Abstract
Composed of 35 amino acids, O3 is the smallest characterized protein encoded by vaccinia virus (VACV) and is an integral component of the entry-fusion complex (EFC). O3 is conserved with 100% identity in all orthopoxviruses except for monkeypox viruses, whose O3 homologs have 2 to 3 amino acid substitutions. Since O3 is part of the EFC, high conservation could suggest an immutable requirement for interaction with multiple proteins. Chordopoxviruses of other genera also encode small proteins with a characteristic predicted N-terminal α-helical hydrophobic domain followed by basic amino acids and proline in the same relative genome location as that of VACV O3. However, the statistical significance of their similarity to VACV O3 is low due to the large contribution of the transmembrane domain, their small size, and their sequence diversity. Nevertheless, trans-complementation experiments demonstrated the ability of a representative O3-like protein from each chordopoxvirus genus to rescue the infectivity of a VACV mutant that was unable to express endogenous O3. Moreover, recombinant viruses expressing O3 homologs in place of O3 replicated and formed plaques as well or nearly as well as wild-type VACV. The O3 homologs expressed by the recombinant VACVs were incorporated into the membranes of mature virions and, with one exception, remained stably associated with the detergent-extracted and affinity-purified EFC. The ability of the sequence-divergent O3 homologs to coordinate function with VACV entry proteins suggests the conservation of structural motifs. Analysis of chimeras formed by swapping domains of O3 with those of other proteins indicated that the N-terminal transmembrane segment was responsible for EFC interactions and for the complementation of infectivity.
INTRODUCTION
Vaccinia virus (VACV), the best-studied member of the Poxviridae, is a large DNA virus that encodes about 200 proteins and replicates entirely in the cytoplasm of infected cells (17). The mature virion (MV) enters cells via the plasma membrane at a neutral pH or by a low-pH-dependent endosomal route following macropinocytosis or fluid-phase uptake (1, 11, 16, 18, 27). Four proteins participate in cell attachment via glycosaminoglycans and laminin (6, 7, 10, 15), but no specific cell receptor has been identified. At least 12 MV proteins participate in membrane fusion and core entry; with one possible exception, these proteins are physically associated to form the entry-fusion complex (EFC) (3, 4, 13, 19–26). The roles of the entry proteins have been determined mainly by analysis of inducible conditional lethal mutants. Under nonpermissive conditions, normal-appearing but noninfectious virions form. These defective virions can bind to cells but are unable to mediate membrane fusion and core entry. In addition, the absence of individual EFC proteins destabilizes the entire complex.
The protein requirements for the entry of poxviruses into cells have been studied exclusively with VACV. However, homologs of 11 of the 12 EFC proteins are highly conserved in all chordopoxviruses, making it likely that they use the same entry mechanism. A possible exception is the VACV O3 protein. The O3L open reading frame (ORF), encoding the O3 protein, was not annotated originally because of its small size (35 amino acids). Nevertheless, we demonstrated that the protein was expressed and that the phenotype of an inducible mutant under nonpermissive conditions was similar to that of other EFC mutants (22). Although a VACV O3L deletion mutant was constructed, it replicated poorly, and entry was very inefficient. A BLAST search against the translated nucleotide database (tblastn) revealed homologous proteins with 100% sequence identity in other orthopoxviruses; the exception was the monkeypox virus homolog, with 2 to 3 amino acid substitutions. However, BLAST hits were not found in all chordopoxvirus genera. Moreover, the statistical significance of the hits was weakened because of the small size of the proteins and the bias caused by the contribution of the transmembrane (TM) domain (30). Nevertheless, an ORF predicting a small hydrophobic protein is located in the same relative position and orientation in all chordopoxvirus genomes (22). Since the O3 protein is required to stabilize the VACV EFC, presumably through multiple protein interactions (22), it seemed possible that the sequence-divergent O3-like proteins might exhibit genus specificity. Remarkably, we found that the related proteins of other chordopoxviruses can complement a VACV O3 deficiency despite extensive sequence diversity. Furthermore, we demonstrate the importance of the N-terminal hydrophobic domain in EFC interactions and the function of the O3 homologs.
MATERIALS AND METHODS
Cells and viruses.
BS-C-1 cells were maintained in minimal essential medium with Earle's salts supplemented with 100 U of penicillin and streptomycin, 10% fetal bovine serum, and 2 mM l-glutamine (Quality Biologicals, Gaithersburg, MD). The VACV Western Reserve (WR) strain, vO3-HAi (an O3-inducible virus), and vO3Δ (a virus lacking the O3L ORF) have been described previously (22). The vO3-HAi virus expressing firefly luciferase (LUC) under the control of the strong synthetic early/late promoter (vO3-HAi-Luc) (5) was generated by inserting the gene cassette between the VACV F12L and F13L gene loci. Cyan fluorescent protein (CFP) expressed by the late P11 promoter (2) was used as a reporter to pick recombinant virus. VACV MVs were purified by sedimentation through a 36% sucrose cushion, followed by banding in a 25-to-40% sucrose density gradient (8).
Gene synthesis.
O3L ORFs from chordopoxviruses were synthesized by GeneArt (Carlsbad, CA) and were cloned into its pMK vector.
Plaque assay and determination of virus yields.
Plaque assays were performed in BS-C-1 cell monolayers in 12-well culture plates with 10-fold serial dilutions of virus. The virus was adsorbed for 1 h at room temperature; unbound virus was removed; and the cells were washed and incubated with a medium containing a 0.5% methylcellulose solution in the presence of 100 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 48 h. Plaques were stained with crystal violet and were counted.
For trans-complementation assays, the plasmid concentration was estimated using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE). BS-C-1 cells were infected with the O3-inducible virus in the absence of IPTG, and equivalent amounts of plasmid were transfected using the Lipofectamine 2000 reagent (Invitrogen). After 24 h, cells were harvested and were lysed by freeze-thawing. The virus yield was determined either by a plaque assay in the presence of IPTG or by an entry assay based on LUC expression at 90 min after infection.
Generation of O3L replacement viruses.
Replacement viruses were generated from the vO3Δ deletion mutant. O3L genes or chimeras from other chordopoxviruses were inserted with DsRED in the place of green fluorescent protein (GFP) by homologous recombination. The recombinant viruses were isolated based on DsRED expression and were clonally purified. The identity of the final virus was confirmed by sequencing the mutated genome segment. Replacement viruses expressing hemagglutinin (HA)-tagged VACV or ORFV O3 (VACV O3-HA or ORFV O3-HA) were further modified by incorporating the VACV A28 protein expressing tandem affinity calmodulin- and streptavidin-binding peptide tags (A28-TAP) as described previously (22).
Western blot analysis.
Proteins of whole-cell lysates and purified virions were separated in 4 to 12% Novex NuPAGE acrylamide gels with 2-(N-morpholino)ethanesulfonic acid buffer and were transferred to nitrocellulose membranes using the iBlot system (Invitrogen). The membrane was blocked with 5% nonfat milk in phosphate-buffered saline with 0.05% Tween 20. The membrane was incubated for 1 h at room temperature or overnight at 4°C in the same solution with primary antibodies at appropriate dilutions. Excess antibodies were removed by washing with phosphate-buffered saline containing Tween 20 followed by phosphate-buffered saline without detergent. IRDye 680- or IRDye 800-conjugated secondary antibodies against mouse and rabbit antibodies were added, and the mixture was incubated for 1 h at room temperature, washed, and developed using an Odyssey infrared imager (LI-COR Biosciences, Lincoln, NE). A mouse monoclonal antibody (MAb) against the human influenza virus HA epitope (Covance, Princeton, NJ) and a rabbit anti-calmodulin-binding peptide antibody (Immunology Consultants Laboratory, Portland, OR) were used. Antibodies against VACV entry-fusion proteins have been described previously (22).
Affinity purification of the EFC.
BS-C-1 cells were infected with recombinant viruses containing an HA-tagged protein or a streptavidin- and calmodulin-binding peptide tandem affinity purification (TAP)-tagged protein. Cells were lysed in 50 mM Tris, 200 mM NaCl, 1% Triton X-100, 5 mM EDTA, and a protease inhibitor cocktail (Roche, Indianapolis, IN) for 1 h at 4°C. Lysates were centrifuged at 15,000 × g to obtain a soluble extract and were bound either to an anti-HA affinity matrix (Roche) or to streptavidin-agarose conjugate beads (Millipore, Billerica, MA) for 12 h at 4°C. The beads were washed in the same buffer, and bound proteins were either eluted with biotin (for TAP proteins) or boiled directly with a lithium dodecyl sulfate loading buffer. Affinity-purified proteins were identified by Western blotting with specific antibodies.
Extraction of membrane protein from virions.
Purified virions were incubated with 50 mM Tris, 200 mM NaCl, 50 mM dithiothreitol, and 1% NP-40 buffer for 1 h at 37°C. Soluble membrane proteins were detected in the detergent phase after centrifugation at 15,000 × g for 30 min at 4°C. Equivalent amounts of soluble and insoluble extracts were subjected to Western blot analysis in order to determine protein distribution.
Low-pH-induced syncytium formation.
BS-C-1 cells were infected with viruses at a multiplicity of infection of 2 PFU per cell. After 16 h, cells were treated with a pH 5.5 or pH 7.4 buffer for 2 min. The cells were washed and were incubated with fresh medium for an additional 3 h. The live cells were viewed by phase-contrast microscopy to visualize syncytia.
RESULTS
Comparison of O3L-like genes of chordopoxviruses.
The O3 protein of VACV is conserved with 100% identity in all orthopoxvirus species except monkeypox viruses, which have only 2 to 3 amino acid substitutions. Since the O3 protein is part of the EFC, such high conservation could suggest an immutable requirement for interaction with multiple proteins. In contrast to the results for other EFC proteins, tblast searches did not detect O3 homologs in all chordopoxviruses, and the statistical significances of those found were low due to their small size and sequence variability. In addition, the hydrophobic transmembrane domain imposes an amino acid bias that inflates the percentage of identity to VACV O3 and the apparent significance of the match (30). In fact, very few amino acids in O3 are highly conserved throughout the genera (Table 1). Nevertheless, common features, including the location in the genome, the presence of isoleucine- or leucine-valine immediately after the initiating methionine, the presence of a predicted N-terminal membrane-spanning α-helix followed by a positively charged amino acid residue (mostly arginine or lysine) that may determine membrane orientation (28) and a helix-breaking proline that may provide flexibility for the C-terminal segment, suggested that all of the proteins are bona fide homologs (Table 1). We therefore set out to test whether the predicted O3 homologs could functionally replace VACV O3.
Table 1.
Comparison of O3-like proteins of chordopoxviruses
| Genusa | Species | Sequenceb | % IDc | Ed |
|---|---|---|---|---|
| OP | Variola virus Congo-1970 | MLVVIMFFIAFAFCSWLSYSYLRPYISTKELNKSR | 100 | 2e−14 |
| OP | Monkeypox virus Zaire-96-I-16 | MLVVIMFFIAFVFCSWLSYNYLCPYISTKELNKSR | 91.4 | 3e−12 |
| CPe | Lumpy skin disease virus NI-2490 | MIVLVFFFIAFSFCVWISYSFLKPY | 48.6 | 0.003 |
| SP | Swinepox virus 17077-99 | MIVFIMFVIAFSFAVWLSYTFLRPYMINENI | 48.6 | 0.005 |
| LP | Myxoma virus 6918 | MIVFVIFIIAFVFCGWISYGFLKPYMFLNRKH | 45.7 | 0.022 |
| YP | Yaba monkey tumor virus | MIVFTIFIIAFGFCIWLSYLSLKPYVVIT | 42.9 | 0.04 |
| AP | Fowlpox virus HP-440 | MIVTVLFLIMFFICTLYSYHYLKPWIFYVEREVT | 37.1 | >0.1 |
| MP | Molluscum contagiosum virus | MLVTLIFYLAFALCAAYAVAFLRPFLLLNSDLDAAPVARRE | 29.5 | >0.1 |
| PP | Orf virus OV-SA00 | MIVVFVWLLALTVGMWMAWARMGPFLRSANAADRHEPPAGPTIRNPRP | 14.0 | >0.1 |
OP, Orthopoxvirus; CP, Capripoxvirus; SP, Suipoxvirus; LP, Leporipoxvirus; YP, Yatapoxvirus; AP, Avipoxvirus; MP, Molluscipoxvirus; PP, Parapoxvirus.
Sequences recognized by a tblast search against VACV O3 are underlined; predicted α-helical domains are italicized; boldface indicates a basic amino acid and proline following a TM domain.
ID, amino acid identity.
E, expectation value.
The sheeppox virus O3 homolog (MIVLFFFFIAFSFCVWISYSFLKPYTYKV>) was used for expression studies.
trans-Complementation of VACV infection with chordopoxvirus O3L-like genes.
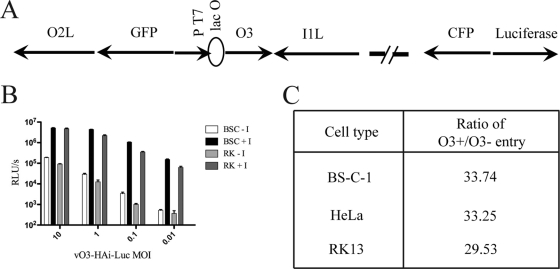
A new recombinant VACV was engineered in order to help determine the roles of O3 homologs in trans-complementation virus entry assays. vO3-HAi-Luc was derived from the previously described epitope-tagged, O3-inducible VACV vO3-HAi (22) by insertion of the firefly LUC ORF controlled by the VACV synthetic early/late promoter (Fig. 1A). The replication of vO3-HAi-Luc was dependent on IPTG at all multiplicities of infection and in various cell types (Fig. 1B). The specific infectivity of virions made in the presence of IPTG was approximately 30-fold greater than that of virions made in the absence of IPTG (Fig. 1C). These results are similar to those obtained previously with the parent virus vO3-HAi (22) and justify the use of vO3-HAi-Luc here.
Fig 1.
Construction and characterization of an O3L-inducible VACV that expresses LUC. (A) Diagram of vO3-HAi-Luc. The O3L ORF is regulated by the phage T7 promoter (PT7) and the Escherichia coli lac operator (lacO). ORFs encoding enhanced green fluorescent protein (GFP), cyan fluorescent protein (CFP), and LUC are regulated by VACV promoters. The direction of transcription is indicated by arrows. The vO3-HAi-Luc genome also contains the T7 RNA polymerase ORF regulated by a VACV promoter and lacO, as well as the E. coli lac repressor regulated by a VACV promoter (not shown). (B) LUC expression. BS-C-1 and RK-13 cells were infected with vO3-HAi-Luc at a range of multiplicities in the presence (+I) or absence (−I) of IPTG for 24 h. The cells were harvested, lysed, and used to infect fresh BS-C-1 cells. LUC was measured after 90 min and was plotted as relative light units (RLU) per second. (C) BS-C-1 cells were infected with vO3-HAi-Luc in the presence or absence of IPTG, and the virus particles produced were purified by sucrose gradient sedimentation. Equivalent numbers of particles were used to infect BS-C-1, RK-13, and HeLa cells. After 90 min, LUC activity was measured and the ratios plotted. The number of particles was determined from the optical density at 260 nm by using the value 1.2 × 1010 particles/unit (14).
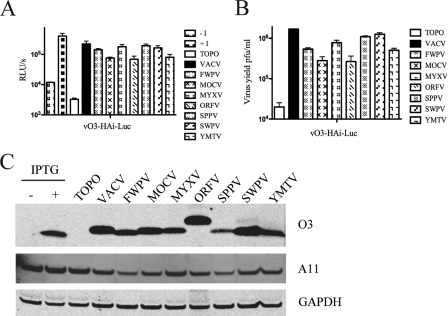
Putative VACV O3 homologs from representatives of eight genera were chemically synthesized with C-terminal Strep(II) and HA epitope tags and were regulated by the natural VACV O3L promoter. The genes were inserted into plasmids and were analyzed in a trans-complementation assay in order to determine whether they possessed a common functionality despite sequence divergence. trans-Complementation assays were performed by infecting cells with vO3-HAi-Luc in the absence of IPTG and transfecting the cells with plasmids. Lysates were prepared after 24 h, and the formation of entry-competent virus presumably containing the O3 ortholog (shown below) was determined by infecting fresh cells and measuring LUC activity after 90 min (Fig. 2A). In all cases, transfection with plasmids containing chordopoxvirus O3L-like genes complemented the O3L-inducible virus relative to the vector alone. The complementation ranged from 30 to 87% relative to VACV O3. The lowest (but still respectable) complementation values, 33% and 30% relative to VACV O3, were obtained with the O3-like proteins derived from molluscum contagiosum virus (MOCV) and orf virus (ORFV), respectively, which had the lowest sequence identities to VACV O3 (Table 1). The abilities of the O3 homologs to complement vO3-HAi-Luc in the absence of IPTG were also measured by plaque assays. The results were similar to those obtained by the LUC assay (Fig. 2B). The lowest complementation values, 16% and 15% relative to VACV
Fig 2.
trans-Complementation with chordopoxvirus O3-like proteins. (A and B) trans-complementation with chordopoxvirus O3-expressing plasmids. BS-C-1 cells were infected with the vO3-HAi-Luc virus in the presence (+I) or absence (−I) of IPTG for positive and negative controls. Additional cells infected in the absence of IPTG were transfected with the vector plasmid (TOPO) or with plasmids containing an O3 homolog from the indicated virus, and virus yields at 24 h postinfection were determined by LUC assays (results expressed as RLU/s) (A) or plaque assays (B). FWPV, fowlpox virus; MYXV, myxoma virus; SPPV, sheeppox virus; SWPV, swinepox virus; YMTV, Yaba monkey tumor virus. (C) Western blot analysis. The O3 proteins of the indicated chordopoxviruses were expressed by transfection as for panels A and B. Cells were infected with the vO3-HAi-Luc virus in the presence or absence of IPTG for positive and negative controls. The expression of O3 was determined by Western blotting with an anti-HA antibody; the expression of the VACV late protein A11 and cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was determined with specific antibodies in the same blot. TOPO denotes the empty vector control. The slower migration of the ORFV homolog is consistent with its larger size.
O3, were again obtained with the MOCV and ORFV ORFs, respectively. The expression of proteins of the predicted sizes was verified by Western blotting with an anti-HA antibody (Fig. 2C). The MOCV and ORFV proteins were as abundant as, or more abundant than, proteins that had higher complementation levels.
Restoration of the replication of an O3L deletion mutant by homologs.
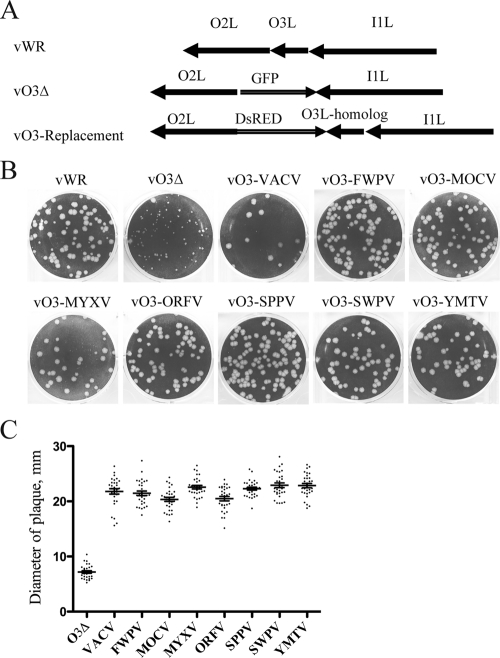
Having demonstrated complementation by chordopoxvirus O3L-like genes expressed from plasmids, we confirmed and extended this result by making O3L replacement viruses. This was accomplished by using the VACV O3L deletion virus (vO3Δ), in which GFP replaced the O3L ORF, as the parent. The VACV O3L gene or an O3L-like gene from another chordopoxvirus was placed adjacent to the DsRED gene and was flanked by the O2L and I1L genes to enable recombination at the O3L locus (Fig. 3A). Recombinant virus plaques were detected by DsRED expression, and the viruses were clonally purified.
Fig 3.
Construction of recombinant VACVs with chordopoxvirus O3L homologs replacing O3L. (A) Diagrams of the parental VACV strain WR and the O3 deletion and O3 homolog replacement viruses. The GFP in the O3 deletion virus was replaced with DsRED and O3L homologs by homologous recombination. (B) Plaque phenotypes of viruses, named for the origin of the O3 homolog, in BS-C-1 cells. (C) The diameters (in millimeters) of the plaques shown in panel B were determined using ImageJ software. The horizontal lines indicate the mean diameters.
The plaques produced by viruses that expressed chordopoxvirus O3L-like genes were all larger than that of vO3Δ and similar to that of the wild-type VACV O3-expressing virus, except for the marginally smaller MOCV and ORFV variants (Fig. 3B and C). These results, together with the transfection data, demonstrated that the chordopoxvirus proteins could functionally replace VACV O3 and provided strong support for the identification of these proteins as orthologs. Nevertheless, the sequence diversity led us to question whether this role required physical interaction with other EFC proteins.
Association of chordopoxvirus O3 homologs with virions and their infectivities.
Our previous studies with recombinant VACV demonstrated the association of O3 with the virion membrane (22). In order to verify that the chordopoxvirus O3 homologs were functioning in a similar manner, their association with virions was determined. The O3L replacement viruses were grown in BS-C-1 cells, and virions were purified by sucrose density gradient centrifugation. The yields of purified virions were measured at 260 nm, and equal amounts were separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels. The O3 homologs were detected by Western blotting using an antibody against the attached HA tag. O3 was found in all cases, although the levels varied relative to that of the loading control, A3 core protein (Fig. 4A). The association of O3 with the virion membrane was confirmed by extraction with the nonionic detergent NP-40 in the presence of the reducing agent dithiothreitol (Fig. 4B). The virion core protein A3, used as a negative control, could not be extracted and remained in the pellet fraction. Thus, the chordopoxvirus O3 protein homologs were virion membrane-associated proteins.
Fig 4.

Virion membrane association of chordopoxvirus O3 homologs. (A) Virus particles were purified from cells infected with the O3 replacement viruses indicated, and equal amounts were separated by SDS-PAGE and were probed by Western blotting with antibodies to HA for the detection of O3 and the VACV core protein A3 as a loading control. A small amount of uncleaved A3, migrating as the higher band, is present in virions. (B) The membrane association of O3 was determined by detergent extraction. Purified virions were incubated with a buffer containing NP-40 detergent and dithiothreitol, and soluble (S) and pellet (P) fractions were obtained by centrifugation. Western blotting was performed as for panel A. (C) Specific infectivities of purified virions. Equal amounts of purified virions determined by optical density (1.2 × 1012 particles) were serially diluted, and the number of PFU was determined by a plaque assay. Particle-to-PFU ratios were calculated and were compared with that of wild-type VACV.
The specific infectivities of the recombinant virions were calculated using the optical density to estimate the number of virus particles and the number of plaques for infectivity. Equal numbers of virus particles were serially diluted and plated on BS-C-1 cells, and the ratios of particles to PFU were determined. Using the value of 1.2 × 1010 particles per optical density unit (14), we calculated a particle/PFU ratio of approximately 70 for VACV WR. Except for ORFV, the values ranged within 1- to 3-fold that for the wild-type virus (Fig. 4C). The particle-to-PFU ratio of ORFV virions was nearly 7-fold higher, indicating lower infectivity.
Interaction of chordopoxvirus O3 homologs with the VACV EFC.
The ability of the chordopoxvirus O3 homologs to associate with the VACV EFC proteins was analyzed by coimmunopurification. BS-C-1 cells infected with the recombinant viruses were lysed with Triton X-100 detergent, and the soluble extracts were incubated with beads containing attached anti-HA antibodies. After washing, the bound proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE), and Western blotting was performed with antibodies to several different EFC components in order to determine the presence of the complex. With VACV O3, each of the six additional EFC proteins analyzed was detected as a strong band on the Western blot (Fig. 5A). EFC proteins were also associated with most of the other O3 homologs, although the amounts detected differed. Lesser amounts of EFC proteins were pulled down with the MOCV homolog and little or none with the ORFV homolog (Fig. 5A).
Fig 5.
Interaction of chordopoxvirus O3 homologs with VACV EFC. (A) BS-C-1 cells were infected with wild-type VACV or the indicated chordopoxvirus O3L replacement virus. O3 and associated proteins were affinity purified from soluble cell extracts by using anti-HA beads. The bound proteins were eluted and identified by Western blotting with antibodies to six different EFC proteins in addition to O3-HA, as indicated on the right. Infection with wild-type VACV WR, which does not express an HA-tagged protein, was used as a control to show specificity. (B) Cells were infected with recombinant viruses expressing VACV or ORFV O3 proteins with tandem streptavidin- and calmodulin-binding peptides attached to their C termini. The TAP-tagged VACV O3 and ORFV O3 proteins in the soluble extract were pulled down using streptavidin-agarose beads. The bound proteins were analyzed by Western blotting as for panel A. (C) The A28R gene expressing tandem affinity tags (A28-TAP) was inserted into O3-HA-expressing VACV and ORFV. After infection with these viruses, A28 was affinity purified using streptavidin-agarose beads, and the interacting EFC proteins were determined by Western blot analysis with the antibodies indicated on the right.
To enhance the efficiency of the EFC purification, replacement viruses were constructed with tandem streptavidin- and calmodulin-binding peptides attached to the C termini of VACV and ORFV O3 proteins. However, the ORFV O3 homolog still failed to pull down the VACV EFC efficiently (Fig. 5B).
The interactions of the VACV and ORFV O3 proteins with EFC proteins were further analyzed by “reverse pulldown.” The A28R gene expressing tandem affinity tags (A28-TAP) was inserted into O3-HA-expressing VACV and ORFV. After infection with these viruses, A28 was affinity purified using streptavidin-agarose beads, and the interacting EFC proteins were determined by Western blot analysis. A28 pulled down VACV O3 and other EFC proteins efficiently (Fig. 5C). However, A28 associated poorly with ORFV O3 and other EFC proteins but relatively better with H2, which interacts directly with A28 (Fig. 5C). These results confirmed that VACV O3 is necessary to stabilize the EFC for affinity purification but that the ORFV homolog was unable to do this. Nevertheless, this outcome does not preclude an important but weak association of the ORFV O3 homolog with the VACV EFC in the viral membrane.
Ability of chordopoxvirus O3 homologs to mediate syncytium formation.
The proteins involved in the entry of VACV are also required for the fusion of infected cells upon low-pH treatment. To evaluate the abilities of O3L replacement viruses to mediate fusion, BS-C-1 cells that had been infected for 16 h were treated with a pH 5.5 or pH 7.4 buffer for 2 min and were then incubated for an additional 3 h in fresh normal medium. Importantly, all O3L replacement viruses mediated fusion upon low-pH treatment, although the syncytia were smaller in cells infected with ORFV or MOCV O3-expressing viruses than in cells infected with viruses expressing other O3 homologs (Fig. 6). Thus, the relative plaque sizes, specific infectivities, interactions with EFC proteins, and fusion activities are consistent for the various O3L replacement viruses.
Fig 6.
Abilities of O3L chimeric viruses to mediate low-pH-induced syncytium formation. BS-C-1 cells were infected with viruses at a multiplicity of 2 PFU per cell. After 16 h, cells were treated with a pH 5.5 or pH 7.4 buffer for 2 min. The cells were washed, incubated with fresh medium for an additional 3 h, and then viewed by phase-contrast microscopy.
O3 domain swapping.
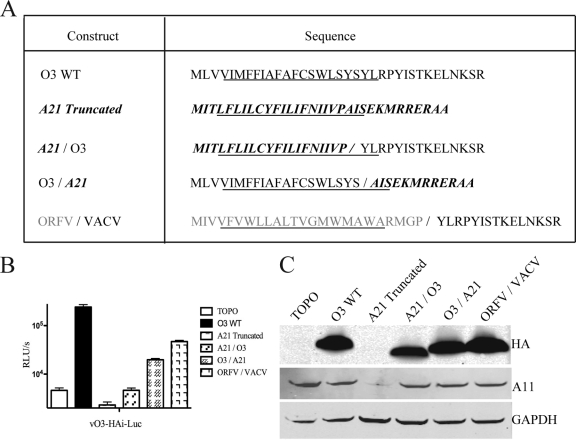
Each of the chordopoxvirus O3 homologs has a predicted N-terminal TM domain and a variable-length nonhomologous C-terminal segment. This arrangement suggested the importance of the TM domain, although Clustal analysis failed to show strong sequence identities even in this part of the protein. The absence of strong sequence conservation prompted us to investigate the abilities of other TM domains to perform the function of O3. The VACV-encoded A21 membrane protein, also part of the EFC, has a topology similar to that of O3. Like that of O3, the predicted α-helical TM domain of A21 starts from amino acid 4 and includes predominantly hydrophobic residues (Fig. 7A). We shortened the C-terminal domain of A21 to make it similar in length to that of O3 and swapped the TM domains between O3 and A21 and between the ORFV O3 homolog and VACV O3 (Fig. 7A). The truncated A21 was expressed poorly or was unstable (Fig. 7C) and appeared to have a dominant negative effect, since complementation was poorer than that with the vector-alone control (Fig. 7B). Addition of the C-terminal domain of O3 to the A21 TM domain enhanced expression (Fig. 7C), but the chimeric protein failed to complement O3 function better than the vector control (Fig. 7B). In contrast, a chimeric construct that expressed the O3 TM domain and the truncated C terminus of A21 complemented the O3-inducible virus, as did the ORFV O3 homolog TM domain with the C-terminal domain of VACV O3, though not as well as the wild-type virus (Fig. 7B). As additional specificity controls, we tested a modified 27-amino-acid version of the VACV A9 protein with the TM domain at the N terminus that had previously been shown to be incorporated into the MV membrane (12) and the 44-amino-acid hydrophobic E5 protein encoded by bovine papillomavirus (9). Neither complemented the O3-inducible virus (data not shown). Thus, the segment of O3 providing specific functionality is the TM domain, which can be replaced by the corresponding region of an O3 ortholog but not by hydrophobic sequences of similar length from other proteins tested.
Fig 7.
trans-Complementation of O3L chimeric constructs. (A) Sequences of proteins. The constructs are named according to the origins of the N- and C-terminal domains, which are on the left and right sides of the slash, respectively. VACV O3 sequences are shown in black type; VACV A21 sequences are in boldface italics; ORFV O3 homolog sequences are shown in gray type. The predicted α-helical regions are underlined. (B) trans-Complementation of O3 chimeric constructs. BS-C-1 cells infected with the vO3-HAi-Luc virus in the absence of IPTG were transfected with the vector plasmid (TOPO) or plasmids containing the constructs shown in panel A. Virus yields at 24 h postinfection were determined by LUC assays; results are expressed as RLU/s. (C) O3 chimeric proteins were analyzed by Western blotting of cell lysates with an antibody to the HA tag on O3 proteins, with a VACV A11-specific antibody as a control for virus infection, and with an antibody to cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a cell loading control.
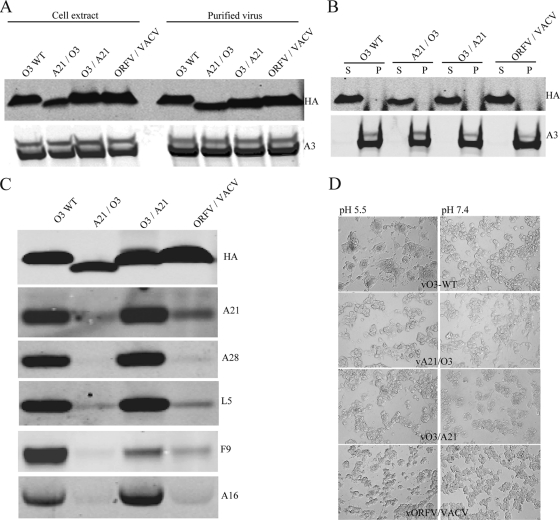
To confirm the complementation results described above, recombinant viruses expressing chimeric constructs A21/O3 (in which the A21 N terminus was attached to the O3 C terminus), O3/A21, and ORFV/VACV O3, as well as the modified form of VACV A9 and a construct with papillomavirus E5 in place of O3, were constructed. The plaque size of the virus expressing A21/O3 was small and comparable to that of the vO3Δ virus (Fig. 8A and B), as were those of the A9 and E5 constructs (not shown). On the other hand, plaques formed by viruses expressing the O3/A21 or ORFV/VACV O3 chimeric protein resembled those of the wild-type virus (Fig. 8A and B). When purified chimeric O3 viruses were analyzed by Western blotting, all three proteins (A21/O3, O3/A21, and ORFV/VACV O3) were detected in virions, and detergent extraction confirmed membrane association (Fig. 9A and B). Whereas O3/A21 was able to interact stably with the EFC, A21/O3 did not, as determined by affinity purification and Western blotting (Fig. 9C). The association of ORFV/VACV chimeric O3 protein with EFC proteins was weak and similar to that of full-length ORFV O3 (Fig. 9C). Since the chimeric protein has the ORFV O3 TM domain, it demonstrates the role of the TM domain in mediating a protein-protein interaction with EFC proteins.
Fig 8.
Plaque phenotypes of viruses expressing chimeric O3. (A) Plaques formed in BS-C-1 cells by recombinant wild-type VACV (vO3-WT), a VACV deletion mutant (vO3Δ), and viruses with chimeric O3L ORFs (vA21/O3, vO3/A21, vORFV/VACV) replacing the endogenous O3L ORF. (B) The plaque sizes (in millimeters) of the viruses shown in panel A were determined using ImageJ software tools. The horizontal lines indicate the mean diameters.
Fig 9.
Virion and EFC association of chimeric O3 proteins encoded by VACV. (A) Association of chimeric O3 proteins with virions. BS-C-1 cells were infected with the viruses described in the legend to Fig. 8. Cell extracts and purified virions were analyzed by Western blotting using antibodies to the HA tag on O3 and the A3 core protein. (B) Association of chimeric O3 proteins with the virion membrane. Purified virions were extracted with NP-40 detergent and dithiothreitol, and soluble (S) and pellet (P) fractions were analyzed by Western blotting as for panel A. (C) Interaction of chimeric O3 proteins with the EFC. Membrane proteins from infected cells were solubilized with NP-40, immunopurified with an antibody to HA, immobilized on beads, and analyzed by Western blotting using antibodies to HA and to the five EFC proteins given on the right. (D) Abilities of O3 chimeric proteins to mediate low-pH-induced syncytium formation. BS-C-1 cells were infected with the wild-type virus or with recombinant viruses expressing chimeric O3 proteins. After 16 h, cells were treated with a pH 5.5 or pH 7.4 buffer for 2 min. The cells were washed, incubated with fresh medium for an additional 3 h, and then viewed by phase-contrast microscopy.
The abilities of chimeric viruses to mediate cell-cell fusion upon low-pH treatment were determined as described above. It was somewhat surprising that none of the chimeric viruses were able to mediate fusion (Fig. 9D), even though vO3/A21 and vORFV/VACV O3 complemented infectivity. However, this result is consistent with other data suggesting that more-active fusion complexes are required for cell-cell fusion than for virus-cell fusion and entry (29).
DISCUSSION
With only 35 amino acids, O3 is the smallest characterized protein encoded by VACV. The significance of this small ORF was established by our finding that O3 is a functional component of the EFC (22). The O3L ORF is highly conserved in orthopoxviruses, with identical sequences in all species except for monkeypox viruses, with 2 to 3 amino acid substitutions, whereas the O3L genes of other chordopoxviruses exhibit considerable divergence. In view of the sequence differences, it was unclear whether these ORFs could functionally replace O3 in the context of other VACV EFC proteins. To investigate this question, we carried out trans-complementation and recombination experiments with the corresponding genes of other chordopoxviruses.
Remarkably, every chordopoxvirus O3L-like gene could substitute functionally for VACV O3L, indicating that these genes are true homologs and have conserved important structural elements. In most cases, replication of the O3L replacement viruses was efficient, and the plaques were indistinguishable from those of wild-type VACV except for the slightly smaller plaque sizes of recombinants containing the ORFV and MOCV homologs, which have the greatest sequence divergence. VACV entry proteins assemble into a complex that can be isolated even after detergent extraction (24). Moreover, this complex must be held together by multiple protein interactions, since repression of individual components, including O3, destabilizes the complex, which can no longer be affinity purified (22). One might anticipate that such interactions would be sequence specific. It was surprising that despite diversity, each tagged O3 homolog except for that of ORFV could pull down the EFC. In this regard, the 48-amino-acid ORFV O3 homolog is longer than the others, but perhaps more importantly, its predicted TM domain is 2 amino acids shorter. Nevertheless, the ORFV homolog was incorporated into MVs and could be extracted with a nonionic detergent, indicating that it was a membrane component. In view of the ability of the ORFV O3 to complement VACV, it seems likely that the homolog has weak interactions with other VACV EFC components within the membrane that are abrogated by detergent extraction or by washing of the affinity beads. It would be informative to determine whether ORFV O3 interacts more strongly with an EFC composed of ORFV proteins rather than VACV proteins.
We considered that the TM segment was the important functional domain of O3 and its orthologs for several reasons. First, each O3 homolog had a predicted N-terminal helical domain of the same length (except for ORFV, noted above) followed by a basic amino acid and a proline. Second, there was no motif or sequence conservation in the C-terminal domain, which varied in length. Moreover, the C-terminal domains of two orthologs were only 5 amino acids long. Swapping experiments were carried out to investigate the relative roles of the two domains of the O3 proteins. A chimera composed of the N-terminal domain of VACV O3 and a segment of the C-terminal portion of the VACV A21 EFC protein (O3/A21) was able to complement wild-type O3, whereas the reverse construct, composed of the N-terminal domain of A21 and the C-terminal domain of O3 (A21/O3), was unable to do so, even though the latter was incorporated into the MV membrane. As expected from the functional ability of the O3/A21 chimera, it interacted stably with the EFC. However, while a chimera that was formed from the ORFV O3 N-terminal domain and the VACV O3 C-terminal domain was able to complement VACV replication, stable interaction with the EFC was not observed. The latter property was similar to that of the full-length ORFV O3 protein, reinforcing the idea that EFC component interactions are determined by the TM domain. Other constructs containing heterologous TM domains were unable to complement O3 or to interact with the EFC. Taken together, these results suggested that the TM domain of O3 is responsible for its function and for interaction with the TM domains of other EFC proteins. Future studies will be directed toward understanding the functionally important sequence and structural features of the O3 TM domain and of the TM domains of other EFC proteins.
ACKNOWLEDGMENTS
We thank Catherine Cotter for help with cell cultures and Karl Erlandson, Jason Laliberte, other members of our laboratory, and Eugene Koonin for useful discussions.
This research was supported by the Division of Intramural Research, NIAID, NIH.
Footnotes
Published ahead of print 23 November 2011
REFERENCES
- 1. Bengali Z, Townsley AC, Moss B. 2009. Vaccinia virus strain differences in cell attachment and entry. Virology 389: 132–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bertholet C, Drillien R, Wittek R. 1985. One hundred base pairs of 5′ flanking sequence of a vaccinia virus late gene are sufficient to temporally regulate late transcription. Proc. Natl. Acad. Sci. U. S. A. 82: 2096–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bisht H, Weisberg AS, Moss B. 2008. Vaccinia virus L1 protein is required for cell entry and membrane fusion. J. Virol. 82: 8687–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown E, Senkevich TG, Moss B. 2006. Vaccinia virus F9 virion membrane protein is required for entry but not virus assembly, in contrast to the related L1 protein. J. Virol. 80: 9455–9464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chakrabarti S, Sisler JR, Moss B. 1997. Compact, synthetic, vaccinia virus early/late promoter for protein expression. Biotechniques 23: 1094–1097 [DOI] [PubMed] [Google Scholar]
- 6. Chiu WL, Lin CL, Yang MH, Tzou DLM, Chang W. 2007. Vaccinia virus 4c (A26L) protein on intracellular mature virus binds to the extracellular cellular matrix laminin. J. Virol. 81: 2149–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chung C-S, Hsiao J-C, Chang Y-S, Chang W. 1998. A27L protein mediates vaccinia virus interaction with cell surface heparin sulfate. J. Virol. 72: 1577–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Earl PL, Moss B, Wyatt LS, Carroll MW. 1998. Generation of recombinant vaccinia viruses, p 16.17.1–16.17.19 In Ausubel FM, et al. (ed), Current protocols in molecular biology, vol 2 Greene Publishing Associates & Wiley Interscience, New York, NY [Google Scholar]
- 9. Horwitz BH, Burkhardt AL, Schlegel R, DiMaio D. 1988. 44-amino-acid E5 transforming protein of bovine papillomavirus requires a hydrophobic core and specific carboxyl-terminal amino acids. Mol. Cell. Biol. 8: 4071–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hsiao JC, Chung CS, Chang W. 1999. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 73: 8750–8761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang CY, et al. 2008. A novel cellular protein, VPEF, facilitates vaccinia virus penetration into HeLa cells through fluid phase endocytosis. J. Virol. 82: 7988–7999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Husain M, Weisberg AS, Moss B. 2007. Sequence-independent targeting of transmembrane proteins synthesized within vaccinia virus factories to nascent viral membranes. J. Virol. 81: 2646–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Izmailyan RA, Huang CY, Mohammad S, Isaacs SN, Chang W. 2006. The envelope G3L protein is essential for entry of vaccinia virus into host cells. J. Virol. 80: 8402–8410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Joklik WK. 1962. The purification of four strains of poxvirus. Virology 18: 9–18 [DOI] [PubMed] [Google Scholar]
- 15. Lin CL, Chung CS, Heine HG, Chang W. 2000. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 74: 3353–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mercer J, Helenius A. 2008. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science 320: 531–535 [DOI] [PubMed] [Google Scholar]
- 17. Moss B. 2007. Poxviridae: the viruses and their replication, p 2905–2946 In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 18. Moss B. 2006. Poxvirus entry and membrane fusion. Virology 344: 48–54 [DOI] [PubMed] [Google Scholar]
- 19. Nichols RJ, Stanitsa E, Unger B, Traktman P. 2008. The vaccinia I2L gene encodes a membrane protein with an essential role in virion entry. J. Virol. 82: 10247–10261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ojeda S, Domi A, Moss B. 2006. Vaccinia virus G9 protein is an essential component of the poxvirus entry-fusion complex. J. Virol. 80: 9822–9830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ojeda S, Senkevich TG, Moss B. 2006. Entry of vaccinia virus and cell-cell fusion require a highly conserved cysteine-rich membrane protein encoded by the A16L gene. J. Virol. 80: 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Satheshkumar PS, Moss B. 2009. Characterization of a newly identified 35-amino-acid component of the vaccinia virus entry/fusion complex conserved in all chordopoxviruses. J. Virol. 83: 12822–12832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Senkevich TG, Moss B. 2005. Vaccinia virus H2 protein is an essential component of a complex involved in virus entry and cell-cell fusion. J. Virol. 79: 4744–4754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Senkevich TG, Ojeda S, Townsley A, Nelson GE, Moss B. 2005. Poxvirus multiprotein entry-fusion complex. Proc. Natl. Acad. Sci. U. S. A. 102: 18572–18577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Senkevich TG, Ward BM, Moss B. 2004. Vaccinia virus entry into cells is dependent on a virion surface protein encoded by the A28L gene. J. Virol. 78: 2357–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Townsley A, Senkevich TG, Moss B. 2005. The product of the vaccinia virus L5R gene is a fourth membrane protein encoded by all poxviruses that is required for cell entry and cell-cell fusion. J. Virol. 79: 10988–10998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Townsley AC, Weisberg AS, Wagenaar TR, Moss B. 2006. Vaccinia virus entry into cells via a low-pH-dependent endosomal pathway. J. Virol. 80: 8899–8908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. von Heijne G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225: 487–494 [DOI] [PubMed] [Google Scholar]
- 29. Wolfe CL, Ojeda S, Moss B. 19 October 2011. Transcriptional repression and RNA silencing act synergistically to demonstrate the function of the eleventh component of the vaccinia virus entry-fusion complex. J. Virol doi:10.1128/JVI.05935-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wong WC, Maurer-Stroh S, Eisenhaber F. 2010. More than 1,001 problems with protein domain databases: transmembrane regions, signal peptides and the issue of sequence homology. PLoS Comput. Biol. 6: e1000867. [DOI] [PMC free article] [PubMed] [Google Scholar]