Abstract
Meq is the major Marek's disease virus (MDV)-encoded oncoprotein and is essential for T-cell lymphomagenesis. Meq and several noncoding RNAs, including three microRNA (MiR) clusters, are expressed from the repeats of the MDV genome during latent infection of T cells. To investigate the state of the chromatin in this and flanking regions, we carried out chromatin immunoprecipitation (ChIP) analysis of covalent histone modifications and associated bound proteins. T-cell lines and a lymphoma were compared. The chromatin around the promoters for Meq and the noncoding RNAs in both cell lines and the lymphoma were associated with H3K9 acetylation and H3K4 trimethylation, which are marks of transcriptionally active chromatin. These correlated with bound Meq–c-Jun heterodimers. The only binding site for Meq homodimers is located at the lytic origin of replication (OriLyt), next to the lytic gene pp38. This region lacked active marks and was associated with repressive histone modifications (H3K27 and H3K9 trimethylation). DNA CpG methylation was investigated using methylated DNA precipitation (MeDP). In cell lines, DNA methylation was abundant across the repeats but noticeably reduced or absent around the active promoters. In primary tumors, CpG methylation occurred less than 2 months after infection, focused within the ICP4 gene. These data suggest that nonrandom de novo DNA methylation occurs early in lymphomagenesis. In addition, the histone data indicate a role for Meq in the epigenetic regulation of the MDV genome repeats in transformed T cells and suggest that the OriLyt region and the Meq/MiR region might be separated by chromatin boundary elements, and preliminary data on CTCF binding are consistent with this.
INTRODUCTION
Marek's disease (MD) is a common lymphoproliferative and neurological disease of poultry caused by the highly contagious alphaherpesvirus Marek's disease virus (MDV). Because of its contagious nature, rapid disease onset, and persistence in both the host and environment, MDV is arguably one of the most economically significant pathogens of poultry. More than 5 billion doses of MDV vaccine are used annually in an attempt to control the disease (31).
The pathogenesis of MD is complex. Infection is via the respiratory route and is followed shortly by a cytolytic infection of mainly B cells in lymphoid organs. It is thought that activated T cells (largely of the CD4+ phenotype) are recruited to the site of cytolytic infection and become latently infected with MDV and then transformed. MDV tumors and their derived cell lines contain MDV genomes integrated into the telomeres of the host chromosome. While integration appears to be associated intrinsically with transformation and latency, integration into the telomeres is not essential; however, it is crucial for efficient reactivation (15, 16, 24, 53). This leads to neoplastic T-cell lesions in visceral organs, and infiltrating lymphocytes can cause edema in peripheral nerves and produce paralysis (5, 54). The virus also replicates in feather follicle epithelium, the site of a productive infection that allows shedding and horizontal spread. Although MDV is an alphaherpesvirus, biologically it more closely resembles the lymphotropic oncogenic gammaherpesviruses, such as Epstein-Barr virus (EBV), Kaposi's sarcoma-associated herpesvirus (KSHV), and herpesvirus saimiri (HVS) (18).
The MDV genome contains two unique regions flanked by repeats; all identified proteins and RNAs expressed during latency are encoded in these repeat regions. The long repeat adjacent to the unique long (UL) region contains the MDV origin of lytic replication (OriLyt) flanked by genes associated with lytic replication. These include the genes expressing pp14, pp38, and the related pp24 protein, which are expressed only during lytic replication of MDV (13, 44). Deletion of the pp38 gene from the Md5 strain by use of both cosmid and bacterial artificial chromosome (BAC) technology has shown its importance in lytic replication but that it is dispensable for the formation of tumors (21, 43). The pp38 protein can act as a transcriptional regulator of its own promoter when it is dimerized with pp24 (17).
Adjacent to the pp38 gene is the gene encoding Meq. This is the major MDV oncoprotein that is expressed during both latent and lytic replication and closely resembles a B-ZIP transcription factor. Meq can homodimerize or heterodimerize with c-Jun, and the dimerization state determines its DNA binding affinity (25, 41). Heterodimers bind with high affinity to DNAs resembling 12-D-tetradecanoylphorbol 13-acetate (TPA) and cyclic AMP (cAMP) response elements (AP-1 sites)—these are termed Meq-responsive elements I (MERE-I) (TGACA/GTCA). Meq–c-Jun heterodimers activate transcription in transient reporter assays. In contrast, Meq homodimers bind MERE-II (ACACA) sites and appear to act as repressors of transcription (25, 29, 42). The only homodimer binding site that has been identified in the MDV genome is at OriLyt, but several MERE-1 (AP-1) sites are found in the latency-associated region (27, 34). Meq homo- and heterodimerization and Meq binding to the cellular corepressor CtBP are all required for its oncogenic activity (8, 9, 30, 49, 50).
Located 3′ of Meq is the gene encoding vIL8, a viral CXC chemokine suggested, based on its expression kinetics, to be a late protein associated with lytic replication (28, 35). Deletion of vIL8 from MDV by cosmid recombination showed that it is important in early cytolytic infection but dispensable for T-cell transformation and lymphomagenesis (14). Also described for this region are spliced transcripts encoding both Meq and vIL8 which have been detected in some cell lines, but their significance is currently unclear (2).
The lytic ICP4 gene is also found within the short repeat of the genome, encoding a protein product of 155 kDa that has conserved domains with and amino acid sequence similarity to ICP4 proteins of other alphaherpesviruses, such as herpes simplex virus (HSV) (1). A monoclonal antibody against MDV ICP4 detects a protein in lytically infected chicken embryo fibroblasts (CEF) (55), and overexpression of ICP4 in the MSB-1 cell line resulted in increased expression of pp38, suggesting a role in MDV replication or reactivation (40). Running antisense to ICP4 is a 10-kb latency-associated transcript (LAT) that appears to be expressed during latent infection. LATs have been detected in MDV-transformed cell lines and lymphomas, but transcripts from the ICP4 gene have been detected only during lytic infection (11, 12, 26).
The MDV-encoded microRNAs (MiRs) are found in clusters on either side of the Meq gene and at the beginning of the LAT. They are expressed during both lytic replication and latency (10, 56). As with Meq, deletion of the MiRs completely abrogates oncogenesis, and specific deletion of the MiR-155 ortholog MiR-M4 has the same effect (57). The long repeat also encodes the MDV viral telomerase subunit (vTR) (19), which has been shown to be expressed both in lytic infection and in MDV-transformed T-cell lines. Deletion analysis has shown that it is dispensable for lytic replication and important but not absolutely essential for tumor formation (53).
Here we have examined histone modifications and DNA methylation on the chromatin of and adjacent to the latency-associated region of MDV and related this to patterns of transcription and transcription factor binding.
MATERIALS AND METHODS
Cell culture.
MDV-carrying T-cell lines 265L and RPL1 were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal calf serum, 10% tryptose phosphate broth, penicillin, streptomycin, 1 mM sodium pyruvate (Sigma), and 50 μM 2-mercaptoethanol (Invitrogen) at 38.5°C with 5% CO2. The 265L cell line was established from a lymphoma from the liver of a line P chicken infected with the wild-type RB1B strain of MDV (57). The RPL1 cell line was established from a transplantable T-cell tumor induced by the JM strain of MDV (32).
Cell treatments with chromatin-modifying drugs.
Trichostatin A (TSA), 5-azacytidine (AZA), and sodium butyrate (NaB) were all purchased from Sigma and were used at concentrations of 500 nM, 5 μM, and 2.5 mM, respectively.
Western blotting.
Western blotting was performed essentially as previously described (52). Briefly, proteins were extracted with RIPA buffer, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to Protran nitrocellulose membranes (Schleicher and Schuell). Membranes were blocked with 5% milk powder in phosphate-buffered saline (PBS)–0.05% Tween 20 and probed with appropriate primary and horseradish peroxidase (HRP)-conjugated secondary antibodies. An ECL kit (Amersham Biosciences) was used for visualization. The following primary antibodies were used: mouse monoclonal anti-γ-tubulin (T6557; Sigma) and mouse monoclonal anti-pp38 (Institute for Animal Health [IAH], Compton, United Kingdom).
qPCR.
Quantitative PCR (qPCR) was carried out using Platinum SYBR green qPCR SuperMix (Invitrogen) on an ABI 7900 384-well real-time PCR machine. Sequences of primers used are shown in Table 1. Cycling conditions used were 2 min at 50°C, 2 min at 95°C, and then 40 cycles of 15 s at 95°C and 1 min at 60°C.
Table 1.
Primers used for qPCR
| Primer no. | Primer name | Sequence (5′-3′) | Primer no. | Primer name | Sequence (5′-3′) |
|---|---|---|---|---|---|
| 1 | UL2-1_For | TCCCTCTCACTTCAACGGAC | 45 | IRS10_For | GTCAAGAATGGCTAAAGGGATGATA |
| 2 | UL2-1_Rev | CCTAACACATATAACGGTAAATCTT | 46 | IRS10_Rev | CGGTGTACAGAGTTTCAGCAGT |
| 3 | UL4-For | CGTAGGTTCCATAAATGGTTTCA | 47 | IRS12_For | GTTGTCGGCCCATTAACTCT |
| 4 | UL4-Rev | GCAAGCATCTGTCAAATAGCAA | 48 | IRS12_Rev | GGTACTGTCCTCTATCCCAATT |
| 5 | UL6-1_For | GTGTGATCTCTTAGCAGTATCAGG | 49 | IRS14_For | CCCTTCTGATATTGGTGGAGT |
| 6 | UL6-1_Rev | CCGTAATATGTGTTGTTCGTTGTAG | 50 | IRS14_Rev | GGCATTGGGAGTGGTGTCTG |
| 7 | UL8-For | ACCACATAGCTCAAATACTAGAACA | 51 | IRS16_For | CAGACCCTTGGAAGAACTCG |
| 8 | UL8-Rev | GGGTTCTAAATTTTCACTCGCT | 52 | IRS16_Rev | AAAAAGATAACATTTTAGCTACTCT |
| 9 | Ori0-For | AGAAAAAAGGTGGATTGTAATAGGC | 53 | IRS18_For | CCGCAGTCTAAGGGGAGAAA |
| 10 | Ori0-Rev | CGGATTTGATGGCAATCTTTTG | 54 | IRS18_Rev | CGGTTTCTTGGGGTAAGTAGT |
| 11 | Ori-2-For-1 | ATTAATGCTGGCCGAAAGAC | 55 | IRS20_For | TTATTGCCCCGTACTCACCG |
| 12 | Ori-2-Rev-1 | CGGATAGAATAGTACGGGGT | 56 | IRS20_Rev | CATTTAAAGTCTTTCCATGCCAAAC |
| 13 | Ori4A-For-1 | GCGGTATAGGATAAGAGATCAC | 57 | IRS22_For | GTCCCCACCTCCTTTGTCTC |
| 14 | Ori4A-Rev-1 | GCGAAGAAGTTCCAAACGAT | 58 | IRS22_Rev | GCTCCGACCTAAACTAGGGAC |
| 15 | Ori6a-For | CCCCTTATTCCTCCATAGCAC | 59 | IRS24_For | CCCACCACCGATACTTCCAG |
| 16 | Ori6a-Rev | TGTGGGAGAAAGTATGTCGATTT | 60 | IRS24_Rev | ATAAGAAGGGGAACGGTCAC |
| 17 | Ori8-For | AGAGATTGCTAGGTCGAAAGC | 61 | IRS26_For | CCTTTTGTGCTTAGTCGATGG |
| 18 | Ori8-Rev | TCCTTTGAGTGACGGGAAGC | 62 | IRS26_Rev | AATTGTGTCCTGCTGAACGA |
| 19 | Mir2-For | CGTTCGAAGCAGTGATTATTCTT | 63 | IRS28_For | CCCATAGCTTCGTTAACACTT |
| 20 | Mir2-Rev | TGAATTTCACTCTCCCCTCAAA | 64 | IRS28_Rev | GGGAGTGTGTTATCTTGTCGC |
| 21 | Mir4-For | AGTTCCATTTTTCTTACATGCTCA | 65 | IRS30_For | CCAGGAATACGATTTGGCTAAT |
| 22 | Mir4-Rev | GATTGGCCGTTGTAGCATAC | 66 | IRS30_Rev | ACGGTTCAGGGGATATGAGA |
| 23 | Mir6-For | TCAGGGCCCCAGACATCAAA | 67 | IRS32_For | GCCCCTAATGTACTACTTGCT |
| 24 | Mir6-Rev | CCCATGAGAAATTTTTGATAACTGA | 68 | IRS32_Rev | GGAGTGAAATCTTTAGGGAGGG |
| 25 | AP1-2-For-1 | ACGGTTCATGACAGTTGATAAAC | 69 | UL30-1_For | CACACATCCTCACAATCCTCA |
| 26 | AP1-2-Rev-1 | GAGCAGTCGGTTGTAACCAA | 70 | UL30-1_Rev | CCCAAATGCCTAGATGACAA |
| 27 | AP1-4-For-1 | CCCGAACCATTAGATATCAGT | 71 | GAPDH_For | GTCAACGGATTTGGCCGTAT |
| 28 | AP1-4-Rev-1 | ACCGTTGAAGTGATGTAATATTGAA | 72 | GAPDH_Rev | CCACTTGGACTTTGCCAGAGA |
| 29 | AP1-6a-2-For | ATCTAATACTTCGGAACTCCT | 73 | MDV072_For | AACCCAGAGATCCAAACACA |
| 30 | AP1-6a-2-Rev | AAATTTTCCTTACCGTGTAGC | 74 | MDV072_Rev | CGGAACCATCAAGCTCCATTTTA |
| 31 | AP1-8-For | CGACTGTAGCACTTAGAATCGC | 75 | pp38_For | ATTAATGCTGGCCGAAAGAC |
| 32 | AP1-8-Rev | TCCGCGTCCCAGCAATCAGG | 76 | pp38_Rev | CGGATAGAATAGTACGGGGT |
| 33 | AP1-10-For | ACAGGAGTTTGCATCAAAAGG | 77 | Meq_For | GGAGTTTGTCTACATAGTCCGT |
| 34 | AP1-10-Rev | GGTCGTGGGAATTTTCTCATATACA | 78 | Meq_Rev | GCCCCTCCAAACACCCCTTC |
| 35 | AP1-12-For | CCACCATCTTCTCCAACCAT | 79 | vIL8_For | CCACCATCTTCTCCAACCAT |
| 36 | AP1-12-Rev | GCCTTGGGTACAGCAGTTTATTAAA | 80 | vIL8_Rev | GCCTTGGGTACAGCAGTTTATTAAA |
| 37 | IRS2-For | CCCTAATCGGAGGTATTGATGG | 81 | vTR_For | CCCTAATCGGAGGTATTGATGG |
| 38 | IRS2-Rev | TAAACAGCGGGCGGAGGGAG | 82 | vTR_Rev | TAAACAGCGGGCGGAGGGAG |
| 39 | IRS4-For | GGGCGATAAGACACTTTCCC | 83 | MIR#3_For | GCGACAATCAGGACGATGTAAT |
| 40 | IRS4-Rev | TATGTGCCGGTTCCAGTGTG | 84 | MIR#3_Rev | GGGAAAATCTGTTGTTCCGTAG |
| 41 | IRS6_For | ACAAAACACTTACCCTCTCAACT | 85 | LAT_For | TTATTGCCCCGTACTCACCG |
| 42 | IRS6_Rev | CGTTCCTGATTTCCTTTCGC | 86 | LAT_Rev | CATTTAAAGTCTTTCCATGCCAAAC |
| 43 | IRS8_For | GCGACAATCAGGACGATGTAAT | 87 | UL1_For | GCCCCTAATGTACTACTTGCT |
| 44 | IRS8_Rev | GGGAAAATCTGTTGTTCCGTAG | 88 | UL1_Rev | GGAGTGAAATCTTTAGGGAGGG |
qRT-PCR.
For quantitative reverse transcriptase real-time PCR (qRT-PCR), RNA was extracted from approximately 5 × 106 cells for each cell line by using an RNeasy minikit from Qiagen, with a DNase digestion step, following the manufacturer's instructions. For quantification, 1 μl of cDNA (generated from total RNA by using SuperScript III first-strand synthesis SuperMix [Invitrogen] per the manufacturer's instructions) was used for qPCR. PCR products were quantified by standard curve analysis in SDS2.3 and then normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The calculated errors in the graphs are the standard deviations for three replicate qRT-PCRs for each mRNA.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were carried out essentially as described previously (38), using a ChIP assay kit (Millipore) according to the manufacturer's instructions. Chromatin was sheared to 200 to 1,000 bp for 1 × 106 cells per ChIP in 200 μl of lysis buffer, using a Bioruptor sonicator (UCD-200; Diagenode) on the high setting for a total of 12 min (applying 30-s “on” and 30-s “off” intermittent sonication). Chromatin was precipitated using antibodies raised against various histone modifications and transcription factors (Table 2); 5 μg per 1 × 106 cells was used for each precipitation, unless otherwise indicated by the manufacturer's protocol. For precipitation of proteins other than histones, reaction mixtures were scaled up 10-fold (1 × 107 cells) until elution. DNA was purified using a Qiagen PCR purification kit and eluted in a 300-μl volume. Isolated DNA was assayed by qPCR. Using standard curves, 10% of the input was compared to the IP sample, and the values from the IgG negative control were subtracted as background. The calculated errors in all graphs presenting ChIP data are the standard deviations for three replicate qPCRs for precipitated chromatin, input chromatin, and background (chromatin precipitated with nonspecific rabbit IgG).
Table 2.
Rabbit antibodies used in ChIP assays
| Antibody target | Manufacturer | Antibody no. |
|---|---|---|
| H3K9Ac | Millipore | 17-658 |
| H3K9me3 | Millipore | 17-625 |
| H3K27me3 | Millipore | 17-622 |
| H3K4me3 | Millipore | 17-614 |
| H3Ac | Millipore | 06-599 |
| H4Ac | Millipore | 06-866 |
| RNA polymerase II pS5 | Abcam | ab5131 |
| c-Jun | Abcam | ab31419 |
| CTCF | Millipore | 07-729 |
| Rad21 | Abcam | ab992 |
| Smc1 | Abcam | ab9262 |
| Smc3 | Abcam | ab9263 |
| Meq | IAH Compton | See reference 8 |
MeDP with His-tagged MBD2b.
Methylated DNA precipitation (MeDP) assays were carried out essentially as described previously (38). After isolating genomic DNA by using a Qiagen blood and tissue minikit as described in the manufacturer's protocol, 2 μg of DNA was sonicated in 200 μl of H2O for five 20-s sonication rounds, using a Bioruptor sonicator (UCD-200; Diagenode) on the high setting for a total of 12 min (30-s “on” and 30-s “off” intermittent sonication). Methylated DNA was precipitated from 1 μg of sheared DNA by using a MethylCollector kit (Active Motif) according to the manufacturer's instructions. Precipitated DNA was quantified by qPCR. As a background control, precipitations with magnetic beads only were performed, without adding His-MBD2b. As additional controls, precipitations were performed using purified MDV-BAC DNA (unmethylated) and artificially methylated MDV-BAC DNA (fully methylated in vitro). The calculated errors in all graphs presenting methylated DNA precipitation data are the standard deviations for three replicate qPCRs for precipitated DNA and input DNA.
Animal experiments.
All animal experiments were carried out in accordance with United Kingdom Home Office guidelines by trained staff holding a Home Office Personal License for the various procedures, using the specific-pathogen-free inbred line P (B19/19), obtained from the Poultry Production Unit of the Institute for Animal Health. For pathogenicity studies, 1-day-old birds were infected with 1,000 PFU of the cell-associated virus stocks by the intra-abdominal route. The MDV-infected birds were maintained in separate HEPA-filtered rooms. Blood (in 3% sodium citrate) collected at the indicated times after infection with MDV was used to purify peripheral blood lymphocytes (PBL) for MeDP and ChIP.
RESULTS
Analyses of RNA transcripts within and flanking the MDV repeat regions.
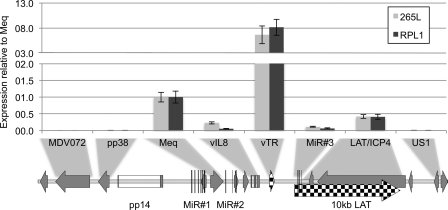
In order to establish the pattern of latency-associated transcription in the MDV-carrying T-cell lines used in this study, the expression of several genes and noncoding RNAs from within the two repeat regions (short and long) of the MDV genome was investigated by qRT-PCR, using two independent and distinct MDV cell lines, RPL1 and 265L. These cell lines were chosen because they have very different histories; RPL1 is a cell line first described in 1977 (32), and 265L is a recent line generated from a tumor from a line P bird that was infected with MDV strain RB1B (57). RNA was reverse transcribed using a mixture of oligo(dT) primers and random hexamers, and specific transcripts were quantified using primers (69 to 82) shown in Table 1 and then normalized against GAPDH controls. The qRT-PCR analysis of RNA levels in T-cell lines RPL1 and 265L demonstrated that they had remarkably similar expression profiles for the pp38, Meq, and vIL8 genes and the vTR, MiR cluster 3, and LAT/ICP4 regions (Fig. 1), despite their distinct origins and long-term culture of these cells. The expression profiles for both cell lines closely matched that described in the literature for genes across this region whose transcripts were previously shown to be highly expressed in MDV-transformed cell lines: Meq, the vTR region, and the LAT/ICP4 region all showed high levels of expression in both RPL1 and 265L cells (19, 26, 28). The extremely high levels of transcripts corresponding to vTR were probably because these form a stable RNA subunit of the telomerase enzyme (20). Expression of the third microRNA cluster (MiR3) was also detectable, but at a much lower level, as is often the case for MiRs. Two genes that flank the repeat regions, MDV072 and US1, in the UL and US regions, respectively, showed no detectable expression. The region within the long repeat that flanks the UL contains the OriLyt plus associated lytic genes pp14 and pp38 and showed no detectable expression. This demonstrated that latent gene expression was restricted to a core region within the repeats surrounded by silenced regions in both the long repeat and surrounding unique regions. The results suggest that for latency to be maintained, the lytic infection-associated genes are repressed through a stable, possibly epigenetic mechanism that is highly conserved in distinct MDV cell lines, and these patterns of expression are likely to occur early during the development of MDV-associated tumors.
Fig 1.
Expression of the genes in the MDV repeat regions. Quantitative RT-PCR was carried out on mRNAs extracted from two MDV-transformed tumor-derived T-cell lines: a historically established line (RPL1) and a newly established line (265L). Expression of the genes within the long and short repeats was detected. This included genes encoding the viral oncoprotein Meq, the CXC chemokine vIL8, and the viral telomerase subunit vTR, the third microRNA cluster, and transcripts across the LAT/ICP4 region. Expression from the vTR region appeared to be significantly higher; this may be because, as an RNA subunit of telomerase, it is more stable. Adjacent genes encoding lytic proteins, namely, the MDV072 gene in the unique long region and the gene for the phosphoprotein pp38 at the origin of lytic replication, and the unique short region gene US1 showed no detectable expression. Expression is shown relative to that of Meq and normalized to that of GAPDH.
Profiles of histone 3 modifications across the repeats of MDV in T-cell lines.
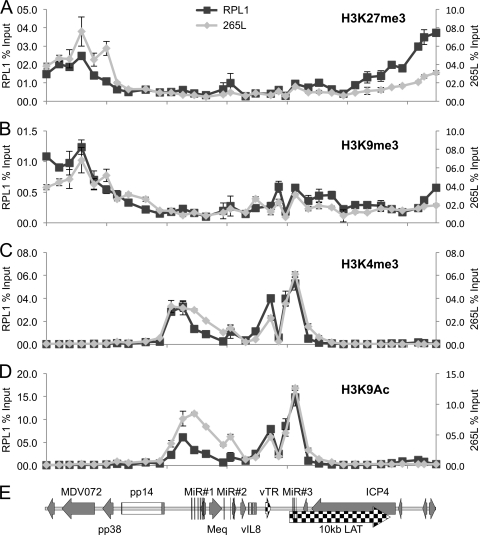
To investigate whether the pattern of transcriptional repression and activation was related to modification of the histones located at the promoters of the relevant genes, ChIP followed by qPCR was performed. Antibodies against trimethylation modifications of histone 3 at lysine 27 (H3K27me3) and lysine 9 (H3K9me3), both of which are associated with repressed chromatin, were used to identify silent regions. To understand the core active region, antibodies against activation-associated modifications were used, specifically for trimethylation of histone 3 at lysine 4 (H3K4me3) and acetylation of histone 3 at lysine 9 (H3K9ac). Multiple (n = 34) primer pairs (Table 1) distributed approximately 1 kb apart across the whole region were used for qPCR.
This analysis revealed that the repressive marks H3K27me3 and H3K9me3 were both associated with the part of the repeat region around the OriLyt from which gene expression was absent in both cell lines (Fig. 2). This is consistent with the repressed nature of the chromatin maintaining the inactive state of this part of the genome. In contrast, the active marks H3K4me3 and H3K9Ac were restricted to the core region that contained the MiR clusters, Meq, the vTR region, and the 10-kb LAT. There were no detectable active marks outside this region, and both cell lines showed remarkably similar patterns of active marks. Close inspection suggests the occurrence of four peaks of H3K9Ac and H3K4me3 across the region: the first corresponds to the first MiR cluster and the Meq promoter, the second includes MiR2, the third contains the promoter for the vTR region, and the final peak clearly maps to the third MiR cluster and the promoter for the LAT. Very similar profiles were seen for both cell lines with two other commercial antibodies raised against other marks of active chromatin, i.e., H3Ac and H4Ac (data not shown).
Fig 2.
Profile of histone 3 (H3) modifications across the repeats of MDV in T-cell lines. Chromatin immunoprecipitation was carried out with two MDV-transformed T-cell lines, RPL1 and 265L. Results are shown as percentages of total input, with RPL1 and 265L cell data shown on independent y axes and the x axis indicating positions (bp) across the MDV genome. Antibodies against specific histone modifications were used, with two against modifications related to repression (H3K27me3 [A] and H3K9me3 [B]) and two against modifications associated with activation (H3K4me3 [C] and H3K9Ac [D]). Thirty-four sets of primers were used for qPCR analysis. (E) Gene map of the region for reference.
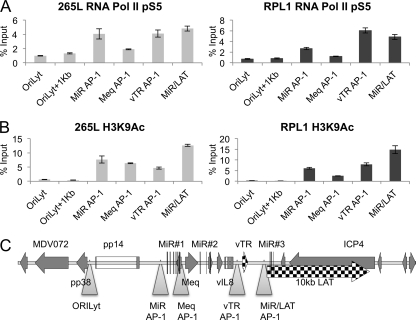
Binding and transcription by RNA Pol II across the MDV repeat regions.
The patterns of active chromatin and expressed transcripts showed a good correlation, so in order to determine the location of bound RNA polymerase II (Pol II) in an active state of transcript elongation, ChIP assays were performed. Initiation of transcription by RNA Pol II requires it to be phosphorylated at the appropriate serine (7). ChIP assays were therefore carried out with an antibody recognizing RNA Pol II phosphorylated at serine 5 (pS5), a marker for transcription initiation and early elongation. The profile of RNA Pol II pS5 at sites of active chromatin is shown in Fig. 3A, and again, a remarkable similarity between the two cell lines (RPL1 and 265L) can be seen. The profile of bound RNA Pol II closely matches that of active chromatin, as shown by an antibody that recognizes N-terminal acetylation of histone 3 (H3K9Ac) (Fig. 3B). There are several peaks shown, corresponding to the region around the first MiR cluster, Meq, the vTR region, and the last MiR cluster. There is also evidence of a significant amount of transcription across the LAT/ICP4 region (Fig. 3A). This most likely corresponds to initiation and elongation of the LAT and its spliced products.
Fig 3.
RNA polymerase II binding is associated with regions of active chromatin. (A) To correlate RNA Pol II binding in the region with active chromatin, chromatin immunoprecipitation was carried out with antibodies against RNA Pol II phosphorylated at serine 5 (an indicator of transcript initiation occurring) and two MDV-transformed T-cell lines, 265L and RPL1. Results are shown as percentages of total input. (B) Active chromatin as revealed by H3K9Ac ChIP, correlated with RNA Pol II binding across the region. (C) Map of the region and locations of qPCR primers (triangles).
MDV chromatin in primary tumors.
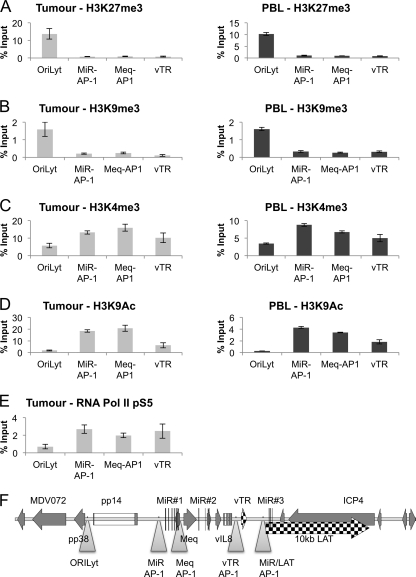
The data presented above show active and repressed regions of the chromatinized MDV genome from the two tumor-derived T-cell lines. Although the profiles of histone marks and RNA Pol II transcription in these cells are highly conserved—despite the different histories of the lines—there is the possibility of selection in culture leading to a profile that would not necessarily be representative of a primary lymphoma. Therefore, we analyzed the viral chromatin state of MDV-transformed T cells in vivo, firstly in a primary tumor removed from the liver of a line P chicken sacrificed 28 days after infection with MDV strain RB1B and then in the PBL from an RB1B-infected line P chicken at day 32 postinfection. This bird went on to develop multiple T-cell lymphomas. Samples were fixed in formaldehyde and analyzed by ChIP and qPCR. Primer pairs corresponding to several distinct regions of the MDV latency region (suggested by the cell line histone data) were used. This revealed that both H3K27me3 and H3K9me3 (Fig. 4A and B) were present around the lytic origin and at the genes encoding pp14 and pp38. In contrast, the two markers of active chromatin, H3K4me3 and H3K9Ac, were increased around the first MiR cluster, Meq, and the vTR region (Fig. 4C and D). This was similar to the data from the T-cell lines, with distinct active and repressed regions of chromatin. However, unlike the case in the cell lines, there was evidence of active chromatin surrounding OriLyt in the tumor. This was probably due to activation of the lytic genes in a subset of cells in the tumor and to the lack of selection for a completely latent population, as would occur during long-term cell culture. Finally, RNA Pol II pS5 distribution was also investigated at the same four regions of MDV in the primary tumor. The binding was consistent with and largely corroborated the histone modification data (Fig. 4E). Unfortunately, insufficient MDV-positive cells were present in the PBL for reliable detection of Pol II on viral chromatin (our unpublished observations).
Fig 4.
Histone modifications and binding of RNA Pol II in primary MDV lymphoma and PBL. Chromatin immunoprecipitation was carried out with antibodies against histone modifications and RNA Pol II phosphorylated at serine 5 (pS5) on chromatin extracted from a primary MDV lymphoma. (A) H3K27me3; (B) H3K9me3; (C) H3K4me3; (D) H3K9Ac; (E) RNA Pol II pS5. Results are shown as percentages of total input. The chromatin profile and regions of active transcription were very similar to those seen for the tumor-derived cell lines. (F) Map of the region and locations of qPCR primers (triangles).
DNA methylation analysis across the repeats of MDV in T-cell lines.
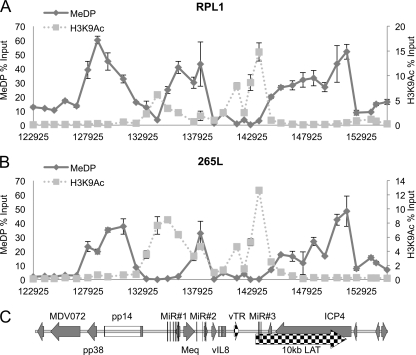
Long-term repression of the chromatin around promoters can be associated with CpG DNA methylation (45). Therefore, the role of CpG DNA methylation was investigated using the MeDP technique on purified genomic DNAs from the two cell lines, followed by qPCR analysis. The CpG DNA methylation profiles of the two cell lines (RPL1 and 265L) (Fig. 5A and B) both showed that methylation was present across the repeat region; however, it was conspicuously absent at the active promoter regions around the MiR cluster, Meq, the vTR region, and LATs. In contrast, the lytic promoters either side of the repeat region and surrounding OriLyt showed peaks of methylated CpG DNA. This is clearly illustrated in Fig. 5 by the overlays showing comparisons of H3K9Ac (dotted lines) with methylation (solid lines), with peaks of active chromatin occurring in the regions of reduced or no methylation. Control MeDP was carried out using in vitro-methylated RB1B-BAC, which showed the effectiveness of the protocol and gave an indication of the density of CpGs distributed across the region (data not shown).
Fig 5.
CpG methylation analysis of MDV DNA from T-cell lines. Fragmented DNAs from RPL1 (A) and 265L (B) cells were subjected to methylated DNA precipitation as described in Materials and Methods. Again, 34 primer sets were used for qPCR, and the methylated DNA profile is shown as an unbroken line; for comparison, the H3K9Ac data from Fig. 2 are shown as a dotted line. Results are shown as percentages of total input, with H3K9Ac and methylation shown on independent y axes and the x axis indicating positions (bp) across the MDV genome. (C) Map of the region.
DNA methylation of primary tumor material and PBL from recently infected chickens.
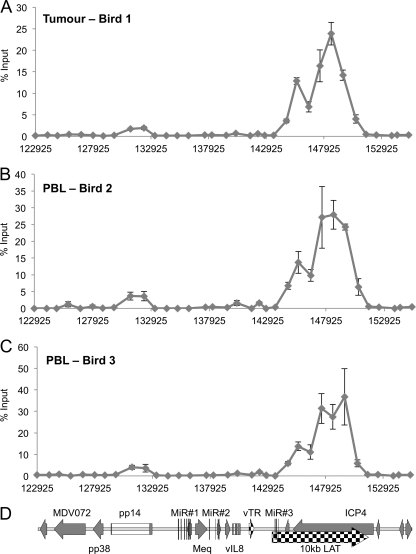
To investigate the DNA methylation status of the viral genome in a primary lymphoma and the PBL of infected chickens, genomic DNAs were extracted from a tumor removed from a line P chicken at 43 days and from the PBL taken from two chickens at 32 days postinfection, using the RB1B strain of MDV for infection. The MeDP protocol was followed by qPCR analysis to determine the amount of methylated DNA compared to the input DNA for all three samples. This demonstrated a distinct but related profile compared to that found in the cell lines (compare Fig. 6 with Fig. 5A and B). The tumor material showed the presence of intragenic CpG methylation, specifically in the ICP4/LAT region and, to a much lesser extent, in pp14. However, there was a distinct absence of methylation in and around the nearby actively transcribed promoters, as found in the cell lines. It should be noted that there are CpGs distributed throughout the repeat region. It was particularly striking that all three DNA methylation profiles (for the tumor and the two independent samples of PBL) were very similar, although the DNAs came from the infection of different birds at different times with different batches of virus. This suggests that the MDV genome was modified by the host DNA methylation machinery in the same way on these three separate occasions and that there must be some nonrandom underlying guiding principle.
Fig 6.
CpG methylation analysis of DNAs from primary MDV lymphoma and PBL. (A) Methylated DNA precipitation was carried out on fragmented DNAs from a primary MDV lymphoma isolated from an RB1B-infected line P chicken. Again, 34 primer sets were used for the qPCR. (B and C) Similar analysis of PBL from two RB1B-infected line P chickens that developed lymphomas. Results are shown as percentages of total input on the y axis, with the x axis indicating locations (bp) across the MDV genome. All three methylation profiles are remarkably similar, with methylation largely within the body of the pp14 gene and the LAT/ICP4 region. (D) Map of the region.
CpG DNA methylation in a gene in the UL region.
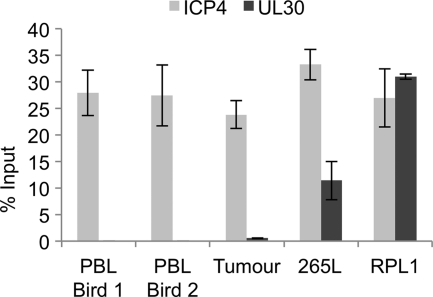
With the presence of methylated MDV DNA in the repeat region in both the T-cell lines and the primary tumor and PBL material, the methylation status of another region of the viral genome was investigated in the tumor, PBL, and cell lines. Methylation in the body of the ICP4 gene was compared to methylation within the body of UL30 (Fig. 7). There was no detectable methylation in UL30 from the PBL and tumor, although there was detectable methylation in ICP4. However, the cell lines had detectable methylation of both ICP4 and UL30. This again suggests that CpG methylation of ICP4 in tumor development is nonrandom and that methylation in the cell lines is more extensive.
Fig 7.
CpG methylation of a gene (UL30) in the UL region of the MDV genome. Methylated DNA precipitation was carried out on fragmented DNAs from PBL isolated from two RB1B-infected line P chickens that developed lymphomas, from a primary MDV lymphoma isolated from an RB1B-infected line P chicken, and from two T-cell lines (265L and RPL1). Results are shown as percentages of total input. Methylation within the ICP4 gene was compared to methylation within the intragenic region of UL30. There was no detectable methylation in the UL30 gene within the PBL and tumor, but both cell lines had detectable methylation.
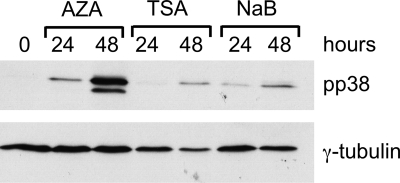
Reactivation of MDV by treatment with chromatin-modifying drugs.
To assess whether the histone modifications and methylation marks were functional in maintaining the latency state of the 265L cell line, the cells were subjected to drug treatment. This was carried out using the histone deacetylase (HDAC) inhibitor TSA, the methylation inhibitor AZA, and NaB. This revealed that all three drugs have the ability to cause initiation of the lytic cycle, as indicated by expression of pp38 (Fig. 8). This is consistent with histone modification and DNA methylation being involved in the maintenance of the viral genome in its latent state.
Fig 8.
Initiation of MDV lytic gene expression by chromatin-modifying drugs. Western blotting was carried out on the 265L cell line treated with AZA, TSA, and NaB. Samples were taken 0, 24, and 48 h after drug treatment, separated by SDS-PAGE, Western blotted, and probed with antibodies against γ-tubulin and pp38. All three treatments induced the expression of pp38, a lytic MDV antigen.
Binding of Meq and c-Jun proteins in the repeat region of the cell lines.
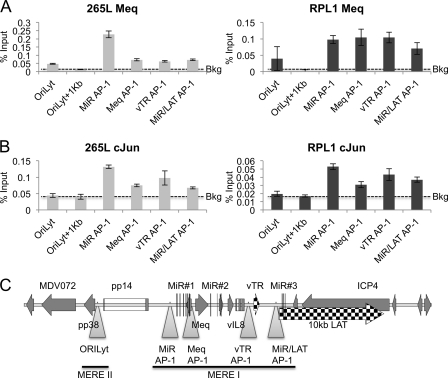
Previous low-resolution studies have shown that Meq and c-Jun can bind within the MDV repeat region (25). To obtain higher-resolution data, we used the same approach of ChIP followed by qPCR across the repeat region of the viral genome, using antibodies directed against Meq and c-Jun. Binding of Meq and c-Jun to specific sites across this region of the MDV genome from the cell lines is shown in Fig. 9. Meq binding could be seen at the OriLyt, where the only known Meq homodimer (MERE-II) binding site is located, and at several of the AP-1 (MERE-I) sites across the repeat region (Fig. 9A). At the homodimer site, the binding of Meq is associated with the presence of the repressive histone marks, and at the heterodimer binding sites it is associated with the presence of active chromatin (compare Fig. 9 with Fig. 2 and 3). As expected, binding of c-Jun was found only at the heterodimer binding sites (Fig. 9B), with no c-Jun binding above the control nonspecific antibody background found at the OriLyt binding site.
Fig 9.
Meq and c-Jun binding on the repeat region of the MDV genome. ChIP was carried out using antibodies against the Meq or c-Jun protein with the T-cell lines 265L and RPL1. Results are shown as percentages of total input. (A) Bound Meq was detectable above the background at all predicted MERE sites (OriLyt+1kb, where there is no Meq binding site) in both RPL1 and 265L cells. (B) Bound c-Jun was detectable above the control antibody background (Bkg) only at the heterodimer binding (AP-1, MERE-1) sites. (C) Map of the region and locations of qPCR primers (triangles).
Boundary elements in the repeat region of MDV.
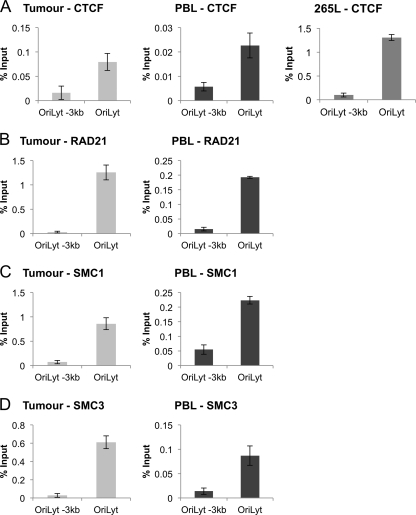
With the distinct loci of active and repressed chromatin in the repeat region, it is likely that boundary elements exist to keep the distinct chromatin regions functionally separate. To investigate this possibility, ChIP assays were performed using an antibody directed against the boundary element protein CTCF. This was performed on the primary tumor-derived chromatin, PBL-derived chromatin, and chromatin from the 265L cell line and showed the presence of CTCF binding at or near OriLyt (Fig. 10A), but not 3 kb upstream. Since recruitment of CTCF can result in the formation of higher-order chromatin structures through the recruitment of cohesins (36), we investigated the presence of cohesin subunits Rad21, SMC1, and SMC3 at the same sites on the tumor- and PBL-derived MDV chromatin. All three factors showed binding at OriLyt, but not 3 kb upstream (Fig. 10B, C, and D), consistent with the CTCF binding and with the possibility that chromatin looping and/or higher-order chromatin structures may be established (33, 36, 39, 48). For reasons we do not understand, it was not possible to consistently show binding of the cohesion factors at either site in extracts from the 265L cells.
Fig 10.
The insulator protein CTCF binds to a region around the origin of lytic replication in a primary MDV lymphoma, PBL, and the 265L cell line. (A) Chromatin immunoprecipitation was carried out using an antibody against CTCF and was quantified using qPCR. Binding was strongest at the OriLyt. (B, C, and D) Similar ChIP assays were then carried out using antibodies against the cohesion factors Rad21, SMC1, and SMC3. Results are shown as percentages of total input. All three molecules were found bound at the same region as CTCF. A site 3 kb upstream of OriLyt showed no detectable binding for CTCF, Rad21, SMC1, or SMC3. Binding of these factors was not detected consistently in 265L cells (data not shown).
DISCUSSION
During latency, MDV, like other herpesviruses, expresses a very limited subset of transcripts, including microRNAs, protein-encoding genes, and other noncoding RNAs, all of which have a direct or indirect role in the maintenance of the latent state. For latency to be achieved, there must be controlled shutdown of the lytic genes in concert with the expression of transcripts needed to establish and maintain the latent state. Therefore, chromatinization of the viral genome and use of the host epigenetic machinery to allow regulatory modifications on histones are likely to be key events (for example, see references 6, 23, and 51). Here we investigated the types of epigenetic marks that are incorporated into MDV chromatin during latency and how these marks correspond to binding sites for the best candidate regulator of latency, Meq. Furthermore, we established the similarities and differences of these histone modifications between T-cell lines and primary lymphomas from infected chickens in order to understand how cell selection in culture may affect the accumulation of these marks on latent MDV chromatin.
Initially, transcript levels in two tumor-derived T-cell lines with distinct histories were analyzed. The profiles from both lines were very similar and matched well with previous studies of the different transcripts within the repeat region (10, 12, 14, 19, 26, 27). The most abundant transcript was the vTR region, probably because of its stability as an RNA subunit in the telomerase complex (20). The similarity in the expression profiles was striking and clearly indicated the presence of an active core region within the repeats surrounded by silenced chromatin. It also suggested that the control of active and repressed chromatin states is maintained during cell culture selection.
To reveal how this pattern of transcription is maintained, we investigated the histone modifications that are distributed throughout the repeat region of the genome. ChIP-qPCR data revealed two distinct domains within the repeat regions of the viral genome in the T-cell lines. The first is associated with the repressive marks H3K9me3 and H3K27me3 and localized primarily around the origin of lytic replication (OriLyt). The second region is associated with active chromatin marks H3K9Ac and H3K4me3 and is found in the region from Meq and the first MiR cluster through to the final MiR cluster and the LAT promoter. More specifically, the active chromatin is closely associated with active promoters within the repeat regions, with clear peaks at the promoter for the first MiR cluster, the Meq promoter, the vTR region promoter, and the final MiR cluster and LAT promoter. These regions of active chromatin are indicated by multiple peaks of not only H3K9Ac and H3K4me3 but also acetylated histones 3 and 4 (H3Ac and H4Ac) (data not shown).
It was confirmed that the active chromatin-bound RNA Pol II was in a phosphorylation state (with pS5) indicative of transcription initiation and early elongation. Again, despite their distinct origins, both cell lines showed almost identical patterns of bound RNA polymerase II pS5, again suggesting the conservation of this activity profile.
To further understand the effect of cell culture selection, we investigated the nature of these profiles in a primary tumor and explanted cells. This material was analyzed using the same protocols used for the T-cell lines. The same two chromatin domains were identified, with H3K9me3 and H3K27me3 present at OriLyt and H3K9Ac and H3K4me3 present at the promoters for MiRs, Meq, and the vTR region within the repeat. This demonstrated that the profile was established early during development of the tumor and then maintained during subsequent cell culture. The main difference revealed was that some H3K4me3 was detected at OriLyt in the tumor and PBL of an RB1B-infected line P chicken (Fig. 4C). There are two possible explanations for this. Firstly, and perhaps most likely, reactivating lytic virus in a subpopulation of cells within the tumor could account for the presence of active marks around OriLyt. Alternatively, the presence of both H3K27me3 and H3K4me3 could indicate the presence of a bivalent or “poised” chromatin structure (4). Recent studies of the KSHV epigenome have shown the presence of such bivalent chromatin on the RTA promoter (23, 51). Although data from the primary tumor suggested the presence of bivalent chromatin at the MDV OriLyt, the epigenetic profiles of the T-cell lines showed no obvious bivalent marks. Moreover, H3K9me3 was present at OriLyt in both the tumor and the cell lines, and this mark suggests a more stable heterochromatin-associated form of silencing (3).
Since H3K27me3 and H3K9me3 can lead to the recruitment of DNA methyltransferases (DNMTs) and to accumulation of CpG DNA methylation (45), we next investigated the amount of CpG methylation across the repeat region. We first investigated the two cell lines that were used for histone analysis, and these showed extensive methylation across the repeat region. The conspicuous absence of CpG methylation at several of the active promoters in the region suggests that the presence of transcription factors, for instance, Meq–c-Jun or factors that they recruit, might block the access of DNMTs. The exclusion of methylation and transcription factor binding correlates with the presence of active chromatin and RNA Pol II occupancy at these regions. The same technique was used to investigate the CpG DNA methylation of MDV in a primary tumor and in the PBL of RB1B-infected line P birds. These each demonstrated a strikingly similar pattern of CpG DNA methylation (Fig. 6), suggesting that the mechanism of DNMT recruitment follows the same nonrandom pattern. In comparison to the profile of the cell lines, it is quite clear that methylation was restricted to intragenic regions of the genome and still absent from the active promoters. The methylated regions of MDV found in the tumor and PBL were a subset of those in the cell lines, suggesting that the spread of CpG methylation occurs during outgrowth of cell lines. To determine whether methylation appeared preferentially within the repeat region or was distributed equally throughout the genome, we examined the UL30 gene, which would normally be repressed during latency. Surprisingly, the intragenic region of UL30 showed no methylation in the primary tumor or PBL, although ICP4 was clearly methylated (Fig. 7). This was in contrast to the cell lines, which carried methylated MDV DNA in both regions, suggesting that methylation occurs preferentially at ICP4 in T cells in vivo. It should be noted that the methylated DNA in the ICP4 gene is probably adjacent to the site of integration of MDV DNA into the chicken genome, at a telomeric locus (24). MDV genome methylation might be linked to integration, suggesting an additional role for integration in the development of a latent state. The repressive histone marks and DNA methylation detected were consistent with the demonstration that MDV lytic activity was initiated in 265L cells by chromatin-modifying drugs (Fig. 8).
The best candidate for an MDV protein involved in histone modification is Meq, since it is one of the few proteins expressed during latency, has a nuclear distribution, binds DNA, and regulates transcription (9, 49, 50). The ability to repress transcription as a homodimer probably relates to its ability to interact with the cellular corepressor CtBP (8), and this provides a potential link to the polycomb complexes responsible for H3K27me3 (46). In addition, complexes have been identified that contain CtBP and the histone methyltransferase G9A that is responsible for H3K9me3 and heterochromatin formation (47). Unfortunately, with the anti-CtBP antibodies available, we have been unable to perform ChIP assays with chicken CtBP. Within the repeat region, Meq–c-Jun heterodimers maintain active transcription during latency, probably through their ability to recruit histone acetyltransferases such as p300/CBP to AP-1 sites (22). Where Meq–c-Jun heterodimers were detected (Fig. 9), there was little or no DNA methylation detected, consistent with transcription factor binding and CpG methylation being mutually exclusive.
Because of the presence of two adjacent domains, one active and one repressed, we investigated the binding of the chromatin boundary element factor CTCF around OriLyt. ChIP assays revealed a distinct CTCF peak near OriLyt that coincided with binding of the cohesion factors Rad21, SMC1, and SMC3. This suggests that the presence of a higher-order chromatin structure within the repeat region may play an important role in maintaining the active chromatin. It is likely that there are additional CTCF-binding sites within the MDV genome and that these may facilitate chromosome looping (33, 39, 48). Further studies to identify these should lead to a much greater understanding of the maintenance of the MDV genome in its latent state.
In summary, we have shown that the MDV genome is chromatinized and have described the types of epigenetic mark associated with the establishment and maintenance of MDV latency. Independent infections of T cells appear to lead to strikingly similar patterns of histone modifications, DNA methylation, and transcription across the latency-associated repeats. It is possible that the Meq protein plays a role in the establishment of this latency profile, through its ability to interact with CtBP and c-Jun and to bind DNA. Furthermore, since Meq probably binds to chromatin in the host genome during MDV latency, changes to the host epigenome may contribute to MDV-mediated T-cell transformation and lymphomagenesis, as has been reported for other tumor viruses (e.g., see reference 37).
ACKNOWLEDGMENTS
We thank Lorraine P. Smith and Lydia Kgosana (IAH, Compton, United Kingdom) for help in obtaining the tumor and peripheral blood leukocyte material used in the analysis.
This work was supported by the Biotechnology and Biological Sciences Research Council, United Kingdom (grant BB/G001197/1).
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Anderson AS, Francesconi A, Morgan RW. 1992. Complete nucleotide sequence of the Marek's disease virus ICP4 gene. Virology 189: 657–667 [DOI] [PubMed] [Google Scholar]
- 2. Anobile JM, et al. 2006. Nuclear localization and dynamic properties of the Marek's disease virus oncogene products Meq and Meq/vIL8. J. Virol. 80: 1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beisel C, Paro R. 2011. Silencing chromatin: comparing modes and mechanisms. Nat. Rev. Genet. 12: 123. [DOI] [PubMed] [Google Scholar]
- 4. Bernstein BE, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- 5. Biggs PM. 1997. The Leeuwenhoek Lecture, 1997. Marek's disease herpesvirus: oncogenesis and prevention. Philos. Trans. R. Soc. Lond. B Biol. Sci. 352: 1951–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bloom DC, Giordani NV, Kwiatkowski DL. 2010. Epigenetic regulation of latent HSV-1 gene expression. Biochim. Biophys. Acta 1799: 246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brookes E, Pombo A. 2009. Modifications of RNA polymerase II are pivotal in regulating gene expression states. EMBO Rep. 10: 1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown AC, et al. 2006. Interaction of MEQ protein and C-terminal-binding protein is critical for induction of lymphomas by Marek's disease virus. Proc. Natl. Acad. Sci. U. S. A. 103: 1687–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown AC, et al. 2009. Homodimerization of the Meq viral oncoprotein is necessary for induction of T-cell lymphoma by Marek's disease virus. J. Virol. 83: 11142–11151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burnside J, et al. 2006. Marek's disease virus encodes microRNAs that map to meq and the latency-associated transcript. J. Virol. 80: 8778–8786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cantello JL, Anderson AS, Morgan RW. 1994. Identification of latency-associated transcripts that map antisense to the ICP4 homolog gene of Marek's disease virus. J. Virol. 68: 6280–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cantello JL, Parcells MS, Anderson AS, Morgan RW. 1997. Marek's disease virus latency-associated transcripts belong to a family of spliced RNAs that are antisense to the ICP4 homolog gene. J. Virol. 71: 1353–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen XB, Sondermeijer PJ, Velicer LF. 1992. Identification of a unique Marek's disease virus gene which encodes a 38-kilodalton phosphoprotein and is expressed in both lytically infected cells and latently infected lymphoblastoid tumor cells. J. Virol. 66: 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cui X, Lee LF, Reed WM, Kung H-J, Reddy SM. 2004. Marek's disease virus-encoded vIL-8 gene is involved in early cytolytic infection but dispensable for establishment of latency. J. Virol. 78: 4753–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delecluse HJ, Hammerschmidt W. 1993. Status of Marek's disease virus in established lymphoma cell lines: herpesvirus integration is common. J. Virol. 67: 82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Delecluse HJ, Schüller S, Hammerschmidt W. 1993. Latent Marek's disease virus can be activated from its chromosomally integrated state in herpesvirus-transformed lymphoma cells. EMBO J. 12: 3277–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ding J, Cui Z, Lee LF. 2007. Marek's disease virus unique genes pp38 and pp24 are essential for transactivating the bi-directional promoters for the 1.8 kb mRNA transcripts. Virus Genes 35: 643–650 [DOI] [PubMed] [Google Scholar]
- 18. Epstein MA. 2001. Historical background. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356: 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fragnet L, Blasco MA, Klapper W, Rasschaert D. 2003. The RNA subunit of telomerase is encoded by Marek's disease virus. J. Virol. 77: 5985–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gilley D, Blackburn EH. 1999. The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc. Natl. Acad. Sci. U. S. A. 96: 6621–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gimeno IM, et al. 2005. The pp38 gene of Marek's disease virus (MDV) is necessary for cytolytic infection of B cells and maintenance of the transformed state but not for cytolytic infection of the feather follicle epithelium and horizontal spread of MDV. J. Virol. 79: 4545–4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goodman RH, Smolik S. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14: 1553–1577 [PubMed] [Google Scholar]
- 23. Günther T, Grundhoff A. 2010. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 6: e1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaufer BB, Jarosinski KW, Osterrieder N. 2011. Herpesvirus telomeric repeats facilitate genomic integration into host telomeres and mobilization of viral DNA during reactivation. J. Exp. Med. 208: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Levy AM, et al. 2003. Characterization of the chromosomal binding sites and dimerization partners of the viral oncoprotein Meq in Marek's disease virus-transformed T cells. J. Virol. 77: 12841–12851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li DS, Pastorek J, Zelník V, Smith GD, Ross LJ. 1994. Identification of novel transcripts complementary to the Marek's disease virus homologue of the ICP4 gene of herpes simplex virus. J. Gen. Virol. 75: 1713–1722 [DOI] [PubMed] [Google Scholar]
- 27. Liu JL, Kung HJ. 2000. Marek's disease herpesvirus transforming protein MEQ: a c-Jun analogue with an alternative life style. Virus Genes 21: 51–64 [PubMed] [Google Scholar]
- 28. Liu JL, et al. 1999. MEQ and V-IL8: cellular genes in disguise? Acta Virol. 43: 94–101 [PubMed] [Google Scholar]
- 29. Liu JL, Ye Y, Lee LF, Kung HJ. 1998. Transforming potential of the herpesvirus oncoprotein MEQ: morphological transformation, serum-independent growth, and inhibition of apoptosis. J. Virol. 72: 388–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lupiani B, et al. 2004. Marek's disease virus-encoded Meq gene is involved in transformation of lymphocytes but is dispensable for replication. Proc. Natl. Acad. Sci. U. S. A. 101: 11815–11820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nair V. 2004. Successful control of Marek's disease by vaccination. Dev. Biol. (Basel) 119: 147–154 [PubMed] [Google Scholar]
- 32. Nazerian K, et al. 1977. A nonproducer T lymphoblastoid cell line from Marek's disease transplantable tumor (JMV). Avian Dis. 21: 69–76 [PubMed] [Google Scholar]
- 33. Ohlsson R, Lobanenkov V, Klenova E. 2010. Does CTCF mediate between nuclear organization and gene expression? Bioessays 32: 37–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Parcells MS, Arumugaswami V, Prigge JT, Pandya K, Dienglewicz RL. 2003. Marek's disease virus reactivation from latency: changes in gene expression at the origin of replication. Poult. Sci. 82: 893–898 [DOI] [PubMed] [Google Scholar]
- 35. Parcells MS, et al. 2001. Marek's disease virus (MDV) encodes an interleukin-8 homolog (vIL-8): characterization of the vIL-8 protein and a vIL-8 deletion mutant MDV. J. Virol. 75: 5159–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parelho V, et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- 37. Paschos K, Allday MJ. 2010. Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends Microbiol. 18: 439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paschos K, et al. 2009. Epstein-Barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim. PLoS Pathog. 5: e1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Phillips JE, Corces VG. 2009. CTCF: master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pratt WD, Cantello J, Morgan RW, Schat KA. 1994. Enhanced expression of the Marek's disease virus-specific phosphoproteins after stable transfection of MSB-1 cells with the Marek's disease virus homologue of ICP4. Virology 201: 132–136 [DOI] [PubMed] [Google Scholar]
- 41. Qian Z, Brunovskis P, Lee L, Vogt PK, Kung HJ. 1996. Novel DNA binding specificities of a putative herpesvirus bZIP oncoprotein. J. Virol. 70: 7161–7170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qian Z, Brunovskis P, Rauscher F, Lee L, Kung HJ. 1995. Transactivation activity of Meq, a Marek's disease herpesvirus bZIP protein persistently expressed in latently infected transformed T cells. J. Virol. 69: 4037–4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reddy SM, et al. 2002. Rescue of a pathogenic Marek's disease virus with overlapping cosmid DNAs: use of a pp38 mutant to validate the technology for the study of gene function. Proc. Natl. Acad. Sci. U. S. A. 99: 7054–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ross N, et al. 1997. Marek's disease virus EcoRI-Q gene (meq) and a small RNA antisense to ICP4 are abundantly expressed in CD4+ cells and cells carrying a novel lymphoid marker, AV37, in Marek's disease lymphomas. J. Gen. Virol. 78: 2191–2198 [DOI] [PubMed] [Google Scholar]
- 45. Schlesinger Y, et al. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 39: 232–236 [DOI] [PubMed] [Google Scholar]
- 46. Sewalt RG, Gunster MJ, van der Vlag J, Satijn DP, Otte AP. 1999. C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol. Cell. Biol. 19: 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shi Y, et al. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422: 735–738 [DOI] [PubMed] [Google Scholar]
- 48. Splinter E, et al. 2006. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev. 20: 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Suchodolski PF, et al. 2009. Homodimerization of Marek's disease virus-encoded Meq protein is not sufficient for transformation of lymphocytes in chickens. J. Virol. 83: 859–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Suchodolski PF, et al. 2010. Both homo and heterodimers of Marek's disease virus encoded Meq protein contribute to transformation of lymphocytes in chickens. Virology 399: 312–321 [DOI] [PubMed] [Google Scholar]
- 51. Toth Z, et al. 2010. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog. 6: e1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Touitou R, et al. 2001. Physical and functional interactions between the corepressor CtBP and the Epstein-Barr virus nuclear antigen EBNA3C. J. Virol. 75: 7749–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Trapp S, et al. 2006. A virus-encoded telomerase RNA promotes malignant T cell lymphomagenesis. J. Exp. Med. 203: 1307–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Venugopal K. 2000. Marek's disease: an update on oncogenic mechanisms and control. Res. Vet. Sci. 69: 17–23 [DOI] [PubMed] [Google Scholar]
- 55. Xing Z, Xie Q, Morgan RW, Schat KA. 1999. A monoclonal antibody to ICP4 of MDV recognizing ICP4 of serotype 1 and 3 MDV strains. Acta Virol. 43: 113–120 [PubMed] [Google Scholar]
- 56. Yao Y, et al. 2007. Marek's disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation with those encoded by MDV-1. J. Virol. 81: 7164–7170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhao Y, et al. 2011. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek's disease lymphomas. PLoS Pathog. 7: e1001305. [DOI] [PMC free article] [PubMed] [Google Scholar]