell line DG-75. Virology 308: 83–91 [DOI] [PubMed] [Google Scholar]
- 49. Robinson MJ, Erlwein O, McClure MO. 2011. Xenotropic murine leukaemia virus-related virus (XMRV) does not cause chronic fatigue. Trends Microbiol. 19: 525–529 [DOI] [PubMed] [Google Scholar]
- 50. Rumlova-Klikova M, Hunter E, Nermut MV, Pichova I, Ruml T. 2000. Analysis of Mason-Pfizer monkey virus Gag domains required for capsid assembly in bacteria: role of the N-terminal proline residue of CA in directing particle shape. J. Virol. 74: 8452–8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Satterfield BC, et al. 2011. Serologic and PCR testing of persons with chronic fatigue syndrome in the United States shows no association with xenotropic or polytropic murine leukemia virus-related viruses. Retrovirology 8: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schutzer SE, Rounds MA, Natelson BH, Ecker DJ, Eshoo MW. 2011. Analysis of cerebrospinal fluid from chronic fatigue syndrome patients for multiple human ubiquitous viruses and xenotropic murine leukemia-related virus. Ann. Neurol. 69: 735–738 [DOI] [PubMed] [Google Scholar]
- 53. Sfanos KS, et al. 2008. A molecular analysis of prokaryotic and viral DNA sequences in prostate tissue from patients with prostate cancer indicates the presence of multiple and diverse microorganisms. Prostate 68: 306–320 [DOI] [PubMed] [Google Scholar]
- 54. Shin CH, et al. 2011. Absence of XMRV retrovirus and other murine leukemia virus-related viruses in patients with chronic fatigue syndrome. J. Virol. 85: 7195–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Still A, Huseby D, Barklis E. 2011. Analysis of the N-terminal region of the murine leukemia virus nucleocapsid protein. Virus Res. 155: 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Switzer WM, et al. 2010. Absence of evidence of xenotropic murine leukemia virus-related virus infection in persons with chronic fatigue syndrome and healthy controls in the United States. Retrovirology 7: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ulbrich P, et al. 2006. Distinct roles for nucleic acid in in vitro assembly of purified Mason-Pfizer monkey virus CANC proteins. J. Virol. 80: 7089–7099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Urisman A, et al. 2006. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2: e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59. van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. 1996. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116: 17–24 [DOI] [PubMed] [Google Scholar]
- 60. Verhaegh GW, et al. 2011. Prevalence of human xenotropic murine leukemia virus-related gammaretrovirus (XMRV) in Dutch prostate cancer patients. Prostate 71: 415–420 [DOI] [PubMed] [Google Scholar]
- 61. von Schwedler UK, et al. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17: 1555–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77: 5439–5450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang MQ, Goff SP. 2003. Defects in virion production caused by mutations affecting the C-terminal portion of the Moloney murine leukemia virus capsid protein. J. Virol. 77: 3339–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wildova M, et al. 2008. The effect of point mutations within the N-terminal domain of Mason-Pfizer monkey virus capsid protein on virus core assembly and infectivity. Virology 380: 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wilk T, et al. 2001. Organization of immature human immunodeficiency virus type 1. J. Virol. 75: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wright ER, et al. 2007. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 26: 2218–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yeager M, Wilson-Kubalek EM, Weiner SG, Brown PO, Rein A. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. U. S. A. 95: 7299–7304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yu F, et al. 2001. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J. Virol. 75: 2753–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]