Abstract
The complete nucleotide sequences of the chloroplast (cp) and mitochondrial (mt) genomes of resurrection plant Boea hygrometrica (Bh, Gesneriaceae) have been determined with the lengths of 153,493 bp and 510,519 bp, respectively. The smaller chloroplast genome contains more genes (147) with a 72% coding sequence, and the larger mitochondrial genome have less genes (65) with a coding faction of 12%. Similar to other seed plants, the Bh cp genome has a typical quadripartite organization with a conserved gene in each region. The Bh mt genome has three recombinant sequence repeats of 222 bp, 843 bp, and 1474 bp in length, which divide the genome into a single master circle (MC) and four isomeric molecules. Compared to other angiosperms, one remarkable feature of the Bh mt genome is the frequent transfer of genetic material from the cp genome during recent Bh evolution. We also analyzed organellar genome evolution in general regarding genome features as well as compositional dynamics of sequence and gene structure/organization, providing clues for the understanding of the evolution of organellar genomes in plants. The cp-derived sequences including tRNAs found in angiosperm mt genomes support the conclusion that frequent gene transfer events may have begun early in the land plant lineage.
Introduction
Plastid and mitochondria are essential organelles in plant cells. Chloroplasts conduct photosynthesis in the presence of sunlight and mitochondria indirectly supply energy within plant cells; together they form the powerhouses of the cell. Both chloroplasts and mitochondria possess their own genomes. The chloroplast (cp) genome and mitochondrial (mt) genomes are often used for the study of plant evolution [1], [2]. From the information of all sequenced cp genomes, most of them range from 120 to 160 kb in length and have GC contents of 30 to 40%. The quadripartite organization is shared by almost all cp genomes, consisting of a large-single-copy region (LSC; 80–90 kb) and a small-single-copy region (SSC; 16–27 kb), as well as two copies of inverted repeats (IRs) of ∼20 to 28 kb in size. The gene content and structure of angiosperm cp genome is highly conserved [3], [4]. Expansion and contraction of the IR as well as gene and intron losses have been documented in a wide range of angiosperms [5], [6].
The mt genome plays crucial roles in plant development and productivity [7]. In comparison to other non-plant eukaryotes, plants have large and complex mt genomes [8], [9]. Mt genomes of seed plants are unusually large and vary in size at least in an order of magnitude. Much of these variations occur within a single family [10]. Seed plant mitochondrial genomes are characteristic for their very low mutation rate [11], frequent uptake of foreign DNA by intracellular and horizontal gene transfer [12], and dynamic structure [13]. The evolving land plants have gained new mechanisms to facilitate more frequent gene exchanges between mt and cp genomes as well as between mt and nuclear genomes, which make mt genomes increase their sizes. [14].
In the past several years, we have witnessed a dramatic increase in the number of complete organellar genomes, especially those of plants. Until now, there are 206 complete cp genomes and 47 mt genomes having been deposited in GenBank Organelle Genome Resoures. With the emergence of next-generation sequencers, new approaches for genome sequencing have been gradually applied due to their high-throughput, time-saving, and low-cost advantages. With a new gene-based strategy and combining data from the two next-generation sequence platforms, pyrosequencing (Roche GS FLX) and ligation-based sequencing (Life Technologies SOLiD), we successfully assembled cp and mt genomes of resurrection plant B. hygrometrica (Bunge) R. Br [15]. B. hygrometrica or Bh is a small dicotyledenous, homiochlorophyllous resurrection plant in the Gesneriaceae family, and it is widespread in China, inhabiting shallow rock crevices from humid tropical regions to arid temperate zones [16], [17]. In this study, we analyze genomic features and structures of both cp and mt genomes of B. hygrometrica. Through organellar genome comparison with other lower plants and angiosperms, we provide information for the better understanding of organellar genome evolution in land plants.
Results and Discussion
Features of B. hygrometrica cp genome and mt genome
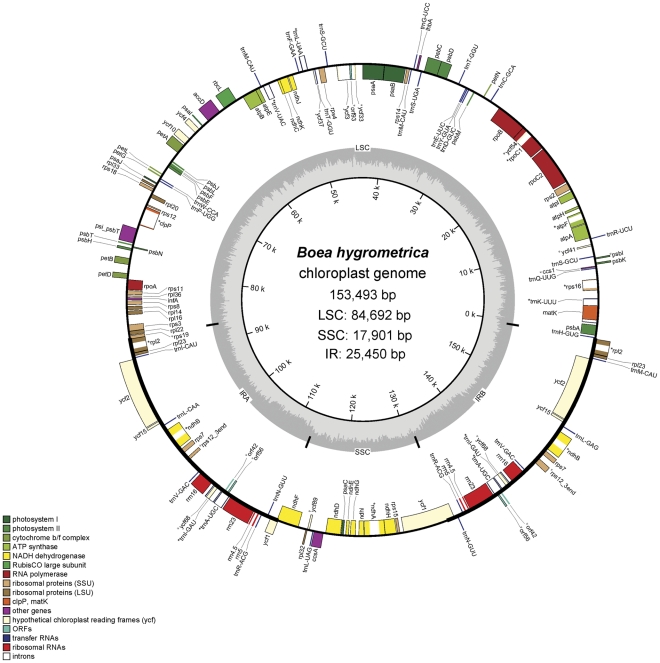
The Bh cp genome is 153,493 bp in length and has a GC content of 37.59%. Similar to those of other angiosperms [4], [18], [19], the Bh cp genome maps as a circular molecule with the typical quadripartite structure: a pair of IRs (25,450 bp, covering 16.6%) separated by the LSC (84,692 bp, covering 55.1%) and SSC (17,901 bp, 11.7%) regions ( Figure 1 ). It encodes 147 predicted functional genes and 19 of them are duplicated in the IR regions. Among these 147 genes, we identified 103, 36, and 8 protein-coding, tRNA, and rRNA genes, respectively ( Table 1 and Table 2 ). 38% of the genome is non-coding, including introns, intergenic spacers, and pseudogenes. All the rRNA genes (rrn16, rrn23, rrn5 and rrn4.5) and 7 tRNA (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU) genes are located in IR regions. Similar to other dicot species, Bh has two genes (rps19 and trnH) located in the position of IR/LSC junctions. This is different in monocots, such as rice and maize, whose cpDNAs have a fully duplicated rps19 gene in the IR/LSC junctions [18]. The average length of intergenic regions is 385 bp, varying from 1 to 2,221 bp. There are 4 cases of overlapping genes (psbD-psbC, ndhK-ndhC, atpE-atpB, and ycf1-ndhF), resulting in an average coding density (including conserved genes, unique ORFs and introns) of 1/1,058 bp. The cp genome has 19 intron-containing genes with 12 in protein-coding genes and 7 in tRNA genes. In terms of size, gene content, and intron composition, Bh cpDNA closely is mapped to Olea europaea cpDNA (155,872 bp, GC 37%) [20] among all angiosperms. Sequence alignment shows that 93% (142,189 bp) of the Bh cp genome sequence are covered by that of O. europaea with 95.4% identity (Figure S1). 36 tRNAs are detected, enabling B. hygrometrica cp genome to decode all 61 codons. All of 3 stop codons are present with UAA being the most frequently used (UAA 40%, UAG 33% and UGA 27%) (Table S1). Phylogenetic analyses, which were constructed by 63 protein-coding sequences from 12 cp genomes (one green algae as outgroup, one land and 10 seed plants) (Table S2), indicates that the phylogenetic position of B. hygrometrica is closer and older than V. vinifera among the analyzed dicots (Figure S2).
Figure 1. The circular-mapping chloroplast genome of B. hygrometrica.
Features on transcriptionally clockwise and counter-clockwise strands are drawn on the inside and outside of the outer circle, respectively. Genes belonging to different groups are color-coded (Ψ, pseudogene). The genome coordinate and GC content are shown in the inner circle. The thick lines indicate inverted repeats (IRA and IRB), which separate the genome into small (SSC) and large (LSC) single copy regions. The map was drawn by using OGDRAW [47].
Table 1. General features of B. hygrometrica cp and mt genomes.
| Feature | Chloroplast | Mitochondrion |
| Genome size (bp) | 153,493 | 510,519 |
| GC content (%) | 37.59 | 43.27 |
| Coding sequences (%)a | 72 | 12.19 |
| Gene content (%)b | 61.21 | 7.89 |
| No. of protein-coding gene | 103 | 33 |
| No. of intron | 19 | 23 |
| No. of tRNA genes | 36 | 28 |
| No. of rRNA operons | 8 | 4 |
| Repeat sequence (%)c | 2.36 | 1.45 |
| Chloroplast-derived (%) | \ | 10.52 |
Conserved genes, unique ORFs, introns, and intron ORFs are considered as coding sequences.
Unique ORFs and intron ORFs are not taken into account.
Predicted by using RepeatMasker Web Server.
Table 2. Gene content of B. hygrometrica cp and mt genomes.
| cpDNA | Photosystem I | psaA,psaB,psaC,psaI,psaJ |
| Photosystem II | psbA,psbB,psbC,psbD,psbE,psbF,psbH,psbI,psbJ,psbK,psbL,psbM,psbN,psbT,psi_psbT | |
| Cytochrome b/f complex | petA,petB,petD,petG,petL,petN | |
| ATP synthase | atpA,atpB,atpE,atpF,atpH,atpI | |
| NADH dehydrogenase | ndhA,ndhB,ndhC,ndhD,ndhE,ndhF,ndhG,ndhH,ndhI,ndhJ,ndhK | |
| RubisCO large subunit | rbcL | |
| RNA polymerase | rpoA,rpoB,rpoC1,rpoC2 | |
| Ribosomal proteins (SSU) | rps2,rps3,rps4,rps7(×2),rps8,rps11,rps12,rps12_3end(×2),rps14,rps15,rps16,rps18,rps19 | |
| Ribosomal proteins (LSU) | rpl2(×2),rpl14,rpl16,rpl20,rpl22,rpl23(×2),rpl32,rpl33,rpl36 | |
| Other genes | clpP,matK,accD,ccs1,ccsA,infA,lhbA,cemA | |
| hypothetical chloplast reading frames | ycf1(×2),ycf2(×2),ycf3,ycf4,ycf10,ycf15(×2),ycf33,ycf37,ycf41,ycf54,ycf68(×2),ycf89 | |
| ORFs | orf42(×2),orf56(×2),orf93 | |
| Transfer RNAs | trnA-UGC(×2),trnC-GCA,trnD-GUC,trnE-UUC,trnF-GAA,trnG-UCC,trnH-GUG,trnI-CAU,trnI-GAU(×2),trnK-UUU,trnL-CAA,trnL-GAG,trnL-UAA,trnL-UAG,trnM-CAU(×3),trnN-GUU(×2),trnP-UGG,trnQ-UUG,trnR-ACG(×2),trnR-UCU,trnS-GCU(×2),trnS-UGA,trnT-GGU(×2),trnV-GAC(×2),trnV-UAC,trnW-CCA,trnY-GUA | |
| Ribosomal RNAs | rrn4.5(×2),rrn5(×2),rrn16(×2),rrn2(×2) | |
| mtDNA | Genes of Mitochondrial Origin | |
| Complex I (NADH dehydrogenase) | nad1,nad2,nad3,nad4,nad4L,nad5,nad6,nad7,nad9 | |
| Complex II (succinate dehydrogenase) | sdh3 | |
| complex III (ubichinol cytochrome c reductase) | Cob | |
| Complex IV (cytochrome c oxidase) | cox1,cox2,cox3 | |
| Complex V (ATP synthase) | atp1,atp4,atp6,atp8,atp9 | |
| Cytochrome c biogenesis | ccmB,ccmC,ccmFc,ccmFn | |
| Ribosomal proteins (SSU) | rps3,rps4,rps7,rps12,rps13 | |
| Ribosomal proteins (LSU) | rpl16 | |
| Maturases | matR | |
| Other genes a | BohyM-orf1,BohyM-orf2,BohyM-orf3 | |
| Transfer RNAs | trnC-GCA, trnE-UUC(×2), trnG-GCC,trnI-CAU,trnK-UUU,trnL-UAA,trnM-CAU(×2), trnP-UGG, trnS-GCU,trnS-UGA,trnT-UGU, trnY-GUA | |
| Ribosomal RNAs | rrn5(×2),rrn18,rrn26 | |
| Genes of Chloroplast Origin | ||
| Genes with intact ORFs | atpA,atpI,ndhB,petD,petN,psaJ,psbC,psbM,rpl20,rpl33,rpl36,rpoB,rps14,rps18,rps2,rps7,rrn16,rrn4.5,rrn5,ycf15 | |
| Pseudogenes b | accD(×2),atpA,atpE,atpF(×2),clpP,lhbA,matK(×2),ndhC,ndhJ,ndhK,orf42,orf56,petB,psaA,psaB(×3),psbD,psi_psbT,rpl16,rpl2,rpl22,rpoA,rpoC1(×2),rpoC2(×5),rps11,rps12_3end(×2),rps16,rps3(×3),rrn23(×2),ycf2(×2),ycf41,ycf54,trnE-UUC,trnF-GAA,trnI-GAU,trnI-GAU,trnL-UAA,trnP-UGG | |
| Transfer RNAs | trnD-GUC,trnF-GAA(×2),trnH-GUG,trnL-CAA,trnM-CAU(×2),trnN-GUU,trnQ-UUG,trnR-ACG,trnS-GGA,trnV-GAC,trnW-CC(×2) | |
Genes that are conserve among other function unknown mitochondrial genes or ORFs.
Genes that are transferred to mtDNA from cpDNA in fragments.
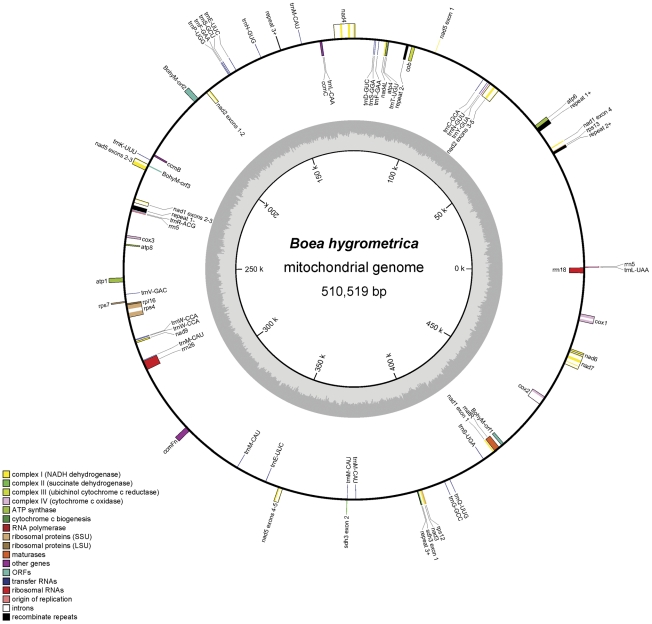
The Bh master mt genome is assembled into a circular molecule of 510,519 bp in length ( Figure 2 ) and has an average GC content of 43.27%. This size is bigger than the mt genome of A. thaliana (366,924 bp) [21], but smaller than Vitis vinifera (773,279 bp) [12] among dicots. With only 12% of coding sequences, the largest part (88%) of this genome is non-coding, containing 1.45% repeat and 10.52% cp-derived sequences ( Table 1 ). The mt genome has 65 genes, including 33 protein-coding, 4 rRNA, and 28 tRNA genes, and 10 genes have exon-intron structure. Similar to other angiosperms, the Bh mitochondrion uses the canonical genetic code. All 61 codons are present in mt genome, and UAA (46%) is the most frequently-used stop codon (Table S3). The known 33 protein-coding genes in mt genome are similar to other published angiosperm mt genomes ( Table 2 ), such as 9 subunits of NADH dehydrogenase (complex I), 5 subunits of ATP synthase (Complex V), and 3 subunits of cytochrome c oxidase (complex IV). Compared to other angiosperms, we observed that there is one sdh3 and no sdh4 in Bh mt genome. There are 3 copies of sdh3 in mt genome of Nicotiana tabacum [22], and both sdh3 and sdh4 are present in that of V. vinifera [12]. The Bh cox1 has an intron/exon structure that is unlike other higher seed plants (Table S4). There are two 5S rRNA (rrn5) copies and one copy of rrn5 from its cp genome. The best sequence alignment score belongs to V. vinifera mt genome, with 23% (119,377 bp) of Bh mt genome being alignable to that of V. vinifera with 94.2% identity (Figure S3).
Figure 2. The circular-mapping mitochondrial genome of B. hygrometrica.
Features on transcriptionally clockwise and counter-clockwise strands are drawn on the inside and outside of the outer circle, respectively. The genome coordinate and GC content are shown in the inner circle. Genes belonging to different groups are color-coded. The map was drawn by using OGDRAW [47].
Plant cells often contain multiple clones or copies of cp and genomes, and thus the organellar genomes can be regarded as a population with genetic heterogeneity [4]. Polymorphic sites can be detected by aligning thousands of high quality reads to assembling of the cp or mt genome [23]. Our SNP analysis shows that there is no intervarietal SNPs (intraSNPs) found in Bh cp genome. However, we identified 729 SNPs in Bh mt genome (Table S5) and SNPs in mt genome occurred at a rate of 1 in 700 bp. We only detected 9 SNPs in gene regions with 2, 1, 1, and 5 in rrn5, rrn26, trnM-CAU, and rrn18, respectively ( Table 3 ). There are no intraSNPs detected in known protein-coding gene regions. The intraSNPs have been demonstrated to be present in both cp and mt genomes of rice [23], [24]. As an indicator for the heterogeneous nature of cp and mt populations, the intravarietal polymorphisms provide us useful markers for the future genetic studies on B. hygrometrica.
Table 3. Intravarietal polymorphisms (IntraSNPs) in the gene regions of B. hygrometrica mt genome.
| Gene | Position | SNP | Read Coverage |
| rrn5 | 234508 | C/T | 273/212 |
| rrn5 | 234560 | G/A | 320/152 |
| rrn26 | 289003 | A/G | 366/86 |
| trnM-CAU | 339634 | A/T | 415/868 |
| rrn18 | 509408 | G/A | 193/80 |
| rrn18 | 509565 | C/T | 191/39 |
| rrn18 | 509941 | G/T | 107/24 |
| rrn18 | 509978 | T/C | 148/90 |
| rrn18 | 510096 | T/G | 182/57 |
Structural dynamics of Bh mitochondrial genome in ontogeny
All previous studies on complete sequencing of flowering plant mt genomes are based on the master circle (MC) hypothesis [21], [22], [25]–[28]. Arrieta-Montiel et al. reported on the structural dynamics of the common bean mt genome [29]. The analysis of 10 recombinant clones supports existence of the MC molecule in wheat mt genome [7]. However, the result of field-inversion electrophoresis suggests that Physcombitrella mt genome does not consist of a multipartite structure, as seen in angiosperms [30]. As mitochondrial gene orders are significantly different between lower plants and higher flowering plants, the multipartite structures as seen among angiosperms may originate during the evolution of pteridophytes or seed plants [9], [30].
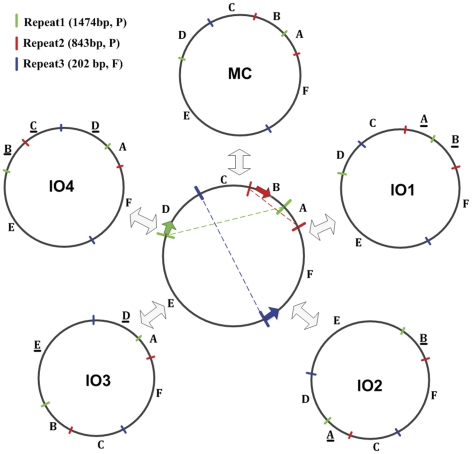
Repeat prediction by REPuter shows that there are 14 repeat pairs in Bh mt genome ( Table 4 ). Since not all the repeats are involved in recombination, from the mt assembly [15] we detected 3 repeat-specific contigs that are candidates for the recombination among the MC and other isomeric (IO) and subgenomic molecules, which have been confirmed by SOLiD sequencing [15]. Those 3 repeat contigs are the repeat pairs of two palindromic matches (1,474 bp and 843 bp) and one forward match (222 bp). By aligning all SOLiD long mate-pair reads to both ends of the repeats, we constructed the MC and 4 isomeric molecules ( Figure 3 ). These subgenomic molecules are not discussed further because we have yet to find significant differences among the sequence reads.
Table 4. Repeat pairs predicted based on REPuter in B. hygrometrica mt genome.
| Repeat length(bp) | Start Position | Match direction* | Repeat length (bp) | Start Position | E-value |
| 1474 | 51709 | P | 1474 | 232397 | 0.00E+00 |
| 843 | 42273 | P | 843 | 108092 | 0.00E+00 |
| 222 | 153742 | F | 222 | 405380 | 3.56E-118 |
| 202 | 44767 | F | 202 | 395750 | 1.77E-111 |
| 180 | 449174 | F | 180 | 504438 | 4.53E-93 |
| 165 | 156474 | P | 165 | 272697 | 4.08E-84 |
| 131 | 25864 | P | 131 | 291976 | 3.89E-66 |
| 119 | 449235 | F | 119 | 504499 | 5.92E-59 |
| 116 | 42870 | P | 116 | 246903 | 3.70E-57 |
| 116 | 108222 | F | 116 | 246903 | 3.70E-57 |
| 109 | 300367 | F | 109 | 308910 | 1.74E-55 |
| 120 | 403184 | P | 120 | 496901 | 3.15E-55 |
| 102 | 126580 | P | 102 | 462677 | 1.32E-44 |
| 101 | 521 | F | 101 | 173834 | 5.13E-44 |
Note: the 3 recombination of repeat pairs for mt genomes are shown in bold.
*F and P stand for forward and palindromic matches, respectively.
Figure 3. Production of various molecular structures from the MC molecule by intra-molecular recombination between 3 different repeat pairs.
A, B, C, D, E, and F are 6 segments separated by repeat pairs. Underlined segments are the negative strand as compared to the MC molecule. Three repeat pairs are shown in different colors. The keys show repeat name, length, and matching direction. F and P stand for forward and palindromic matches, respectively.
The length of recombinant repeats (222 bp, 843 bp, and 1,474 bp) of Bh is different from that of V. radiata, and demonstrates the recombination across short mt repeats (38–297 bp) [31]. The longer repeats are reminiscent of those found in other angiosperm mitochondrial genomes, which are involved in mt genome rearrangements and can result in stoichiometric shifting of subgenomic mt genome topologies, occasionally beyond detection level for one (or more) of alternative DNA topologies [29], [32]. The 3 recombinant repeats are located in gene-rich regions and split mt genome into 6 segments. Each segment has some essential conserved genes, such as nad1 e4 and rps13 in segment A, and atp6, nad2 e3-5 and coxb in segment B. From this point, it is possible that the mt genome of Bh do not have subgenomic molecules. There are 4 genes (nad1, nad2, nad5 and sdh3) with exon-intron structure separated by the recombinant repeats ( Table 5 ). The nad1 gene in MC molecule is cross-strand gene with exon 4 in positive strand and exon 1–3 in negative strand. Recombination involving introns might lead to rearranged molecules without loss of essential genes [30]. In all rearranged 4 isomeric molecules, the Trans-splicing genes are different, and the IO3 molecule has all 4 Trans-splicing genes. Trans-splicing status of group II intron widely distributes in the mt genome of higher plant [33]–[35]. We compared gene structures of 15 mt genomes from lower to higher plants, and found 3 conserved genes (nad1, nad2, and nad5) as well as other higher plants contain trans-splicing intron ( Table 6 ), while there is no intron in those genes of Chara vulgaris. The 3 genes structure supported the multipartite structures formed by multiple recombination may arise with the earliest tracheophytes [30], [32], and can be a molecular signature of plant evolution.
Table 5. The segments and organization of multipartite structures in B. hygrometrica mt genome.
| Mt segment | Start position | End position | Length (bp) | Exon of cross-strand gene |
| A | 43116 | 51708 | 8592 | nad1 e4 |
| B | 53183 | 108091 | 54908 | nad2 e3-5; nad5 e1 |
| C | 108935 | 153741 | 44806 | |
| D | 153964 | 232396 | 78432 | nad2 e1-2; nad5 e2-3; nad1 e2-3 |
| E | 233871 | 405379 | 171508 | nad5 e4-5; sdh3 e2 |
| F | 405602 | 42272 | 147189 | sdh3 e1; nad1 e1 |
Note: segments or exons in negative strand are underlined.
Table 6. Structural comparison of nad1, nad2, and nad5 genes in plant mt genomes.
| Species | nad1 | nad2 | nad5 |
| Chara vulgaris | −* | +* | +* |
| Marchantia polymorpha | +* | ++ | ++ |
| Megaceros aenigmaticus | ++++ | +++ | ++++ |
| Cycas taitungensis | +−−−− | −−+++ | +++++ |
| Triticum aestivum | −−−−+ | ++−−− | +++++ |
| Oryza sativa | +−−−− | +++−− | ++−−− |
| Sorghum bicolor | +−−+− | −−+++ | ++−++ |
| Tripsacum dactyloides | −−−−+ | ++−−− | +++++ |
| Zea mays | ++++− | −−+++ | ++−−− |
| Beta vulgaris | −−−+− | ++−−− | +++−− |
| Brassica napus | −++++ | −−+++ | ++−−− |
| Arabidopsis thaliana | −−−−− | −−−−− | −−−−− |
| Nicotiana tabacum | +−−−− | −−+++ | ++−++ |
| Vitis vinifera | +++−− | +++++ | −−−−− |
| Boea hygrometrica | −−−++ | −−−−− | +++++ |
Note: “+” or “−” shows exons located in positive or negative strands, respectively.
“*” indicates genes that lack exon-intron structures.
The number of “+” or “−” strands for the number of exons in a gene.
Comparative analysis of cp genome organization
We compared 12 cp genomes ranging from green algae to angiosperm (Table S6). The GC contents of cp genome are lowest in lower plants (Charophyta and Bryophyta) but highest in Cycads. Monocots seem to have slightly higher GC contents than dicots among their cp genomes. The genome size and structure of cp genomes are also different in those cp genomes. For example, C. vulgaris (184,933 bp; Charophyta) has the largest genome while the smallest genome is found in lower plant Marchantia polymorpha (121,024 bp). The genome size of angiosperms is more stable than lower plants with dicots larger than monocots. Compared to lower pants, the most variable portions of angiosperm cp genomes are percentages of IRs (34% in A. thaliana) and LSC (54.5% in A. thaliana) regions. This is the result of IR expansion into the LSC region from green algae to angiosperm [4].
The cp genome contains genes that encode structural and functional components of the organelle. Although some genes and gene clusters are well conserved among all plants, the overall structure of cp genomes show remarkable differences (Table S6). First, there are 63 core protein-coding genes, shared among all plants, whereas there are 3 additional core genes (chlB, chlL and ycf12) only found in the lower plant lineage (Charophyta, Bryophyta, and Cycads). The 63 core cp genes are involved in photosynthesis, energy metabolism, and other housekeeping functions. Second, there are 10 genes (psaM, rpl5, rpl12, rpl19, tufA, ycf20, ycf62, ycf66, odpB, and ftsH) are unique to green algae. All of them reside in the LSC region except ycf20 gene that is duplicated in IR regions. Compared to seed plants, there is only one gene, ribosomal protein L21 (rpl21) is conserved in both green algae and liverwort. Third, all four ribosomal RNA genes (rrn4.5, rrn5, rrn16, and rrn23) have 2 copies in IR regions except the 2 copies of rrn4.5 that is lost in Charophyta. Fourth, gene loss and transfer to the nucleus is a common feature of cp genomes [36]. We detected 3 genes (petL, petN, and ycf3) that are lost at the base of the Bryophyta lineage and 2 genes (accD and ycf2) are lost in monocots as compared to dicots. There are also some species-specific gene lost events, such as psaJ in O. sativa and nadJ-ccsA lost in O. europaea [20]. The unique loss of psbZ in LSC region testifies the convergent evolution of B. hygrometrica and O. europaea.
The order of cp genes in plants is not constant, changing among different regions of the genome as large gene clustering become rare. Among 63 core protein-coding genes, 50 are always reside in LSC region, 5 (psaC, ndhD, ndhE, ndhG, and ndhI) in SSC region, and 8 (ndhA, ndhB, ndhF, ndhH, rpl2, rpl23, rpl32 and rps7) in variable positions among 12 examined cp genomes. No conserved protein-coding genes are found constant in IR regions. These mobile genes may serve as an indication of lineage markers, since 4 of them (ndhB, rpl2, rpl23, and rps7) locate on LSC region in lower plants and migrated to IR regions in higher plants. Genes residing in the boundary of LSC/IRA or IRB/LSC are usually ribosomal proteins S12 (rps12) in higher plants and the position-conserved ycf1 is more likely present in the boundary of IRA/SSC and SSC/IRB in dicots.
Comparative analysis of plant mt genomes
The plant mt genomes are exceptionally variable in size, structure, and sequence content and the accumulation of repetitive sequences contributes the most to such variation [31]. From the feature comparison of 15 plant mt genomes ( Table 7 ), we noticed that their genome sizes vary from 67,737 bp in C. vulgaris to 773,279 bp in V. vinifera. Recently, the large mt genome have been reported in Cucurbita pepo with 982,833 bp [10]. The GC contents of these genomes are also variable from 40 to 47%. There is a massive difference of coding sequences between lower and higher plants. The coding sequence in C. vulgaris is 90.7%, whereas it is 4.94% in Tripsacum dactyloides. Repeat content ranges from 1 to 41% among seed plants and are smallest in B. hygrometrica, composed of only 1.45% of the genome. Both large (>1,000 nt) and small (<50 nt) repeats affect recombination in seed plants [7], [31], [37]. The protein-coding genes and tRNAs in mt genomes also vary largely because of the large number of function-unknown proteins or ORFs in mt genomes and frequent plastid DNA insertions including cp tRNA genes [26], [38].
Table 7. Comparison of basic features among 15 mt genomes.
| Scientific Name | Size (bp) | GC (%) | Coding (%) | Repeats (%) | Protein-coding genes | tRNAs | rRNAs |
| Chara vulgaris | 67,737 | 40.9 | 90.7 | 3.2 | 46 | 27 | 3 |
| Marchantia polymorpha | 186,609 | 42.41 | 20.3 | 10.1 | 76 | 29 | 3 |
| Megaceros aenigmaticus | 184,908 | 46.01 | 17.9 | 2.6 | 48 | 18 | 3 |
| Cycas taitungensis | 414,903 | 46.92 | 10.1 | 15.1 | 39 | 26 | 3 |
| Triticum aestivum | 452,528 | 44.35 | 8.6 | 10.1 | 39 | 25 | 9 |
| Oryza sativa | 490,520 | 43.85 | 11.1 | 28.8 | 53 | 22 | 3 |
| Sorghum bicolor | 468,628 | 43.73 | 6.69 | 13.1 | 32 | 18 | 3 |
| Tripsacum dactyloides | 704,100 | 43.93 | 4.94 | 40.6 | 33 | 18 | 3 |
| Zea mays | 569,630 | 43.93 | 6.2 | 11.4 | 163 | 29 | 4 |
| Beta vulgaris | 368,801 | 43.86 | 10.3 | 12.5 | 140 | 26 | 5 |
| Brassica napus | 221,853 | 45.19 | 17.3 | 5.5 | 79 | 17 | 3 |
| Arabidopsis thaliana | 366,924 | 44.77 | 10.6 | 7.8 | 117 | 21 | 3 |
| Nicotiana tabacum | 430,597 | 44.96 | 9.9 | 10.8 | 156 | 23 | 4 |
| Vitis vinifera | 773,279 | 44.14 | 4.98 | 1.71 | 74 | 31 | 3 |
| Boea hygrometrica | 510,519 | 43.27 | 12.19 | 1.45 | 33 | 28 | 4 |
Note: Parts of the dataset are available from published data [53].
We also carefully examined conserved genes in different plant lineages (Table S4). First, there are 14 conserved core protein-coding genes shared among all lineages, including seven subunits of NADH dehydrogenase (Complex I), one subunit of ubichinol cytochrome c reductase (Complex III), three subunits of cytochrome c oxidase (Complex IV), and three subunits of ATP synthase (Complex V). All these genes play important roles either in proton movement across the inner membrance of the mitochondrion or electron transfer reactions in the respiratory chain. However, the gene structures are not conserved among them, and only two genes (nad4 and cox2) have exon-intron structures in all mt genomes. For comparison, there are 9 genes (nad3, nad4L, nad6, nad9, cob, cox3, atp1, atp9, and ccmFN) without exon-intron structure in both seed and early land plants and with exon-intron structure at least in one lower plant. Intron structure in mt genes is common as we only detected 6 genes (sdh3, sdh4, atp4, atp8, ccmB, and ccmFN) have no introns among all plants. Second, gene loss is more frequent in dicots than monocots, as genes in cytochrome c biogenesis are lost in both B. vulgaris and A. thaliana. Three species (Nicotiana tabacum, V. vinifera and B. hygrometrica) gained sdh3 as in this study. The number of ribosomal protein genes is different in various plant mt genomes. Most ribosomal proteins (23) are present in V. vinifera genome. Contrast to higher plants, there is no matR detected in liverwort and green algae in this study. However, it is reported that in mosses, Takakia and Sphagnum have part of matR [35], [39]. Most of mt genomes in plants have 3 ribosomal RNAs (rrn5, rrn18, and rrn26), but there are multiple copies found in angiosperms (such as T. aestivum and B. vulgaris). Copy number-variable mt genes are reported in wheat, rice, and maize [7]. In summary, since the gene coding fraction is much less among mt genomes as compared to cp genomes, even conserved genes are also variable in gene content, structure, and intron positioning [30].
Plastid DNA insertions in mt genome
One of the important events in determining mt genome size in angiosperms is the frequent capture of sequences from the cp genome [10], [26], [28], [40]. A recently study demonstrates frequent DNA transfer from cp to mt genomes occur as far back as the common ancestor of the extant gymnosperms and angiosperms, about 300 MYA and the frequency of cp-derived sequence transfer is positively correlated with variations in mt genome size [41]. For instance, B. hygrometrica mt genome contains fragments of cp origin, ranging from 50 to 5,146 bp in length (Table S7). The total fraction of cp-derived sequences present in Bh mt genome is 53,440 bp, 10.5% of the whole mt genome. Most of these insertions are conserved, as evidenced from the observation that 45 out 80 insertions (over 50 bp) are identified in mt genomes of other plants. The average GC content between old insertions (at least found one homolog in the mt genomes of other plants) and new insertions (no homologs found in other mt genomes) is obviously different (Figure S4). The average GC content of old insertions (41.59%) is distinct from that of mt genomes (43.27%) as well as the average GC content of new insertions (35.32%) is close to the GC content of cp genomes (37.59%; Table 8 ). The result suggests positive correlations between the GC content of cp genomes and new insertions (coefficient value r2 = 0.69) and between the GC content of mt genomes and old insertions (coefficient value r2 = 0.64). There is a significant difference between the GC content of old and new insertions (T test: P<0.01). All of cp-derived sequences in lower plants are old insertion, and it is strange to see there are no new insertions from cp genomes during the evolution of A. thaliana. Compared to other angiosperms, the most striking feature of Bh mt genome is thefrequent sequence transfer from the cp genome in its recent evolution. Cp-derived sequence analyses may provide clues for the understanding functional indications of DNA insertions from cp in mt genomes.
Table 8. Comparison of cp-derived sequences in mt genomes among plants.
| Species | Chara | Marchantia | Cycas | Triticum | Oryza | Sorghum | Zea | Arabidopsis | Nicotiana | Vitis | Boea |
| Cp-GC (%) | 26.19 | 28.81 | 39.45 | 38.31 | 38.99 | 38.49 | 38.46 | 36.29 | 37.85 | 37.4 | 37.59 |
| * Mt-GC (%) | 40.9 | 42.41 | 46.92 | 43.93 | 43.85 | 43.73 | 43.93 | 44.77 | 44.96 | 44.14 | 43.27 |
| *Mtpt No | 11 | 16 | 40 | 57 | 71 | 49 | 41 | 30 | 43 | 51 | 80 |
| *Mtpt length (bp) | 915 | 1379 | 17608 | 15089 | 34969 | 33006 | 24859 | 5150 | 11415 | 68925 | 53440 |
| Old mtpt | 11 | 16 | 21 | 54 | 63 | 44 | 38 | 30 | 39 | 33 | 45 |
| New mtpt | 0 | 0 | 19 | 3 | 8 | 5 | 3 | 0 | 4 | 18 | 35 |
| Old-GC (%) | 54.54 | 52.79 | 45.85 | 44.63 | 40.62 | 41.23 | 44.55 | 46.06 | 46.2 | 40.52 | 41.59 |
| New-GC (%) | \ | \ | 40.75 | 34.49 | 37.72 | 39.47 | 42.62 | \ | 34.09 | 34.08 | 35.32 |
*Mtpt: cp-derived sequences from chloroplast to mitochondrial genomes. Only the genus names are indicated.
We also analyzed the inserted cp genomic sequences to mt genomes. First, there are 85 cp-derived fragments in the Bh mt genome. Most of them, especially protein-coding sequences, have no intact gene structure or have frameshifts/indels and the fact suggests that theses cp-derived sequences are degenerated and lack functional constraints [41]. However, protein-coding genes such as ribosomal proteins and tRNAs originated from cp genomes appear still functional in Bh mt genome. Similar genes are also seen in other angiosperms [26], [28]. Second, it is curious to investigate the locations of cp-derived sequences in cp DNAs to see if any particular regions in cp genomes are hotspots of DNA transfer. Among 80 transferred sequence fragments in Bh cp genome, 45, 7, and 28 are from LSC, SSC, and IR regions, respectively. The numbers of fragments appear correlating to the lengths of LSC (84,692 bp), SSC (17,901 bp) and IRs (50,900 bp), and such correlations are also seen in other angiosperms, including maize, rice, wheat, and tobacco [41]. In conclusion, DNA transfer from cp genomes to mt genomes in angiosperms occurs randomly as it has been proposed earlier by Mastsuo et al in rice [42].
tRNAs transfer between cp and mt genomes
To investigate whether mt genomes encodes a full set of tRNAs species necessary for protein synthesis in the organelle, we identified 28 tRNA genes from the complete Bh assembly based on tRNA structures and realized that all 61 codons are used by Bh mitochondria (Table S3). However, the tRNA genes encoded by the mt genome alone are not sufficient to decode all codons; for instance, trnA is missing in Bh, and it suggests that the role of the missing tRNA is supplied by either cp or nuclear genomes [22], [26], [41]. tRNAs originated from plastids are called cp-derived tRNAs and their counterparts are native mt tRNAs. Half of the 28 mt tRNAs in B.hygrometrica are identified as cp-derived tRNAs (Table S8) and 19 amino acids are encoded by only one codon except for leucine (UAA and CAA) and serine (GCU, UGA, and GGA).
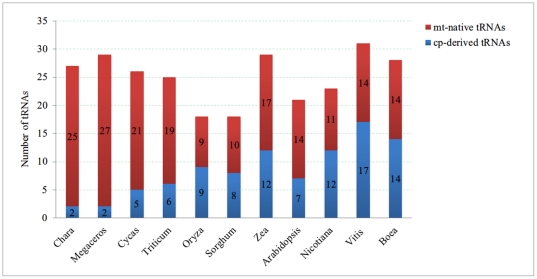
In contrast to the protein coding genes, mt tRNA genes appear constantly being transferred from cp genomes during the evolution of angiosperms ( Figure 4 and Table S8), and the proportion of cp-derived tRNAs in mt genomes increases from 8% in Charophyta to 55% in dicotyledonous plants. There are 17 cp-derived tRNAs and 14 mt native tRNAs in mt genome of V. vinifera, which has the most cp-derived tRNAs among dicotyledonous plants. Seven mt-native tRNA genes (trnD-GUC, trnE-UUC, trnI-CAU, trnK-UUU, trnM-CAU, trnS-GCU, trnS-UGA and trnW-GUA) and one cp-derived tRNA gene (trnF-GAA) are common to all 15 species. Compared to the mt native tRNA genes in lower plants, there are three tRNA genes (trnH-GUG, trnN-GUU, and trnW-GUA) integrated as part of large cp genomic fragments into mt genomes among angiosperms [26]. This indicates that frequent DNA transfer from cp to mt genomes occur as far back as the emergence of seed plants [41]. We detected two different tRNAs transfer events in seed plants. One is trnC-GCA transfer in monocots and the other involves two (trnD-GUC and trnQ-UUG) gene transfer events in dicots. Cp-derived tRNA genes replace their mt counterparts were identified in all sequenced angiosperms, even in gymnosperm Cyas mtDNA. But these replacement not occurred in Marchantia, Reclinomonas, Cyanidioschyzon, Nephroselmis, Chara, and Physcomitrella [43].The mt-native tRNA gene (trnG-GCC) had all been lost in monocots and six mt-native genes (trnA-UGC, trnG-UCC, trnL-UAG, trnR-UCU, trnR-ACG, and trnT-GGU) are mostly lost in all angiosperms.
Figure 4. Distribution of tRNA genes in 15 plant mt genomes.
Gene gain and loss in plant organelle genome
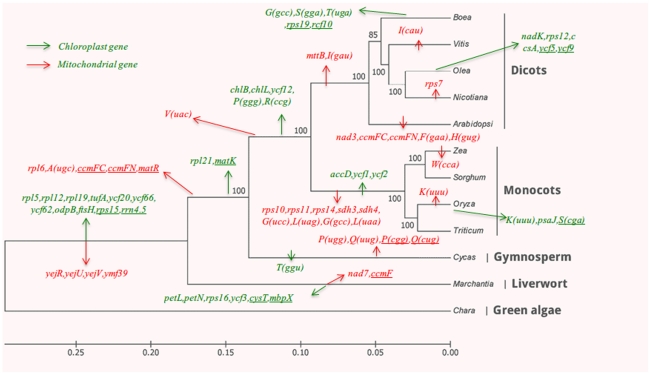
Starting from Bh organellar genomes, we have analyzed in a systematic way representative cp and mt genomes of various lineages and our results provide information for a better understanding of organellar genome evolution and function. Sequence-based phylogenetic analysis clearly supports the conclusion that Bh is much close to V. vinifera. Structural dynamics of Bh mt genome suggest that the multipartite structures may have started during the evolution of seed plants [30]. However, mechanisms for rapid mt genome rearrangement and expansion among plant lineages remain enigmatic. Based on eleven known cp and mt genomes of different lineages, we showed a strong relationship between the changing organellar genomes among angiosperms, and some of the lineage-associated gene gain and loss may provide excellent markers for phylogenetic studies ( Figure 5 ). For instance, there are 9 cp and 4 mt genes lost during the evolution from green algae to lower land plants. It seems that monocots have a faster rate of evolution than dicots in organellar genomes in our study, because 3 cp and 9 mt genes are lost in monocots and only 2 mt genes are lost in dicots. In additional, gene structures and positioning of cp and mt genomes are also very informative for the understanding of land plant evolution. In agreement with the results of several previous studies, most of the transferred angiosperm sequences from cp to mt genomes become degenerated and are regarded as junk sequences, whereas some of the cp-derived tRNAs are still functional in mt genomes [26], [28], [41]. As more plant organellar genome sequences become available, the evolution of plant organellar genomes will unveil its details and mechanisms.
Figure 5. Phylogenetic distribution of gene gain and loss from chloroplast and mitochondrial genomes in plant lineages.
The genes that are gained independently in different lineages are underlined. Genes of chloroplast and mitochondrial origins are indicated with green and red, respectively. Phylogenetic relationships of the plants are determined based on the conserved genes of chloroplast genomes described in Figure S2.
Materials and Methods
Genome sequencing and assembly
We developed an efficient procedure for Bh organellar genome sequencing and assembly using whole genome data from 454 GS FLX sequencing platform [15]. Briefly, we collected fresh leaves and extracted genomic DNA for 454 GS FLX sequencing (see manuals of GS FLX Titanium for detail). In order to validate genome assembly and to make sure for the assembly of the master circle or MC, we construed two mate-pair libraries (2×50 bp) for SOLiD 4.0 sequencing platform with insert sizes of 1–2 kb and 3–4 kb by following the SOLiD Library Preparation Guide. The method for assembling organellar genome was based on correlation between contig read depth and copy number in the genome [44]. We first filtered cp reads from the raw data according known plant cp genome sequences and then assembled the “clean” read into cp genome into the major segments: large-single-copy (SSC), small-single-copy (SSC) and inverted repeats (IRs) regions. The mt genome assembly is more complicated than that of cp genomes. We filtered the contigs including mt conserve genes (such as NADH dehydrogenase and succinate dehydrogenase) and removed the contamination of cp sequences. The gene-based method for assembling mt genome has been reported earlier [7]. Mapping all the SOLiD mate-pair reads to mt contigs with Bioscope, we obtained the major contig relationship map in the repeat regions to assemble the MC.
Genome annotation
The cp genome was annotated by using the program DOGMA (Dual Organellar GenoMe Annotator) [45] coupled with manual corrections for start and stop codons. Protein-coding genes are identified by using the plastid/bacterial genetic code. Codon usage is predicted by using CodonW (http://codonw.sourceforge.net/). We construct a custom-designed amino acid database for protein-coding genes and nucleotide databases for rRNA and tRNA genes, compiled from all previously annotated plant mt genomes available at the organelle genomic biology website at NCBI (http://www.ncbi.nlm.nih.gov/genomes/ORGANELLES/organelles.html). NCBI BlastX and BlastN searches of the mt genome against the databases allow us to find protein and RNA genes, respectively. All BlastN and BlastX searches are carried out by using the default settings with e-value 1e-10. Putative RNA editing sites are inferred to create proper start and stop codons as well as to remove internal stop codons. We also used tRNAscan-SE [46] to corroborate tRNA boundaries identified by BlastN. The annotated GenBank files of the cp and mt genomes of Bh are used to draw gene maps using OrganellarGenome DRAW tool (OGDRAW) [47]. The maps were then examined for further comparison of gene order and content.
Analyses on SNPs, repeats, and cp-derived sequences
We identified intra-specific SNPs in both cp and mt genomes. Using BioScope, we mapped two runs of SOLiD mate-pair reads to both cp and mt genomes (BioScope Software User Guide). We carried out repeat sequence analysis using the REPuter web-based interface (http://bibiserv.techfak.uni-bielefeld.de/reputer/) [48], including forward, palindromic, reverse and complemented repeats with a minimal length of 50 bp. Transposable elements and other repeated elements were mapped with RepeatMasker Web Server (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) running under the cross_match search engine. Cp-derived sequences are identified with BlastN search of mt genomes against Bh annotated cp genomes (Identity ≥80%, E-value≤1e-5, and Length ≥50 bp). The cp-derived sequences were then aligned to all known plant mt genomes by using BlastN (Identity ≥80%, E-value≤1e-5, and Coverage ≥50%). tRNAs transferred to the mt genome were identified by aligning to all tRNAs in the cp genome of the same species by using BlastN (Identity ≥80%, E-value≤1e-5, and Coverage ≥50%).
Phylogenetic Analysis
We compare the Bh cp genome with other plant organellar genomes, and use the homologous protein-coding sequences to construct phylogenetic tree. Sixty-three cp protein sequences (psaA, psaB, psaC, psaI, psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, petA, petB, petD, petG, atpA, atpB, atpE, atpF, atpH, atpI, rbcL, rpoA, rpoB, rpoC1, rpoC2, ndhA, ndhB, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, rpl2, rpl14, rpl16, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36, rps2, rps3, rps4, rps7, rps8, rps11, rps14, rps18, clpP, cemA, and ycf4) from 12 different organisms (Table S2) are aligned and concatenated into a dataset of 196,313 amino acids.
We align amino acid sequences from individual genes using MUSCLE v3.8.31 [49], remove ambiguously aligned regions in each alignment using GBLOCKS 0.91b [50], and concatenate the aligned sequences. We use maximum likelihood method and PhyML v3.0 [51] under Jones-Taylor-Thornton (JTT and gamma distribution of rates across sites with four categories) model of sequence evolution to construct phylogenetic trees. Confidence of branch points is estimated based on 100 bootstrap replications. We obtained the best tree after heuristic search with the help of Modelgenerator [52].
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are as follows: Chara vulgaris, NC_008097 and NC_005255; Marchantia polymorpha, NC_001319 and NC_001660; Megaceros aenigmaticus, NC_012651; Cycas taitungensis, NC_009618 and NC_010303; Triticum aestivum, NC_002762 and NC_007579; Oryza sativa, NC_001320 and NC_011033; Sorghum bicolor, NC_008602 and NC_008360; Tripsacum dactyloides, NC_008362; Zea mays, NC_001666 and NC_007982; Beta vulgaris, NC_002511; Brassica napus, NC_008285; Arabidopsis thaliana, NC_000932 and NC_001284; Nicotiana tabacum, NC_001879 and NC_006581; Vitis vinifera, NC_007957 and NC_012119; Olea europaea, NC_013707; Boea hygrometrica, JN107811 and JN107812.
Supporting Information
Chloroplast genomic alignment between Boea hygrometrica and Olea europaea. Alignments with direct match are shown in red and reverse match are shown in blue. Obviously, alignments of two IR regions are indicated in blue.
(DOC)
Molecular phylogenetic analysis based on Maximum Likelihood method by using JTT matrix-based model (Chara vulgaris as outgroup). The bootstrap consensus tree inferred from 100 replicates is taken to represent the evolutionary history of selected plants. The dataset is composed of sixty-three conserved cp proteins concatenated to 14,894 positions from 12 plant cp genomes. Nodes receive over 80% bootstrap replicates are indicated at phylogenetic positions and B. hygrometrica is next to Vitis vinifera among dicots.
(DOC)
Mitochondrial genomic alignment between Boea hygrometrica and Vitis vinifera. Alignments with direct match are shown in red and reverse match are shown in blue.
(DOC)
The GC distribution between new and old cp-derived sequences in mitochondrial genome of Boea hygrometrica. Hits stand for the results of homologous alignments with other known mitochondrial genomes.
(DOC)
Table Codon usage table of the chloroplast genes.
(DOC)
cp and mt genomes of 15 plants comparison in this study.
(DOC)
Codon usage table of the mitochondrial genes.
(DOC)
Gene content and characteristic comparison of 15 mt genomes.
(XLS)
Intravarietal single nucleotide polymorphisms (intraSNPs) in mt genome.
(XLS)
Gene content and characteristic comparison of 12 cp genomes.
(XLS)
Blast result of chloroplast-derived DNA segment in mitochondrial genome of Boea hygrometrica.
(XLS)
tRNAs comparison of both cp and mt genomes among 11 pant.
(XLS)
Acknowledgments
We thank Beijing Institute of Genomics, Chinese Academy of Sciences for technical support from. We also wish to thank Liancheng Huang for his preparation of the B. hygrometrica materials for this project.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work is supported by grants from Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-R-01-04), Natural Science Foundation of China (90919024), Natural Science Foundation of China (30900831), from the Ministry of Science and Technology as the National Science and Technology Key Project (2008ZX10004-013), and the National Basic Research Program (973 Program) from the Ministry of Science and Technology of the People's Republic of China (2011CB944100). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Olmstead R, Palmer J. Chloroplast DNA systematics: A review of methods and data analysis. American journal of botany. 1994;81:1205–1224. [Google Scholar]
- 2.Qiu Y-L, Li L, Wang B, Xue J-Y, Hendry TA, et al. Angiosperm phylogeny inferred from sequences of four mitochondrial genes. Journal of Systematics and Evolution. 2010;48:391–425. [Google Scholar]
- 3.Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, et al. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Molecular Biology and Evolution. 2006;23:2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- 4.Yang M, Zhang XW, Liu GM, Yin YX, Chen KF, et al. The Complete Chloroplast Genome Sequence of Date Palm (Phoenix dactylifera L.). Plos One. 2010;5 doi: 10.1371/journal.pone.0012762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen DR, Dastidar SG, Cai Z, Penaflor C, Kuehl JV, et al. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Mol Phylogenet Evol. 2007;45:547–563. doi: 10.1016/j.ympev.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Chang CC, Lin HC, Lin IP, Chow TY, Chen HH, et al. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Molecular Biology and Evolution. 2006;23:279–291. doi: 10.1093/molbev/msj029. [DOI] [PubMed] [Google Scholar]
- 7.Ogihara Y, Yamazaki Y, Murai K, Kanno A, Terachi T, et al. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic acids research. 2005;33:6235–6250. doi: 10.1093/nar/gki925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li LB, Wang B, Liu Y, Qiu YL. The Complete Mitochondrial Genome Sequence of the Hornwort Megaceros aenigmaticus Shows a Mixed Mode of Conservative Yet Dynamic Evolution in Early Land Plant Mitochondrial Genomes. Journal of Molecular Evolution. 2009;68:665–678. doi: 10.1007/s00239-009-9240-7. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Xue JY, Wang B, Li L, Qiu YL. The mitochondrial genomes of the early land plants Treubia lacunosa and Anomodon rugelii: dynamic and conservative evolution. Plos One. 2011;6:e25836. doi: 10.1371/journal.pone.0025836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alverson AJ, Wei XX, Rice DW, Stern DB, Barry K, et al. Insights into the Evolution of Mitochondrial Genome Size from Complete Sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Molecular Biology and Evolution. 2010;27:1436–1448. doi: 10.1093/molbev/msq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer JD, Herbon LA. Plant mitochondrial DNA evolves rapidly in structure, but slowly in sequence. Journal of Molecular Evolution. 1988;28:87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- 12.Goremykin VV, Salamini F, Velasco R, Viola R. Mitochondrial DNA of Vitis vinifera and the Issue of Rampant Horizontal Gene Transfer. Molecular Biology and Evolution. 2009;26:99–110. doi: 10.1093/molbev/msn226. [DOI] [PubMed] [Google Scholar]
- 13.Lonsdale DM, Brears T, Hodge TP, Melville SE, Rottmann WH. The Plant Mitochondrial Genome: Homologous Recombination as a Mechanism for Generating Heterogeneity. Philosophical Transactions of the Royal Society of London B, Biological Sciences. 1988;319:149–163. [Google Scholar]
- 14.Fujii S, Kazama T, Yamada M, Toriyama K. Discovery of global genomic re-organization based on comparison of two newly sequenced rice mitochondrial genomes with cytoplasmic male sterility-related genes. Bmc Genomics. 2010;11 doi: 10.1186/1471-2164-11-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang TW, Zhang XW, Hu SN, Yu J. An efficient procedure for plant organellar genome assembly, based on whole genome data from the 454 GS FLX sequencing platform. Plant methods. 2011 doi: 10.1186/1746-4811-7-38. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng X, Hu ZA, Wang HX. mRNA differential display visualized by silver staining tested on gene expression in resurrection plant Boea hygrometrica. Plant Molecular Biology Reporter. 1999;17:279–279. [Google Scholar]
- 17.Jiang GQ, Wang Z, Shang HH, Yang WL, Hu Z, et al. Proteome analysis of leaves from the resurrection plant Boea hygrometrica in response to dehydration and rehydration. Planta. 2007;225:1405–1420. doi: 10.1007/s00425-006-0449-z. [DOI] [PubMed] [Google Scholar]
- 18.Goulding S, Wolfe K, Olmstead R, Morden C. Ebb and flow of the chloroplast inverted repeat. Molecular and General Genetics MGG. 1996;252:195–206. doi: 10.1007/BF02173220. [DOI] [PubMed] [Google Scholar]
- 19.Palmer J. Plastid chromosomes: structure and evolution. Cell Culture and Somatic Cell Genetics of Plants, vol 7A, The Molecular Biology of Plastids. 1991:5–53. [Google Scholar]
- 20.Besnard G, Hernandez P, Khadari B, Dorado G, Savolainen V. Genomic profiling of plastid DNA variation in the Mediterranean olive tree. Bmc Plant Biology. 2011;11 doi: 10.1186/1471-2229-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unseld M, Marienfeld JR, Brandt P, Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nature Genetics. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 22.Sugiyama Y, Watase Y, Nagase M, Makita N, Yagura S, et al. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Molecular Genetics and Genomics. 2005;272:603–615. doi: 10.1007/s00438-004-1075-8. [DOI] [PubMed] [Google Scholar]
- 23.Tian XJ, Zheng J, Hu SN, Yu J. The rice mitochondrial genomes and their variations. Plant Physiology. 2006;140:401–410. doi: 10.1104/pp.105.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang J, Xia H, Cao M, Zhang X, Zeng W, et al. A comparison of rice chloroplast genomes. Plant Physiology. 2004;135:412–420. doi: 10.1104/pp.103.031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubo T, Nishizawa S, Sugawara A, Itchoda N, Estiati A, et al. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA). Nucleic acids research. 2000;28:2571–2576. doi: 10.1093/nar/28.13.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, et al. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Molecular Genetics and Genomics. 2002;268:434–445. doi: 10.1007/s00438-002-0767-1. [DOI] [PubMed] [Google Scholar]
- 27.Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic acids research. 2003;31:5907–5916. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clifton SW, Minx P, Fauron CMR, Gibson M, Allen JO, et al. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiology. 2004;136:3486–3503. doi: 10.1104/pp.104.044602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arrieta-Montiel M, Lyznik A, Woloszynska M, Janska H, Tohme J, et al. Tracing evolutionary and developmental implications of mitochondrial stoichiometric shifting in the common bean. Genetics. 2001;158:851–864. doi: 10.1093/genetics/158.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terasawa K, Odahara M, Kabeya Y, Kikugawa T, Sekine Y, et al. The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Molecular Biology and Evolution. 2007;24:699–709. doi: 10.1093/molbev/msl198. [DOI] [PubMed] [Google Scholar]
- 31.Alverson AJ, Zhuo S, Rice DW, Sloan DB, Palmer JD. The Mitochondrial Genome of the Legume Vigna radiata and the Analysis of Recombination across Short Mitochondrial Repeats. Plos One. 2011;6 doi: 10.1371/journal.pone.0016404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hecht J, Grewe F, Knoop V. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biology and Evolution. 2011 doi: 10.1093/gbe/evr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malek O, Knoop V. Trans-splicing group II introns in plant mitochondria: the complete set of cis-arranged homologs in ferns, fern allies, and a hornwort. RNA. 1998;4:1599–1609. doi: 10.1017/s1355838298981262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linda B. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion. 2008;8:26–34. doi: 10.1016/j.mito.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Qiu YL, Palmer JD. Many independent origins of trans splicing of a plant mitochondrial group II intron. Journal of Molecular Evolution. 2004;59:80–89. doi: 10.1007/s00239-004-2606-y. [DOI] [PubMed] [Google Scholar]
- 36.Robbens S, Derelle E, Ferraz C, Wuyts J, Moreau H, et al. The complete chloroplast and mitochondrial DNA sequence of Ostreococcus tauri: Organelle genomes of the smallest eukaryote are examples of compaction. Molecular Biology and Evolution. 2007;24:956–968. doi: 10.1093/molbev/msm012. [DOI] [PubMed] [Google Scholar]
- 37.Small I, Suffolk R, Leaver CJ. Evolution of plant mitochondrial genomes via substoichiometric intermediates. Cell. 1989;58:69–76. doi: 10.1016/0092-8674(89)90403-0. [DOI] [PubMed] [Google Scholar]
- 38.Marechal-Drouard L, Guillemaut P, Cosset A, Arbogast M, Weber F, et al. Transfer RNAs of potato (Solanum tuberosum) mitochondria have different genetic origins. Nucleic acids research. 1990;18:3689–3696. doi: 10.1093/nar/18.13.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dombrovska O, Qiu Y-L. Distribution of introns in the mitochondrial gene nad1 in land plants: phylogenetic and molecular evolutionary implications. Mol Phylogenet Evol. 2004;32:246–263. doi: 10.1016/j.ympev.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 40.Alverson AJ, Rice DW, Dickinson S, Barry K, Palmer JD. Origins and Recombination of the Bacterial-Sized Multichromosomal Mitochondrial Genome of Cucumber. The Plant Cell Online. 2011 doi: 10.1105/tpc.111.087189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang D, Wu Y-W, Shih AC-C, Wu C-S, Wang Y-N, et al. Transfer of Chloroplast Genomic DNA to Mitochondrial Genome Occurred At Least 300 MYA. Molecular Biology and Evolution. 2007;24:2040–2048. doi: 10.1093/molbev/msm133. [DOI] [PubMed] [Google Scholar]
- 42.Matsuo M, Ito Y, Yamauchi R, Obokata J. The Rice Nuclear Genome Continuously Integrates, Shuffles, and Eliminates the Chloroplast Genome to Cause Chloroplast–Nuclear DNA Flux. The Plant Cell Online. 2005;17:665–675. doi: 10.1105/tpc.104.027706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L, Wang B, Liu Y, Qiu YL. The complete mitochondrial genome sequence of the hornwort Megaceros aenigmaticus shows a mixed mode of conservative yet dynamic evolution in early land plant mitochondrial genomes. J Mol Evol. 2009;68:665–678. doi: 10.1007/s00239-009-9240-7. [DOI] [PubMed] [Google Scholar]
- 44.Alexander J. Identification and quantification of genomic repeats and sample contamination in assemblies of 454 pyrosequencing reads. 2010 [Google Scholar]
- 45.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 46.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic acids research. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bock R, Lohse M, Drechsel O. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Current Genetics. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 48.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic acids research. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 51.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, et al. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Systematic Biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 52.Keane T, Creevey C, Pentony M, Naughton T, Mclnerney J. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. Bmc Evolutionary Biology. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaw SM, Shih ACC, Wang D, Wu YW, Liu SM, et al. The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Molecular Biology and Evolution. 2008;25:603–615. doi: 10.1093/molbev/msn009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chloroplast genomic alignment between Boea hygrometrica and Olea europaea. Alignments with direct match are shown in red and reverse match are shown in blue. Obviously, alignments of two IR regions are indicated in blue.
(DOC)
Molecular phylogenetic analysis based on Maximum Likelihood method by using JTT matrix-based model (Chara vulgaris as outgroup). The bootstrap consensus tree inferred from 100 replicates is taken to represent the evolutionary history of selected plants. The dataset is composed of sixty-three conserved cp proteins concatenated to 14,894 positions from 12 plant cp genomes. Nodes receive over 80% bootstrap replicates are indicated at phylogenetic positions and B. hygrometrica is next to Vitis vinifera among dicots.
(DOC)
Mitochondrial genomic alignment between Boea hygrometrica and Vitis vinifera. Alignments with direct match are shown in red and reverse match are shown in blue.
(DOC)
The GC distribution between new and old cp-derived sequences in mitochondrial genome of Boea hygrometrica. Hits stand for the results of homologous alignments with other known mitochondrial genomes.
(DOC)
Table Codon usage table of the chloroplast genes.
(DOC)
cp and mt genomes of 15 plants comparison in this study.
(DOC)
Codon usage table of the mitochondrial genes.
(DOC)
Gene content and characteristic comparison of 15 mt genomes.
(XLS)
Intravarietal single nucleotide polymorphisms (intraSNPs) in mt genome.
(XLS)
Gene content and characteristic comparison of 12 cp genomes.
(XLS)
Blast result of chloroplast-derived DNA segment in mitochondrial genome of Boea hygrometrica.
(XLS)
tRNAs comparison of both cp and mt genomes among 11 pant.
(XLS)