Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) encodes 12 pre-microRNAs (pre-miRNAs). Current studies have shown that these miRNAs are involved in regulation of viral and host gene expression, implicating a role in the maintenance of viral latency and suppression of antiviral innate immunity. However, the functions of these miRNAs remain largely unknown. On the basis of the sequence homology between oncogenic miR-155 and KSHV-encoded miR-K12-11, we hypothesized that miR-K12-11 could attenuate transforming growth factor β (TGF-β) signaling, facilitating viral infection and tumorigenesis. In the present study, we demonstrated that ectopic expression of miR-K12-11 in Ramos, a TGF-β-sensitive cell line, downregulated TGF-β signaling and facilitated cell proliferation upon TGF-β treatment by directly targeting SMAD5, an important mediator in TGF-β signaling. In addition, the downregulation of SMAD5 by miR-K12-11 was further confirmed in a de novo KSHV infection system or latently infected KSHV-positive B-lymphoma cell lines. More importantly, repression of miR-K12-11 by a specific sponge inhibitor restored the expression of SMAD5 in both de novo-infected and latently infected cells. Finally, we found that restoration of SMAD5, in addition to the TGF-β type II receptor, which was epigenetically silenced by the latent viral protein latency-associated nuclear antigen, sensitized BC3 cells to the cytostatic effect of TGF-β signaling. Taken together, our findings highlight a novel mechanism in which miR-K12-11 downregulates TGF-β signaling and suggest that viral miRNAs and proteins may exert a dichotomy regulation in virus-induced oncogenesis by targeting the same signaling pathway.
INTRODUCTION
Transforming growth factor β (TGF-β) and related growth factors are multifunctional cytokines involved in diverse biological processes, including embryonic development, regulation of cell growth, differentiation, hematopoiesis, immunity, and apoptosis (26, 28). The transductive networks involve the receptor serine/threonine kinase at the cell surface and their substrates, the SMAD proteins, which migrate into the nucleus, where they activate or repress target gene expression in association with DNA-binding partners (31). Distinct repertoires of receptors, SMAD proteins, and DNA-binding partners seemingly underlie, in a cell-specific manner, the multifunctional nature of TGF-β and related factors. The frequent mutations in these pathways often result in various forms of human cancer and developmental disorders (28), and it is now accepted that loss of TGF-β signaling is a key event in the development and progression of these diseases (5, 27, 28).
Recently, there were several studies that revealed a close relationship of microRNA (miRNA) hsa-miR-155 with TGF-β and bone morphogenetic protein (BMP) signaling (25, 35, 47, 48). miR-155 is a human oncogenic miRNA in which aberrant expression in a transgenic mouse (Eμ-miR-155) was associated with the development of lymphoblastic leukemia/high-grade lymphoma (10). In diffuse large-B-cell-lymphoma (DLBCL) cells, miR-155 has been reported to directly target the BMP-responsive transcriptional factor SMAD5, contributing to the development of lymphoid and myeloid malignancies (35). In addition, Flemington and colleagues (2008 and 2010) have shown that miR-155 inhibits BMP signaling by targeting SMAD1/5 (47, 48), which is important for the survival of Epstein-Barr virus (EBV) type III latency-associated tumors, through the inhibition of BMP-mediated viral reactivation and cell death. Finally, miR-155 has been implicated in the modulation of TGF-β signaling in myeloid cells by targeting SMAD2, with a significant effect on fibrosis, angiogenesis, and immunity (25). Interestingly, there is a viral ortholog of miR-155, miR-K12-11, which shares a 100% seed sequence homology and has been reported in Kaposi's sarcoma-associated herpesvirus (KSHV) (15, 39), a human lymphotropic gammaherpesvirus associated with Kaposi's sarcoma (KS) and B-cell lymphoma. On the basis of the seed sequence homology between miR-K12-11 and miR-155, it is reasonable to speculate that both miRNAs regulate a common set of target genes and, as a result, could have similar biological activities in TGF-β or BMP signaling.
Previously, several studies have described an interfering effect of KSHV-encoded proteins on TGF-β or BMP signaling. In a viral lytic infection, viral interferon regulatory factor 1 (vIRF-1) reportedly inhibited TGF-β signaling via direct interaction with both SMAD3 and SMAD4, where this interaction disrupted SMAD3-SMAD4 complex formation and DNA binding (38). K-bZIP, on the contrary, through its binding to CREB-binding protein (CBP), disrupted TGF-β signaling by interfering with the recruitment of CBP into transcription initiation complexes of TGF-β-responsive elements (43). Moreover, another KSHV lytic product, K5, was involved in the ubiquitination of the BMP type II receptor (BMPRII) by targeting the lysine residue (K180) in the cytoplasmic domain, leading to subsequent lysosomal degradation (13). In latent infections, however, it has been found that latency-associated nuclear antigens (LANAs), products of viral latent genes, can epigenetically silence TGF-β type II receptor (TβRII) gene expression, which contributes to the antiproliferative effects of TGF-β signaling (12). These studies have shown that in KSHV latent or lytic infections, TGF-β or BMP signaling, which has a wide range of effects in physiological processes (28), must be under stringent regulation to control its expression and activity.
Interestingly, KSHV-infected B-cell lines displayed decreased or nondetectable levels of miR-155 (39), suggesting that expression of viral miR-K12-11 may be substituted for expression of miR-155, which has been shown to modulate TGF-β or BMP signaling. On the basis of the similarity between miR-155 and miR-K12-11, we hypothesized that miR-K12-11 could attenuate TGF-β or BMP signaling, facilitating viral infection and tumorigenesis. Through the study described herein, we demonstrated that ectopic expression of miR-K12-11 in Ramos, a TGF-β-sensitive cell line, downregulated TGF-β signaling and facilitated cell proliferation upon TGF-β1 treatment by directly targeting SMAD5, which affected no obvious change in SMAD1 or SMAD2/3. Furthermore, the expression levels of SMAD5 were much lower in de novo-infected human embryonic kidney 293T (HEK293T) cells or latently infected KSHV-positive B-lymphoma cell lines than an uninfected control. Finally, inhibition of the effect of miR-K12-11 by the sponge inhibitor derepressed the expression of SMAD5, which then sensitized KSHV-positive PEL cells to TGF-β signaling. Altogether, our findings highlight a general phenomenon in which viral miRNAs and proteins may exert a dichotomy regulation in virus-induced oncogenesis by targeting the same signaling pathway.
MATERIALS AND METHODS
Plasmids and cell lines.
Wild-type or mutated 3′ untranslated region (UTR) reporters were constructed by cloning the appropriate 3′ UTR sequence into pGL3 cM (Promega, Madison, WI) downstream of the firefly luciferase gene. The primers used for amplification and mutagenesis are listed in Table 1. pCDH-miR-K12-11 and pCDH-sponge/K12-11 were constructed as reported previously (22). The cDNA clone of TβRII (NM_003242.5) was purchased from Sino Biological (Beijing, China) and further subcloned into the expression vector pCDH (System Biosciences, CA). The construct of SMAD5 was amplified from a cDNA library and inserted into pCMV-myc (Clontech, CA). KSHV-positive (BCBL1 and BC3) and -negative (BJAB, Ramos, and DG75) cell lines, generously provided by Erle S. Robertson (University of Pennsylvania), were grown in RPMI 1640 medium (HyClone, Beijing, China) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. The HEK293T cell line and Vero rKSHV.219 cells, a gift from Jeffrey Vieira (University of Washington), were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (HyClone, Beijing, China), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 μg/ml puromycin.
Table 1.
Sequences of primers used for PCR and qRT-PCR
| Name | Sequencea |
|---|---|
| SMAD1UTR-256KpnIF | TTTggtaccGCAGGACTTTGTGTACAG |
| SMAD1UTR-563XhoIR | TCCctcgagCAGCAAGTATGGTCAGCAT |
| SMAD2UTR-8050KpnIF | TGCggtaccAGCCAGAATGTGTTGTTAAC |
| SMAD2UTR-8700XhoIR | AACctcgagCATGGTAAACAACTCAAATGG |
| SMAD3UTR-4168KpnIF | TTTggtaccCAGCATCCAGAAACACCAAAC |
| SMAD3UTR-4388XhoIR | TCCctcgagACACATCAGAGTCCAGAACAG |
| SMAD5UTR-580KpnIF | TGAggtaccACTATGCACTGCTGTAACTGG |
| SMAD5UTR-930XhoIR | AAGctcgagAGGACGTAAGGATAAGCATC |
| SMAD5UTR-MRE1F | CAAGTATATGTCATTTACTCAAAATCTGTCATAAcgtaatCTTTATAGCTAGTGACAG |
| SMAD5UTR-MRE1R | CTGTCACTAGCTATAAAGattacgTTATGACAGATTTTGAGTAAATGACATATACTTG |
| SMAD5UTR-MRE2F | GACAATGTTGCAAGAACTCTATTTTTGACATGgtaattTCTTTTATTTTGCACTTTTAT |
| SMAD5utr-MRE2R | ATAAAAGTGCAAAATAAAAGAaattacCATGTCAAAAATAGAGTTCTTGCAACATTGTC |
| Tgfbr2-1EcoRIF | AATgaattcGCCACCATGGGTCGGGGGCTGCTCAG |
| Tgfbr2-1704BamHIR | AATggatccCTATTTGGTAGTGTTTAGGGAG |
| SMAD5qPCR-F | CCAGCAGCTGCAGCCTCAAAAT |
| SMAD5qPCR-R | TGCCGGTGATATTCTGCTCCCCAA |
| SMAD5-1SalIF | CGCgtcgacCATGACGTCAATGGCCAGCTT |
| SMAD5-1395NotIR | CGCgcggccgcCTTATGAAACAGAAGATATGG |
Lowercase nucleotides represent restriction enzyme recognition or PCR-based mutagenesis sites.
Antibodies and reagents.
Antibodies recognizing SMAD1, SMAD2/3, SMAD5, phosphorylated SMAD1/5 (p-SMAD1/5), p-SMAD2, and TβRII were purchased from Cell Signaling Technology (Danvers, MA). Recombinant human TGF-β1 and BMP2 were purchased from R&D Systems (Minneapolis, MN). β-Actin antibody, 12-O-tetradecanoylphorbol-13-acetate (TPA), valproate (VPA), and doxycycline hyclate (Dox) were obtained from Sigma (St. Louis, MO).
Dual luciferase reporter assay.
The dual luciferase reporter assay was comprised of two reporters: a Renilla luciferase expression construct, pRL-TK (Promega, Madison, WI), which was cotransfected as an internal control to correct for the differences in both transfection and harvest, and a firefly luciferase expression construct in pGL3 cM containing the assayed 3′ UTR sequences. For the luciferase reporter assay, HEK293T cells were plated in a 12-well plate and then cotransfected with 1 μg of either miR-K12-11 or an empty vector, 100 ng of either wild-type or a mutated 3′ UTR reporter, and 4 ng of pRL-TK, using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells were collected 48 h after transfection and analyzed using a dual luciferase reporter assay system (Promega, Madison, WI). Luciferase activity was detected using a Veritas luminometer (Turner BioSystems, Sunnyvale, CA). Firefly luciferase activity of each sample was normalized to Renilla luciferase activity. Transfection assays were done in triplicate in independent experiments.
RNA extraction and qRT-PCR.
The extraction of total RNA and quantification of SMAD5, miR-K12-11, and miR-155 were performed following a previously reported protocol (23). The primers used to detect SMAD5 are listed in Table 1, and bulge-loop miRNA quantitative real-time (qRT)-PCR primer sets (one reverse transcription primer and a pair of quantitative PCR primers for each set) specific for miR-K12-11 and miR-155 were designed by RiboBio (Guangzhou, China).
rKSHV.219 virus preparation and de novo infection.
Recombinant KSHV.219 (rKSHV.219) stocks were prepared from stable Vero cells as described previously, with minor modifications (45). Briefly, well-grown Vero cells (50 to 60% confluent) were synchronized by serum starvation for 24 h and then supplemented with fetal bovine serum to a final concentration of 10%. After culturing for an additional 12 to 16 h, cells were induced for 24 h with 25 ng/ml TPA and 3 mM VPA and then cultured in complete medium. At 72 h postinduction, the medium was collected, cells were removed by centrifugation (4,000 rpm for 20 min), and the supernatant was passed through a 0.45-μm-pore-size filter and then pelleted at 20,000 rpm for 2 h (Beckman SW28 rotor). Viral pellets were resuspended in medium and then aliquoted and stored at −80°C. The rKSHV.219 de novo infection of HEK293T cells was performed as described previously (32). Briefly, 70 to 80% confluent 293T cells were infected with viral stock in serum-free medium in a 12-well plate at a multiplicity of infection (MOI) of 10. Centrifugation enhancement was used for infection by centrifuging the culture plate at 450 × g for 120 min, with replacement of medium 2 h after centrifugation. After 48 h, the infection efficiency could be determined by counting the percentage of green fluorescent protein (GFP)-positive cells.
Lentiviral transduction.
To generate the stable and inducible cell line expressing miR-K12-11 upon Dox induction, pre-miR-K12-11 was subcloned into pLVX-tight-puro (Clontech, CA). HEK293T cells were transfected with packaging vectors and pLVX-Tet-On or pLVX-miR-K12-11 as reported previously (24). The viruses were collected from the culture supernatants on days 2 and 3 posttransfection, concentrated by ultracentrifugation for 1.5 h at 25,000 rpm, and resuspended in phosphate-buffered saline (PBS). Ramos cells in a 12-well plate were sequentially infected with lentiviral particles in the presence of 8 μg/ml Polybrene (Sigma, St. Louis, MO) and then incubated with 2 μg/ml puromycin and 1 mg/ml hygromycin for 2 weeks.
Cell proliferation and cell cycle assays.
To measure the effect of miR-K12-11 on growth rate, stable Ramos cells were initially treated with 2 μg/ml Dox for 48 h and then starved in serum-free medium overnight. Aliquots of cells (1 × 105) were spun down, resuspended in 2 ml reduced serum medium (1% fetal bovine serum), and treated with either 10 ng/ml TGF-β1 or vehicle over 3 days. Cell densities were measured every 24 h. For cell cycle profile studies, cells were harvested 72 h after exposure to 10 ng/ml TGF-β1 or vehicle and then identified by propidium iodide staining (Sigma, St. Louis, MO), followed by fluorescence-activated cell sorter analyses as previously described (11). To determine the effect of endogenous miR-K12-11 on cell proliferation, BC3 cells, after transduction with sponge construct or vector control, were electroporated with TβRII as mentioned above. An equal number of cells (1 × 105) was spun down, resuspended in 2 ml reduced serum medium (1% fetal bovine serum), and treated with either 100 ng/ml TGF-β1 or vehicle over 4 days. Cell density was determined every 24 h.
RESULTS
Ectopic expression of miR-K12-11 attenuates the cytostatic effect of TGF-β1 in Ramos cells.
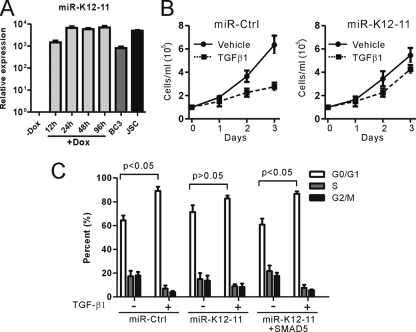
In order to study whether KSHV-encoded miR-K12-11 is involved in modulation of TGF-β signaling, we transduced TGF-β-sensitive Ramos cells, as described previously (2, 12, 40), with a lentiviral vector containing doxycycline-inducible miR-K12-11 or scrambled control miRNA and then selected with puromycin and blasticidin. In this inducible system, miR-K12-11 expression was induced over a period of 96 h after Dox treatment, which facilitated the determination of the effects of increased levels of miR-K12-11 on TGF-β signaling. First, we carried out bulge-loop qRT-PCR to confirm miR-K12-11 expression. After treatment with Dox, as expected, the expression of miR-K12-11 was highly upregulated to a level comparable to that of KSHV-positive cell line BC3 or JSC (Fig. 1A).
Fig 1.
Ectopic expression of miR-K12-11 attenuated the cytostatic effect of TGF-β1 in Ramos cells. (A) Ramos-teton-miR-K12-11 cells were treated with doxycycline to upregulate the expression of miR-K12-11. RNA from these samples was reverse-transcribed and then used to quantify the mature miR-K12-11 and housekeeping U6 levels with a bulge-loop qRT-PCR. KSHV-positive cell lines, BC3 or JSC, were used as a positive control. All values are expressed as fold induction of the untreated Ramos cells. (B) Growth of Ramos cells overexpressing miR-K12-11 or control miRNA treated with either TGF-β1 or vehicle over 3 days. Error bars represent SDs. (C) Cell cycle analyses show G0/G1 arrest after TGF-β1 exposure in Ramos cells overexpressing control miRNA, miR-K12-11, or miR-K12-11 in combination with SMAD5. The corresponding P value was calculated for each group, and P values are shown at the top (Student's t test).
To measure the effect of miR-K12-11 on cell growth or survival upon TGF-β stimulation, equal numbers of Ramos cells, after antibiotic selections and Dox induction for 48 h, were treated with either TGF-β1 or vehicle over 3 days. In line with previous published data (2), TGF-β treatment inhibited the proliferation of Ramos transduction with control miRNA by 50% compared to vehicle treatment (Fig. 1B, left). In contrast, treatment of Ramos cells transduced with miR-K12-11 resulted in no significant reduction in the proliferation rate (Fig. 1B, right). Next, we determined the influence of miR-K12-11 expression on TGF-β1-mediated cell cycle arrest and found that Ramos cell lines expressing miR-K12-11 either became refractory or had a diminished response to TGF-β1-mediated G0/G1 arrest, in contrast to controls (Fig. 1C). Taken together, these results demonstrated that aberrant expression of miR-K12-11 downregulated TGF-β signaling in Ramos cells, thereby promoting cell growth upon TGF-β1 treatment.
SMAD5 was the predominant target of miR-K12-11 in TGF-β signaling.
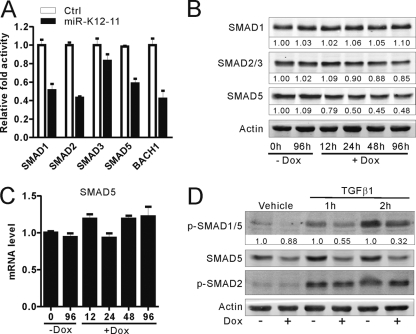
To identify the cellular targets of miR-K12-11 involved in modulation of TGF-β signaling, the TargetScan (http://www.targetscan.org/vert_42/seedmatch.html) and RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/welcome.html) tools were used to predict potential miRNA targets. On the basis of complementarities with the seed region of miR-K12-11, putative binding sites were predicted in the 3′ UTR of SMAD1/2/3/5. Interestingly, SMAD1, SMAD2, and SMAD5 have previously been reported to be targets of miR-155, the human ortholog of miR-K12-11, in different cell lines (25, 35, 47, 48). The 3′ UTR of SMAD1/2/3/5, including the putative miR-K12-11 responsive element (MRE), was cloned into the pGL3 control vector downstream of the luciferase gene. Using a dual luciferase reporter system, we found that overexpression of miR-K12-11 in HEK293T cells was able to repress SMAD1, SMAD2, or SMAD5 3′ UTR-driven luciferase activity by about 40 to 50% (Fig. 2A) compared to that of the control vector but had no significant effect on SMAD3 3′ UTR reporter activity. To validate these findings at the protein level, we utilized the stable miR-K12-11-inducible Ramos cell line mentioned above. After treatment with or without Dox for the indicated times, protein extracts were subjected to Western blotting. We found that ectopic expression of miR-K12-11 reduced the protein levels of SMAD5 significantly to 40 to 50% after 24 h induction but did not affect SMAD1 expression and had only a modest effect on SMAD2/3 (Fig. 2B). Taken together, these results indicated that SMAD5 is the predominant target of miR-K12-11 in the negative regulation of TGF-β signaling. Therefore, SMAD5 was selected for further study.
Fig 2.
SMAD5 was the predominant target of miR-K12-11. (A) miR-K12-11 downregulates SMAD1/2/3/5 3′ UTR reporter activity in HEK293T cells. One hundred nanograms pGL3-SMAD1- 3′ UTR, pGL3-SMAD2- 3′ UTR, pGL3-SMAD3- 3′ UTR, or pGL3-SMAD5- 3′ UTR was cotransfected with either pCDH-miR-K12-11 or pCDH-copGFP (1 μg) into HEK293T cells. The pGL3-BACH1 3′ UTR was used as a positive control. (B) Western blotting of SMAD1/2/3/5 in Ramos-teton-miR-K12-11 cells after induction with or without doxycycline for the indicated times. As a loading control, β-actin was also detected in the same blotting. Values represent percentages of SMAD1/2/3/5 normalized against β-actin and compared with an untreated control. (C) Total RNA of the same samples was reverse-transcribed and then used as a template to determine the SMAD5 mRNA levels by standard qRT-PCR. Values were normalized against β-actin. (D) Ramos-teton-miR-K12-11 cells were cultured with or without doxycycline for 48 h and then starved for 12 h before the addition of TGF-β1 (10 ng/ml). Lysates of these cells were taken 1 or 2 h after TGF-β1 stimulation to quantify the levels of phospho-SMAD1/5 and SMAD5 proteins by Western blotting. As a normalizing control, β-actin was also detected in the same blotting. Densitometry values represent the percentage of p-SMAD1/5 normalized against β-actin and compared with an uninduced control.
In order to determine whether miR-K12-11 regulates SMAD5 expression at the transcriptional or translational level, we carried out qRT-PCR to quantify the mRNA levels of SMAD5. We found that although SMAD5 protein levels were decreased by about 40 to 50% in miR-K12-11-expressing Ramos cells, its mRNA was slightly increased compared to that for untreated controls (Fig. 2C). These data suggested that miR-K12-11 could target endogenous SMAD5 expression by translational inhibition instead of direct mRNA degradation.
Since phosphorylated SMAD5 (p-SMAD5) is the functional form, we addressed whether miR-K12-11 affects its phosphorylation and determined the levels of p-SMAD5 in the presence of miR-K12-11 after TGF-β1 treatment. Typically, TGF-β1 signals are transduced via the heteromeric TβRII and TβRI complex to activate SMAD2/3 (26). However, noncanonical signals linking TGF-β1 to SMAD1/5 have recently been reported not only in endothelial and epithelial tissues but also in primary B cells and some B-lymphoma cell lines, such as diffuse large-B-cell-lymphoma (DLBCL) cells (2, 35). In Ramos cells, exposure to TGF-β1 resulted in phosphorylation of SMAD2 and also SMAD1/5 (Fig. 2D). After treatment of Ramos cells with or without Dox for 48 h, cells were starved overnight and then treated with vehicle or 10 ng/ml TGF-β1. p-SMAD1/5 levels were quantified by Western blotting at different time points spanning the first 2 h of induction. Over this period, upregulation of miR-K12-11 did affect the expression of SMAD5 as well as its phosphorylated form, p-SMAD1/5, the levels of which were reduced by 50 and 60%, respectively, whereas no significant changes in the levels of p-SMAD2 were observed (Fig. 2D). In addition, transfection of Ramos cells stably expressing miR-K12-11 with SMAD5 expression constructs rescued their sensitivity to TGF-β1-mediated G0/G1 arrest (Fig. 1C). On the basis of these experiments, we concluded that SMAD5 is the major target of miR-K12-11 in modulating TGF-β signaling in Ramos cells.
SMAD5 is a direct target of miR-K12-11.
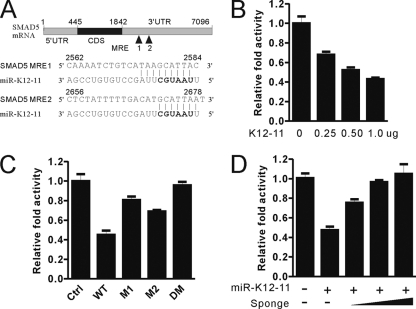
After showing the downregulation of SMAD5 in miR-K12-11-transduced Ramos cells, we then attempted to explore whether SMAD5 is a direct target of miR-K12-11. First, we found that SMAD5- 3′ UTR luciferase activity was inhibited by miR-K12-11 expression plasmids in a dose-dependent manner (Fig. 3B). Second, as SMAD5 was predicted to have two perfect matches with the seed sequence of miR-K12-11 (Fig. 3A), we speculated that the downregulation of SMAD5 by miR-K12-11 would be seed sequence dependent. We then constructed several reporter mutants: M1 with a mutation in MRE1, M2 with a mutation in MRE2, and DM with mutations in both MRE1 and MRE2. In the presence of the SMAD5- 3′ UTR, reporter M1 or M2 was able to partially resist the miR-K12-11 repression effect, whereas reporter DM totally abolished the miR-K12-11 repression effect compared to a wild-type (WT) reporter (Fig. 3C). Finally, we designed a specific miR-K12-11 sponge construct containing six tandem binding sites of miR-K12-11 immediately downstream of the enhanced GFP (EGFP) stop codon. Cotransfection of the sponge/K12-11 construct with WT reporter SMAD5 3′ UTR abrogated the repression effect of miR-K12-11 in a dose-dependent manner (Fig. 3D). These data collectively suggested that SMAD5 is a direct target of miR-K12-11.
Fig 3.
SMAD5 is the direct target of miR-K12-11 in TGF-β signaling pathway. (A) Diagram of two MREs within the 3′ UTR of SMAD5. The seed match sites of miR-K12-11 in the 3′ UTR were predicted by use of the TargetScan and RNAhybrid tools. (B) miR-K12-11 downregulates SMAD5 3′ UTR reporter activity in a dose-dependent manner. pGL3-SMAD5- 3′ UTR (100 ng) and pRL-SV40 (5 ng) were cotransfected with 0, 100, 200, or 500 ng of pCDH-miR-K12-11 into HEK293T cells, and the total mass of plasmids was compensated to 500 ng with the empty vector pCDH-copGFP. (C) Mutations of MREs in the SMAD5 3′ UTR reporter abolish miR-K12-11 downregulation effect. WT, wild type; M1, mutated MRE1; M2, mutated MRE2; DM, mutated MRE1 and MRE2. (D) Sponge reverses the miR-K12-11 repression effect on the SMAD5 3′ UTR reporter in a dose-dependent manner. HEK293T cells were cotransfected with pRL-SV40 (5 ng), pCDH-miR-K12-11 (500 ng), pGL3-SMAD5- 3′ UTR (100 ng), and 0, 100, 200, or 500 ng pCDH-sponge/K12-11, and the total mass of plasmids was compensated to 500 ng with an empty vector. All the reporter luciferase activity was normalized to Renilla luciferase activity. Data are presented as means ± SDs (n = 3).
SMAD5 was downregulated by miR-K12-11 in KSHV de novo-infected HEK293T cells.
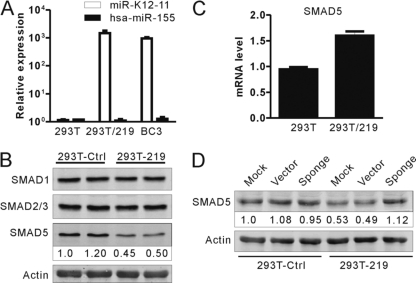
As expression of miR-K12-11 can downregulate endogenous SMAD5 expression in Ramos cells, we sought to determine whether miR-K12-11 could also regulate the expression of SMAD5 in a KSHV de novo infection system using the recombinant virus rKSHV.219. Concentrated rKSHV.219 viruses expressing GFP were used to infect HEK293T cells (MOI = 10). More than 95% of the HEK293T cells were found to be GFP positive at 72 h postinfection and were designated HEK293T/219. Then, we carried out bulge-loop qRT-PCR to confirm miRNA expression. We found that mature miR-K12-11 was highly expressed in HEK293T/219 cells and that expression was comparable to that in KSHV-positive BC3 cells but was completely absent in HEK293T control cells (Fig. 4A). Importantly, miR-155 was barely detectable in either HEK293T/219 or HEK293T cells (Fig. 4A). We found that SMAD5 protein levels were dramatically decreased in HEK293T/219 cells in comparison to the control cells, whereas no obvious changes in the protein levels of SMAD1 and SMAD2/3 were observed (Fig. 4B). In contrast, the mRNA levels of SMAD5 were not reduced but, rather, were upregulated in the HEK293T/219 cells (Fig. 4C). To determine whether inhibition of miR-K12-11 could rescue SMAD5 expression, HEK293T/219 cells were infected with lentiviruses through the sponge/K12-11 or an empty control vector. We found that the expression of SMAD5 was rescued in sponge/K12-11-expressing cells compared to control cells (Fig. 4D). These data indicated that miR-K12-11 can also downregulate SMAD5 expression in a KSHV de novo infection system.
Fig 4.
SMAD5 was downregulated by miR-K12-11 in de novo KSHV-infected HEK293T cells. (A) The expression levels of miR-K155 or miR-K12-11 were detected 72 h after rKSHV.219 infection (MOI = 10) in HEK293T cells and control cells. (B and C) SMAD5 expression was downregulated in response to rKSHV.219 de novo infection. Western blotting and qRT-PCR were used to detect SMAD5 expression at the protein and mRNA levels, respectively. Densitometry values represent the percentage of SMAD5 normalized against β-actin compared with control cells. (D) Sponge/K12-11 can rescue SMAD5 expression in HEK293T/219 cells. HEK293T/219 cells were transduced with a lentiviral miR-K12-11-specific sponge inhibitor or a control vector, and then the expression levels of SMAD5 and β-actin were determined by Western blotting.
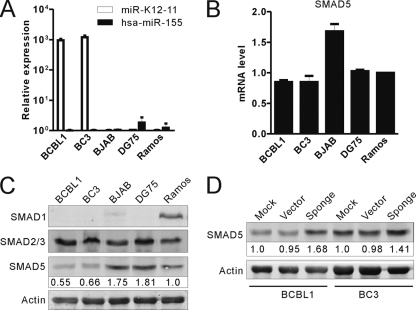
SMAD5 was significantly underexpressed in KSHV-positive PEL cells.
The experiments described above showed that expression of miR-K12-11 can regulate endogenous SMAD5 expression in either transduced Ramos cells or de novo-infected HEK293T cells. Next, we sought to determine whether SMAD5 was similarly regulated by miR-K12-11 in KSHV naturally infected B cells. First, we checked the endogenous expression level of SMAD5 in KSHV-infected PEL cells and compared it with that in KSHV-negative Burkitt's lymphoma cells. As shown in Fig. 5C, the protein levels of SMAD5 in KSHV-infected PEL cells are remarkably lower than those in KSHV-negative lymphoma cells, whereas the mRNA levels of SMAD5 varied (Fig. 5B). Based on these data, we inferred that SMAD5 was downregulated in PEL cells by miR-K12-11 at a posttranscriptional level. However, previous data showed that SMAD5 was also targeted by miR-155. We then sought to determine the expression levels of miR-155 in KSHV-positive or -negative lymphoma cells. Using bulge-loop qRT-PCR, we found that Ramos and DG75 cells have very low levels of miR-155 expression, while BJAB cells and the other two KSHV-positive cells, BCBL1 and BC3, had barely any miR-155 expression (Fig. 5A). These data suggested that miR-K12-11 played an important role in regulating SMAD5 expression in PEL cells through translational inhibition. In order to further confirm this hypothesis, we transduced BCBL1 or BC3 cells using sponge/K12-11–GFP or a control empty vector. At 48 h postinfection, more than 90% of the PEL cells were found to be GFP positive. In cells overexpressing sponge/K12-11–GFP, the protein levels of SMAD5 were rescued compared to vector control cells (Fig. 5D). Therefore, these data suggest that miR-K12-11 was likely responsible for the regulation of SMAD5 expression in KSHV-infected PEL cells.
Fig 5.
SMAD5 was significantly underexpressed in KSHV-positive PEL cells. (A) miR-K155 or miR-K12-11 expression levels were determined in the indicated cells by bulge-loop qRT-PCR. The percentage of SMAD5 normalized against β-actin and compared with the untreated control is shown. (B) The mRNA levels of SMAD5 in the indicated cells were detected by qRT-PCR. (C) Western blotting was used to monitor the expression levels of SMAD1/2/3/5 in KSHV-positive cells in contrast to KSHV-negative cells. Densitometry values denote the percentage of SMAD5 normalized against β-actin and compared with Ramos cells. (D) Sponge/K12-11 rescued SMAD5 expression in BCBL1 or BC3 cells.
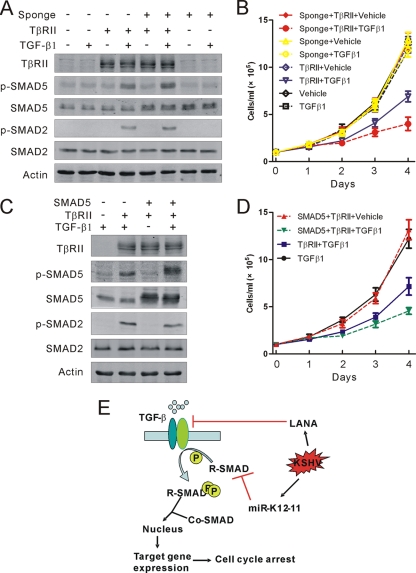
Restoration of SMAD5, in addition to TβRII, sensitizes KSHV-positive B-lymphoma cells to TGF-β signaling.
The above-described experiments showed that SMAD5 was an authentic target of miR-K12-11; however, the functional significance of the downregulation of SMAD5 by miR-K12-11 remained uncharacterized. As the data presented above showed that SMAD5 is essential for relaying signals initiated by TGF-β1 in Ramos cells, it was worthwhile to test whether it was also the case in KSHV-positive PEL cells, where TGF-β signaling has been reported to be blocked by LANA through the downregulation of TβRII by epigenetic modification (12). In line with these data, treatment of BC3 cells with TGF-β1 resulted in no phosphorylation of SMAD2 or SMAD5. To reconstitute TGF-β signaling in PEL cell lines, BC3 cells were transfected with TβRII expression constructs using Nucleofector (Lonza, Cologne, Germany). Treatment of TβRII-transfected BC3 cells with TGF-β1 resulted in phosphorylation of SMAD2 and SMAD5, leading to an efficient inhibition of the growth rate (Fig. 6A and B). However, inhibition of miR-K12-11 with the sponge inhibitor sensitized BC3 cells to TGF-β1 signaling and led to more SMAD5 phosphorylation and a further decrease in growth rate compared to the control vector (Fig. 6A and B). In addition, ectopic expression of exogenous SMAD5 in combination with TβRII in BC3 cells resulted in a similar but modest inhibition of cell growth upon TGF-β1 treatment (Fig. 6C and D). Collectively, these data further supported a functional link between miR-K12-11 and SMAD5 in KSHV-positive PEL cells, indicating that, in addition to LANA, miR-K12-11 also plays an important role in modulating the antiproliferative effect of TGF-β1 signaling.
Fig 6.
Restoration of SMAD5-sensitized, in addition to TβRII-sensitized, BC3 cells to TGF-β signaling. (A and C) Western blotting analyses of the TGF-β signaling cascade on the indicated treatment. (B and D) Growth curve of BC3 cells in response to the indicated treatment. (E) Working model of dichotomy regulation of KSHV on TGF-β signaling. On the one hand, the LANA protein silenced the expression of TβRII by epigenetic modification; on the other hand, miR-12-11 decreased the protein level of SMAD5. Both of them collectively lead to the abolition of TGF-β signaling in KSHV-infected B cells.
DISCUSSION
TGF-β1 is the prototypical member of a large family of pleiotropic cytokines that play an important role in the regulation of survival, proliferation, and differentiation of various cell types (5, 26). In most epithelial, endothelial, and hematopoietic cells, including B lymphocytes, TGF-β1 is a potent inhibitor of cell proliferation; hence, TGF-β1 may suppress tumor progression in the early stages of tumorigenesis (6, 21). Viruses cause about 15% of all human cancers and have developed various strategies to evade the inhibitory effect of TGF-β signaling. In this study, we demonstrated that viral miRNA-mediated gene regulation is used by KSHV to control TGF-β signaling. Ectopic expression of miR-K12-11 attenuated TGF-β signaling in a TGF-β1-sensitive cell line (Ramos), thereby facilitating cell proliferation. In addition, SMAD5, rather than SMAD1 or SMAD2/3, is found to be a direct and predominant target of miR-K12-11 in the TGF-β signaling pathway. More importantly, the expression of SMAD5 is much lower in de novo rKSHV.219-infected HEK293T and latently infected PEL cells than negative cells. Finally, inhibition of miR-K12-11 resulted in derepression of SMAD5, which then sensitized PEL cells to TGF-β signaling. During the preparation of the manuscript, Renne and colleagues validated that miR-K12-11 is a functional ortholog of miR-155 in hematopoiesis in vivo and its overexpression led to a significant expansion of the CD19+ B-cell population in the spleen by targeting CCAAT enhancer-binding protein beta (CEBPβ) (7). This result complemented our novel finding that expression of miR-K12-11 in B lymphocytes latently infected with KSHV may be substituted for miR-155 in the modulation of TGF-β signaling. Since miR-K12-11 is largely expressed during viral latency, we believe that miR-K12-11 is important for the modulation of TGF-β signaling during KSHV latency; however, further studies are still needed to clarify its roles in vivo.
The effect of oncogenic viral infection on TGF-β signaling has been described for several other viruses, including human T-cell lymphotropic virus type 1 (HTLV-1), adenovirus, human papillomavirus (HPV), hepatitis C virus (HCV), Marek's disease virus type 1 (MDV1), and Epstein-Barr virus (EBV) (1, 9, 18, 19, 29, 30, 33, 41, 46). In HTLV-1-infected adult T-cell leukemia (ATL) cells, the oncoprotein Tax induced the production of TGF-β1, which is an inhibitor of T-cell proliferation and cytotoxicity and confers resistance to TGF-β1-induced growth inhibition through JNK/c-Jun constitutive activation (1, 18, 29). Adenovirus E1A blocks TGF-β responses through its interaction with CBP/p300, preventing the transcriptional activity of SMADs and downregulation of TβRII (33, 41). In HCV infection, core and NS3 physically interact with the MH1 region of the SMAD3 and block TGF-β/SMAD3-mediated transcriptional activation through interference with the DNA-binding ability but not the nuclear translocation ability of SMAD3 (9). In addition to virus-encoded proteins, MDV1-miR-M3 has been reported to suppress cisplatin-induced apoptosis by directly downregulating the expression of SMAD2 (46). Here, we showed that KSHV has evolved multidimensional mechanisms for blocking TGF-β signaling. In addition to LANA-mediated epigenetic silencing of TβRII expression (12), miR-K12-11 also played an important role in inhibiting this pathway by downregulating SMAD5. PEL cell lines are defective in TGF-β signaling because of a lack of TβRII expression. Restoration of TβRII expression establishes pathway function and sensitivity to its growth-inhibitory effects. However, inhibition of miR-K12-11 represses the expression of SMAD5, leading to a further inhibition of its growth rate. Our findings highlighted a general phenomenon of virus-induced tumorigenesis, which can be summarized as shown in Fig. 6E. Interestingly, through immunohistochemical staining for primary cases of Kaposi's sarcoma (KS) and multicentric Castleman disease (MCD) (3, 12), previous studies have shown that TβRII is still present in some cells expressing LANA protein. These data indicated that in addition to LANA, other factors, such as miRNAs, also played an important role in the modulation of the TGF-β signaling pathway. During latency, both virus-encoded proteins and miRNAs will function synergistically to modulate the same signaling pathway, proactively creating a cellular environment beneficial to viral latency and oncogenesis.
Viral miRNAs have recently become an emerging hot spot in the study of host-virus interaction. In total, 12 genes were identified within the major latency-associated region of the KSHV genome, giving rise to at least 17 mature miRNAs (8, 16, 34, 36, 44). To date, a few studies have shown that KSHV-encoded miRNAs are involved in the regulation of viral and host gene expression, playing an important role in the maintenance of viral latency and suppression of antiviral innate immunity (4, 17, 20, 22, 23, 49). However, the molecular mechanisms by which these miRNAs modulate the process of viral pathogenesis and the behavior of host cells remain largely unknown. We were especially interested in miR-K12-11, which has a 100% seed sequence homology to human miR-155, shown to be the first oncogenic miRNA and a critical regulator of B-cell development and oncogenesis (15, 39). Increasing evidence pointed out that miR-155 was involved in numerous biological processes, including hematopoiesis, inflammation, and immunity (14). Deregulation of miR-155 has been found to be associated with different kinds of cancer, cardiovascular diseases, and viral infections (42). Thus far, there are more than 40 predicted target genes that have been confirmed experimentally, including ARID2, BACH1, CEBPβ, ETS1, IKBKE, and PU.1 (14). However, in previous studies, only a few common target genes have been identified using reporter assays and gene expression profiles, including BACH1, FOS, TM6SF1, and IKBKE (15, 22, 39). One explanation for this phenomenon is the contribution of sequences outside the seed region to mRNA targeting and target-site selection, although both miR-155 and miR-K12-11 share the same seed sequence. In addition, most target genes are downregulated by miRNAs through translational inhibition rather than mRNA degradation. These targets will be ignored when using gene expression profile assays. Lastly, although some targets, such as SMAD1, can be repressed by miR-K12-11 and miR-155 in reporter assays (Fig. 2), they may not be downregulated by endogenous miR-K12-11 at the protein level. However, the reason for this inconsistency is complicated and needs to be further analyzed. Hence, a detailed analysis of additional targets will aid in providing a better understanding of the function of this viral ortholog in B-cell expansion and oncogenesis and the contribution of sequences outside the seed region in miRNA targeting.
KSHV has evolved multiple strategies to prevent infected cells from growth inhibition by TGF-β signaling (12, 37, 43). Latent infections are presumably requisites for cancer development. Our data have shown that in addition to LANA and several miRNAs previously found to target THBS1 (37), miR-K12-11, the viral ortholog of miR-155, attenuates TGF-β signaling by directly targeting SMAD5, suggesting a novel mechanism by which miRNAs contribute to lymphomagenesis.
ACKNOWLEDGMENTS
We are grateful to Erle S. Robertson (University of Pennsylvania) for critical reading of the manuscript.
This work was supported by grants from the National Basic Research Program of China (2011CB504805 and 2012CB519002), the Natural Science Foundation of China (30770098 and 30970154), and the 100 Talent Program of the Chinese Academy of Sciences, to K.L.
Footnotes
Published ahead of print 19 October 2011
REFERENCES
- 1. Arnulf B, et al. 2002. Human T-cell lymphotropic virus oncoprotein Tax represses TGF-beta 1 signaling in human T cells via c-Jun activation: a potential mechanism of HTLV-I leukemogenesis. Blood 100:4129–4138 [DOI] [PubMed] [Google Scholar]
- 2. Bakkebo M, Huse K, Hilden VI, Smeland EB, Oksvold MP. 2010. TGF-beta-induced growth inhibition in B-cell lymphoma correlates with Smad1/5 signalling and constitutively active p38 MAPK. BMC Immunol. 11:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barbera AJ, Kaye KM. 2008. KSHV LANA's expanding bag of tricks. Blood 111:4425–4426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bellare P, Ganem D. 2009. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 6:570–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bierie B, Moses HL. 2006. TGF-beta and cancer. Cytokine Growth Factor Rev. 17:29–40 [DOI] [PubMed] [Google Scholar]
- 6. Blobe GC, Schiemann WP, Lodish HF. 2000. Role of transforming growth factor beta in human disease. N. Engl. J. Med. 342:1350–1358 [DOI] [PubMed] [Google Scholar]
- 7. Boss IW, et al. 2011. A Kaposi's sarcoma-associated herpesvirus-encoded ortholog of microRNA miR-155 induces human splenic B-cell expansion in NOD/LtSz-scid IL2Rγ null mice. J. Virol. 85:9877–9886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cai X, et al. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. U. S. A. 102:5570–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng PL, Chang MH, Chao CH, Lee YH. 2004. Hepatitis C viral proteins interact with Smad3 and differentially regulate TGF-beta/Smad3-mediated transcriptional activation. Oncogene 23:7821–7838 [DOI] [PubMed] [Google Scholar]
- 10. Costinean S, et al. 2009. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood 114:1374–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Darzynkiewicz Z, Juan G, Bedner E. 2001. Determining cell cycle stages by flow cytometry. Curr. Protoc. Cell Biol., May, Unit 8.4. [DOI] [PubMed] [Google Scholar]
- 12. Di Bartolo DL, et al. 2008. KSHV LANA inhibits TGF-beta signaling through epigenetic silencing of the TGF-beta type II receptor. Blood 111:4731–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Durrington HJ, et al. 2010. Identification of a lysosomal pathway regulating degradation of the bone morphogenetic protein receptor type II. J. Biol. Chem. 285:37641–37649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Faraoni I, Antonetti FR, Cardone J, Bonmassar E. 2009. miR-155 gene: a typical multifunctional microRNA. Biochim. Biophys. Acta 1792:497–505 [DOI] [PubMed] [Google Scholar]
- 15. Gottwein E, et al. 2007. A viral microRNA functions as an orthologue of cellular miR-155. Nature 450:1096–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grundhoff A, Sullivan CS, Ganem D. 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12:733–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lagos D, et al. 2010. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat. Cell Biol. 12:513–519 [DOI] [PubMed] [Google Scholar]
- 18. Lee DK, Kim BC, Brady JN, Jeang KT, Kim SJ. 2002. Human T-cell lymphotropic virus type 1 tax inhibits transforming growth factor-beta signaling by blocking the association of Smad proteins with Smad-binding element. J. Biol. Chem. 277:33766–33775 [DOI] [PubMed] [Google Scholar]
- 19. Lee DK, et al. 2002. The human papilloma virus E7 oncoprotein inhibits transforming growth factor-beta signaling by blocking binding of the Smad complex to its target sequence. J. Biol. Chem. 277:38557–38564 [DOI] [PubMed] [Google Scholar]
- 20. Lei X, et al. 2010. Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA. Nat. Cell Biol. 12:193–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Levy L, Hill CS. 2006. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 17:41–58 [DOI] [PubMed] [Google Scholar]
- 22. Liang D, et al. 2011. A human herpesvirus miRNA attenuates interferon signaling and contributes to maintenance of viral latency by targeting IKK ε. Cell Res. 21:793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin X, et al. 2011. miR-K12-7-5p encoded by Kaposi's sarcoma-associated herpesvirus stabilizes the latent state by targeting viral ORF50/RTA. PLoS One 6:e16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu Y, et al. 2008. Kaposi's sarcoma-associated herpesvirus RTA activates the processivity factor ORF59 through interaction with RBP-Jkappa and a cis-acting RTA responsive element. Virology 380:264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Louafi F, Martinez-Nunez RT, Sanchez-Elsner T. 2010. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-{beta}. J. Biol. Chem. 285:41328–41336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Massague J. 1998. TGF-beta signal transduction. Annu. Rev. Biochem. 67:753–791 [DOI] [PubMed] [Google Scholar]
- 27. Massague J. 2008. TGF[beta] in cancer. Cell 134:215–23018662538 [Google Scholar]
- 28. Massague J. 1990. The transforming growth factor-beta family. Annu. Rev. Cell Biol. 6:597–641 [DOI] [PubMed] [Google Scholar]
- 29. Mori N, et al. 2001. Human T-cell leukemia virus type I oncoprotein Tax represses Smad-dependent transforming growth factor beta signaling through interaction with CREB-binding protein/p300. Blood 97:2137–2144 [DOI] [PubMed] [Google Scholar]
- 30. Mori N, Morishita M, Tsukazaki T, Yamamoto N. 2003. Repression of Smad-dependent transforming growth factor-beta signaling by Epstein-Barr virus latent membrane protein 1 through nuclear factor-kappaB. Int. J. Cancer 105:661–668 [DOI] [PubMed] [Google Scholar]
- 31. Moustakas A, Heldin CH. 2009. The regulation of TGFbeta signal transduction. Development 136:3699–3714 [DOI] [PubMed] [Google Scholar]
- 32. Myoung J, Ganem D. 2011. Infection of primary human tonsillar lymphoid cells by KSHV reveals frequent but abortive infection of T cells. Virology 413:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nishihara A, Hanai J, Imamura T, Miyazono K, Kawabata M. 1999. E1A inhibits transforming growth factor-beta signaling through binding to Smad proteins. J. Biol. Chem. 274:28716–28723 [DOI] [PubMed] [Google Scholar]
- 34. Pfeffer S, et al. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269–276 [DOI] [PubMed] [Google Scholar]
- 35. Rai D, Kim SW, McKeller MR, Dahia PL, Aguiar RC. 2010. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc. Natl. Acad. Sci. U. S. A. 107:3111–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Samols MA, Hu J, Skalsky RL, Renne R. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:9301–9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Samols MA, et al. 2007. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 3:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seo T, Park J, Choe J. 2005. Kaposi's sarcoma-associated herpesvirus viral IFN regulatory factor 1 inhibits transforming growth factor-beta signaling. Cancer Res. 65:1738–1747 [DOI] [PubMed] [Google Scholar]
- 39. Skalsky RL, et al. 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 81:12836–12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Spender LC, et al. 2009. TGF-beta induces apoptosis in human B cells by transcriptional regulation of BIK and BCL-XL. Cell Death Differ. 16:593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tarakanova VL, Wold WS. 2003. Transforming growth factor beta1 receptor II is downregulated by E1A in adenovirus-infected cells. J. Virol. 77:9324–9336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tili E, Croce CM, Michaille JJ. 2009. miR-155: on the crosstalk between inflammation and cancer. Int. Rev. Immunol. 28:264–284 [DOI] [PubMed] [Google Scholar]
- 43. Tomita M, Choe J, Tsukazaki T, Mori N. 2004. The Kaposi's sarcoma-associated herpesvirus K-bZIP protein represses transforming growth factor beta signaling through interaction with CREB-binding protein. Oncogene 23:8272–8281 [DOI] [PubMed] [Google Scholar]
- 44. Umbach JL, Cullen BR. 2010. In-depth analysis of Kaposi's sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J. Virol. 84:695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vieira J, O'Hearn PM. 2004. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology 325:225–240 [DOI] [PubMed] [Google Scholar]
- 46. Xu S, Xue C, Li J, Bi Y, Cao Y. 2011. Marek's disease virus type 1 microRNA miR-M3 suppresses cisplatin-induced apoptosis by targeting Smad2 of the transforming growth factor beta signal pathway. J. Virol. 85:276–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yin Q, et al. 2008. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J. Virol. 82:5295–5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yin Q, et al. 2010. MicroRNA miR-155 inhibits bone morphogenetic protein (BMP) signaling and BMP-mediated Epstein-Barr virus reactivation. J. Virol. 84:6318–6327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ziegelbauer JM. 2011. Functions of Kaposi's sarcoma-associated herpesvirus microRNAs. Biochim. Biophys. Acta 1809:623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]