Abstract
Tissue engineering approaches fabricate and subsequently implant cell-seeded and unseeded scaffold biomaterials. Once in the body, these biomaterials are repopulated with somatic cells of various phenotypes whose identification upon explantation can be expensive and time-consuming. We show that imaging time-of-flight secondary ion mass spectrometry (ToF-SIMS) can be used to distinguish mammalian cell types in heterogeneous cultures. Primary rat esophageal epithelial cells (REEC) were cultured with NIH 3T3 mouse fibroblasts on tissue culture polystyrene and freeze dried before ToF-SIMS imaging. Results show that a short etching sequence with C60+ ions can be used to clean the sample surface and improve the ToF-SIMS image quality. Principal component analysis (PCA) and partial least-squares discriminant analysis (PLS-DA) were used to identify peaks whose contributions to the total variance in the multivariate model were due to either of the two cell types or the substrate. Using PLS-DA, unknown regions of cellularity that were otherwise unidentifiable by SIMS could be classified. From the loadings in the PLS-DA model, peaks were selected that were indicative of the two cell types and ToF-SIMS images were created and overlaid that showed the ability of this method to distinguish features visually.
Keywords: secondary ion mass spectrometry (SIMS), time-of-flight secondary ion mass spectrometry (ToF-SIMS) multivariate analysis (MVA), principal component analysis (PCA), partial least-squares discriminant analysis (PLS-DA), mass spectrometry imaging, cell phenotyping
INTRODUCTION
A common tissue engineering approach is to fabricate and subsequently implant both seeded and unseeded scaffolding biomaterials1–9. Often, autologous cells are harvested by biopsy, cultured in vitro to expand cell numbers, seeded on scaffolds and then reimplanted. However, the phenotypic purity and identity of these cells is often difficult to assess. The creation of phenotypically homogenous cell populations is especially important where stem cells are differentiated to create specific cell populations for implantation.
Current approaches to identify specific cells in heterogeneous cultures often employ a panel of primary antibodies, a time-consuming and costly approach. Additionally, scanning electron microscopy (SEM) can be used to visualize gross morphological differences, but specific cell type identification is challenging. Fluorescent activated cell sorting (FACS) can be used to identify cells in suspensions and sort these cells, but the labeling schemes employed for FACS analysis can also be time-consuming and expensive and do not provide information on cell spatial distributions and colony formation. Here, we show that time-of-flight secondary ion mass spectrometry (ToF-SIMS) in the imaging mode, in conjunction with multivariate analysis (MVA), can be used to identify cell types in a heterogeneous culture of epithelial cells and fibroblasts with simple sample preparation.
ToF-SIMS is a surface-sensitive mass spectrometric technique providing molecular information about the outermost 1–2 nm of a sample. It has been used for biological applications such as characterization of adsorbed protein films on biomaterial surfaces based on spectral analysis10–13, the study of lipid membrane characteristics using the imaging mode14–16, the analysis of explanted biomaterials17, and the identification of cell types including yeast strains18, thyroid tumor cells19, and prostate cancer cells20,21 and breast cancer cells22. In the ToF-SIMS imaging mode employed in this study, a series of two-dimensional images is created by rastering the primary ion beam over the sample surface. Each of these images represents spatially defined intensity gradients of chosen ionized atoms or molecules (molecular or combined fragments). For each image pixel analyzed, an entire mass spectrum is obtained (typically from 1 to 860 a.m.u., for experiments described here). Then for each selected mass, these intensity images can be created and used to spatially identify molecular features and characterize samples with sub-micrometer lateral resolution (down to 60 nm)23–27.
For cell specimens, there are a number of potential preparation methods intended to preserve membrane chemistry (or intracellular chemistry) while dehydrating the samples sufficiently for the ultra-high vacuum environment of the ToF-SIMS analysis chamber. Cells are often cultured on silicon wafers followed by air drying, flash frozen with liquid nitrogen or liquid propane prior to lyophilization, freeze fractured, or analyzed in a frozen hydrated state28–36. Additionally, chemical treatments are often applied to increase ionization of desired ions or preserve surface structure30 or increase ionization of particular fragments37,38. Etching with C60 primary ion sources minimizes surface artifacts associated with deposited remnant media or other contaminants39. This approach increases the signal to noise ratio for optimal 2D imaging27. It also simplifies sample preparation since less than pristine surfaces can be cleaned with little or no damage.
In this study, we show that it is possible to use mass spectral information to identify individual cell types within ToF-SIMS images and to categorize unidentified cellular regions using the MVA classification technique called partial least-squares discriminant analysis (PLS-DA). PLS-DA, a multivariate classification strategy, is a robust mathematical prediction tool for discriminating ToF-SIMS data40. For studies employing engineered biomaterials designed to direct cell fate and improve regeneration and healing, this one-pass analysis technique could prove useful in both identifying and spatially locating cell types while simultaneously identifying new or unexpected cell phenotypes41,42. Additionally, this technique could be a useful tool for determining phenotypic purity of populations of differentiated stem cells.
EXPERIMENTAL SECTION
Please see the Supplemental Information for experimental details.
RESULTS AND DISCUSSION
Determination of C60+ Etching Parameters
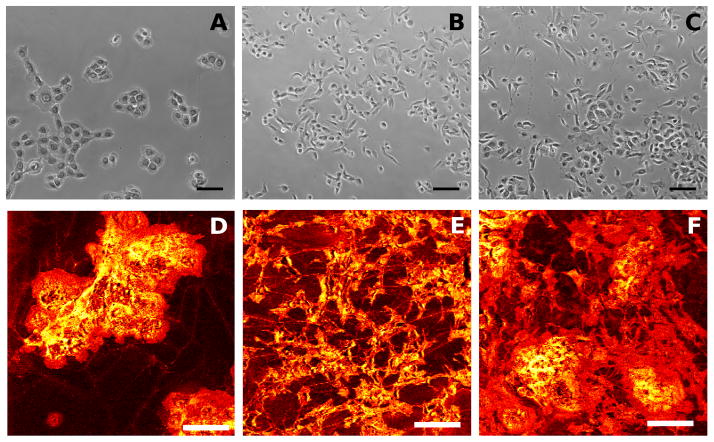
It was hypothesized that ToF-SIMS could be a useful tool for differentiating phenotypically heterogeneous populations of cells using a simple sample preparation method and a single pass analysis. To test this hypothesis, primary rat esophageal epithelial cells (REECs) were cultured with NIH 3T3 fibroblasts and dried for analysis. Figure 1(A–C) contains optical microscope phase contrast images of in vitro populations of REECs, 3T3s, and the heterogeneous mixture of the two. These two cell types were chosen for this study for their visually obvious morphological dissimilarities when grown in culture. In Figure 1A, the typical connected cuboidal colonies associated with primary epithelial cultures can be seen. This is in contrast to the 3T3 fibroblast cultures seen in Figure 1B, which grow in isolated, elongated spindle shapes and can be less than half the total area of an individual REEC from Figure 1A. In Figure 1C, the heterogeneous culture of the two cell types can be seen. Individual spindle-shaped cells, assumedly 3T3s, can be observed in isolation in Figure 1C. Areas of higher cell density are likely REEC colonies, but accurate and definitive identification is not possible by simple visual inspection. In addition, it is possible that these areas could have 3T3 cells within and/or on top of colonies of REECs. ToF-SIMS total ion intensity images for analogous samples can be seen as follows in Figure 1: REECs (Fig 1D), 3T3s (Fig 1E), and heterogeneous mixture (Fig 1F). The ToF-SIMS images are shown with a “hot” color scheme where yellow indicates high relative intensity regions, red indicates intermediate intensity regions, and black represents low or zero intensity regions. The C60 etching parameters used in Fig. 1D through Fig 1F are discussed in Figure 2 (30 seconds etching, corresponding to a primary ion dose of 4.0×1013 C60+ ions). Within the total ion images seen in Fig. 1D through Fig. 1F, it is observed that the cell regions are clearly identified above the substrate background and that the heterogeneous cell sample in Figure 1F has overlapping regions within the image field making visual identification difficult.
Figure 1.
Phase contrast images (A–C) and ToF-SIMS total ion images (D–F) of isolated and heterogeneous cultures. The images are as follows: Rat esophageal epithelial cells (A and D); NIH 3T3 mouse fibroblasts (B and E); Heterogeneous mixture of rat esophageal epithelial cells and 3T3 fibroblasts (C and F). Scale bars represent 100 μm.
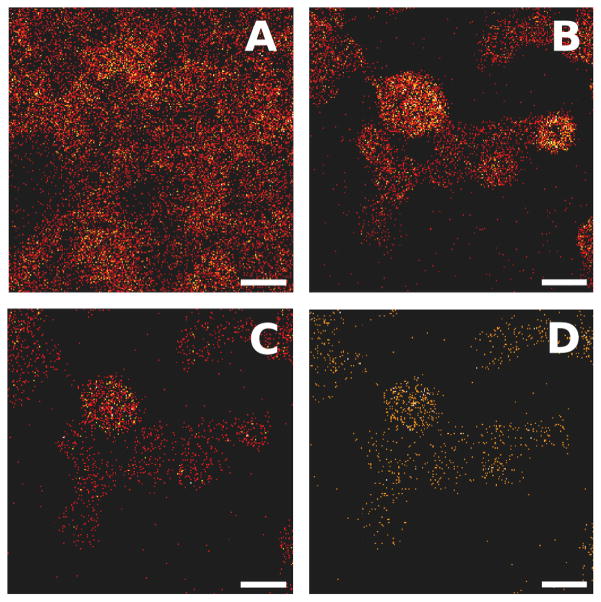
Figure 2.
C60 etching time optimization. The cholesterol fragment at m/z C27H45+ is imaged here for rat esophageal epithelial cells. A) no etching; B) 30 second etching; C) 60 second etching; D) 90 second etching. Scale bars represent 50 μm.
Figure 2 contains a collection of ToF-SIMS images taken from the REEC control samples, before and after C60 etching. Etching with C60 was investigated to increase the relevance of the ToF-SIMS image data by removing surface contaminants associated with residual media components. Several studies have shown that information-rich, molecular depth profiling of entire cells and of biological films is possible using C60 clusters16,27,34,46. In addition to media residues, it is likely that sections of the underlying cells are also removed during etching. The goal for C60 etching in this study is to retain as much of the surface molecular structure as possible while removing biochemical “noise” not directly associated with the cells being analyzed. Each of the images in Figure 2 represents the same location on the REEC sample (seen in Figure 1A). Ion images were acquired for this area after no etching, 30, 60, and 90 seconds of etching (details of C60 etching are listed in the Experimental Section). The ion images in Figure 2 are from the C27H45+ fragment (m/z 369) which has been shown in previous studies to be associated with cholesterol from cell membranes16,45.
In Figure 2A, before C60 etching, the cholesterol signal is diffuse and difficult to differentiate from the background. As shown in Figure 2B, the identification of cholesterol localized to the REECs is improved after etching with C60+ for 30 seconds. In Figure 2C and Figure 2D, the longer etching times (i.e., 60 and 90 seconds) do not significantly improve the quality of the cell images and, in fact, have likely etched through portions of the cells that contain useful information. It is also possible that ion-induced chemical damage might be responsible for the observed signal decrease with increasing etching time. For this reason, 30 seconds of C60 etching with the 10 keV source (corresponding to a C60+ ion dose of 4.0×1013 and an estimated removal of 16 nm of material) was chosen. This was shown to be sufficient to remove media residues while preserving the underlying molecular information44.
Principal Component Analysis Modeling and Classification of Heterogeneous Culture Regions
To test the ability of our models to classify unknown cellular regions within the mixed culture samples, regions from the mixed cultures were projected into the PCA models built from the calibration data (Figures 3 and 4). Figure 3 represents the projection of the mixed culture regions onto the first two principal components within the PCA models created from the control regions (calibration data) for the non-etched and etched surfaces. In all of the PCA plots in Figure 3 and Figure 4, the colored sample labeled with 95% confidence intervals represent the calibration data) while the grey samples represent the projected mixed culture regions data. It should be noted that in all of the multivariate analyses in this study, the data was normalized to the sum of the selected peaks. If this normalization were not performed, the majority of observed variations would be due to ionization differences. However, by normalizing to the sum of the selected peaks, we are removing these ionization differences from our analyses and focusing on the differences between variations in peak intensities only.
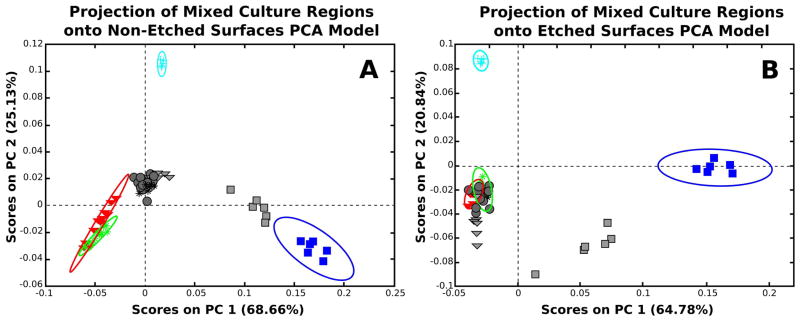
Figure 3.
Principal component analysis models depicting PC 1 vs. PC 2 calculated from the calibration data set for the non-etched (A) and etched (B) surfaces. The following samples represent the calibration data: NIH 3T3 fibroblasts (red triangle), REECs (green asterisk), media coated tissue culture polystyrene (light blue cross), and the clean tissue culture polystyrene (blue squares). The test regions from the mixed culture samples are projected onto the PCA models built from the calibration data. The test regions are represented in grey with analogous shapes to the calibration data with the exception of the unknown cellular regions that are represented with grey circles.
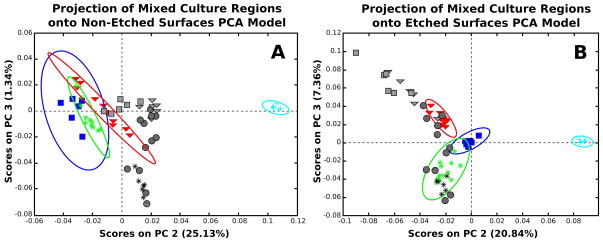
Figure 4.
Principal component analysis models depicting PC 2 vs. PC 3 calculated from the calibration data set for the non-etched (A) and etched (B) surfaces. The following samples represent the calibration data: NIH 3T3 fibroblasts (red triangle), REECs (green asterisk), media coated tissue culture polystyrene (light blue cross), and the clean tissue culture polystyrene (blue squares). The test regions from the mixed culture samples are projected onto the PCA models built from the calibration data. The test regions are represented in grey with analogous shapes to the calibration data with the exception of the unknown cellular regions that are represented with grey circles.
In Figure 3, PC 1 shows separation between the TCPS and biologic material surfaces (i.e. media-coated TCPS and cell-based samples) for the non-etched (Fig 3A) and etched (Fig 3B) surfaces. Then, PC 2 separates the media-coated TCPS surface from the two cell types in both figures. It is not unexpected that the majority of the variance in these models separates the TCPS and media coated TCPS areas from the cell regions since the chemistry of these regions should be very different. The model for the non-etched surfaces captured a combined variance of 93.79% of the total model in the first two PCs (Fig 3A) while the model for the etched surfaces captured a combined variance of 85.62% in the first two PCs (Fig 3B). In both Figure 3 and Figure 4, the grey symbols represent selected regions within the mixed culture ToF-SIMS images that were projected onto the calibration data set PCA models. For this, analogously shaped grey data points represent cells of each type that were visually identified within the mixed cultures. These visually identified regions were then used to confirm that the regions within the mixed cultures could be matched with analogous cell type regions from the calibration data. In Fig 3A, the non-etched, projected, mixed culture regions in grey did not align with their analogous counterparts in the PCA model as these regions project onto this model close to the origin and do not appear to correlate well with any of the control regions. Conversely, in Fig 3B, the etched projected mixed culture cellular regions were classified within or near their analogous cell region counterparts from the calibration data, though it is noted that there is significant overlap between the two cell types (red triangles and green asterisk).
For Figure 3, it is noteworthy that the TCPS regions from the mixed cultures (grey squares) did not project onto the model with the TCPS regions from the calibration data (blue squares). This is likely due to the known problems in etching TCPS with C60+ clusters which results in heavy cross-linking of the target and thus in low ion signals47. This low ion yield is the likely cause of the poor fit associated with the etched TCPS regions. This did indeed appear to be the case when the Q-residual values were calculated (Supplemental Figures) as the etched TCPS regions clearly do not fit within the model as represented by their high Q-residual values.
Figure 4 shows PC 2 vs. PC 3 for the non-etched (Fig 4A) and etched (Fig 4B) surfaces. In Figure 4A, PC 3 captures only 1.34% of the overall variance within the PCA model for the non-etched surfaces. The confidence intervals in Figure 4A for the two cell regions are clearly overlapping with each other and with the TCPS regions in the calibration data. Additionally, the projected cellular regions in grey cannot be clearly identified with any of the regions from the calibration data. In Figure 4B, we see that PC 3 captures a larger percentage of the overall variance in the PCA model for the etched surfaces. Graphically, it can be seen that the differences in the two cell types are the major contributors to the variance in PC 3 (7.36% of the variance in the total model). The projected data from the selected regions from the mixed culture surfaces correlate well with their respective cell types (grey regions in Fig 4B). In addition, the semi-random cellular areas which are used here to demonstrate the predictive ability of the model (grey circles) are shown to all fall within the confidence limits of the regions associated with either the REEC areas or the 3T3 areas in Figure 4B. The fact that clear separation between the two cell types was not seen until PC 3 suggests that the differences between the cell types is smaller than the differences between TCPS, media coated TCPS and the cell surfaces as would be expected.
Partial Least-Squares Discriminant Analysis (PLS-DA) Classification of Projected Heterogeneous Culture Regions
PLS-DA is a multivariate classification technique used to classify test data with a set of calibration data. Similar to PCA, PLS-DA is an axis rotation that creates a new set of axes, which in the case of PLS-DA, maximize the variance between groups. Scores and loadings are calculated like PCA that can be used to identify the relationship between the original data and the captured variance from the model. Additionally in PLS-DA, test data (in this case regions from the heterogeneous cultures) can be projected onto the calibration data. A discrimination line is created (seen in red in Figure 5) that graphically represents the area of maximum separation between groups in the calibration data. Then, regions from the test data that fall above or below the discrimination line represent data points that are classified (or not classified) with the particular region from the calibration data that is graphically represented above the discrimination line in that particular graph. In Figure 5A, the classification of the 3T3 regions from the mixed cultures are seen to fall above the red discrimination line with the 3T3 regions from the calibration data for the etched surfaces (non-etched data not shown). The discrimination line represents the minimization of the intersection of the probability distributions of the calibration group and the predicted groups such that the samples that fall above the discrimination line in Figure 5 (grey samples) are classified with the calibration group used for the discrimination calculation (fibroblasts in Figure 5A and REECs in Figure 5B). The PLS-DA method has been described in a paper by Perez et. al.48 and this method has bee suggested as a more robust choice for discriminating biological surface regions40. A number of the unknown cellular regions (dark grey circles) fall with this class as well suggesting that these regions are predicted as 3T3 regions. As discussed earlier, Figure 5A shows that the TCPS regions appear misclassified by the model. Since the unknown regions were selected as cellular regions from the total ion images, it is unlikely that their classification in Figure 5A is related to the TCPS misclassification problem.
Figure 5.
Predicted classification on the etched cell and TCPS samples of the following: (A) the NIH 3T3 regions and (B) the REEC regions of the mixed culture samples with the calibration data set. The labels for samples are as follows: NIH 3T3 fibroblasts (red triangles), REECs (green asterisks), media coated tissue culture polystyrene (light blue crosses), and the clean tissue culture polystyrene (blue squares). The test regions are represented in grey with analogous shapes to the calibration data with the exception of the unknown cellular regions that are represented with dark grey circles and labeled sequentially.
In Figure 5B, the classification of the REEC regions in the mixed cultures is seen to fall above the discrimination line with the REEC regions in the calibration data. There was clear distinction between the groups on the etched surfaces and there was no significant misclassification in the model associated with the REEC regions. The classification trends in Figure 5 are similar to the results shown in Figure 3 and Figure 4 (from the PCA models). The application of PCA for ToF-SIMS data sets has been used numerous times previously. PLS-DA has not been used for this application, so the similarity in the results obtained in Figure 4 with those found in Figure 5 validates the accuracy of the PLS-DA results. Additionally, the classification of the unknown cellular regions is more distinct in the PLS-DA routine based on the graphical identification of regions associated with the projected test data onto the calibration data. This suggests that PLS-DA is a potentially more robust and sensitive routine for the application of identifying cell types in heterogeneous cultures using ToF-SIMS imaging.
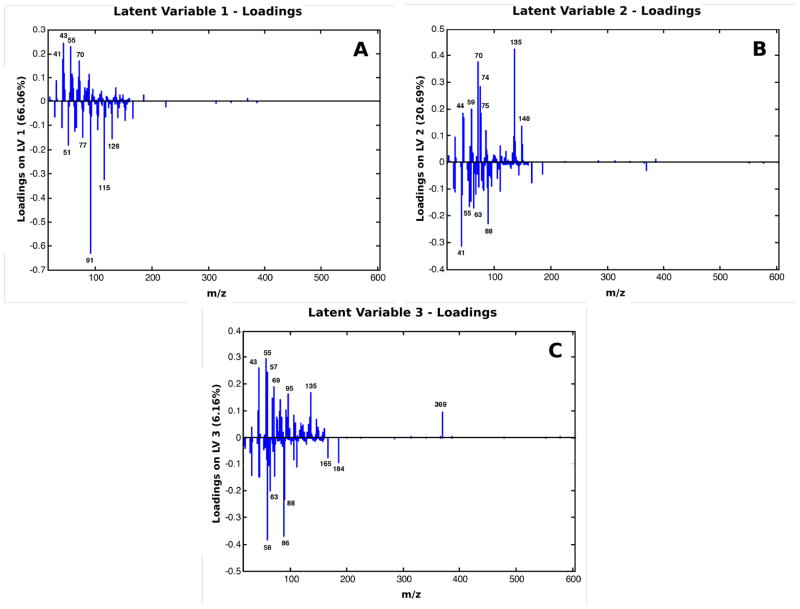
In Figure 6, the loadings for the PLS-DA model are shown for the first three latent variables. Like the PCA data in Figure 3 and Figure 4, the first three latent variables (LV) captured the majority of the overall variance in the analysis (data not shown). Like the PC model, the first LV separated the TCPS from the biologic surfaces, the second LV separated the media coated TCPS from the two cell types and the third LV distinguished between the two cell types. Subsequently, the loadings on LV3 in Figure 6C are the fragments associated with either the 3T3s or the REECs. In Figure 6C, it can be seen that fragments at m/z 86 (C5H12N+), 165 (C8H10NO+)16 and 184 (C5H15NO4P+ phosphocholine head group)45 contribute to the negative location of the 3T3 scores on the LV3 axis. The fragment at m/z 369 (C27H45 cholesterol [M - OH]+) is a major contributor to the positive location of the REEC scores on the LV3 axis. The two higher mass fragments (m/z 184 and 369) have been attributed to cell membranes in previous studies16,45. The two fragments at m/z 86 and 165 have previously been related to amino acid fragmentation patterns16. However in a ToF-SIMS study analyzing model lipid surfaces produced using a Langmuir-Blodgett (LB) film deposition method, the fragments at m/z 86 and 165 were also seen associated with the phosphocholine head group16. It is difficult to assign molecular fragment identities to the majority of fragments in the lower mass regions (below m/z 100) due to the resolution of the spectra associated with the ToF-SIMS images and the potential for overlap between the fragmentation patterns of the large macromolecules associated with a cell surface. Ideally, all of the fragments would be counted equally in the classification strategy. Using the PLS-DA multivariate classification routine, all of the lipid and amino acid fragments are taken into account equally and discrimination between classes can be calculated in an unbiased manner.
Figure 6.
Loadings associated with the PLS-DA model created in Figure 6 for the etched surfaces. (A) Latent variable 1 capturing 66.06% of the total variance in the model, (B) Latent variable 2 capturing 20.69% of the total variance in the model, (C) Latent variable 3 capturing 6.16% of the total variance in the model. The scores plots for the three latent variables can be seen in the Supplemental Figures.
Combined Images Derived from Loadings Results
The goal of this study was to identify individual cell types in heterogeneous cultures using ToF-SIMS images. In order to achieve this, we have shown that PCA can be used to enhance our ability to classify unknown cell regions within these heterogeneous cultures. PLS-DA appears to have a more distinct class assignment (cell type assignment) capacity. To visually identify cell types with the ToF-SIMS images acquired from the heterogeneous cultures, it would be possible to manually decide to plot various fragments based on their visual prominence. In Figure 7, we used the loadings associated with the PLS-DA classification in order to create combined images that could be overlaid for more precise discrimination of regions. For this, mass fragments from the loadings in LV 3 that were associated with the REEC cell type were combined and plotted together (Figure 7A). Additionally, mass fragments from the loadings in LV 3 that were associated with the 3T3 cell type were combined and plotted together (Figure 7B). Because a fragment is more prevalent in one cell type versus another does not preclude that fragment from existing within the other cell type. However, using this method, the signal intensity and region specificity is enhanced in Figure 7A and Figure 7B. For the 3T3 regions associated with Figure 7B, it is seen that good definition of individual 3T3 cells can be seen in portions of the image, but in some of the more dense regions, cellular definition is not as good. However, in Figure 7A, the REEC regions have more region specificity and appear to grow in colonies as expected (see Figure 1). However, in Figure 7B, there does appear to be a low level of signal associated with the 3T3s. The overlap in both of the first two images in Figure 7 was expected because these combination images contain fragments associated with both the lipid membrane and protein and carbohydrate structures (among others). The multivariate classification works by monitoring changes in all of the included fragments simultaneously and is generally based on relative “amounts” of each of the fragments. A simple overlay of the two images will increase the potential for visual cell discrimination. In Figure 7C, the two images from Figure 7A and Figure 7B are normalized to the total ion intensity and overlaid with a threshold value of 0.25. The numerical threshold was chosen empirically as the value that appeared to minimize noise while retaining signal indicative of one cell type or the other. For normalized images such as these, all of the pixels have intensities that range from 0 to 1. By setting a threshold value, pixels above the set ratio will be set to 1 and pixels below the set ratio will be set to 0. This is done for both of the images in the overlay. Using this method, it can be seen in Figure 7C that the identification of the REEC regions (green) is obvious, but also the thresholding appears to discriminate the 3T3 regions from the REEC regions more clearly.
Figure 7.
Two-dimensional ToF-SIMS images of the combined ion fragments selected using the loadings from LV3 in Figure 6. Images represent REEC fragments (A), NIH 3T3 fragments (B), and the normalized overlay of A and B with a threshold value of 0.25.
CONCLUSIONS
The identification of individual cell phenotypes within heterogeneous cultures generally requires antibody staining paired with microscopy. In this study, we showed that it is possible to use imaging time-of-flight secondary ion mass spectrometry (ToF-SIMS) to identify individual cell types when cultured together based upon their emitted surface mass fragments. The cell differentiation abilities of this method were increased by etching the surface with C60 and the discrimination of cellular regions in the mixed cultures was aided by multivariate analysis (MVA). Using MVA in this study, principal components analysis (PCA) showed that C60 etching improved the cell specificity in the collected data and that regions from the two cell types were clearly separated in the scores plot. Then, partial least squares - discriminant analysis (PLS-DA) was used to classify unknown cellular regions from within the heterogeneous cultures. The heterogeneous cell culture model system is presented here as a proof of concept for the hypothesis that ToF-SIMS images can be used to distinguish cell types. A limitation to studying cells with ToF-SIMS is the preparation of dried cell samples suitable for the ultrahigh vacuum conditions necessary for analysis. It is difficult to preserve the integrity of the cell membranes during the drying process without rupture and spillage of the cytoplasmic constituents. This is a possible limitation to the studies presented here and may have contributed to the difficulty in identification of the 3T3 regions in the PCA (Figures 3 and 4). However, in our analyses we found numerous mass fragments seen in previous studies to be attributed to cell membranes and our identifications were improved in the PLS-DA (Figures 5 and 6) despite this possible interference. Lastly, we were able to clearly differentiate regions in the combined ion images created from the PLS-DA loadings in Figure 7 showing that despite the possible need for future improvements to cell preservation/drying methods, this study represents a successfully proof of principle that the technique is a feasible method for differentiating cell phenotypes using ion images. As surface sensitive mass spectrometers such as the ToF-SIMS instrument used here evolve into smaller less expensive analytical tools with simplified sample preparation requirements, the implementation of topical mass spectrometry as a simple characterization method for cell phenotyping may become routine. The unbiased acquisition of data associated with ToF-SIMS sampling makes the method ideal for identifying surface features associated with specific cell phenotypes based on their surface chemistries (i.e., differences in cell surface proteins, sugars and lipids).
Supplementary Material
Acknowledgments
The authors acknowledge the funding and facilities provided by the National ESCA and Surface Analysis Center for Biomedical Problems (NESAC/BIO) through grant EB-002027 from the National Institutes of Health at the University of Washington. Additional funding and facilities were also provided by the University of Washington Engineered Biomaterials 21st Century (UWEB21) center, an Engineering Research Center. Jeremy Brison is currently a postdoctoral researcher of the Belgian Fund for Scientific Research (FNRS-FRS) and Christopher A. Barnes is currently a postdoctoral researcher at the Institute of Molecular Systems Biology at ETH Zürich in Zürich, Switzerland.
Footnotes
Supporting Information Available
All of the experimental details along with three supplemental Figures are included in the Supporting Information: (S1) Eigenvalues for PCA model; (S2) Q-residuals for PCA model; (S3) Scores for the Latent Variables associated with Figure 5 and Figure 6.
References
- 1.Ratner BD, Bryant SJ. Annu Rev Biomed Eng. 2004;6:41–75. doi: 10.1146/annurev.bioeng.6.040803.140027. [DOI] [PubMed] [Google Scholar]
- 2.Oberpenning F, Meng J, Yoo JJ, Atala A. Nat Biotechnol. 1999;17:149–155. doi: 10.1038/6146. [DOI] [PubMed] [Google Scholar]
- 3.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, Taylor DA. Nat Med. 2008;14:213–221. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 4.Griffith LG, Naughton G. Science. 2002;295:1009. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- 5.Langer R, Tirrell DA. Nature. 2004;428:487–492. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- 6.Lutolf MP, Hubbell JA. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 7.Stevens MM, George JH. Science. 2005;310:1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 8.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaushal S, Amiel GE, Guleserian KJ, Shapira OM, Perry T, Sutherland TW, Rabkin E, Moran AM, Schoen FJ, Atala A, Soker S, Bischoff J, Mayer JE. Nat Med. 2001;7:1035–1040. doi: 10.1038/nm0901-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner MS, Castner DG. Langmuir. 2001;17:4649–4660. [Google Scholar]
- 11.Canavan HE, Graham DJ, Cheng XH, Ratner BD, Castner DG. Langmuir. 2007;23:50–56. doi: 10.1021/la062330o. [DOI] [PubMed] [Google Scholar]
- 12.Lhoest JB, Wagner MS, Tidwell CD, Castner DG. J Biomed Mater Res. 2001;57:432–440. doi: 10.1002/1097-4636(20011205)57:3<432::aid-jbm1186>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 13.Brown BN, Barnes CA, Kasick RT, Michel R, Gilbert TW, Beer-Stolz D, Castner DG, Ratner B, Badylak SF. Biomaterials. 2010;31:428–437. doi: 10.1016/j.biomaterials.2009.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Science. 2004;305:71–73. doi: 10.1126/science.1099791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostrowski SG, Kurczy ME, Roddy TP, Winograd N, Ewing AG. Anal Chem. 2007;79:3554–3560. doi: 10.1021/ac061825f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker MJ, Zheng N, Winograd N, Lockyer NP, Vickerman JC. Langmuir. 2008;24:11803–11810. doi: 10.1021/la802582f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klerk LA, Dankers PYW, Popa ER, Bosman AW, Sanders ME, Reedquist KA, Heeren RMA. Anal Chem. 2010;82:4337–4343. doi: 10.1021/ac100837n. [DOI] [PubMed] [Google Scholar]
- 18.Jungnickel H, Jones EA, Lockyer NP, Oliver SG, Stephens GM, Vickerman JC. Anal Chem. 2005;77:1740–1745. doi: 10.1021/ac048792t. [DOI] [PubMed] [Google Scholar]
- 19.Nygren H, Malmberg P, Nilsson M, Kriegeskotte C, Arlinghaus HF. Appl Surf Sci. 2008;255:1285–1288. [Google Scholar]
- 20.Baker MJ, Gazi E, Brown MD, Clarke NW, Vickerman JC, Lockyer NP. Appl Surf Sci. 2008;255:1084–1087. doi: 10.1016/j.apsusc.2008.05.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker MJ, Brown MD, Gazi E, Clarke NW, Vickerman JC, Lockyer NP. Analyst. 2008;133:175–179. doi: 10.1039/b712853c. [DOI] [PubMed] [Google Scholar]
- 22.Kulp KS, Berman ESF, Knize MG, Shattuck DL, Nelson EJ, Wu LG, Montgomery JL, Felton JS, Wu KJ. Anal Chem. 2006;78:3651–3658. doi: 10.1021/ac060054c. [DOI] [PubMed] [Google Scholar]
- 23.Cannon DM, Winograd N, Ewing AG. Annu Rev Biophys Biomol Struct. 2000;29:239–263. doi: 10.1146/annurev.biophys.29.1.239. [DOI] [PubMed] [Google Scholar]
- 24.Lockyer NP, Vickerman JC. Appl Surf Sci. 2004;231:377–384. doi: 10.1016/j.apsusc.2008.05.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunnarsson A, Sjovall P, Hook F. Nano Letters. 2010;10:732–737. doi: 10.1021/nl904208y. [DOI] [PubMed] [Google Scholar]
- 26.Gunnarsson A, Kollmer F, Sohn S, Hook F, Sjovall P. Anal Chem. 2010;82:2426–2433. doi: 10.1021/ac902744u. [DOI] [PubMed] [Google Scholar]
- 27.Jones EA, Lockyer NP, Vickerman JC. Int J Mass Spectrom. 2007;260:146–157. [Google Scholar]
- 28.Berman ESF, Fortson SL, Checchi KD, Wu L, Felton JS, Wu KJJ, Kulp KS. J Am Soc Mass Spectrom. 2008;19:1230–1236. doi: 10.1016/j.jasms.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Lanekoff I, Kurczy ME, Hill R, Fletcher JS, Vickerman JC, Winograd N, Sjovall P, Ewing AG. Anal Chem. 2010;82:6652–6659. doi: 10.1021/ac101243b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parry SA, Kurczy ME, Fan X, Halleck MS, Schlegel RA, Winograd N. Appl Surf Sci. 2008;255:929–933. doi: 10.1016/j.apsusc.2008.05.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parry S, Winograd N. Anal Chem. 2005;77:7950–7957. doi: 10.1021/ac051263k. [DOI] [PubMed] [Google Scholar]
- 32.Chandra S. Appl Surf Sci. 2008;255:1273–1284. [Google Scholar]
- 33.Chandra S, Smith DR, Morrison GH. Anal Chem. 2000;72:104A–114A. doi: 10.1021/ac002716i. [DOI] [PubMed] [Google Scholar]
- 34.Fletcher JS, Lockyer NP, Vaidyanathan S, Vickerman JC. Anal Chem. 2007;79:2199–2206. doi: 10.1021/ac061370u. [DOI] [PubMed] [Google Scholar]
- 35.Piehowski PD, Davey AM, Kurczy ME, Sheets ED, Winograd N, Ewing AG, Heien ML. Anal Chem. 2009;81:5593–5602. doi: 10.1021/ac901065s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malm J, Giannaras D, Riehle MO, Gadegaard N, Sjovall P. Anal Chem. 2009;81:7197–7205. doi: 10.1021/ac900636v. [DOI] [PubMed] [Google Scholar]
- 37.Arlinghaus HF, Kriegeskotte C, Fartmann M, Wittig A, Sauerwein W, Lipinsky D. Appl Surf Sci. 2006;252:6941–6948. [Google Scholar]
- 38.Wehbe N, Delcorte A, Heile A, Arlinghaus HF, Bertrand P. Appl Surf Sci. 2008;255:824–827. [Google Scholar]
- 39.Kurczy ME, Piehowsky PD, Willingham D, Molyneaux KA, Heien ML, Winograd N, Ewing AG. J Am Soc Mass Spectrom. 2010;21:833–836. doi: 10.1016/j.jasms.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lutz U, Lutz RW, Lutz WK. Anal Chem. 2006;78:4564–4571. doi: 10.1021/ac0522299. [DOI] [PubMed] [Google Scholar]
- 41.Belu AM, Graham DJ, Castner DG. Biomaterials. 2003;24:3635–3653. doi: 10.1016/s0142-9612(03)00159-5. [DOI] [PubMed] [Google Scholar]
- 42.Beckstead BL, Pan S, Bhrany AD, Bratt-Leal AM, Ratner BD, Giachelli CM. Biomaterials. 2005;26:6217–6228. doi: 10.1016/j.biomaterials.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 43.Brison J, Muramoto S, Castner DG. J Phys Chem C. 2010;114:5565–5573. doi: 10.1021/jp9066179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brison J, Benoit DSW, Muramato S, Robinson M, Stayton PS, Castner DG. Surf Interface Anal. 2010;43:354–357. doi: 10.1002/sia.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magnusson U, Friber P, Sjovall P, Dangardt F, Malmberg P, Chen Y. Clin Physiol Funct Imaging. 2008;28:202–209. doi: 10.1111/j.1475-097X.2008.00796.x. [DOI] [PubMed] [Google Scholar]
- 46.Breitenstein D, Rommel CE, Mollers R, Wegener J, Hagenhoff B. Angew Chem, Int Ed Engl. 2007;46:5332–5335. doi: 10.1002/anie.200604468. [DOI] [PubMed] [Google Scholar]
- 47.Mahoney CM. Mass Spectrom Rev. 2010;29:247–293. doi: 10.1002/mas.20233. [DOI] [PubMed] [Google Scholar]
- 48.Perez NF, Ferre J, Boque R. Chemom Intell Lab Syst. 2009;95:122–128. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.