Abstract
The species B human adenoviruses (HAdVs) infect cells upon attaching to CD46 or desmoglein 2 (DSG-2) by one or several of their 12 fiber knob trimers (FKs). To test whether DSG-2 and CD46 simultaneously serve as virus receptors for adenovirus type 3 (Ad3), we performed individual and combined CD46/DSG-2 loss-of-function studies in human lung A549 and 16HBE14o cells. Our results suggest that in these cells, DSG-2 functions as a major attachment receptor for Ad3, whereas CD46 exerts a minor contribution to virus attachment and uptake in the range of ∼10%. However, in other cells the role of CD46 may be more pronounced depending on, e.g., the expression levels of the receptors. To test if avidity allows Ad3/7 to use CD46 as a receptor, we performed gain-of-function studies. The cell surface levels of ectopically expressed CD46 in CHO or human M010119 melanoma cells lacking DSG-2 positively correlated with Ad3/7 infections, while Ad11/35 infections depended on CD46 but less on CD46 levels. Antibody-cross-linked soluble CD46 blocked Ad3/7/11/35 infections, while soluble CD46 alone blocked Ad11/35 but not Ad3/7. Soluble Ad3/7-FKs poorly inhibited Ad3/7 infection of CHO-CD46 cells, illustrating that Ad3/7-FKs bind with low affinity to CD46. This was confirmed by Biacore studies. Ad3/7-FK binding to immobilized CD46 at low density was not detected, unlike that of Ad11/35-FK. At higher CD46 densities, however, Ad3/7-FK bound to CD46 with only 15-fold-higher dissociation constants than those of Ad11/35-FK. These data show that an avidity mechanism for Ad3/7 binding to CD46 leads to infection of CD46-positive cells.
INTRODUCTION
Human Adenoviridae comprise 55 types, classified into seven species, A to G (http://www.vmri.hu/∼harrach/AdVtaxlong.htm), based on genome sequence comparison, hemagglutination, and additional features. The B1 viruses adenovirus type 3 (Ad3), Ad7, Ad16, Ad21, and Ad50 (Ad3/7/16/21/50) predominantly infect the upper respiratory tract, whereas the B2 viruses Ad11/14/34/35 are associated with kidney and urinary tract infections with fatal outcomes in immunocompromised patients (30, 54, 68). Recent epidemiological reports described the reemergence of several of these virus types associated with outbreaks of respiratory disease (7, 32, 39, 77). The tropism of species B viruses is broader than that of the C species and includes cancer cells, dendritic cells, and hematopoietic stem cells. This feature makes the B species interesting vectors for gene therapy and vaccination approaches (52).
Ads attach to their host cells by binding of the trimeric fiber protein to a cellular surface receptor. The fiber protein consists of a tail for anchorage to the penton base, a shaft of variable length, and a globular fiber knob (FK). The latter is responsible for the binding of the virus particle (vp) to a primary attachment receptor (43). Species B Ads bind a different cell surface receptor(s) than do most of the other species members (76). Two receptors have been identified, CD46 for Ad11 (57), Ad35 (15), Ad3 (60), and species D Ad37 and Ad49 (31, 74), and desmoglein 2 (DSG-2) for Ad3/7/11/14 (69, 70). Whether CD46 functions as an attachment receptor for all species B types has been controversial. Virus competition, CD46 antibody blocking, and small interfering RNA (siRNA) knockdown of CD46 experiments suggested that more than one receptor exists for species B Ads (15, 19, 37, 56, 57, 60, 67). It was suggested that all species B Ads except Ad3/7 would utilize CD46 and that all serotypes, including Ad3/7, would bind to a second, common receptor (sBAR) (37, 56). Another group proposed an alternative classification, where group I members (Ad16/21/35/50) would almost exclusively use CD46 while group II members (Ad3/7/14) would use not CD46 but DSG-2 and the only member of group III (Ad11p) would be able to use both receptors (67, 70). Both classifications contrast, however, with findings by others, who reported functional utilization of CD46 by Ad3 and Ad7 in rodent cells ectopically expressing CD46 (13, 14, 20, 40, 60, 61).
Analysis of monovalent interactions of different species B FKs with CD46 short consensus repeat (SCR) I-II revealed a broad range of affinities, with similar dissociation constants (Kd) for Ad11- and Ad35-FK (Ad11/35-FK) in the range of 5 to 19 nM but strongly increased Kd values of 284 nM for Ad21-FK and 437 nM for Ad16-FK and an approximately 2,000-fold-reduced affinity of both Ad7-FK and Ad14-FK for CD46 SCR I-II, compared to Ad11-FK (10, 47, 48). The crystal structures of FKs for Ad3 (11), Ad35 (46, 71), Ad16 (47), and Ad7/14 (48) have revealed a generally conserved overall fold and trimeric organization. Interestingly, the different FKs have low sequence identity, especially at the surface loops, which mediate binding to CD46, as indicated by cocrystal structures of CD46 SCR I-II with Ad11-FK (49) or Ad21-FK (10). These crystal structures also suggested interactions of the trimeric fiber molecule with three CD46 molecules, albeit involving substantial differences in the number and types of contacts. The binding surface on CD46 SCR I-II for Ad11-FK comprises a large continuous area of 1,681 Å2, with three main contact points composed of fiber knob DG, HI, and IJ loop residues. A more recent cocrystal structure of Ad11-FK in complex with an extended CD46 SCR I-IV confirmed the involvement of the DG and HI loops but not of the IJ loop (50). Together, this wealth of structural evidence indicates that there are no central binding motifs among species B Ads, which would explain the macroscopic observations that there is a wide range of affinities for CD46 between the different FKs (10, 47, 71).
Here we tested two hypotheses: first, that DSG-2 and CD46 simultaneously serve as receptors for Ad3, and second, that avidity effects allow the low-affinity CD46 binders Ad3/7-FK to attach to CD46 and thereby allow these viruses to use CD46 as an entry receptor. We performed loss-of-function studies in human A549 and 16HBE14o lung cells and showed that DSG-2 functions as a major attachment receptor for Ad3, whereas CD46 exerts a minor contribution in the range of ∼10% to virus attachment and uptake. Gain-of-function experiments in CD46-negative CHO or CD46-low human melanoma cells both negative for DSG-2 demonstrated that Ad3/7 infections increased with increasing levels of ectopic CD46. We further showed that multimerized soluble CD46 blocked Ad3/7 infections and that Ad3/7-FK reduced Ad3/7 infections in A549 cells expressing high levels of CD46. Finally, biosensor measurements demonstrated that affinities of Ad3/7-FKs for immobilized CD46 increased at high receptor densities. These data argue that CD46 is a receptor for all species B adenoviruses if present at sufficient levels and may function together with or separately from DSG-2.
MATERIALS AND METHODS
Viruses and cells.
Ad3CMV-eGFP, Ad7CMV-eGFP, Ad11CMV-eGFP, and Ad35CMV-eGPF containing the cytomegalovirus-enhanced green fluorescent protein (CMV-eGFP) expression cassette in the deleted E1 region were prepared at concentrations of 7.1 × 1011, 1.6 × 1011, 6.6 × 1011, and 1.3 × 1012 virus particles (vp)/ml and have been described previously (13, 14). Ad3 (prototype strain GB), Ad7 (prototype strain Gomen), Ad11p, and Ad35 (Holden strain) were radiolabeled as described previously (14). Specific activities were in the range of 2.6 × 10−5 to 4.9 × 10−5 cpm/vp. Likewise, Ad2, Ad3, and Ad35 were labeled with atto488 (Atto-tec; Germany) in a manner similar to that described previously (65).
Chinese hamster cell lines CHO-K1 and CHO-15B6 (containing a mutation in the N-acetylglucosamininyltransferase 1), the human 293T and 911 embryonic kidney cell lines, and A549 lung carcinoma cells were grown in Dulbecco's modified Eagle's medium (DMEM) plus 8% fetal bovine serum (FBS). The human 16HBE14o bronchial epithelial cells (17) and the M010119 primary human melanoma cell cultures (55) were grown in RPMI 1640 plus 8% FBS as described earlier. CHO-CD46 cell lines stably expressing the BC1 splice isoform were generated as described for the BHK-CD46 cells (60). For stable transfection of CHO-15B6 and M010119 cells with the N-terminal eGFP-tagged CD46-encoding cDNA (eGFP-CD46), a PCR-amplified CD46 sequence was cloned into pcDNA3.1-CARSP-eGFP-CAR (5), replacing the mature coxsackie virus B and Ad receptor (CAR) cDNA. In the resulting construct, the 19 amino acid residues of the human CAR signal peptide sequence were followed by two spacer residues, 239 residues of eGFP, 10 additional spacer residues, and 344 residues of the mature CD46 BC1 isoform. To avoid eGFP-induced dimerization of the tagged CD46, an A206K mutant of eGFP was utilized (75). Following selection in DMEM plus 8% FBS and 0.8 mg/ml G418, individual clones were picked and screened, giving rise to the limiting dilution clone CHO-eGFP-CD46#33.5. To generate human melanoma M010119 and lung carcinoma A549 cells expressing eGFP-CD46, a lentiviral expression system was used. For this, the eGFP-CD46 cDNA was subcloned into the lentiviral pBlasti vector (27), and lentiviral vectors were generated by transient transfection in 293T cells. M010119 and A549 cells transduced with lentiviral vector pBlasti-eGFP-CD46 were selected in blasticidin-containing medium at 4 μg/ml. Resulting bulk cultures M010119-eGFP-CD46#8 and A549-eGFP-CD46#2 revealed homogenous CD46 expression.
Immune reagents.
Cytofluorometric analysis, CD46-specific antibodies, and secondary fluorochrome conjugates have been described previously (14, 60). The hybridoma cell line secreting the MCI20.6 anti-CD46 antibody was a generous gift from Denis Gerlier (42). Monoclonal 8E5- and 6D8-antidesmoglein 2 were purchased from Santa Cruz Inc., and goat anti-human IgG-Fc was from Bethyl Laboratories Inc.
Virus binding and transduction assays.
Binding experiments including radiolabeled Ads were performed as described previously (14). For binding experiments using atto488-labeled viruses, the same procedure was used, except that cells were analyzed by cytofluorometric means. For eGFP expression analysis, triplicates of 105 cells were seeded in 12-well plates. After incubation for 3 h at 37°C and 5% CO2, the cells were infected with recombinant eGFP-expressing Ads at virus concentrations of 10, 100, or 1,000 vp/cell. Medium was replaced 5 h postinfection (p.i.), and cells were analyzed 2 days p.i. by flow cytometric analysis (Cytomics FC 500; Beckman Coulter).
For FK competition experiments, cells were seeded in triplicate 12-well plates and were allowed to adhere as described above. After 3 h, the medium was removed and the cells were washed with cold phosphate-buffered saline (PBS). For blocking with FK proteins, serial 5-fold dilutions of the different FKs were prepared in PBS, resulting in concentrations of 5,000, 1,000, 200, 40, and 8 ng/ml. Five hundred microliters of FK dilutions was added to the cells and incubated for 1 h on ice under constant shaking. Subsequently, eGFP-encoding Ad vectors were added and incubated for 1 h. The amount of virus added was optimized in preceding experiments and was adjusted such that for all five eGFP-expressing vectors, similar reporter expression levels in the range of 100 to 500 were obtained by fluorescence-activated cell sorting (FACS) measurement. Thus, for A549 cell transduction, Ad3-, Ad5-, Ad7-, Ad11-, and Ad35-eGFP vectors were used at 14,800, 2,825, 8,200, 1,314, and 2,540 vp/cell, respectively. For CHO-CD46#2, the virus input was adjusted to 29,600, 2,825, 8,200, 657, and 1,088 vp/cell, respectively. The cells were washed twice with cold PBS and were returned to standard cell culture conditions. Analysis of cells was performed 2 days p.i., as described above. For competition experiments, including recombinant CD46ex-huFc, FK proteins were replaced with the indicated concentrations of CD46ex-huFc or CARex-huFc. In experiments that included cross-linking of CD46ex-huFc and CARex-huFc, the adapter proteins were preincubated on ice with goat anti-human IgG antibody for 30 min before being added to the cells. Specific siRNAs for CD46 (15) and DSG-2 (70) were synthesized by Microsynth (Switzerland), and scrambled siRNA control was obtained from Qiagen. siRNA transfections were performed with Lipofectamine 2000 (Invitrogen) for A549 cells and Interferin (Polyplus) for 16HBE14o cells. Knockdown cells were infected with recombinant eGFP-expressing Ads at virus concentrations of 500 vp/cell. Statistical evaluation was performed using Student's t test.
Generation of recombinant soluble Ad receptor and fiber knob proteins.
Production of the soluble CD46ex-huFc and CARex-huFc has been described earlier (12, 60). CD46ex-huFc comprises 295 amino acids of the mature extracellular domain, including all SCR I-IV domains and the STP of BC1, plus the short region of unknown function, fused to the 232 amino acids of the human IgG1-Fc domain, including hinge, CH2, and CH3 regions. Production of CD46-SCR I-II consisting of 145 amino acids, including an N-terminal His tag and spacer, has been described in reference 14. CARex-huFc protein comprises the extracellular domain of CAR fused to the human IgG1-Fc domain. The five recombinant FKs derived from Ad3, Ad5, Ad7, Ad11, and Ad35 were produced using the Bac-to-Bac baculovirus expression system (pFastBac; Stratagene). All FKs contained an N-terminal 6×His tag used for single-step nickel-nitrilotriacetic acid (Ni-NTA)-agarose affinity chromatography, followed by the recognition site for tobacco etch virus (TEV) protease. FK comprised the fiber sequence residues 114 to 319 for Ad3 (NCBI GenBank identification [GI]: 78059423), 384 to 581 for Ad5 (GI: 56160559), 126 to 325 for Ad7 (GI: 51173336), 126 to 325 for Ad11 (GI: 56160807), and 126 to 323 for Ad35 (GI: 56160945). Calculated molecular masses of the FK monomer/trimers were 26.28/78.85, 24.98/74.95, 25.63/76.89, 25.90/77.71, and 25.50/76.50 kDa for FK of Ad3, Ad5, Ad7, Ad11, and Ad35, respectively.
SPR analysis for kinetics/affinity between CD46 and Ad-FKs.
Surface plasmon resonance (SPR) experiments were performed on a Biacore T100 system (GE Healthcare) at 25°C equipped with CM5 sensor chips. All reagents, including the amine-coupling kit, HBS-P+, and CM5 chips, were purchased from Biacore (GE Healthcare). Buffer HBS-P+ (10 mM HEPES, pH 7.4, 150 mM NaCl, 0.05% [vol/vol] surfactant P20) was used as running buffer for the entire measurement. Each CM5 chip contained four flow cells, designated Fc1 to Fc4. Fc2 and Fc4 were used to immobilize the CD46ex-huFc ligand, while Fc1 and Fc3 were used to immobilize the CARex-huFc ligand. The immobilization procedure included three steps: surface activation by using a 1:1 mix of 0.4 M 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide in water (EDC) and 0.1 M N-hydroxysuccinimide (NHS) in water for 420 s with a flow rate of 5 μl/min, immobilization of 0.12 μM ligands in 10 mM sodium acetate (pH 4.0) buffer, and deactivation of excess reactive groups by using 1 M ethanolamine-HCl (pH 8.5) for 420 s with a flow rate of 5 μl/min. Following completion of immobilization, the sensor chip was conditioned three times with 50 mM NaOH to stabilize the surface. For initial binding assays, Ad3-, Ad11-, and Ad35-FK analytes were injected at 18.75 and 150 nM and Ad7-FK analyte was injected at 11.07 and 88.59 nM in HBS-P+ buffer at either 30 or 50 μl/min using a contact time of 240 or 280 s. For kinetic studies, Ad3-, Ad7-, Ad11-, and Ad35-FK analytes were injected at 0.27, 0.82, 2.47, 7.41, 22.22, and 66.67 nM in HBS-P+ buffer at either 30 or 50 μl/min using a contact time of 300 or 360 s and a dissociation time of 3,600 s. After each binding cycle, the CM5 surface chip was regenerated by injection of 3 M MgCl2 for 30 to 45 s, followed by NaOH in a range of 4 to 50 mM for 20 s. All analyte concentrations were repeated twice, and the obtained sensorgrams were corrected by subtracting the data from the reference flow cell and from blank buffer injections (double referencing). For details of the kinetic and affinity analysis as well as the fitting procedure, see the supplemental methods.
RESULTS
CD46 and desmoglein 2 simultaneously serve as Ad3 receptor.
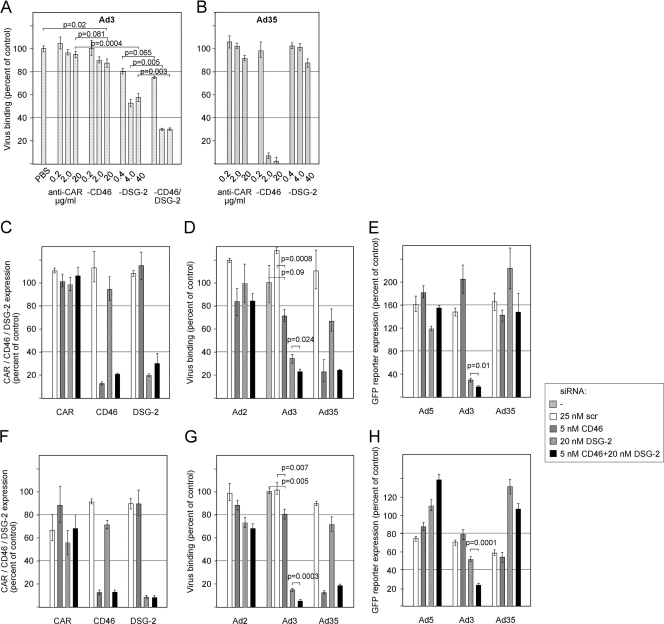
To test whether CD46 and DSG-2 simultaneously or additively serve as attachment receptors, we performed loss-of-function studies in human lung carcinoma A549 cells. These cells stained positively for CD46 (see Fig. S1A in the supplemental material) and DSG-2 (see Fig. S1B) expression, revealing arbitrary mean fluorescence intensities (MFI) of 74 and 29, respectively. We tested antibodies against CD46, DSG-2, and a mix of both and determined their effects on Ad3 and Ad35 binding. For this, cells were incubated with 1,000 vp of atto488-labeled wild-type Ad3/35 followed by determination of cell-associated viruses by cytofluorometric analysis. Compared to PBS and anti-CAR antibody controls, preincubation of A549 cells with the known CD46-blocking antibody MEM258 (14) gave 13 and 8% reduction of Ad3 binding at the highest concentration of 20 μg/ml, respectively (Fig. 1A). This is in agreement with earlier results reporting inhibition of Ad3 binding to human cells: for example, a 40% reduction of binding to human K562 cells (60) or up to 48% inhibition for human hepatoblastoma HuH-7 cells (67). Preincubation with increasing concentrations of DSG-2 antibodies on the other side resulted in 47 and 42% reduction of Ad3 binding at 4 and 40 μg/ml, respectively, which is comparable to the approximately 60% maximal inhibition reported by Wang et al. using twice the concentration of the same mix of antibodies used here (70). Interestingly, when CD46 and DSG-2-specific antibodies were combined, we noticed a significant additive inhibition resulting in 70% inhibition of Ad3 binding. In comparison, the high concentrations of CD46 antibody reduced Ad35 binding by 93 and 98%, but the DSG-2 antibody mix had no significant effects (Fig. 1B). These results are consistent with the notion that Ad35 uses CD46 but not DSG-2 as an attachment receptor (13, 67).
Fig 1.
Loss-of-function studies for individual contribution of CD46 and DSG-2 to Ad3 infection. (A and B) Inhibition of Ad3 (A) and Ad35 (B) binding to A549 cells by anti-CD46 and DSG-2 antibodies. A549 cells were incubated with the indicated concentrations of CD46 (MEM258), DSG-2 (6D8 and 8E5), a mix of the two types of antibodies, control CAR antibodies, or PBS, followed by the addition of atto488-labeled Ad3 or Ad35 at 4°C. Virus binding was assessed by cytofluorometric analysis. The data were normalized to the amounts of virus bound when using PBS. Mean values and standard deviations of triplicates from one representative experiment are shown. (C to H) Effect of siRNA-mediated downregulation on virus infection in A549 (C to E) and 16HBE14o (F to H) cells. (C and F) Cells were transfected with the indicated siRNAs, resulting in specific downregulation of CD46 and DSG-2 but not of unrelated CAR. Shown is one representative analysis. (D and G) Binding of atto488-labeled Ad2 (control), Ad3, and Ad35 to siRNA-transfected cells was assessed as described above. (E and H) Transduction of siRNA-transfected cells with eGFP-expressing control Ad5, Ad3, and Ad35 vectors at 500 vp/cell. eGFP expression values were analyzed 2 days p.i. by flow cytometry and are expressed as MFI.
We next performed siRNA-mediated knockdown of CD46, DSG-2, and the two receptors together and analyzed these cells for virus binding and transduction. Initial tests for optimizing siRNA concentrations confirmed that the best CD46 downregulation was obtained using a single CD46-specific siRNA at 5 nM, whereas a pool of four DSG-2-specific siRNAs each at 5 nM was optimal for DSG-2 protein downregulation when measured by cytofluorometric analyses (15, 70). In five independent experiments, siRNA-mediated reduction of CD46 was in the range of 74 to 88% with a mean of 82%, using the CD46-specific siRNA alone, and 67 to 86% with a mean of 77%, using the CD46-specific siRNA in combination with DSG-2-specific siRNAs (Fig. 1C shows a representative experiment). Similarly, siRNA-mediated reduction of DSG-2 was in the range of 68 to 80% with a mean of 73%, using the DSG-2 siRNAs alone, and 63 to 82% with a mean of 71%, using the DSG-2 siRNAs in combination with the CD46 siRNA. When binding of atto488-labeled Ad3 to these cells was analyzed, we noticed that in CD46 siRNA-only cells binding was reduced by 28% in the experiment shown in Fig. 1D, whereas binding of Ad3 to DSG-2 siRNA-only cells was reduced by 66%. Similarly to the antibody-blocking experiments, combination of the two siRNAs additively reduced Ad3 binding by 77%. Of note, the additive Ad3 binding reduction was not paralleled by a reduction of the DSG-2 expression level in the siRNA combination cells, indicating specificity for CD46. The range of reduction varied from 3 to 23%, with a mean of 11%. As for the antibody inhibition experiment, Ad35 attachment was mainly affected by CD46 but not by DSG-2 interference, resulting in 76 and 77% reductions in binding for CD46-only siRNA and CD46 plus DSG-2 siRNAs, respectively. Attachment of the CAR-binding Ad2 was not affected in these cells, demonstrating the specificity of the observed virus binding reduction for Ad3/35.
We then used the four different siRNA cells to analyze transduction by Ad5-, Ad3-, and Ad35-eGFP reporter viruses. CD46 interference gave no decrease in eGFP expression for Ad3/5 (Fig. 1E). However, Ad3-eGFP-mediated expression was reduced by 71% in DSG-2 siRNA-treated cells and by 82% in DSG-2 plus CD46 siRNA combination cells, revealing a small additive effect of 11% (Fig. 1E). The additive reduction in Ad3 infection by siRNA combination cells was reproducibly significant and varied from 11 to 17%, with a mean of 13% in 4 of 5 experiments. It should be noted here that CD46 downregulation had a very small effect on the transduction by Ad35-eGFP. We therefore repeated this experiment using a chimeric Ad5 containing the fiber knob and shaft from Ad35 (Ad5-eGFP-FK35) and found a 60% reduction of transgene expression upon CD46 siRNA silencing (data not shown). This was similar to data from others using a similar fiber chimeric vector (15). As suggested earlier, a possible explanation for these observations includes the idea that fiber-knob-swapped vectors have altered intracellular trafficking or stability (63).
Using the same approach, we also tested human 16HBE14o bronchial epithelial cells, which are a commonly used model for respiratory epithelial cell infections (17, 35). These cells revealed about 2-fold-lower CD46 (see Fig. S1C in the supplemental material) and 3- to 4-fold-lower DSG-2 (see Fig. S1D) expression, compared to A549 cells. siRNA-mediated reduction of CD46 and DSG-2 was on average 89 and 91%, respectively, when using the single-specific siRNA alone or in combination (Fig. 1F). Binding of atto488-labeled Ad3 was reduced by 20 and 85% in CD46 and DSG-2 siRNA-only cells, respectively, and the combination of the two siRNAs additively reduced Ad3 binding by 95% (Fig. 1G). When using the four different siRNA cells to analyze transduction by eGFP reporter viruses, Ad3-eGFP-mediated expression was reduced by 48% in DSG-2 siRNA-treated cells and by 76% in DSG-2 plus CD46 siRNA combination cells, revealing an additive effect of 28% (Fig. 1H).
Taken together, our results suggest that in A549 and 16HBE14o cells expressing both DSG-2 and CD46, DSG-2 functions as a major attachment receptor for Ad3, whereas CD46 exerts an accessory role for binding and uptake. In other human cells, however, the CD46 contribution may be more pronounced depending, e.g., on the expression levels of the receptors.
Increasing ectopic expression of CD46 leads to enhanced transduction of Ad3/7/11/35 in rodent and human cells.
Avidity effects allow viruses to infect susceptible cells despite somewhat low affinity for cell surface receptors (22, 73). We therefore tested if avidity effects occur during interaction of the Ad3/7 serotypes in CD46-gain-of-function experiments. We and others had shown earlier that rodent cells such as hamster BHK and CHO cells or mouse Ltk− and B16 cells stably expressing CD46 become sensitive to infection with various species B human Ads (HAdVs), including Ad3/7/11/35 (13, 14, 20, 40, 60, 61).
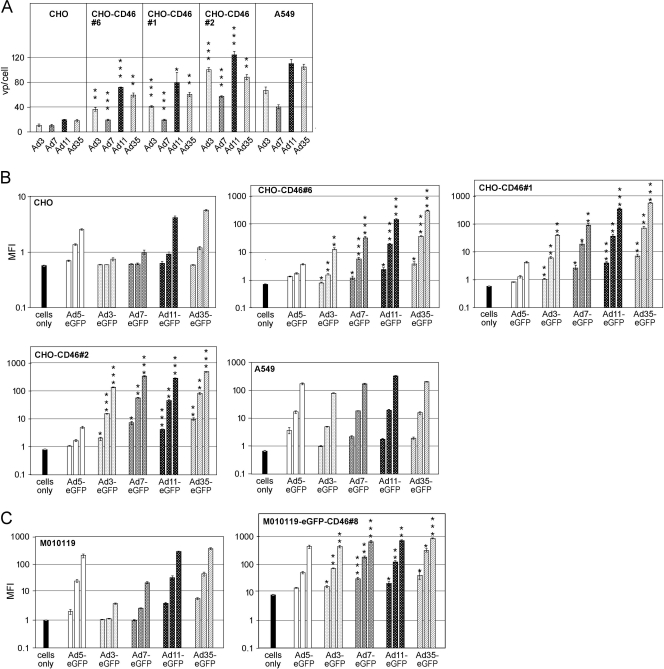
To address if CD46 needs to be expressed at a certain threshold level to be an Ad3/7 receptor, we generated clonal CHO cells stably expressing different levels of the CD46 BC1 isoform and compared their binding and transduction levels for species B serotypes. Of note, rodent cells, including unmodified CHO cells, were found to be negative for Ad3 binding (15, 20, 37, 67, 70) (Fig. 2B), and they did not stain with a DSG-2 antibody (data not shown), suggesting that they lack expression of DSG-2 or that hamster DSG-2 is not able to bind human Ad3. The highest levels of CD46 were measured in CHO-CD46#2 cells with an arbitrary MFI of 161, followed by CHO-CD46#1 (MFI of 45) and CHO-CD46#6 (MFI of 33) (see Fig. S1A in the supplemental material). All three CHO clones were derived from CHO-15B6 cells. Additional CHO-CD46#RC cells, which were derived from CHO-K1 cells (4) and expressed CD46 levels similar to those of CHO-CD46#1, were included as controls in the transduction study. With these cells, we obtained data similar to those for CHO-CD46#1 (not shown). We noticed, however, that the CD46 stably transfected cells had broader expression ranges than, for example, human lung carcinoma A549 cells, which had an MFI of 74. We therefore used the stable CD46-expressing CHO cells at low passage numbers to ensure comparable expression levels. Cells were incubated with 1,000 vp of [3H]thymidine-labeled wild-type Ad3/7/11/35 followed by determination of cell-associated viruses by liquid scintillation counting (Fig. 2A). CHO-CD46#2 cells, expressing the highest levels of CD46, revealed the highest virus binding for all four viruses, followed by CHO-CD46#1 and CHO-CD46#6 cells, which bound 2- to 4-fold more virus than did parental CHO cells. Binding of Ad3/7/11 to A549 cells was intermediate compared to CHO-CD46#2 and CHO-CD46#1 cells, whereas binding of Ad35 was higher to A549 than to CHO-CD46#2 cells.
Fig 2.
Binding and transduction of adenovirus species B viruses in CD46 gain-of-function cells negative for DSG-2. (A) For Ad binding assays, 5 × 105 human A549 cells and the different rodent CHO-CD46 cells were incubated on ice with 1,000 vp of the indicated 3H-labeled species B serotypes. After incubation for 2 h, the cells were washed and cell-associated radioactivity was determined. Mean values and standard deviations of triplicates from one representative experiment are shown. Asterisks indicate here and in the experiments described below the level of significance (*, P < 0.05; **, P < 0.005; ***, P < 0.0005, for comparisons of corresponding Ad binding [infection] in parental CHO versus CD46-transfected cells). (B) Transduction assays of human A549, parental CHO, and CHO-CD46-expressing cells. Cells (105) were incubated with eGFP-expressing Ad3, Ad7, Ad11, and Ad35 vectors at increasing virus concentrations of 10, 100, and 1,000 vp/cell. eGFP expression was analyzed 2 days p.i. by flow cytometry and is expressed as MFI. (C) Transduction assays of human parental and stable CD46-transfected M010119 melanoma cells. Cells were transduced and tested as described above. Background fluorescence intensity for uninfected M010191-eGFP-CD46 cells was higher due to the eGFP-tagged CD46 in these cells.
Since binding experiments have a relatively low dynamic range, we performed transduction experiments using eGFP-expressing vectors. Increasing amounts of recombinant Ad3/7/11/35 expressing eGFP gave a robust increase of transgene expression in all four CHO-CD46 cells (including CHO-CD46#RC cells) (Fig. 2B and Table 1). At a virus concentration of 1,000 vp/cell, e.g., Ad3-mediated eGFP expression increased 18-fold in CHO-CD46#6 cells, 55-fold in CHO-CD46#1 cells, and 192-fold in CHO-CD46#2 cells, compared to parental CHO cells. When standardized to A549 cells, Ad3-eGFP-mediated transgene expression levels amounted to 0.24% in parental CHO cells, 19% in CHO-CD46#6 cells (with a 2.2-fold-lower CD46 level than that of A549), 60% in CHO-CD46#1 cells (with about a 1.5-fold-lower CD46 level than that of A549), and 211% in CHO-CD46#2 cells (2.2-fold-higher CD46 level than that of A549). For better comparison of transduction efficiencies, vp/cell concentrations necessary to reach the arbitrary MFI value of 100 were calculated (Table 1). These values showed that the highest input of 8,207 Ad3 vp/cell was needed for CHO-CD46#6 cells, 2,597 vp/cell was needed for CHO-CD46#1 cells, and 717 vp/cell was needed for CHO-CD46#2 cells to reach comparable transduction levels. Transduction with Ad7 was about 2-fold more efficient than that with Ad3 when comparing vp/ml values necessary to reach an MFI of 100. Ad11/35 consistently transduced all tested cells more efficiently than did Ad3/7, including control A549 cells and parental CHO cells. When standardized to Ad35, Ad3 infection of CHO-CD46#6, CHO-CD46#1, CHO-CD46#2, and A549 required 27-, 16-, 4.2-, and 2.7-fold-higher vp input to obtain the MFI value of 100, respectively. For Ad7, these numbers amounted to 9.5-, 7.3-, 1.57-, and 1.1-fold, respectively. In contrast, Ad11 had a transduction efficacy similar to that of Ad35. Of note, when using Ad5-eGFP in these cells, transduction levels were very low, within a factor of 2 for the different CD46-expressing clones, including the parental CHO cells.
Table 1.
Species B serotype-mediated eGFP transgene expression in different CHO-CD46-expressing cells compared to parental CHO and human A549 cells
| eGFP transgene | eGFP expression level for cell line |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold change compared to CHOa |
% of A549 expressionb |
No. of vp/cell needed to reach MFI of 100c |
|||||||||
| CHO-CD46#6 | CHO-CD46#1 | CHO-CD46#2 | CHO | CHO-CD46#6 | CHO-CD46#1 | CHO-CD46#2 | CHO-CD46#6 | CHO-CD46#1 | CHO-CD46#2 | A549 | |
| Ad3-eGFP | 18 | 55 | 192 | 0.24 | 19 | 60 | 211 | 8,207 (27) | 2,597 (16) | 717 (4.2) | 1,513 (2.7) |
| Ad7-eGFP | 37 | 85 | 354 | 0.23 | 21 | 50 | 202 | 2,919 (9.5) | 1,210 (7.3) | 267 (1.57) | 602 (1.1) |
| Ad11-eGFP | 40 | 88 | 79 | 1.03 | 49 | 105 | 95 | 634 (2.1) | 291 (1.8) | 314 (1.8) | 329 (0.6) |
| Ad35-eGFP | 60 | 106 | 96 | 2.66 | 177 | 305 | 284 | 307 | 165 | 170 | 562 |
Ratios of eGFP mean fluorescence intensity levels from CD46-expressing cell line to that from parental CHO cells were determined for a 1,000-vp/cell input.
eGFP mean fluorescence intensity levels of CD46-expressing cells as percentages of those of A549 cells were determined for a 1,000-vp/cell input.
Regression lines were calculated for the eGFP expression levels; numbers in parentheses correspond to fold virus concentration input over that of Ad35-eGFP.
We also tested whether human melanoma M010119 cells with low CD46 expression (MFI of 17.8) (see Fig. S1E in the supplemental material) were increasingly transduced with Ad-eGFP vectors by increasing CD46 levels. Stable transfection of these cells (M010119-eGFP-CD46#8) resulted in about 9-fold-higher CD46 expression levels (MFI of 155) compared to the parental cells. In this case, an N-terminal eGFP-tagged CD46 BC1 isoform was used, where the endogenous signal peptide was replaced by the CAR signal peptide, followed by the eGFP sequence and the sequence of the mature CD46 BC1 isoform. Of note, the receptor function of the eGFP-tagged CD46 form was retained for all four species B serotypes tested in M010119-eGFP-CD46#8 cells (Fig. 2C; Table 2). When using 1,000 vp/cell, M010119-eGFP-CD46#8 cells with the 9-fold-higher CD46 level revealed 86-, 26-, 2.1-, and 2.3-fold enhancements of reporter eGFP for Ad3/7/11/35-mediated transduction, respectively, compared to parental M010119 cells. Determination of vp/cell ratios necessary to reach an MFI value of 100 revealed 127-, 52-, 3.1-, and 2-fold enhancement of Ad3/7/11/35-mediated transduction, respectively. Of note, both the parental M010119 low-CD46 cells and the M010119-eGFP-CD46#8 high-CD46 cells expressed no DSG-2 when analyzed by cytofluorometric means (see Fig. S1B and S1F in the supplemental material), in contrast to A549 cells, which expressed high levels of DSG-2.
Table 2.
eGFP transgene expression analysis in M010119 and M010119-eGFP-CD46#8 cells
| Transgene | eGFP expression level |
||
|---|---|---|---|
| Fold change compared to M010119a | No. of vp/cell needed to reach MFI of 100 in cell lineb |
||
| M010119 | M010119-eGFP-CD46#8 | ||
| Ad3-eGFP | 86 | 34,513 | 271 (127) |
| Ad7-eGFP | 26 | 4,695 | 91 (52) |
| Ad11-eGFP | 2.1 | 344 | 110 (3.1) |
| Ad35-eGFP | 2.3 | 258 | 130 (2.0) |
Ratios of eGFP mean fluorescence intensity levels from M010119-eGFP-CD46#8 cells to that from parental M010119 cells were determined for a 1,000-vp/cell input.
Regression lines were calculated for the eGFP expression levels. Numbers in parentheses correspond to fold enhancement of transduction efficiency in M010119-eGFP-CD46#8 cells over that of M010119 cells, based on MFI values of 100.
In summary, ectopic expression of CD46 in rodent cells or enhanced expression of CD46 in human cells low in CD46 and negative for DSG-2 resulted in increased transduction with Ad3/7/11/35 species B serotypes. Transduction efficacy of Ad3/7 correlated with CD46 density levels in these cells, whereas Ad11 reached a plateau with the medium CHO-CD46#1 cells and Ad35 was largely independent of the CD46 levels. This is compatible with the notion that Ad3/7 exhibit a lower affinity for CD46 than do Ad11/35 but can compensate for the lower affinity by avidity binding.
Cross-linking of soluble CD46 enhances blocking of Ad3 and Ad7 infection of CHO-CD46 and A549 cells.
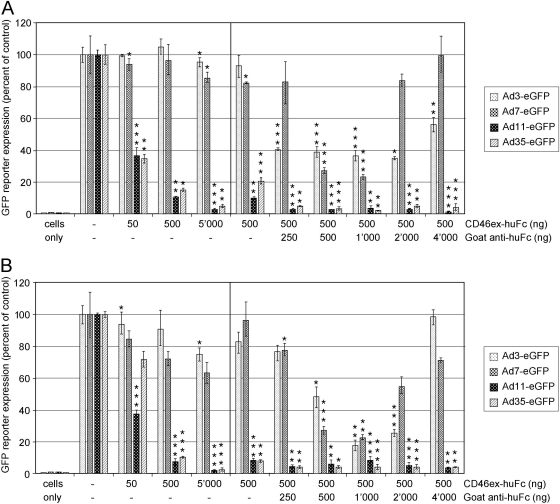
We earlier documented that soluble CD46ex-huFc was at least 100-fold more potent at blocking Ad11/35 binding to rodent cells expressing CD46 than were Ad3 and Ad7 (13). To further test the hypothesis that avidity allows Ad3/7 to use CD46 as a functional receptor, we repeated the virus blocking assays with CD46ex-huFc but included this time cross-linked adapter proteins. We reasoned that antibody-mediated cross-linking of CD46ex-huFc could provide locally more binding sites per molecule, mimicking the situation on the cell membrane where CD46 can diffuse freely and form clusters following multivalent binding (1, 9).
When using 50, 500, and 5,000 ng/ml of CD46ex-huFc protein alone, only Ad11-eGFP- or Ad35-eGFP-mediated transgene expression was significantly reduced in CHO-CD46#2 or A549 cells (Fig. 3A and B). In contrast, CD46ex-huFc did not result in significant reduction of Ad3/7-mediated transgene expression. However, when we combined CD46ex-huFc protein at a concentration of 500 ng/ml with 2-fold-increasing amounts of cross-linking goat anti-human Fc antibody, significant blocking was also achieved for both Ad3- and Ad7-mediated transgene expression. Inhibition followed a precipitin-type curve, reaching a plateau in the range of 500, 1,000, or 2,000 ng/ml of goat anti-human Fc. The highest blocking for Ad3-eGFP amounted to 65% in CHO-CD46#2 cells and 83% in A549 cells, and for Ad7-eGFP, the highest blocking was 77% in both cell types. We conclude that CD46ex-huFc multimerization leads to efficient blocking of Ad3/7. A similarly multimerized complex consisting of CARex-huFc had no significant effects on Ad3/7 infection (see Fig. S2A and S2B in the supplemental material) but CARex-huFc inhibited Ad2/5 transduction (12, 38).
Fig 3.
Cross-linking of CD46ex-huFc strongly increases blocking of Ad3/7 infection in CHO-CD46#2 and A549 cells. CHO-CD46#2 cells (A) or A549 cells (B) were preincubated for 1 h in the cold using the indicated concentrations of adapter CD46ex-huFc alone or in combination with a 2-fold-increasing series of goat anti-human Fc antibody. Following addition of the different eGFP-expressing vectors for another 1 h, cells were washed and analyzed 48 h p.i. Asterisks indicate the levels of significance (*, P < 0.05; **, P < 0.005; ***, P < 0.0005, for comparisons of corresponding Ad infection using CD46ex-huFc versus control CARex-huFc of Fig. S2 in the supplemental material).
Species B adenovirus fiber knobs cross-compete for CHO-CD46 and A549 transductions.
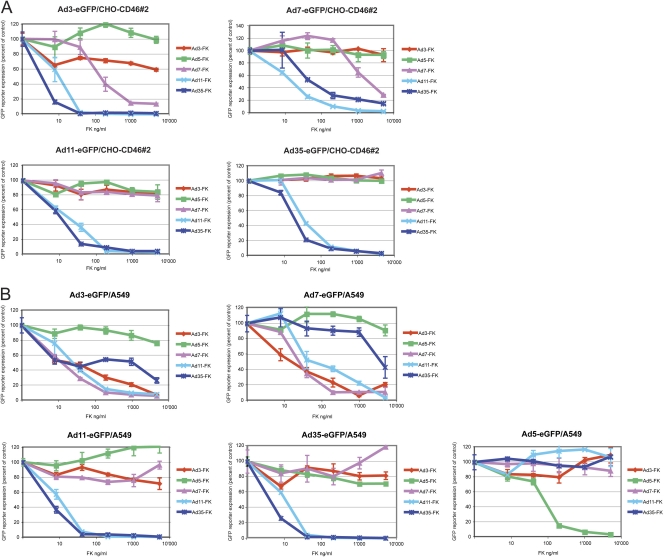
To further characterize affinity differences and receptor usage by species B Ads, we performed blocking studies with species B Ads and soluble FKs in CHO-CD46#2 and A549 cells. CHO-CD46#2 cells do not express DSG-2, unlike human A549 cells. Virus inhibition assays were performed using five different eGFP-expressing vectors as readout for binding and infectivity. For CHO-CD46#2 cells, Ad3/7/11/35-eGFP inputs of 29,600, 8,200, 657, and 1,088 vp/cell, respectively, were used. For A549 cells, the virus inputs amounted to 14,800, 8,200, 1,314, and 2,540 vp/cell, and in addition, for Ad5-eGFP an input of 2,825 was applied. The virus input concentrations were chosen such that the unblocked transgene expression values amounted to fluorescence intensities in the range of about 200, thereby allowing an optimal dynamic range (not shown). We ensured that FKs of Ad3/7/11/35 were predominantly present in trimeric form (see Fig. S2A and S2B in the supplemental material). All five Ad FK proteins were tested in a dose-dependent manner with the highest concentration of 5,000 ng/ml diluted in a 5-fold dilution series to the lowest concentration of 8 ng/ml (Fig. 4A and B and Table 3). The specificity of this assay was confirmed by the finding that the species C Ad5-FK gave rise to inhibition only of Ad5, not of any of the species B serotypes. Likewise, species B FKs inhibited only species B serotypes, but not Ad5-eGFP.
Fig 4.
Inhibition of Ad3-, Ad7-, Ad11-, and Ad35-eGFP transduction in CHO-CD46#2 and A549 cells by recombinant Ad fiber knobs. Cells were preincubated for 1 h in the cold using a 5-fold dilution series of the individual FK proteins, followed by addition of the different eGFP-expressing vectors for another 1 h. (A) For CHO-CD46#2 cells, the viral inputs amounted to 29,600, 8,200, 657, and 1,088 vp/cell for Ad3-, Ad7-, Ad11-, and Ad35-eGFP, respectively. The virus input concentrations had been determined in preceding experiments and were chosen such that the unblocked transgene expression values amounted to fluorescence intensity values of about 200. eGFP analysis was performed 48 h p.i. FKs are color coded as follows: Ad5-FK in green, Ad3-FK in red, Ad7-FK in purple, Ad11-FK in cyan, and Ad35-FK in dark blue. (B) For A549 cells, virus inputs amounted to 14,800, 8,200, 1,314, 2,540, and 2,825 vp/cell for Ad3-, Ad7-, Ad11-, Ad35-, and Ad5-eGFP, respectively. Otherwise, the procedure was the same as that described above.
Table 3.
Inhibition of Ad3/7/11/35-eGFP- and Ad5-eGFP-mediated reporter expression by fiber knob cross-competition
| Competitor | Cell line | % inhibition of eGFP expressiona |
FK concn for 50% inhibition (ng/ml)b |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ad3-eGFP | Ad7-eGFP | Ad11-eGFP | Ad35-eGFP | Ad5-eGFP | Ad3-eGFP | Ad7-eGFP | Ad11-eGFP | Ad35-eGFP | Ad5-eGFP | ||
| Ad3-FK | CHO-CD46 | 40 | 7 | 18 | −3 | NDc | ND | ND | ND | ND | ND |
| A549 | 93 | 78 | 28 | 18 | 17/−9 | 25 | 23 | ND | ND | ND | |
| Ad7-FK | CHO-CD46 | 86 | 81 | 21 | −10 | ND | 169 | 2,690 | ND | ND | ND |
| A549 | 95 | 87 | 4 | −18 | 2/12 | 17 | 31 | ND | ND | ND | |
| Ad11-FK | CHO-CD46 | 99 | 98 | 99 | 97 | ND | 14 | 21 | 22 | 37 | ND |
| A549 | 93 | 95 | 99 | 100 | −10/−6 | 31 | 87 | 13 | 14 | ND | |
| Ad35-FK | CHO-CD46 | 98 | 85 | 97 | 96 | ND | 5 | 63 | 14 | 26 | ND |
| A549 | 74 | 56 | 99 | 99 | −1/−7 | 470 | 4,459 | 33 | 5 | 105 | |
| Ad5-FK | CHO-CD46 | 1 | −35 | 16 | 0 | ND | |||||
| A549 | 24 | 9 | −21 | 29 | 26/97 | ||||||
The cross-competition assays revealed differences in the capacities of the individual FKs to compete with species B viruses. The Ad3/7-FKs on one side and Ad11/35-FKs on the other side showed similar behaviors, which, however, were different in the two cell types tested. In CHO-CD46#2 cells, Ad3/7-FKs elicited at best weak competition for corresponding and noncorresponding Ad3 and Ad7 and no effect on noncorresponding Ad11 and Ad35. This is illustrated by the 40% inhibition of Ad3-eGFP transduction at the highest Ad3-FK concentration of 5,000 ng/ml and a nonsignificant 7% inhibition for Ad7-eGFP transduction. Ad3-FK concentrations necessary for 50% inhibition (FK-50%) were larger than the highest concentration available and could thus not be calculated. Ad7-FK was more efficient than Ad3-FK, and using 5,000 ng/ml, 86 and 81% of Ad3/7-mediated eGFP expression was inhibited, respectively. FK-50% inhibition values for Ad7-FK were determined to be 169 and 2,690 ng/ml for Ad3/7-mediated transgene expression, respectively. In contrast, Ad11/35-FKs led to efficient blocking of all B-type viruses in these cells. Likewise, Ad35-FK blocking efficiency was in the range of 85 to 97% at 5,000 ng/ml, and the corresponding FK-50% values were 5, 63, 14, and 26 ng/ml.
The binding inhibition pattern for A549 cells was different than that for CHO-CD46#2 cells. In A549 cells, Ad3/7-FKs efficiently competed for corresponding and noncorresponding Ad3/7 in A549 cells with FK-50% inhibition values of 25 and 23 ng/ml, respectively (Fig. 4B; Table 3). With the highest concentrations of Ad7-FK, blocking efficiency of 95 and 87% was obtained for Ad3/7-mediated eGFP expression, respectively, with FK-50% values of 17 and 31 ng/ml, respectively. However, as for CHO-CD46#2 cells, Ad3/7-FKs revealed no effect on noncorresponding Ad11/35 in A549 cells. Ad11-FK efficiently blocked all four species B serotypes, with FK-50% inhibition values of 31, 87, 13, and 13 ng/ml, whereas Ad35-FK was less efficient at inhibiting Ad3 and Ad7, with FK-50% values of 470 and 4,459 ng/ml, respectively, but very efficient at inhibiting Ad11 and Ad35, with FK-50% values of 33 and 5 ng/ml, respectively.
In summary, blocking studies with Ad3/7-FKs revealed different effects for corresponding viruses in CHO-CD46 cells or human A549 cells. In CHO-CD46 cells, the observed low blocking efficiency confirms the low affinity of the Ad3/7 soluble FK molecules for CD46. In contrast, the efficient blocking activity of Ad3/7-FK in A549 is compatible with our findings that up to 90% of Ad3 binds with a presumably higher affinity to DSG-2, whereas a minor fraction binds to CD46.
Binding and affinity determinations of CD46 to Ad3/7/11/35-FK by surface plasmon resonance.
To further characterize species B Ad interactions with CD46, we applied biosensor technology using soluble CD46 and the FKs of Ad3/7/11/35, which were predominantly present in trimeric form (see Fig. S3A and S3B in the supplemental material). Initial Biacore experiments using sensor chips had previously been described for immobilized FKs and monovalent CD46-SCR I-II as soluble analyte, thereby avoiding avidity effects of CD46 (10, 47, 48, 71, 72). The SCR I-II domain comprised the binding sites for the FKs (14, 49, 53, 71). Under these conditions, we determined the affinities of Ad11/35-FK to CD46-SCR I-II and found that they were consistent with a 1:1 (monophasic) binding model, where one surface FK monomer interacted with one soluble CD46-SCR I-II (data not shown). Ad3/7-FK, however, did not bind to immobilized CD46-SCR I-II, most likely due to low affinity of their monomers for CD46-SCR I-II, in agreement with earlier data for Ad7-FK (48).
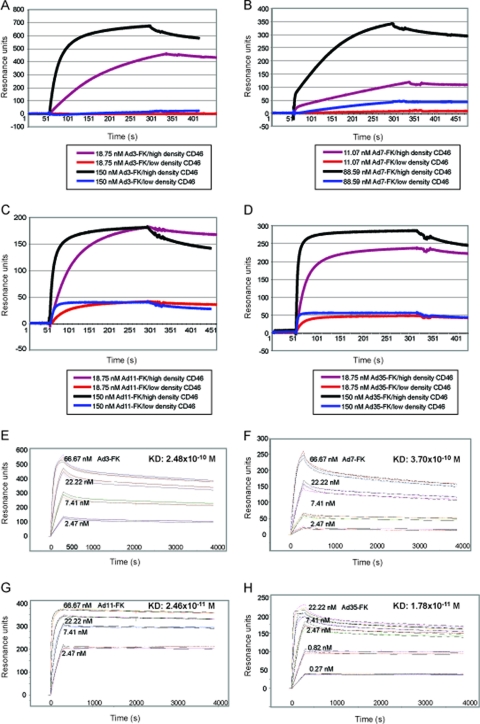
In a reversed setup of the SPR measurements, we immobilized the extracellular domain of CD46 fused to human Fc (CD46ex-huFc; see reference 60) to the flexible dextran matrix of the sensor chip and used FKs as analytes. CD46ex-huFc is a disulfide-linked dimer, as suggested by native gel electrophoresis (see Fig. S3C in the supplemental material). We first tested if different concentrations of immobilized CD46ex-huFc would give different binding responses for the FKs. Therefore, flow cell 2 (Fc2) of the CM5 sensor chip was coated with a high density of CD46ex-huFc (2,630 resonance units [RU]), and the control Fc1 was coated with a high density of CARex-huFc (3,431 RU). In a parallel setup, Fc4 was coated with a low surface density of CD46ex-huFc (345 RU) and Fc3 was coated with a low density of CARex-huFc (278 RU). From the sensorgrams of Ad3/7/11/35-FK binding to CD46ex-huFc, we noticed that higher concentrations of soluble Ad3/7-FK (150 and 89 nM, respectively) were required to reach equilibrium binding at high CD46ex-huFc density levels compared to Ad11/35-FK (18.75 nM) (Fig. 5A to D). At equilibrium binding, both Ad3/7-FK and Ad11/35-FK slowly dissociated, indicative of stable complexes. At low CD46ex-huFc density, the Ad11/35-FKs and to a very small extent also Ad7-FK but not Ad3-FK showed detectable binding. The absolute binding values (Rmax) varied by factors of 2 to 3 between Ad3- and Ad7-FK and between Ad11- and Ad35-FK, as well as different technical replicates of the experiments, possibly reflecting different degrees of FK purities and chip conditions.
Fig 5.
Subtracted SPR sensorgrams for Ad3-, Ad7-, Ad11-, and Ad35-FK interacting with CD46. Soluble receptor CD46ex-huFc was immobilized on a CM5 chip at high density in Fc2 (2,630 RU) and low density in Fc4 (345 RU). Control CARex-huFc was immobilized at high density in Fc1 (3,431 RU) and low density in Fc3 (278 RU). (A to D) FK analytes were injected over the sensor surface at 18.75 and 150 nM concentrations for Ad3-FK (A), Ad11-FK (C), and Ad35-FK (D) and at 11.07 and 88.59 nM for Ad7-FK (B), respectively. Association times were either 240 or 280 s. (E to H) Overlay of analyte responses for Ad3-, Ad7-, Ad11-, and Ad35-FK at different concentrations. Measurements were performed using the CD46 high-density 2,630-RU CM5 chip. Serial concentrations of 0.27, 0.82, 2.47, 7.41, 22.22, and 66.67 nM Ad3-FK (E), Ad7-FK (F), Ad11-FK (G), and Ad35-FK (H) were injected in HBS-P+ buffer at 30 μl/min under a contact time of 300 s and a dissociation time of 3,600 s (for better visibility, only four of the six binding curves are shown). Data evaluation was fitted globally by a two-stage reaction model with Biacore T100 evaluation software, and resulting kinetics/affinity results (listed in Table S1 in the supplemental material and summarized in Table 4).
Combined binding kinetics and affinity measurements were carried out for each of the four FKs. For this, we used three independent measurements in the range of 0.27, 0.82, 2.47, 7.41, 22.22, and 66.67 nM and two different high-density CD46 biosensor chips of 2,630 RU and 1,121 RU. The two biosensor chips gave similar results (Fig. 5E to H) (see Table S1 in the supplemental material). In addition, Ad3-FK binding was found to be independent of the flow rate at the standard flow rate of 30 μl/min and an increased flow rate of 55 μl/min, excluding the possibility that the binding reactions were influenced by mass transfer effects (41). The values for the kinetic rate constants were highly reproducible for the individual FKs, with low standard errors for the association and dissociation constants (Fig. 5E and H) (see Table S1). The kinetics and binding data could be best fitted to a two-stage reaction model rather than monophasic or trivalent binding models (see the supplemental methods). Modeling of the measured association and dissociation rate constants and R values was carried out based on global fittings for ka1, ka2, kd1, and kd2 and local calculations of Rmax and percent χ2/Rmax. All percent χ2/Rmax values were below 5%. The average apparent Kd values (Table 4) were not significantly different for Ad3-FK (2.48 × 10−10 M) and Ad7-FK (3.70 × 10−10 M), whereas the apparent Kds for Ad11-FK and Ad35-FK were 10- to 15-fold lower (P = 0.002/0.0669 for Ad3/7 compared to Ad11, and P = 0.0014/0.063 for Ad3/7 versus Ad35) and amounted to 2.46 × 10−11 and 1.78 × 10−11 M, respectively. Thus, the 10- to 15-fold difference between the apparent Kd values for Ad3/7 and Ad11/35 at high CD46 densities deviated considerably from the 2,000-fold differences of Kd reported for the monovalent CD46 binding to Ad7-FK and Ad11-FK (48). Since the measurements were performed using similar setups, this suggests that at high surface densities of CD46, trimeric FK interacts with two or even three CD46 binding sites and the resulting avidity effects overcome low-affinity restrictions of single FK-CD46 interactions.
Table 4.
Summary kinetics/affinity analyses of Ad-FK binding to immobilized CD46ex-huFca
| Ad-FK | ka1 (M−1 s−1) | kd1 (s−1) | ka2 (s−1) | kd2 (s−1) | Kd ± SEM (M) |
|---|---|---|---|---|---|
| Ad3-FK | 2.30 × 105 | 6.88 × 10−4 | 2.01 × 10−3 | 1.76 × 10−4 | (2.48 ± 0.27) × 10−10 |
| Ad7-FK | 1.44 × 105 | 4.19 × 10−4 | 1.94 × 10−3 | 1.99 × 10−4 | (3.70 ± 1.38) × 10−10 |
| Ad11-FK | 9.27 × 105 | 3.27 × 10−4 | 5.41 × 10−3 | 1.60 × 10−4 | (2.46 ± 1.50) × 10−11 |
| Ad35-FK | 3.08 × 106 | 4.66 × 10−4 | 1.25 × 10−3 | 1.29 × 10−4 | (1.78 ± 1.04) × 10−11 |
Values are averages of three individual measurements.
DISCUSSION
The data obtained in this study suggest that both DSG-2 and CD46 can serve simultaneously as Ad3 receptors and that avidity effects allow the low-affinity CD46 binders Ad3/7-FK to attach to CD46 for infectious entry. Conspicuously, several Ad serotypes have been reported to utilize more than one attachment receptor, which could be part of an evolutionary strategy to increase viral fitness (76). For example, the species D Ad9, with a very short fiber, was found to allow fiber-independent binding of the penton base directly to integrins, if integrins were present in sufficient amounts (51). In integrin-low cells, the virus instead used CAR for productive infection. Our finding that species B1 adenoviruses use avidity to bind to the CD46 receptor has important consequences for tropism and possibly evolution of the species B1 viruses, where viral receptor-binding proteins are under selective pressure and undergo constant variation to evade neutralization by antibodies (23, 47). High-affinity binding could, for example, represent a dead end in evolutionary terms, as such viruses would be constrained to a single receptor. Weak affinity to a receptor could be essential for gaining access to new attachment receptors.
Avidity captures low-affinity interactions.
Viruses use multiple binding strategies, characterized by a broad range of affinity interactions when analyzed for single binding interaction sites (22, 73). Viruses with low affinities include, e.g., influenza virus with millimolar affinity for sialic acid, and there are micromolar affinities for rhinovirus attachment to intercellular adhesion molecule 1, echovirus attachment to decay-accelerating factor, or reovirus attachment to junction adhesion molecule (2, 6, 28, 62). Biologically important binding of viruses to low-affinity receptors occurs when virus particles expose multiple binding sites. This allows for avidity effects and increases overall affinity.
Avidity is characterized by a synergistic (and not additive) combination of individual bond affinities, which largely depends on the structure and geometry of the involved molecules (36). For example, in the case of an IgG with a valency of two, avidity can lead to a strong increase of overall affinity, compared to corresponding monovalent Fab fragments (see, e.g., an early study in reference 16). Particularly strong avidity effects were also reported for low-affinity ErbB2-specific single-chain variable-fragment antibodies, where the antibody fragment with the lowest affinity showed the highest avidity increase upon dimerization (44). Viruses containing trimeric attachment receptors, such as the reovirus sigma 1 protein or the long and short fibers of bacteriophage T4, have been suggested to employ avidity mechanisms for infection (as discussed in reference 33). Interestingly, binding of Ad3 to DSG-2 may engage the avidity mechanism, since multimerization of the trimeric Ad3-FK was required for efficient binding to DSG-2 (69).
Avidity binding of adenoviruses to CD46 and CAR.
Some studies considered Ad3/7 and also Ad14 to be CD46 nonbinders due to low-affinity interactions with CD46 (15, 19, 37, 56, 57, 67). Several lines of evidence, however, support an avidity-based mechanism for Ad3/7 infection of CD46-positive cells. First, Ad3/7 infection correlated with the levels of ectopically expressed CD46, while Ad11/35 infection was saturated at low CD46 expression levels. Second, the binding of species B Ad3/7-FK to immobilized CD46 on a biosensor chip was more dependent on the receptor density than was binding of Ad11/35-FK, and the Ad3/7-FK binding had only 10- to 15-fold-lower apparent affinity than Ad11/35-FK. Third, cross-linking of soluble CD46 increased its blocking effect on Ad3/7 infection in human A549 cells and CHO-CD46 cells. Fourth, soluble Ad3/7-FKs inefficiently blocked Ad3/7 infection of CHO-CD46 cells, unlike Ad11/35-FKs, which efficiently blocked all four tested B serotypes, confirming that Ad3/7-FKs bind with lower affinity to CD46 than do Ad11/35-FKs. These data support and extend earlier binding site mapping studies, which showed that Ad3/7 engage CD46 through similar surfaces as do species B2 Ad11/35 (13). In support of this, the recent SPR studies by Persson et al. revealed a broad variation of CD46 binding affinities, and the authors concluded that Ad7/11/14/35 are able to bind to the same epitopes on CD46 (48). All these data are in accordance with the observations that both the low-affinity CD46 binder Ad3 and the high-affinity binder Ad35 use virtually identical pathways to infect human cells that are CD46 positive (1, 24). Notably, avidity mechanisms may also allow the species B Ad16 to use CD46 for productive infection by increasing the affinity to CD46 by 70-fold compared to Ad11 (47).
Similar to the species B serotypes, also the CAR-tropic species C Ad serotypes have a broad spectrum of affinities to CAR over nearly 3 orders of magnitude, ranging from the low-affinity binder Ad9 to the high-affinity binder Ad5 (25). In addition, avidity effects could be measured by an increase of FK affinity from about 25 nM for monomeric immobilized Ad2 FK to 1 nM for multivalent immobilized CAR in SPR assays employing minimal-size proteins produced in Escherichia coli (33). The avidity-driven multivalent binding reaction to immobilized high-density CAR (200 RU, equivalent to our CD46 high-density chips) could be fitted to a trivalent binding model. We thus consider it likely that many different Ads use avidity binding for attachment and possibly entry into cells.
Avidity effects for Ad-receptor interactions can occur at two levels. The trivalent FK protein can increase the overall affinity compared to a monovalent protein. In addition, some of the 12 fiber trimers on the virus may simultaneously form multiple contact points with the cell surface. Both of these mechanisms can contribute to increase the overall affinities of the virus particle for its receptor. Importantly, in the case of the CAR binders Ad2/5, CAR clustering leads to viral surfing motions on the cell surface and triggers viral uncoating assisted by confined motions of the viral coreceptor alpha v integrin (5).
Apparent Kd values of species B FKs to divalent CD46 differ from Kd values with monovalent soluble CD46.
Avidity effects are strongly dependent on the valency of the ligand, as observed, for example, with binding of dimeric ephrin-A5 ligand to dimerized EphA3 receptor (45). Accordingly, the measured Kd values for FKs to our CD46ex-Fc were about 5 logs lower for the Ad3/7-FKs and 3 logs lower for Ad11/35-FKs compared to the Kd values for the monovalent CD46-SCR I-II interactions published by others (10, 47, 48, 71, 72). Also in regard to the valency difference, the CD46-SCR I-II protein used in other studies was produced in mutant CHO cells and contained homogenous, high-mannose N-linked carbohydrates (48). Our CD46 protein consisted of the entire extracellular domain plus a dimerizing Fc portion and was produced in normal BHK cells allowing for complex O- and N-linked glycosylations (60). Interestingly, the extended CD46-SCR I-IV domain has a somewhat different mode of binding to Ad11-FK, as the contact site did not include the IJ loop described for CD46-SCR I-II interaction (50). We suggest that the dimeric nature of our CD46ex-huFc explains why the binding of the Ad3/7/11/35-FKs fits better to a two-stage reaction model than to trivalent binding models, described earlier for Ad2-FK interaction with CAR (33). This is based on comparison of the different affinities under our standard conditions. We conclude that the observed Kd differences between the monovalent and trivalent affinities of the Ad3/7/11/35-FKs are based on avidity binding.
Virus-receptor affinity versus receptor density.
A recent systematic study with retargeted measles virus (MV) vectors has shown that viruses displaying HER2/neu-specific single-chain variable-fragment antibodies had high avidity and cell-killing ability due to dozens of single-chain molecules in the viral envelope, even if their single affinities were in the low-micromolar range (21). This study noticed a receptor threshold level for productive infection, together with an inverse correlation of attachment receptor affinity and receptor density requirements for infection. In particular, suprathreshold affinities did not further enhance the efficiency of MV infection, which corresponds well with our results for high-affinity CD46 binders Ad11/35. It also fits well with another observation showing that Ad35-FK mutants with either 4-fold- or 60-fold-higher affinity at the monovalent interaction level did not yield higher transduction levels, regardless of the cellular CD46 receptor density (72).
With the FK neutralization experiments in CHO-CD46#2-expressing cells, we noticed that Ad3/7-FKs did not efficiently compete against the binding of corresponding virus particles, in contrast to Ad11/35-FKs, which efficiently blocked Ad3/7/11/35. This is due to low affinity of Ad3/7-FK for CD46, since binding of the different species B Ads is restricted to CD46 in these cells (13, 60). This conclusion was corroborated by the findings that soluble CD46ex-huFc inefficiently inhibited binding of Ad3/7 to the cells, in agreement with weak efficiency of blocking of Ad3/7 by soluble CD46 (13, 67) and also by soluble monovalent CD46 for MV infection (58). Ad3/7 inhibition was, however, strongly enhanced by cross-linking the CD46ex-huFc protein (Fig. 3), suggesting that cross-linked CD46ex-huFc mimics the multiple-interaction situation when the virus contacts the cell surface. Similarly, CD46 cross-linking by genetically fusing the CD46 ectodomain to the octamer oligomerization domain of C4b binding protein resulted in a 2-log-enhanced MV-neutralizing activity in vitro and in vivo due to enhanced virus binding (8).
These data suggest that the receptor density is of key importance not only for interaction of CD46 with trimeric FK but also for the multivalent virus particle. Together or alone, this gives rise to avidity effects, which allow Ad3/7 to infect cells expressing sufficient levels of CD46 (13, 20, 60). The lack of Ad3/7 infection in CHO-CD46 cells reported by other groups is most likely due to low CD46 expression levels and assay systems of low sensitivity or limited dynamic range (15, 18, 37, 57, 67).
Mechanistic implications of desmoglein 2 and CD46 as receptors for Ad3/7.
Our DSG-2/CD46 combinatorial loss-of-function experiments in A549 and 16HBE14o cells significantly enhanced the reduction of cell surface binding and infection of Ad3 by ∼10% compared to DSG-2 interference alone, showing that CD46 has a significant role for infectious Ad3 entry in these cells. We suggest that in nonpolarized A549, 16HBE14o, or HeLa cells (70) expressing both receptors, DSG-2 functions as a major attachment receptor for Ad3. This could be due to a lower affinity/avidity of Ad3 for CD46 compared to that for DSG-2 or to higher levels of DSG-2 than of CD46. In other human cells, the CD46 contribution may be more pronounced depending on the expression levels of the receptors. This is supported by our data from rodent CHO-CD46 and human M010119 melanoma cells lacking DSG-2, where Ad3/7 uses CD46 as a functional receptor.
For oncolytic therapies with human Ads (HAdVs), it will be important to determine the DSG-2 expression levels among human cells and tissues in health and disease and put these data in relation to CD46 expression. In particular, it is conceivable that DSG-2 and CD46 act independently as Ad3/7 receptors. For example, in polarized cells, DSG-2 is a component of the cell-cell adhesion structure of intercellular junctions (26). CD46 is present on apical and basolateral surfaces (3, 59, 64). Apical CD46 may be an initial docking site for species B HAdVs and lead to the activation of DSG-2 from the lateral cell-cell contact sites. Interestingly, a library screening experiment of fiber-pseudotyped Ads revealed relatively high levels of apical transduction of Caco-2 cells by Ad7 and Ad35, despite low levels of virus attachment to the apical membrane (29). Speculatively, apical infections may be exacerbated by cytokine-mediated relocalization of CD46 and DSG-2 to the apical surface upon cytokine signaling from immune cells, as demonstrated for species C receptors CAR and alpha v beta 3 integrins (35). This may lead to a situation where DSG-2 and CD46 complement each other, for example, by physical interactions. Possibly, DSG-2 or another molecule functions as a tethering factor for CD46 and prolongs the half-life of CD46 on the plasma membrane, thereby increasing the CD46 avidity effects. Clustering of CD46 may occur in particular lipid domains (66) or in the vicinity of integrins or tetraspanins (34). In this scenario, lipids, integrins, or tetraspanins could be modulators for CD46 function and might not be required for Ad3 infection, if the CD46 levels were sufficiently high. Clustering of CD46 by antibodies or Ad3/35 facilitates CD46 internalization by clathrin-mediated endocytosis or macropinocytosis, followed by endosome lysis in the case of Ad3/35 and virus particle penetration from endosomes to the cytosol (1, 9, 24).
Supplementary Material
ACKNOWLEDGMENTS
We thank Leta Fuchs for technical assistance, Richard Sutton (Baylor College of Medicine, Houston, TX) for providing the pBlasti lentiviral vector system, Denis Gerlier (Université Lyon, Lyon, France) for providing the MCI20.6 anti-CD46 hybridoma, Dieter Gruenert (California Pacific Medical Center Research Institute, University of California, San Francisco, CA) for providing 16HBE14o cells, and the FACS Core Facility Irchel of the University of Zurich for the cell sorting service. We also thank Johan Hoebeke, Kessel-Lo, Belgium, for providing the equations and algorithms to evaluate the SPR data with the trivalent binding model.
This work was supported by the Kanton Zürich (S.H. and U.F.G.) and by the grant 31003A-116856 of the Swiss National Science Foundation (S.H.).
Footnotes
Published ahead of print 30 November 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Amstutz B, et al. 2008. Subversion of CtBP1-controlled macropinocytosis by human adenovirus serotype 3. EMBO J. 27:956–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barton ES, et al. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441–451 [DOI] [PubMed] [Google Scholar]
- 3. Blau DM, Compans RW. 1995. Entry and release of measles virus are polarized in epithelial cells. Virology 210:91–99 [DOI] [PubMed] [Google Scholar]
- 4. Buchholz CJ, Schneider U, Devaux P, Gerlier D, Cattaneo R. 1996. Cell entry by measles virus: long hybrid receptors uncouple binding from membrane fusion. J. Virol. 70:3716–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burckhardt CJ, et al. 2011. Drifting motions of the adenovirus receptor CAR and immobile integrins initiate virus uncoating and membrane lytic protein exposure. Cell Host Microbe 10:105–117 [DOI] [PubMed] [Google Scholar]
- 6. Casasnovas JM, Springer TA. 1995. Kinetics and thermodynamics of virus binding to receptor. Studies with rhinovirus, intercellular adhesion molecule-1 (ICAM-1), and surface plasmon resonance. J. Biol. Chem. 270:13216–13224 [DOI] [PubMed] [Google Scholar]
- 7. Chmielewicz B, et al. 2005. Respiratory disease caused by a species B2 adenovirus in a military camp in Turkey. J. Med. Virol. 77:232–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Christiansen D, et al. 2000. Octamerization enables soluble CD46 receptor to neutralize measles virus in vitro and in vivo. J. Virol. 74:4672–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crimeen-Irwin B, et al. 2003. Ligand binding determines whether CD46 is internalized by clathrin-coated pits or macropinocytosis. J. Biol. Chem. 278:46927–46937 [DOI] [PubMed] [Google Scholar]
- 10. Cupelli K, et al. 2010. Structure of adenovirus type 21 knob in complex with CD46 reveals key differences in receptor contacts among species B adenoviruses. J. Virol. 84:3189–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Durmort C, et al. 2001. Structure of the fiber head of Ad3, a non-CAR-binding serotype of adenovirus. Virology 285:302–312 [DOI] [PubMed] [Google Scholar]
- 12. Ebbinghaus C, et al. 2001. Functional and selective targeting of adenovirus to high-affinity Fcgamma receptor I-positive cells by using a bispecific hybrid adapter. J. Virol. 75:480–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fleischli C, et al. 2007. Species B adenovirus serotypes 3, 7, 11 and 35 share similar binding sites on the membrane cofactor protein CD46 receptor. J. Gen. Virol. 88:2925–2934 [DOI] [PubMed] [Google Scholar]
- 14. Fleischli C, et al. 2005. The distal short consensus repeats 1 and 2 of the membrane cofactor protein CD46 and their distance from the cell membrane determine productive entry of species B adenovirus serotype 35. J. Virol. 79:10013–10022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaggar A, Shayakhmetov DM, Lieber A. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9:1408–1412 [DOI] [PubMed] [Google Scholar]
- 16. Greenbury CL, Moore DH, Nunn LA. 1965. The reaction with red cells of 7s rabbit antibody, its sub-units and their recombinants. Immunology 8:420–431 [PMC free article] [PubMed] [Google Scholar]
- 17. Gruenert DC, Basbaum CB, Widdicombe JH. 1990. Long-term culture of normal and cystic fibrosis epithelial cells grown under serum-free conditions. In Vitro Cell. Dev. Biol. 26:411–418 [DOI] [PubMed] [Google Scholar]
- 18. Gustafsson B, et al. 2007. Adenovirus DNA is detected at increased frequency in Guthrie cards from children who develop acute lymphoblastic leukaemia. Br. J. Cancer 97:992–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gustafsson DJ, Segerman A, Lindman K, Mei YF, Wadell G. 2006. The Arg279Gln substitution in the adenovirus type 11p (Ad11p) fiber knob abolishes EDTA-resistant binding to A549 and CHO-CD46 cells, converting the phenotype to that of Ad7p. J. Virol. 80:1897–1905 (Author's correction, 80:5101.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hall K, Blair Zajdel ME, Blair GE. 2009. Defining the role of CD46, CD80 and CD86 in mediating adenovirus type 3 fiber interactions with host cells. Virology 392:222–229 [DOI] [PubMed] [Google Scholar]
- 21. Hasegawa K, et al. 2007. Affinity thresholds for membrane fusion triggering by viral glycoproteins. J. Virol. 81:13149–13157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Helenius A. 2007. Virus entry and uncoating, p 99–118 In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 23. Howitt J, Anderson CW, Freimuth P. 2003. Adenovirus interaction with its cellular receptor CAR. Curr. Top. Microbiol. Immunol. 272:331–364 [DOI] [PubMed] [Google Scholar]
- 24. Kalin S, et al. 2010. Macropinocytotic uptake and infection of human epithelial cells with species B2 adenovirus type 35. J. Virol. 84:5336–5350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kirby I, et al. 2001. Adenovirus type 9 fiber knob binds to the coxsackie B virus-adenovirus receptor (CAR) with lower affinity than fiber knobs of other CAR-binding adenovirus serotypes. J. Virol. 75:7210–7214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kowalczyk AP, et al. 1994. Structure and function of desmosomal transmembrane core and plaque molecules. Biophys. Chem. 50:97–112 [DOI] [PubMed] [Google Scholar]
- 27. Kumar M, Keller B, Makalou N, Sutton RE. 2001. Systematic determination of the packaging limit of lentiviral vectors. Hum. Gene Ther. 12:1893–1905 [DOI] [PubMed] [Google Scholar]
- 28. Lea SM, et al. 1998. Determination of the affinity and kinetic constants for the interaction between the human virus echovirus 11 and its cellular receptor, CD55. J. Biol. Chem. 273:30443–30447 [DOI] [PubMed] [Google Scholar]
- 29. Lecollinet S, et al. 2006. Improved gene delivery to intestinal mucosa by adenoviral vectors bearing subgroup B and d fibers. J. Virol. 80:2747–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leen AM, Rooney CM. 2005. Adenovirus as an emerging pathogen in immunocompromised patients. Br. J. Haematol. 128:135–144 [DOI] [PubMed] [Google Scholar]
- 31. Lemckert AA, et al. 2006. Generation of a novel replication-incompetent adenoviral vector derived from human adenovirus type 49: manufacture on PER. C6 cells, tropism and immunogenicity. J. Gen. Virol. 87:2891–2899 [DOI] [PubMed] [Google Scholar]
- 32. Lewis PF, et al. 2009. A community-based outbreak of severe respiratory illness caused by human adenovirus serotype 14. J. Infect. Dis. 199:1427–1434 [DOI] [PubMed] [Google Scholar]
- 33. Lortat-Jacob H, Chouin E, Cusack S, van Raaij MJ. 2001. Kinetic analysis of adenovirus fiber binding to its receptor reveals an avidity mechanism for trimeric receptor-ligand interactions. J. Biol. Chem. 276:9009–9015 [DOI] [PubMed] [Google Scholar]
- 34. Lozahic S, et al. 2000. CD46 (membrane cofactor protein) associates with multiple beta1 integrins and tetraspans. Eur. J. Immunol. 30:900–907 [DOI] [PubMed] [Google Scholar]
- 35. Lutschg V, Boucke K, Hemmi S, Greber UF. 2011. Chemotactic antiviral cytokines promote infectious apical entry of human adenovirus into polarized epithelial cells. Nat. Commun. 2:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mammen M, Choi S, Whitesides GM. 1998. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. 37:2754–2794 [DOI] [PubMed] [Google Scholar]
- 37. Marttila M, et al. 2005. CD46 is a cellular receptor for all species B adenoviruses except types 3 and 7. J. Virol. 79:14429–14436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meier O, Gastaldelli M, Boucke K, Hemmi S, Greber UF. 2005. Early steps of clathrin-mediated endocytosis involved in phagosomal escape of Fcgamma receptor-targeted adenovirus. J. Virol. 79:2604–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Metzgar D, et al. 2007. Abrupt emergence of diverse species B adenoviruses at US military recruit training centers. J. Infect. Dis. 196:1465–1473 [DOI] [PubMed] [Google Scholar]
- 40. Murakami M, et al. 2010. An adenoviral vector expressing human adenovirus 5 and 3 fiber proteins for targeting heterogeneous cell populations. Virology 407:196–205 [DOI] [PubMed] [Google Scholar]
- 41. Myszka DG. 1999. Improving biosensor analysis. J. Mol. Recognit. 12:279–284 [DOI] [PubMed] [Google Scholar]
- 42. Naniche D, et al. 1993. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 67:6025–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nemerow GR, Pache L, Reddy V, Stewart PL. 2009. Insights into adenovirus host cell interactions from structural studies. Virology 384:380–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nielsen UB, Adams GP, Weiner LM, Marks JD. 2000. Targeting of bivalent anti-ErbB2 diabody antibody fragments to tumor cells is independent of the intrinsic antibody affinity. Cancer Res. 60:6434–6440 [PubMed] [Google Scholar]
- 45. Pabbisetty KB, et al. 2007. Kinetic analysis of the binding of monomeric and dimeric ephrins to Eph receptors: correlation to function in a growth cone collapse assay. Protein Sci. 16:355–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pache L, Venkataraman S, Nemerow GR, Reddy VS. 2008. Conservation of fiber structure and CD46 usage by subgroup B2 adenoviruses. Virology 375:573–579 [DOI] [PubMed] [Google Scholar]
- 47. Pache L, Venkataraman S, Reddy VS, Nemerow GR. 2008. Structural variations in species B adenovirus fibers impact CD46 association. J. Virol. 82:7923–7931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Persson BD, et al. 2009. An arginine switch in the species B adenovirus knob determines high-affinity engagement of cellular receptor CD46. J. Virol. 83:673–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Persson BD, et al. 2007. Adenovirus type 11 binding alters the conformation of its receptor CD46. Nat. Struct. Mol. Biol. 14:164–166 [DOI] [PubMed] [Google Scholar]
- 50. Persson BD, et al. 2010. Structure of the extracellular portion of CD46 provides insights into its interactions with complement proteins and pathogens. PLoS Pathog. 6:e1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roelvink PW, Kovesdi I, Wickham TJ. 1996. Comparative analysis of adenovirus fiber-cell interaction: adenovirus type 2 (Ad2) and Ad9 utilize the same cellular fiber receptor but use different binding strategies for attachment. J. Virol. 70:7614–7621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sakurai F, Kawabata K, Mizuguchi H. 2007. Adenovirus vectors composed of subgroup B adenoviruses. Curr. Gene Ther. 7:229–238 [DOI] [PubMed] [Google Scholar]
- 53. Sakurai F, et al. 2006. The short consensus repeats 1 and 2, not the cytoplasmic domain, of human CD46 are crucial for infection of subgroup B adenovirus serotype 35. J. Control. Release 113:271–278 [DOI] [PubMed] [Google Scholar]
- 54. Schmitz H, Wigand R, Heinrich W. 1983. Worldwide epidemiology of human adenovirus infections. Am. J. Epidemiol. 117:455–466 [DOI] [PubMed] [Google Scholar]
- 55. Schmitz M, et al. 2006. Melanoma cultures show different susceptibility towards E1A-, E1B-19 kDa- and fiber-modified replication-competent adenoviruses. Gene Ther. 13:893–905 [DOI] [PubMed] [Google Scholar]
- 56. Segerman A, Arnberg N, Erikson A, Lindman K, Wadell G. 2003. There are two different species B adenovirus receptors: sBAR, common to species B1 and B2 adenoviruses, and sB2AR, exclusively used by species B2 adenoviruses. J. Virol. 77:1157–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Segerman A, et al. 2003. Adenovirus type 11 uses CD46 as a cellular receptor. J. Virol. 77:9183–9191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Seya T, et al. 1995. Blocking measles virus infection with a recombinant soluble form of, or monoclonal antibodies against, membrane cofactor protein of complement (CD46). Immunology 84:619–625 [PMC free article] [PubMed] [Google Scholar]
- 59. Sinn PL, Williams G, Vongpunsawad S, Cattaneo R, McCray PB., Jr 2002. Measles virus preferentially transduces the basolateral surface of well-differentiated human airway epithelia. J. Virol. 76:2403–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sirena D, et al. 2004. The human membrane cofactor CD46 is a receptor for species B adenovirus serotype 3. J. Virol. 78:4454–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sirena D, Ruzsics Z, Schaffner W, Greber UF, Hemmi S. 2005. The nucleotide sequence and a first generation gene transfer vector of species B human adenovirus serotype 3. Virology 343:283–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531–569 [DOI] [PubMed] [Google Scholar]
- 63. Stecher H, Shayakhmetov DM, Stamatoyannopoulos G, Lieber A. 2001. A capsid-modified adenovirus vector devoid of all viral genes: assessment of transduction and toxicity in human hematopoietic cells. Mol. Ther. 4:36–44 [DOI] [PubMed] [Google Scholar]
- 64. Strauss R, et al. 2009. Epithelial phenotype confers resistance of ovarian cancer cells to oncolytic adenoviruses. Cancer Res. 69:5115–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Suomalainen M, et al. 1999. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 144:657–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tang H, Kawabata A, Takemoto M, Yamanishi K, Mori Y. 2008. Human herpesvirus-6 infection induces the reorganization of membrane microdomains in target cells, which are required for virus entry. Virology 378:265–271 [DOI] [PubMed] [Google Scholar]
- 67. Tuve S, et al. 2006. A new group B adenovirus receptor is expressed at high levels on human stem and tumor cells. J. Virol. 80:12109–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wadell G. 2000. Adenoviruses, p 307–327 In Zuckerman AJ, Banatvala JE, Pattison JR. (ed), Principles and practice of clinical virology, 4th ed John Wiley & Sons, Inc, New York, NY. [Google Scholar]
- 69. Wang H, et al. 2011. Multimerization of adenovirus serotype 3 fiber knob domains is required for efficient binding of virus to desmoglein 2 and subsequent opening of epithelial junctions. J. Virol. 85:6390–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang H, et al. 2011. Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14. Nat. Med. 17:96–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang H, et al. 2007. Identification of CD46 binding sites within the adenovirus serotype 35 fiber knob. J. Virol. 81:12785–12792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang H, et al. 2008. In vitro and in vivo properties of adenovirus vectors with increased affinity to CD46. J. Virol. 82:10567–10579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang J. 2002. Protein recognition by cell surface receptors: physiological receptors versus virus interactions. Trends Biochem. Sci. 27:122–126 [DOI] [PubMed] [Google Scholar]
- 74. Wu E, et al. 2004. Membrane cofactor protein is a receptor for adenoviruses associated with epidemic keratoconjunctivitis. J. Virol. 78:3897–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zacharias DA, Violin JD, Newton AC, Tsien RY. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296:913–916 [DOI] [PubMed] [Google Scholar]
- 76. Zhang Y, Bergelson JM. 2005. Adenovirus receptors. J. Virol. 79:12125–12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhu Z, et al. 2009. Outbreak of acute respiratory disease in China caused by B2 species of adenovirus type 11. J. Clin. Microbiol. 47:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.