Abstract
Hox proteins are a metazoan-specific family of transcription factors that are required for developmental patterning. The genomic arrangement of Hox genes into four paralogous clusters is a primitive feature of jawed vertebrates. Using high-throughput sequencing, we demonstrate the absence of all HoxC transcripts from embryos of the shark, Scyliorhinus canicula, and the skate, Leucoraja erinacea, and the absence of all HoxC genes and two HoxC-associated miRNAs from the genome of L. erinacea. These data suggest a loss of the entire HoxC cluster in elasmobranch fishes, and represent the evidence for natural deletion of an entire Hox cluster in vertebrates.
Jawed vertebrates typically possess four Hox clusters with up to fourteen genes each that arose from an ancestral cluster by genome duplication events. An additional round of whole genome duplication in teleost fishes permitted the Hox genes to diversify both structurally and functionally – teleosts possess seven or more clusters, with variable loss or retention of Hox paralogs. The retention of the four Hox clusters appears to be required for viability since deletion of all copies of HoxA, B, C, or D clusters has not been reported in vertebrates (1).
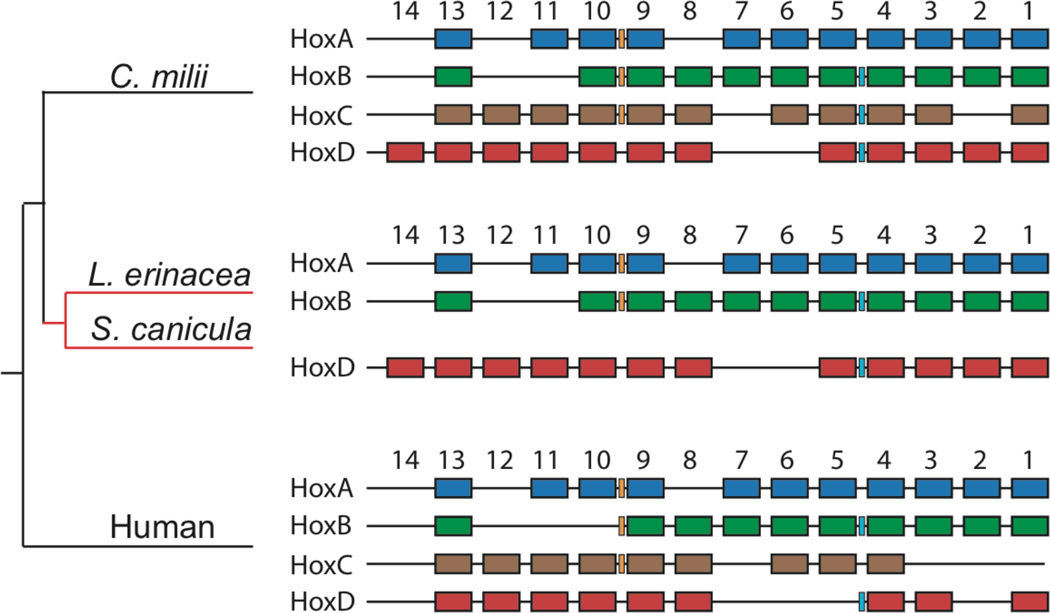
Developmental expression profiling in the three lineages of chondrichthyan fishes (Fig. 1) – holocephalans (Callorhinchus milii), batoids (Leucoraja erinacea), and sharks (Scyliorhinus canicula) – revealed expression of all forty-five members of the HoxA–D clusters in C. milii (average contig length (ACL) = 1214 +/− 565 (SD) base pairs (bp), average coverage (AC) = 25.9 +/− 18.5), but expression of only thirty-four Hox genes encoded by the HoxA, B, and D clusters in L. erinacea and S. canicula (ACL = 1294 +/− 578 bp and 1236 +/− 710 bp, AC = 25.1 +/− 15.1 and 21.0 +/− 21.5, respectively, Table S1). Expression of the eleven HoxC genes was undetectable in S. canicula (consistent with (2)) or L. erinacea (p = 2.60 × 10−7).
Figure 1.
Genomic deletion of the HoxC cluster in elasmobranchs. Jawed vertebrates typically possess four Hox clusters (HoxA–D, shown in blue, green, brown, and red, respectively), each encoding up to fourteen Hox transcription factors and microRNAs belonging to the miR-10 (light blue) and miR-196 (orange) families. Genomic and transcriptomic sequencing reveals the skate (L. erinacea) and shark (S. canicula) retain a full complement of HoxA, B, and D genes, but lack Hox genes and miRNAs encoded by the HoxC cluster. Loss of the HoxC cluster (denoted by red lines) occurred along the elasmobranch stem.
To distinguish between genomic deletion and transcriptional silencing of the HoxC cluster in elasmobranchs (batoids + sharks), we analyzed a 26x coverage assembly of the L. erinacea genome. Contigs encoding thirty-four skate HoxA, B, and D genes were identified (ACL = 5179 +/− 2850 bp, AC = 25.3 +/− 2.8), but no contig or singleton sequence encoding any of the eleven members of the HoxC complex was detectable (p = 6.51 × 10−7). Vertebrate Hox clusters also encode the miR-10 and miR-196 microRNA families (3). The C. milii Hox clusters encode three miR-10 and three miR-196 genes, with miR-10c and miR-196a-2 mapping within the HoxC cluster (Figs. 1 and S1). The L. erinacea genome encodes two members each of the miR-10 and miR-196 families (Fig. S1; ACL = 3008 +/− 1778 bp, AC = 20.5 +/− 1.0). Pre-miRNA sequences for miR-10c and miR-196a-2 were undetectable (p = 6.51 × 10−7). Similarly, of the non-coding sequence elements conserved between orthologous C. milii and human HoxA, B, and D clusters (4), 51% of the 249 elements were identified in the L. erinacea genome, but none of the 25 elements from within the HoxC cluster were detected (p = 1.01 × 10−7, Table S2).
Our data suggest that the lack of HoxC gene expression during L. erinacea and S. canicula development is attributable to a genomic deletion of the entire HoxC cluster in these taxa (Fig. 1). The most likely scenario is that the entire HoxC cluster was lost in a single genomic reduction event following the divergence of holocephalans and elasmobranchs, but prior to the divergence of batoids and sharks (Fig. 1); however, regulated chromosomal diminution cannot be excluded based on embryonic genomic DNA sequence (5). Homozygous mice lacking the HoxC complex exhibit only minor transformations of axial identity, but die perinatally due to pulmonary defects (6). The unique dispensability of the HoxC cluster for body plan development may have enabled elasmobranchs to survive the challenge of a genome reduction abrogating all HoxC cluster function.
Supplementary Material
Acknowledgements
We thank B. Kingham for genome sequencing. B.L.K., H.R.C. and R.D.D. were funded by National Center for Research Resources, NIH (P20RR016463). J.A.G. was funded by a Newton International Fellowship. The skate genome was sequenced by the North East Cyberinfrastructure Consortium. Sequence data are available at the National Center for Biotechnology Information in Gene Expression Omnibus (GSE26235), GenBank (AESE010000000) and Sequence Read Archive (SRA026856).
Footnotes
Publisher's Disclaimer: This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Contributor Information
Benjamin L. King, Email: bking@mdibl.org.
J. Andrew Gillis, Email: jag93@cam.ac.uk.
Heather R. Carlisle, Email: hcarlisl@mdibl.org.
Randall D. Dahn, Email: randalldahn@gmail.com.
References
- 1.Woltering JM, Durston AJ. The zebrafish hoxDb cluster has been reduced to a single microRNA. Nat Genet. 2006 Jun;38:601. doi: 10.1038/ng0606-601. [DOI] [PubMed] [Google Scholar]
- 2.Oulion S, et al. Evolution of Hox gene clusters in gnathostomes: insights from a survey of a shark (Scyliorhinus canicula) transcriptome. Mol Biol Evol. 2010 Dec;27:2829. doi: 10.1093/molbev/msq172. [DOI] [PubMed] [Google Scholar]
- 3.Lund AH. miR-10 in development and cancer. Cell Death Differ. 2010 Feb;17:209. doi: 10.1038/cdd.2009.58. [DOI] [PubMed] [Google Scholar]
- 4.Ravi V, et al. Elephant shark (Callorhinchus milii) provides insights into the evolution of Hox gene clusters in gnathostomes. Proc Natl Acad Sci U S A. 2009 Sep 22;106:16327. doi: 10.1073/pnas.0907914106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith JJ, Stuart AB, Sauka-Spengler T, Clifton SW, Amemiya CT. Development and analysis of a germline BAC resource for the sea lamprey, a vertebrate that undergoes substantial chromatin diminution. Chromosoma. 2010 Aug;119:381. doi: 10.1007/s00412-010-0263-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suemori H, Noguchi S. Hox C cluster genes are dispensable for overall body plan of mouse embryonic development. Dev Biol. 2000 Apr 15;220:333. doi: 10.1006/dbio.2000.9651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.