Abstract
Background
Chronic threat and anxiety are associated with pro-inflammatory transcriptional profiles in circulating leukocytes, but the causal direction of that relationship has not been established. This study tested whether a Cognitive-Behavioral Stress Management (CBSM) intervention targeting negative affect and cognition might counteract anxiety-related transcriptional alterations in people confronting a major medical threat.
Methods
199 women undergoing primary treatment of Stage 0–III breast cancer were randomized to a 10-week CBSM protocol or an active control condition. 79 provided peripheral blood leukocyte samples for genome-wide transcriptional profiling and bioinformatic analyses at baseline, 6-, and 12-month follow-ups.
Results
Baseline negative affect was associated with > 50% differential expression of 201 leukocyte transcripts, including up-regulated expression of pro-inflammatory and metastasis-related genes. CBSM altered leukocyte expression of 91 genes by > 50% at follow-up (Group × Time interaction), including down-regulation of pro-inflammatory and metastasis-related genes and up-regulation of Type I interferon response genes. Promoter-based bioinformatic analyses implicated decreased activity of NF-κB/Rel and GATA family transcription factors and increased activity of Interferon Response Factors and the Glucocorticoid Receptor (GR) as potential mediators of CBSM-induced transcriptional alterations.
Conclusions
In early stage breast cancer patients, a 10-week CBSM intervention can reverse anxiety-related up-regulation of pro-inflammatory gene expression in circulating leukocytes. These findings clarify the molecular signaling pathways by which behavioral interventions can influence physical health and alter peripheral inflammatory processes that may reciprocally affect brain affective and cognitive processes.
Keywords: threat/anxiety, cognitive-behavioral stress management, gene expression, immune system, inflammation, cancer, stress
Introduction
Research in social genomics has linked extended periods of significant life adversity to alterations in the gene transcriptional programs expressed under basal conditions in circulating immune cells (1–4). Across a variety of different threat- and anxiety-related experiences such as imminent bereavement (5), post-traumatic stress disorder (6), chronic loneliness (7, 8), significant life adversity (9), and low SES (10–12), genome-wide transcriptional profiling of leukocytes has shown a common pattern of increased expression of pro-inflammatory genes accompanied by a focal suppression of genes involved in interferon-mediated innate antiviral responses and immunoglobulin G production (1, 4, 8). Promoter-based bioinformatic analyses have implicated several specific transcription factors (TFs) as potential mediators of this transcriptional shift, including activation of pro-inflammatory NF-κB/Rel-family TFs (6, 7, 11) and GATA-family TFs (9), decreased activity of Interferon Response Factors (IRFs) (7), and functional desensitization of the glucocorticoid receptor (GR) (6, 7, 11) which would otherwise antagonize NF-κB/Rel factors and reduce inflammation (13). Several studies suggest that adversity-related alterations in the circulating leukocyte transcriptome are more closely linked to subjective experiences of threat or anxiety than they are to objective external conditions such as social network density or SES (7, 8, 12).
The causal relationship between experienced threat or anxiety and leukocyte transcriptional remodeling remains unclear because existing analyses generally involve correlational study designs. Exogenously triggered leukocyte transcriptional alterations could potentially induce experiences of anxiety, depression, or other negative affective states via cytokine effects on the brain (4, 14–18). However, experimental studies have also shown that negative affective states can alter leukocyte gene expression via signals from the sympathetic nervous system (SNS) and hypothalamic-pituitary-adrenal (HPA)-axis (2, 4, 19, 20). In the present study, we sought to determine whether an experimental intervention that directly targets anxiety-related cognitive and behavioral processes might reverse the pattern of leukocyte transcriptional alterations previously observed in people experiencing significant life adversity (4, 5, 7, 9, 11, 21).
Diagnosis with breast cancer generally evokes substantial health-related anxiety, even among women with early stage non-metastatic disease for whom the odds of survival are high (22, 23). In women undergoing primary treatment for Stage 0–III breast cancer, cognitive-behavioral stress management (CBSM) (24, 25) has been found to reduce anxiety-related symptoms and general negative affect (26) and increase positive affect (27). In these and other studies, CBSM-induced psychological effects have also been associated with peripheral physiologic alterations that may either induce or reflect changes in gene expression, including changes in circulating levels of cortisol and catecholamines (28, 29), circulating numbers of specific leukocyte subsets (28, 30), cytokine production by T lymphocytes (29), and plasma HIV-1 viral load (31). Given the established effects of CBSM in reducing negative affect and cognition, and evidence linking threat/anxiety-related negative affect to immunologic processes, this study tested the hypothesis that CBSM might reverse previously observed anxiety-related transcriptional alterations in circulating immune cells. In particular, we hypothesized that leukocytes from CBSM-treated breast cancer patients would show reduced expression of pro-inflammatory gene programs and enhanced expression of innate antiviral gene programs relative to control group patients (i.e., patterns opposite those previously linked to experienced threat/anxiety) (4). We also tested whether those transcriptional dynamics might be associated with reversal of the specific pattern of bioinformatically-inferred TF activation previously linked to anxiety-related transcriptional alterations (i.e., increased activity of NF-κB/Rel- and GATA-family TFs and decreased activity of IRFs and the GR) (13, 32).
Methods
CBSM Randomized Controlled Trial
Data come from a study of 199 Stage 0–III breast cancer patients (80% Stage I–II) who were recruited 4–8 weeks after primary surgery, but prior to initiation of adjuvant therapy, and randomized to either 1) a 10-week CBSM intervention focusing on anxiety reduction, cognitive restructuring, and coping skills, or 2) an active contact control condition, as previously described (25–27, 29) (NIH Clinical Trial NCT01422551). At baseline, 6, and 12 months post-randomization, 79 participants provided venous blood samples from which 3–10 × 106 peripheral blood mononuclear cells (PBMC) were isolated as previously described (29) and serum cortisol concentrations were measured by ELISA (29) (CONSORT diagram provided as Figure S1 in the Supplement). All research was approved by the Institutional Review Board at the University of Miami.
Demographic, Tumor, and Treatment Characteristics
As previously described (25–27, 29), participant age, annual household income, and ethnicity were assessed by standard self-report instruments, and tumor characteristics (stage, hormone receptor status, number of involved lymph nodes), cancer treatment parameters (surgery type, elapsed time since surgery, radiation therapy, radiation treatment within the 3 weeks prior to each study visit, chemotherapy, chemotherapy treatment within 3 weeks prior to each study visit, hormone therapy), and other medical therapies (including use of pain medications, anxiolytics, and antidepressants prior to each study visit) were derived from patient reports and medical records.
Negative and Positive Affect
Intervention effects on negative and positive affect were assessed by subscales the Affects Balance Scale (ABS) (33) with psychometric properties in this sample previously reported (26, 27) and results summarized for the present analyses by a composite affect balance score computed as the difference between Positive and Negative Affect subscale scores (i.e., Positive – Negative). CBSM effects on time trajectories of ABS Positive Affect, Negative Affect, and composite Affect Balance were analyzed in a 2 (Group: CBSM vs. Control) × 3 (Time: baseline, 6-, and 12-month follow-up) mixed effect linear model analysis treating time as a repeated measure and summarizing time trajectories by a linear trend score (SAS PROC MIXED; SAS Institute, Cary NC) (34). Analyses were conducted on an intent-to-treat basis including all available data (including observations for individuals missing data at other follow-up timepoints).
Gene Expression Profiling and Bioinformatic Analysis
Detailed methods for gene expression profiling and bioinformatic analysis are presented in the Supplement. Briefly, RNA was extracted from PBMC, quality assured for mass and integrity, and subject to genome-wide transcriptional profiling using Illumina Human HT-12 v3 Expression BeadChips (Illumina Inc., San Diego CA) with quantile normalization (35) as previously described (7, 9). Data are deposited as NCBI Gene Expression Omnibus series GSE24079. Initial analysis of baseline data identified genes showing > 50% differential expression across the general range of ABS composite scores (i.e., ± 2 SD relative to the mean value) after control for age, race (white vs. non-), and tumor stage, estrogen receptor (ER)-, and progesterone receptor (PR) status. Subsequent primary analyses identified longitudinal effects of CBSM on expression of each analyzed transcript in a 2 (Group: CBSM vs. Control) × 3 (Time: baseline, 6-, and 12-month follow-up) factorial design treating time as a repeated measure and controlling for individual differences in age, race, tumor stage, ER status, PR status, treatment with chemotherapy, and treatment with radiation. All analyses were conducted on an intent-to-treat basis using mixed effect linear models (34). Genes showing > 50% difference across groups in the magnitude of change over time (contrast: average of 6- and 12-month follow-ups – baseline) were identified as differentially expressed (corresponding to a False Discovery Rate ≤ 5%) (36). Their functional characteristics were identified by GOstat Gene Ontology analysis (37) and their potential regulation by specific transcription factors was inferred from TELiS bioinformatic analysis of transcription factor-binding motifs (TFBMs) in gene promoters (38), using TRANSFAC position-specific weight matrices (39) as previously described (7, 9). Analyses of GR signaling controlled for concurrent serum cortisol concentrations (7) to assess GR signal transduction efficiency above and beyond the effects of CBSM in altering glucocorticoid ligand availability (29). Ancillary analyses also controlled for prevalence of lymphocyte subsets as assessed by flow cytometry (29). Transcript Origin Analysis (8) was employed to identify the specific leukocyte subsets predominately mediating CBSM effects on the overall PBMC pool transcriptome.
Twelve transcripts identified as differentially expressed by microarray analysis at either 6-month follow-up, 12-month follow-up, or on average across both follow-ups were re-verified using quantitative RT-PCR as detailed in the Supplement. Selected genes identified by microarray as differentially expressed only at 6- or 12-month follow-ups were evaluated by RT-PCR to determine whether microarray assays may have underestimated true differences in gene expression (40).
Results
Patient characteristics and CBSM intervention effects on psychological outcomes and cortisol levels have been previously reported for this study (26, 27, 29). Briefly, in this sample of Stage 0–III breast cancer patients recruited after surgery but before the initiation of adjuvant therapy, the 10-week CBSM intervention significantly reduced general anxiety-related symptoms, negative affect, and intrusive thoughts about breast cancer (26), increased positive affect (27), reduced circulating cortisol levels (29), and increased stimulated production of Interleukin 2 and Interferon-γ over a 12-month follow-up in comparison to the active contact control group (29). Within the overall study cohort, 79 patients volunteered for an intensive immunologic sub-study and provided sufficient PBMC samples for genome-wide transcriptional profiling at baseline and at 6- and/or 12-month follow-ups (CBSM N = 45, Control N = 34; CONSORT diagram in Figure S1). CBSM group participants were more likely to provide PBMC samples (48.9%) than were control group participants (31.8%, difference p = .014), but the resulting groups of CBSM and control group patients did not differ in demographic characteristics, tumor characteristics, treatment parameters (surgery type, radiation, chemotherapy, hormone treatment), or baseline affective state (Table 1). PBMC contributors were representative of the total study sample on all demographic, tumor, and treatment-related parameters analyzed (all p ≥ .18) except for exposure to radiation treatment, which was less prevalent among PBMC contributors (45%) than in the total sample (60%, difference p = .043) and CBSM vs. control group assignment as noted above. PBMC contributors showed the same general profile of affective change over time as previously reported for the total study cohort (26, 27) (Group × Time interaction, p = .0042), with the CBSM-treated group showing increased positive affect (linear time trend over 12 months: mean 6.8 ± standard error 2.36 ABS score units, p = .0055), decreased negative affect (−8.22 ± 2.08, p = .0003), and a net positive trend in composite affect balance scores (17.54 ± 4.12, p < .0001), whereas control group participants showed negligible change over time on each dimension (positive affect: −0.16 ± 1.94, p = .936; negative affect: −4.64 ± 3.94, p = .245; overall affect balance: 1.00 ± 3.62, p = .784).
Table 1.
Characteristics of CBSM and control group participants providing PBMC samples for gene expression profiling.
| Control (n=34) | CBSM (n=45) | p | |
|---|---|---|---|
| Age (years)* | 49.2 ± 7.8 | 50.1 ± 7.5 | 0.594 |
| Income ($1,000)* | 80.3 ± 65.4 | 72.8 ± 31.4 | 0.536 |
| Ethnicity (%) | 0.503 | ||
| Non-hispanic white | 67.7 | 79.1 | |
| Hispanic | 23.5 | 14.0 | |
| African American | 8.8 | 7.0 | |
| Stage (%) | 0.449 | ||
| 0 | 8.8 | 16.3 | |
| I | 55.9 | 39.5 | |
| II | 29.4 | 32.6 | |
| III | 5.9 | 11.6 | |
| Lymph nodes+* | 0.4 ± 0.2 | 1.5 ± 3.4 | 0.062 |
| ER+ (%) | 91.3 | 77.8 | 0.194 |
| PR+ (%) | 82.4 | 66.7 | 0.275 |
| Surgery type (%) | 0.109 | ||
| Lumpectomy | 32.4 | 54.6 | |
| Mastectomy | 47.1 | 36.4 | |
| Bilat. mastectomy | 20.6 | 9.1 | |
| Days post-surgery (at study baseline)* | 41.6 ± 22.6 | 38.6 ± 21.6 | 0.561 |
| Chemotherapy | |||
| Ever (%) | 38.2 | 46.7 | 0.454 |
| Within 3 weeks of 6-month follow-up (%) | 20.8 | 10.8 | 0.281 |
| Within 3 weeks of 12-month follow-up (%) | 0.0 | 0.0 | 0.999 |
| Radiation therapy | |||
| Ever (%) | 26.5 | 44.4 | 0.101 |
| Within 3 weeks of 6-month follow-up (%) | 20.8 | 2.8 | 0.020 |
| Within 3 weeks of 12-month follow-up (%) | 0.0 | 0.0 | 0.999 |
| Endocrine therapy (%) | 34.4 | 37.1 | 0.813 |
| Affects Balance Scale (at baseline)* | 31.2 ± 16.3 | 24.3 ± 22.2 | 0.147 |
| Affects Balance Scale (linear trend/follow-up yr)* | +1.0 ± 3.6 | +17.5 ± 4.1 | 0.004 |
Mean ± SD
In analyses relating baseline affective state to PBMC gene expression, genome-wide transcriptional profiling identified 201 named human genes showing > 50% difference in expression across the ± 2-SD range of ABS composite scores at study entry (Table S1 in the Supplement). 177 genes were up-regulated in association with negative affect, including genes encoding pro-inflammatory cytokines (IL1A, IL1B, IL6, TNF), the prostaglandin-synthesis enzyme COX2 (PTGS2), the oxidative stress response (SOD2), inflammatory chemokines and related receptors (CCL3, CCL3L1, CCL4L2, CCL7, CCL20, CXCL9, CXCL10, CXCR6, CXCR7), and transcripts involved in tissue remodeling and epithelial-mesenchymal transition (LMNA, MMP9). Gene Ontology analyses confirmed that negative affect-linked transcripts were disproportionately involved in pro-inflammatory cytokine function (GO:0006954;GO:0005125; both p < .0001) and wound healing (GO:0009611; p < .0001).
To determine whether the CBSM intervention might reverse the pro-inflammatory transcriptional skew associated with significant life adversity in this sample and previous studies (5, 7, 9, 11, 21), we carried out Gene Ontology analyses of all 91 named human genes that showed > 50% difference between CBSM vs. Control groups in the magnitude of change in transcript abundance from baseline to follow-up (i.e., Group × Time interaction, controlling for patient age, race, disease stage (0–III), hormone receptor status (ER+/−, PR+/−) and treatment (chemotherapy, radiation, hormone therapy); genes listed in Table S2 in the Supplement, with additional cross-sectional differences at each follow-up timepoint listed in Tables S3–S4 in the Supplement. 62 transcripts showed significantly greater down-regulation in CBSM-treated patients relative to controls, including genes encoding pro-inflammatory cytokines (IL1A, IL1B, IL6), the prostaglandin-synthesis enzyme COX2 (PTGS2), inflammatory chemokines and their receptors (CCL2, CCL3, CCL3L1, CCL3L3, CCL4L1, CCL4L2, CCL7, CXCL1, CXCL2, CXCR7), and mediators of tissue remodeling and epithelial-mesenchymal transition (G0S2, LMNA, MMP9, OSM). Gene Ontology analyses confirmed that CBSM down-regulated genes were characterized by involvement in pro-inflammatory cytokine activity (GO:0006954;GO:0005125; both p < .0001) and wound healing (GO:0009611; p < .0001). 31 (50%) of the total 62 CBSM-down-regulated transcripts also appeared on the list of genes up-regulated in association with negative affect at baseline (greater than the < 1% overlap expected by chance; binomial p < .0001). Negative affect-related transcripts that were down-regulated by CBSM included pro-inflammatory cytokines (IL1A, IL1B, IL6), COX2 (PTGS2), chemokines and related receptors (CCL3, CCL7, CCL20, CCL3L1, CCL4L2, CXCR7), and mediators of wound healing and epithelial-mesenchymal transition (LMNA, MMP9).
29 genes showed significantly greater up-regulation over time in CBSM-treated patients vs. controls, including transcripts involved in Type I interferon response (IFIT1, IFIT2, IFIT3, IFI44, IFI44L, ISG15, MX2, OAS2, OAS3), Type II interferon signaling (IFNG), and interferon signal transduction (STAT1, STAT2). Gene Ontology analyses confirmed that the most prominent functional characteristic of CBSM up-regulated genes involved their role in antiviral responses (GO:0009615; p < .0001). RT-PCR analysis confirmed microarray-indicated group differences in the relative abundance of 12 of 12 transcripts audited (average 69% difference in expression, all p < .0001; Table S5 in the Supplement).
In CBSM dose-response analyses, only 4 of the 91 differentially expressed genes (4.4%) showed changes in expression that were proportional in magnitude to CBSM group attendance rates. However, CBSM attendance rates were generally high (mean 65.8% ± 4.5% of intervention sessions attended; 80% of participants attending 5 or more of the scheduled 10 sessions), limiting the range of CBSM dose variation available to resolve dose-dependence.
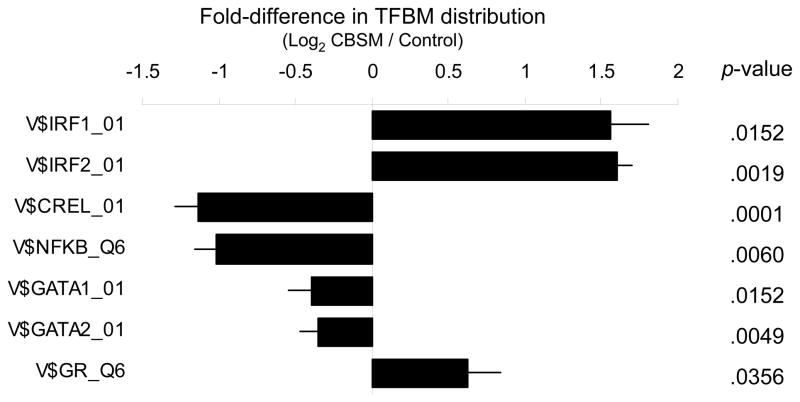
To determine whether CBSM-induced transcriptional alterations might be structured by specific TFs previously implicated in leukocyte transcriptional responses to threat and anxiety, we carried out TELiS bioinformatic analyses of TFBM distributions within the promoters of differentially expressed genes. Promoters of CBSM up-regulated genes showed a significant over-representation of DNA response elements for IRF transcription factors and under-representation of response elements for NF-κB/Rel- and GATA-family TFs (Figure 1). Parallel analyses of gene transcription controlling for concurrent serum cortisol levels showed an over-representation of GR response elements in the promoters of CBSM-up-regulated genes (Figure 1). Differential transcription of genes bearing GR response elements was not attributable to differential expression of the NR3C1 gene encoding the GR, which showed no substantial variation in transcript levels across groups, time-points, or their interaction (all differences ≤ 5%, all p > .20).
Figure 1.
Fold-difference in prevalence of Transcription Factor-Binding Motifs (TFBMs) targeted by the GR and IRF-, NF-κB/Rel-, and GATA-family transcription factors within the promoters of 29 genes found to be up-regulated in PBMC from CBSM-treated breast cancer patients relative to 62 genes up-regulated in active contact controls. V$ TFBM matrix names indicate vertebrate TRANSFAC position-specific weight matrices used in each analysis.
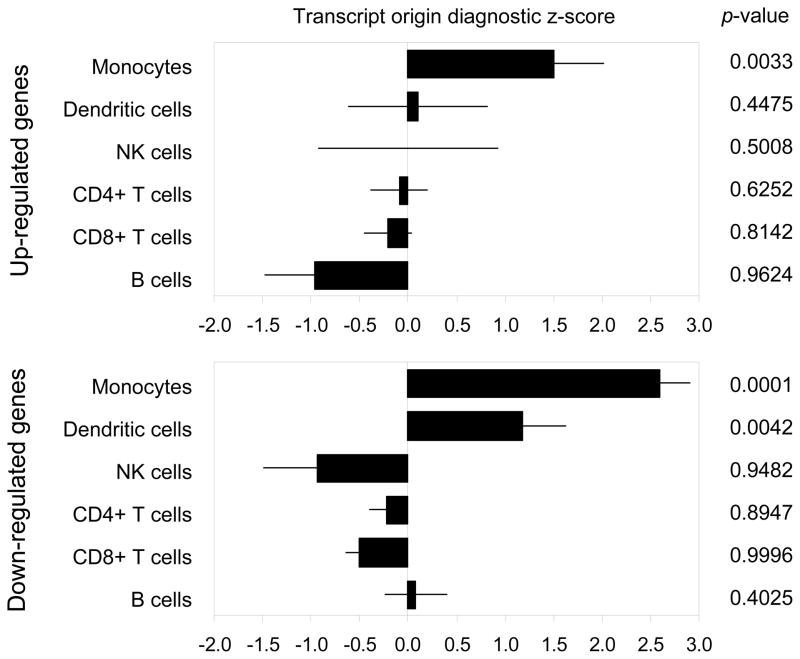
Transcript Origin Analyses (8) identified monocytes and plasmacytoid dendritic cells (pDCs) as cellular contexts for CBSM-induced transcriptional changes (Figure 2), with up-regulated genes deriving predominately from monocytes (p = .0128) and down-regulated transcripts associated with both monocytes (p < .0001) and pDCs (p = .0008).
Figure 2.
Bioinformatically inferred cellular origin of transcripts up- and down-regulated in PBMC from CBSM-treated breast cancer patients. Data represent the mean (± standard error) transcript origin diagnosticity score for the indicated cell type (8), with positive values indicating that differentially expressed genes originate disproportionately from the analyzed cell type and negative values uninformative (i.e., transcripts originate from other cell types, or from indicated cell type as well as other cell types).
Ancillary analyses examined the possibility that CBSM-related differences in the aggregate leukocyte transcriptome might stem from redistribution of leukocyte subsets within the PBMC population (41). Flow cytometry showed no difference in the prevalence of CD19+ B lymphocytes, CD4+ T lymphocytes, CD8+ T lymphocytes, or CD56+ lymphocytes within the assayed PBMC samples (all differences ≤ 5%, all p > .20; no flow cytometry analyses of monocyte or pDC prevalence were available), and controlling for variations in leukocyte subset prevalence continued to show CBSM-induced increases in the expression of interferon-related transcripts and decreases expression of pro-inflammatory genes.
Interferon suppression and pro-inflammatory transcriptional activation also continued to emerge in ancillary analyses that controlled for additional treatment-related variables including recent exposure to chemotherapy or radiation (within 3 weeks prior to each study visit), primary surgery type (lumpectomy, mastectomy, or bilateral mastectomy), and the use of pain medications, anxiolytics, or anti-depressants (down-regulation of pro-inflammatory genes, GO:0006954;GO:0009611; both p < .0001; up-regulation of innate antiviral response genes, GO:0009615; p < .0001).
Discussion
The results of this study link negative affective states to increased leukocyte expression of pro-inflammatory genes in individuals confronting significant life adversity (5–8, 10, 11), and they show that a CBSM intervention targeting anxiety-related affective and behavioral processes can counteract that transcriptional bias by reducing expression of pro-inflammatory and metastasis-related genes and increasing expression of interferon-related genes. Among 79 Stage 0–III breast cancer patients randomized to either a 10-week CBSM intervention or an active contact control condition, 6- and 12-month follow-up assessment showed reduced expression of genes encoding pro-inflammatory cytokines and increased expression of genes encoding both Type I and Type II interferons in PBMC from CBSM-treated patients. These effects emerged from a randomized intervention trial analyzed by intent-to-treat and controlling for any potential confounding effects of patient demographic, tumor, or treatment-related characteristics. These CBSM-induced longitudinal transcriptional alterations provide the first indication that psychological interventions can causally change the basal leukocyte transcriptome, and they implicate threat- and anxiety-related processes as potential CNS mediators of those effects (1, 2). These transcriptional alterations could have significant implications for both cancer-related disease processes (20) and psychological adaptation to the significant threat and anxiety provoked by breast cancer diagnosis and treatment (22, 23).
The common pattern of pro-inflammatory transcriptional bias observed in this study of breast cancer patients and previous studies of different populations confronting other major life adversities (5–8, 10, 11) suggests that there may exist a conserved transcriptional response to adversity (CTRA) that is mediated by the capacity of diverse challenges to activate common psychological reactions (e.g., threat/anxiety) and associated neural and endocrine responses (e.g., SNS and HPA-axis) that ultimately modulate leukocyte gene expression (4). In several previous studies, pro-inflammatory alterations in the basal leukocyte transcriptome were accompanied by selective suppression of Type I interferon-related genes (5, 7, 8, 11). This study found no significant interferon suppression associated with baseline negative affect, but it did identify CBSM-induced up-regulation of Type I interferon-related gene expression over follow-up concurrent with down-regulation of pro-inflammatory cytokines. Thus, experienced threat/anxiety appears to shift the leukocyte basal transcriptional equilibrium away from interferon-related antiviral gene modules in favor of pro-inflammatory cytokines (4). It is unclear why the pro-inflammatory transcriptional skew associated with negative affect at baseline was not accompanied by a detectable suppression of Type I interferon-related genes. It is possible that such a relationship does generally exist, but was not evident due to affective range restriction at baseline (i.e., all participants confronted a significant life adversity in recent breast cancer diagnosis, and thus few or none may have experienced positive/non-anxious affect levels sufficient to reveal associations with Type I interferon signaling) and only emerged after the CBSM intervention induced significant affective differences by 6- and 12-month follow-ups.
Three other features of these data are also consistent with the hypothesis that CBSM effects on the leukocyte transcriptome stem from the reversal of a CNS-mediated conserved transcriptional response to adversity (CTRA). First, the genes down-regulated by CBSM disproportionately included transcripts that were also up-regulated in relationship to negative affect at baseline (enriched > 100-fold relative to the overlap expected by chance). Second, bioinformatic inferences of the specific cell types mediating CBSM transcriptional alterations implicated the same myeloid lineage antigen presenting cells (monocytes and pDCs) linked to CTRA dynamics in previous studies (8). The simultaneous up- and down-regulation of distinct groups of monocyte-related genes is consistent with experimental animal studies documenting effects of chronic threat on monocyte subset differentiation (32, 42). Third, bioinformatic inferences of TF activity associated with CBSM-induced transcriptional alterations mirror those previously linked to CTRA dynamics. In particular, activation of GATA- and NF-κB/Rel-family TFs have been linked to life adversity and SNS signaling (5–7, 9, 11, 21, 43), and the present analyses suggest reductions in their activity following CBSM. The present analyses also indicate CBSM-induced activation of IRF-family TFs and the GR, both of which are inhibited by SNS signaling (42, 44) and were previously implicated in CTRA-related transcriptional down-regulation (5, 7, 9, 11, 21). CBSM increased the expression of genes bearing GR response elements despite the fact that circulating cortisol levels were reduced in CBSM-treated patients relative to controls (29). These effects also emerged despite statistical control for individual differences in circulating cortisol levels, and in the absence of any differential expression of the NR3C1 gene encoding the GR. Such findings are consistent with the hypothesis that CBSM affects GR target gene expression primarily by enhancing GR functional sensitivity (i.e., reversing threat-induced GR desensitization) (5, 11, 32, 45, 46), with such stimulatory effects outweighing the simultaneous effects reduced circulating GR ligand levels (29). Although the present results are consistent with the theorized role of these TFs in mediating the leukocyte CTRA and its reversal by CBSM, it is important to note that the bioinformatic analyses presented here represent indirect inferences of TF activity based on promoter sequence associations and cannot definitively establish that these TFs are causally responsible for the observed transcriptional alterations.
Beyond demonstrating a general influence of cognitive/behavioral processes on the basal transcriptional stance of circulating immune cells in people confronting significant life threat (4), the present results may have specific health implications for women confronting breast cancer (20). CBSM-induced down-regulation of pro-inflammatory cytokine genes (e.g., IL1A, IL1B, IL6) and bioinformatic indications of NF-κB/Rel activity are notable because chronic inflammation has been implicated in breast cancer progression and recurrence (47, 48). CBSM also down-regulated expression of specific genes known to play a role in cancer progression (e.g., those involved in myeloid cell induction of the metastasis-promoting epithelial-mesenchymal transition) (49), while enhancing expression of Type I interferon-related genes which are associated with reduced breast cancer progression (50–52). Those findings provide a molecular framework for understanding both the general link between psychological processes and cancer progression (20, 24) and salutary effects of cognitive-behavioral interventions on breast cancer clinical disease progression (53, 54).
The present findings from a randomized intervention trial show that psychological interventions can exert sustained effects on leukocyte gene expression profiles, but the scope of conclusions that can be drawn from this study are limited in several important respects. In addition to causal effects of cognitive-behavioral processes on leukocyte transcriptional programs, pro-inflammatory cytokines may also signal to the brain to causally affect neural function (14–18), thus inducing a bi-directional regulatory circuit that could propagate associations between inflammation and CNS-mediated threat or anxiety processes (4, 55). Several key gene transcriptional dynamics identified in the present gene expression studies were confirmed in RT-PCR assays of mRNA expression and/or previous studies of protein expression (e.g., IFN-γ production) (29), but future studies will be required to confirm many of the other transcriptional findings and assess their implications within the tumor microenvironment (20). The clinical health impact of the observed leukocyte transcriptional dynamics also requires further definition (e.g., assessing effects on progression-free or total survival times). However, the present molecular findings are consistent with other recent studies documenting improved survival and reduced disease recurrence in non-metastatic breast cancer patients randomized to a broadly similar cognitive-behavioral intervention (53, 54). Although these data document longitudinal changes in leukocyte gene expression for 6 to 12 months after a 10-week CBSM intervention, the ultimate duration of transcriptional impact remains to be established, as do other potential limiting conditions. In particular, CBSM intervention adherence rates were quite high in this study, and it is therefore difficult to determine whether transcriptional effects are dose-dependent on intervention magnitude/adherence. Finally, the present results emerged in a mid- to high-income sample of early stage breast cancer patients, and it is unclear whether similar results would occur in other populations, disease settings, or types of life adversity.
This study demonstrates that a psychologically-targeted intervention delivered in the anxiety-provoking context of primary breast cancer treatment can reverse some of the major changes in immune system gene expression previously observed in people confronting significant life adversity (1, 2). These findings provide a molecular framework for understanding the impact of behaviorally-targeted interventions on human immune function, and they begin the process of mapping specific biological pathways by which those dynamics might potentially alter the course of somatic diseases such as breast cancer (56, 57) and reciprocally feed back to influence threat- and anxiety-related brain processes (4, 15, 55).
Supplementary Material
Acknowledgments
This research was supported by the National Institute of Health (AG010415, AG028748, AG107265, AI052737, CA116778, CA064710, CA140933, DK082344), the Sylvester Cancer Center, and the UCLA Norman Cousins Center. We gratefully acknowledge the patients who participated in this study and the referring physicians, graduate students, and staff at the University of Miami who helped conduct it.
Footnotes
Financial Disclosures
Dr. Antoni reports receiving publication royalties from a book and related training materials that he has authored on CBSM treatments in health psychology. No other author reported any biomedical financial interests or potential conflicts of interest.
Clinical Trial Registration: NIH NCT01422551 (http://clinicaltrials.gov/ct2/show/NCT01422551)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cole SW. Social regulation of human gene expression. Current Directions in Psychological Science. 2009;18:132–137. doi: 10.1111/j.1467-8721.2009.01623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller G, Chen E, Cole SW. Health Psychology: Developing Biologically Plausible Models Linking the Social World and Physical Health. Annu Rev Psychol. 2009;60:501–524. doi: 10.1146/annurev.psych.60.110707.163551. [DOI] [PubMed] [Google Scholar]
- 3.Cole SW. Elevating the perspective on human stress genomics. Psychoneuroendocrinology. 2010;35:955–962. doi: 10.1016/j.psyneuen.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irwin MR, Cole SW. Reciprocal regulation of the neural and innate immune systems. Nat Rev Immunol. 2011;11:625–632. doi: 10.1038/nri3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller GE, Chen E, Sze J, Marin T, Arevalo JM, Doll R, et al. A functional genomic fingerprint of chronic stress in humans: blunted glucocorticoid and increased NF-kappaB signaling. Biol Psychiatry. 2008;64:266–272. doi: 10.1016/j.biopsych.2008.03.017. Epub 2008 Apr 2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Donovan A, Sun B, Cole S, Rempel H, Lenoci M, Pulliam L, et al. Transcriptional control of monocyte gene expression in post-traumatic stress disorder. Dis Markers. 2011;30:123–132. doi: 10.3233/DMA-2011-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole SW, Hawkley LC, Arevalo JM, Sung CY, Rose RM, Cacioppo JT. Social regulation of gene expression in human leukocytes. Genome Biol. 2007;8:1–13. doi: 10.1186/gb-2007-8-9-r189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole SW, Hawkley LC, Arevalo JM, Cacioppo JT. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proc Natl Acad Sci U S A. 2011;108:3080–3085. doi: 10.1073/pnas.1014218108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole S, Arevalo J, Takahashi R, Sloan EK, Lutgendorf S, Sood AK, et al. Computational identification of gene-social environment interaction at the human IL6 locus. Proc Natl Acad Sci U S A. 2010;107:5681–5686. doi: 10.1073/pnas.0911515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen E, Miller GE, Kobor MS, Cole SW. Maternal warmth buffers the effects of low early-life socioeconomic status on pro-inflammatory signaling in adulthood. Mol Psychiatry. 2011;16:729–737. doi: 10.1038/mp.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller GE, Chen E, Fok AK, Walker H, Lim A, Nicholls EF, et al. Low early-life social class leaves a biological residue manifested by decreased glucocorticoid and increased proinflammatory signaling. Proc Natl Acad Sci U S A. 2009;106:14716–14721. doi: 10.1073/pnas.0902971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen E, Miller GE, Walker HA, Arevalo JM, Sung CY, Cole SW. Genome-wide transcriptional profiling linked to social class in asthma. Thorax. 2009;64:38–43. doi: 10.1136/thx.2007.095091. Epub 2008 Nov 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pace TW, Hu F, Miller AH. Cytokine-effects on glucocorticoid receptor function: Relevance to glucocorticoid resistance and the pathophysiology and treatment of major depression. Brain Behav Immun. 2007;21:9–19. doi: 10.1016/j.bbi.2006.08.009. Epub 2006 Oct 2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capuron L, Pagnoni G, Demetrashvili M, Woolwine BJ, Nemeroff CB, Berns GS, et al. Anterior cingulate activation and error processing during interferon-alpha treatment. Biol Psychiatry. 2005;58:190–196. doi: 10.1016/j.biopsych.2005.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. 2008;9:46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison NA, Brydon L, Walker C, Gray MA, Steptoe A, Critchley HD. Inflammation causes mood changes through alterations in subgenual cingulate activity and mesolimbic connectivity. Biol Psychiatry. 2009;66:407–414. doi: 10.1016/j.biopsych.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenberger NI, Inagaki TK, Mashal NM, Irwin MR. Inflammation and social experience: an inflammatory challenge induces feelings of social disconnection in addition to depressed mood. Brain Behav Immun. 2010;24:558–563. doi: 10.1016/j.bbi.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenberger NI, Berkman ET, Inagaki TK, Rameson LT, Mashal NM, Irwin MR. Inflammation-induced anhedonia: endotoxin reduces ventral striatum responses to reward. Biol Psychiatry. 2010;68:748–754. doi: 10.1016/j.biopsych.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ader R, Cohen N, Felten D. Psychoneuroimmunology: interactions between the nervous system and the immune system. Lancet. 1995;345:99–103. doi: 10.1016/s0140-6736(95)90066-7. [DOI] [PubMed] [Google Scholar]
- 20.Antoni MH, Lutgendorf SK, Cole SW, Dhabhar FS, Sephton SE, McDonald PG, et al. The influence of bio-behavioural factors on tumour biology: pathways and mechanisms. Nat Rev Cancer. 2006;6:240–248. doi: 10.1038/nrc1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lutgendorf SK, Degeest K, Sung CY, Arevalo JM, Penedo F, Lucci J, 3rd, et al. Depression, social support, and beta-adrenergic transcription control in human ovarian cancer. Brain Behav Immun. 2009;23:176–183. doi: 10.1016/j.bbi.2008.04.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glanz K, Lerman C. Psychosocial impact of breast cancer: A critical review. Ann Behav Med. 1992;14:204–212. [Google Scholar]
- 23.Moyer A, Salovey P. Psychosocial sequelae of breast cancer and its treatment. Ann Behav Med. 1996;18:110–125. doi: 10.1007/BF02909583. [DOI] [PubMed] [Google Scholar]
- 24.Lutgendorf SK, Sood AK, Antoni MH. Host factors and cancer progression: Biobehavioral signaling pathways and interventions. J Clin Oncol. 2010 doi: 10.1200/JCO.2009.26.9357. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antoni MH. Stress Management Intervention for Women with Breast Cancer. American Psychological Association Press; Washington D.C: 2003. [Google Scholar]
- 26.Antoni MH, Wimberly SR, Lechner SC, Kazi A, Sifre T, Urcuyo KR, et al. Reduction of cancer-specific thought intrusions and anxiety symptoms with a stress management intervention among women undergoing treatment for breast cancer. Am J Psychiatry. 2006;163:1791–1797. doi: 10.1176/ajp.2006.163.10.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antoni MH, Lechner SC, Kazi A, Wimberly SR, Sifre T, Urcuyo KR, et al. How stress management improves quality of life after treatment for breast cancer. J Consult Clin Psychol. 2006;74:1143–1152. doi: 10.1037/0022-006X.74.6.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antoni MH, Cruess DG, Cruess S, Lutgendorf S, Kumar M, Ironson G, et al. Cognitive-behavioral stress management intervention effects on anxiety, 24-hr urinary norepinephrine output, and T-cytotoxic/suppressor cells over time among symptomatic HIV-infected gay men. J Consult Clin Psychol. 2000;68:31–45. doi: 10.1037//0022-006x.68.1.31. [DOI] [PubMed] [Google Scholar]
- 29.Antoni MH, Lechner S, Diaz A, Vargas S, Holley H, Phillips K, et al. Cognitive behavioral stress management effects on psychosocial and physiological adaptation in women undergoing treatment for breast cancer. Brain Behav Immun. 2009;23:580–591. doi: 10.1016/j.bbi.2008.09.005. Epub 2008 Sep 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antoni MH, Cruess DG, Klimas N, Maher K, Cruess S, Kumar M, et al. Stress management and immune system reconstitution in symptomatic HIV-infected gay men over time: effects on transitional naive T cells (CD4(+)CD45RA(+)CD29(+)) Am J Psychiatry. 2002;159:143–145. doi: 10.1176/appi.ajp.159.1.143. [DOI] [PubMed] [Google Scholar]
- 31.Antoni MH, Carrico AW, Duran RE, Spitzer S, Penedo F, Ironson G, et al. Randomized clinical trial of cognitive behavioral stress management on human immunodeficiency virus viral load in gay men treated with highly active antiretroviral therapy. Psychosom Med. 2006;68:143–151. doi: 10.1097/01.psy.0000195749.60049.63. [DOI] [PubMed] [Google Scholar]
- 32.Stark J, Avitsur R, Padgett DA, Sheridan JF. Social stress induces glucocorticoid resistance in macrophages. American J Physiol Reg Int Comp Physiol. 2001;280:R1799–R1805. doi: 10.1152/ajpregu.2001.280.6.R1799. [DOI] [PubMed] [Google Scholar]
- 33.Derogatis LR. The Affects Balance Scale. Clinical Psychometric Research; Baltimore: 1975. [Google Scholar]
- 34.McCulloch CE, Searle SR, Neuhaus JM. Generalized, linear, and mixed models. John Wiley & Sons; Hoboken NJ: 2008. [Google Scholar]
- 35.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B. 1995;57:289–300. [Google Scholar]
- 37.Beissbarth T, Speed TP. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 38.Cole SW, Yan W, Galic Z, Arevalo J, Zack JA. Expression-based monitoring of transcription factor activity: The TELiS database. Bioinformatics. 2005;21:803–810. doi: 10.1093/bioinformatics/bti038. [DOI] [PubMed] [Google Scholar]
- 39.Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cole SW, Galic Z, Zack JA. Controlling false-negative errors in microarray differential expression analysis: a PRIM approach. Bioinformatics. 2003;19:1808–1816. doi: 10.1093/bioinformatics/btg242. [DOI] [PubMed] [Google Scholar]
- 41.Richlin VA, Arevalo JM, Zack JA, Cole SW. Stress-induced enhancement of NF-kappaB DNA-binding in the peripheral blood leukocyte pool: effects of lymphocyte redistribution. Brain Behav Immun. 2004;18:231–237. doi: 10.1016/j.bbi.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Wohleb ES, Hanke ML, Corona AW, Powell ND, Stiner LM, Bailey MT, et al. Beta-Adrenergic Receptor Antagonism Prevents Anxiety-Like Behavior and Microglial Reactivity Induced by Repeated Social Defeat. J Neurosci. 2011;31:6277–6288. doi: 10.1523/JNEUROSCI.0450-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bierhaus A, Wolf J, Andrassy M, Rohleder N, Humpert PM, Petrov D, et al. A mechanism converting psychosocial stress into mononuclear cell activation. Proceedings of the National Academy of Sciences of the USA. 2003;100:1920–1925. doi: 10.1073/pnas.0438019100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collado-Hidalgo A, Sung C, Cole S. Adrenergic inhibition of innate anti-viral response: PKA blockade of Type I interferon gene transcription mediates catecholamine support for HIV-1 replication. Brain Behav Immun. 2006;20:552–563. doi: 10.1016/j.bbi.2006.01.005. Epub 2006 Feb 2028. [DOI] [PubMed] [Google Scholar]
- 45.Miller GE, Cohen S, Ritchey AK. Chronic psychological stress and the regulation of pro-inflammatory cytokines: a glucocorticoid-resistance model. Health Psychol. 2002;21:531–541. doi: 10.1037//0278-6133.21.6.531. [DOI] [PubMed] [Google Scholar]
- 46.Cole SW, Mendoza SP, Capitanio JP. Social stress desensitizes lymphocytes to regulation by endogenous glucocorticoids: insights from in vivo cell trafficking dynamics in rhesus macaques. Psychosom Med. 2009;71:591–597. doi: 10.1097/PSY.0b013e3181aa95a9. Epub 2009 Jun 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cole SW. Chronic inflammation and breast cancer recurrence. J Clin Oncol. 2009;27:3418–3419. doi: 10.1200/JCO.2009.21.9782. Epub 2009 May 3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierce BL, Ballard-Barbash R, Bernstein L, Baumgartner RN, Neuhouser ML, Wener MH, et al. Elevated biomarkers of inflammation are associated with reduced survival among breast cancer patients. J Clin Oncol. 2009;27:3437–3444. doi: 10.1200/JCO.2008.18.9068. Epub 2009 May 3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 50.Tiwari RK, Wong GY, Mukhopadhyay B, Telang NT, Liu J, Hakes TB, et al. Interferon-alpha and gamma mediated gene responses in a human breast carcinoma cell line. Breast Cancer Res Treat. 1991;18:33–41. doi: 10.1007/BF01975441. [DOI] [PubMed] [Google Scholar]
- 51.Recchia F, Sica G, Candeloro G, Necozione S, Bisegna R, Bratta M, et al. Beta-interferon, retinoids and tamoxifen in metastatic breast cancer: long-term follow-up of a phase II study. Oncol Rep. 2009;21:1011–1016. doi: 10.3892/or_00000317. [DOI] [PubMed] [Google Scholar]
- 52.Seth A, Kitching R, Landberg G, Xu J, Zubovits J, Burger AM. Gene expression profiling of ductal carcinomas in situ and invasive breast tumors. Anticancer Res. 2003;23:2043–2051. [PubMed] [Google Scholar]
- 53.Andersen BL, Thornton LM, Shapiro CL, Farrar WB, Mundy BL, Yang HC, et al. Biobehavioral, immune, and health benefits following recurrence for psychological intervention participants. Clinincal Cancer Research. 2010;16:3270–3278. doi: 10.1158/1078-0432.CCR-10-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andersen BL, Yang HC, Farrar WB, Golden-Kreutz DM, Emery CF, Thornton LM, et al. Psychologic intervention improves survival for breast cancer patients: a randomized clinical trial. Cancer. 2008;113:3450–3458. doi: 10.1002/cncr.23969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller AH, Ancoli-Israel S, Bower JE, Capuron L, Irwin MR. Neuroendocrine-immune mechanisms of behavioral comorbidities in patients with cancer. J Clin Oncol. 2008;26:971–982. doi: 10.1200/JCO.2007.10.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chida Y, Hamer M, Wardle J, Steptoe A. Do stress-related psychosocial factors contribute to cancer incidence and survival? Nat Clin Pract Oncol. 2008;5:466–475. doi: 10.1038/ncponc1134. Epub 2008 May 2020. [DOI] [PubMed] [Google Scholar]
- 57.Sloan EK, Priceman SJ, Cox BF, Yu S, Pimentel MA, Tangkanangnukul V, et al. The sympathetic nervous system induces a metastatic switch in primary breast cancer. Cancer Res. 2010;70:7042–7052. doi: 10.1158/0008-5472.CAN-10-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.