Abstract
Radiologists are critically interested in promoting best practices in medical imaging, and to that end, they are actively developing tools that will optimize terminology and reporting practices in radiology. The RadLex® vocabulary, developed by the Radiological Society of North America (RSNA), is intended to create a unifying source for the terminology that is used to describe medical imaging. The RSNA Reporting Initiative has developed a library of reporting templates to integrate reusable knowledge, or meaning, into the clinical reporting process. This report presents the initial analysis of the intersection of these two major efforts. From 70 published radiology reporting templates, we extracted the names of 6,489 reporting elements. These terms were reviewed in conjunction with the RadLex vocabulary and classified as an exact match, a partial match, or unmatched. Of 2,509 unique terms, 1,017 terms (41%) matched exactly to RadLex terms, 660 (26%) were partial matches, and 832 reporting terms (33%) were unmatched to RadLex. There is significant overlap between the terms used in the structured reporting templates and RadLex. The unmatched terms were analyzed using the multidimensional scaling (MDS) visualization technique to reveal semantic relationships among them. The co-occurrence analysis with the MDS visualization technique provided a semantic overview of the investigated reporting terms and gave a metric to determine the strength of association among these terms.
Keywords: Radiology, Structured reporting, Reporting templates, Standardized terminology, RadLex, Mapping, Visualization, Multidimensional scaling
Introduction
The Radiological Society of North America (RSNA) RadLex® vocabulary (http://www.radlex.org) has been designed to create a unifying source for medical imaging terminology [1, 2]. It currently contains more than 32,000 terms, many of which are not found in other medical terminologies. RadLex terms are being acquired to describe the domain of radiology, with the goal of disseminating standardized terminology to facilitate the analysis of radiological information, to permit uniform indexing of image libraries, and to enable new applications for structured capture of medical image information [3, 4]. The latest version has been released on the National Center for Biomedical Ontology (NCBO) BioPortal web site (http://bioportal.bioontology.org/ontologies/1057) with greatly expanded content, particularly in naming anatomy, radiology procedures, and procedure steps.
The intent of RadLex is to reduce variation and improve clarity in radiology reports and image annotations as well as to provide a standardized means of indexing radiological materials in a variety of settings. One application of RadLex is to standardize the vocabulary used in radiology reports. To identify and promote “best practices” radiology reporting, the RSNA has been developing a digital library of XML-encoded, structured radiology reporting templates [5]. The primary goal of this project was to determine how well RadLex covers the elements of templates for structured radiology reports. A secondary goal was to use the reporting templates to provide new terms and relationships to RadLex by analyzing the reporting terms that do not match with the existing RadLex terms. This is facilitated by an information visualization technique called multidimensional scaling (MDS), which reveals the semantic relationships between terms used in the reporting templates.
The concept of information visualization is used to describe 2D and 3D static images or animation to explore information and its structure [6]. It provides a unique way to present complex information visually and allows people to utilize their perceptional capability to understand information and explore knowledge. There are various available information visualization techniques and applications. Among them, MDS has several advantages: (1) the data used in MDS analysis are relatively free of any distributional assumption; (2) MDS can handle various types of data ranging from ordinal data, to interval data, to ratio data; (3) MDS is mature and widely used in many application domains; and (4) many commercial software packages and noncommercial software packages that incorporate MDS are available [7]. MDS facilitates visual clustering analysis for small- to medium-sized datasets, such as the reporting template dataset. The findings of the visual analysis provide a basis for better understanding the nature of unmatched terms. This serves as a rationale for proposing new terms and synonyms for RadLex.
Material and Methods
In the initial phase of the structured reporting initiative, 70 reporting templates were developed by 12 RSNA subcommittees of clinically expert radiologists in collaboration with a variety of subspecialty societies. Text-based templates were encoded into the Extensible Markup Language (XML). The text-based and XML-encoded templates are freely available through the RSNA Reporting Template Library web site (http://www.radreport.org). These 70 reporting templates served as the initial cohort of documents used for analysis.
A semi-automated tool, called RadMap, was developed and applied to assign RadLex IDs to the elements of the 70 reporting templates. RadMap extracted element names from the XML template file and mapped those element names to terms in the RadLex ontology through a web service interface that links to the NCBO BioPortal site. This interface allowed users to select the most appropriate RadLex term [8]. Matching terms were selected by an expert in medical informatics and were verified by two experienced, board-certified radiologists who had extensive experience with RadLex. If no exact match was identified, RadMap listed possible matches based on a keyword search of RadLex terms. The system also allowed users to enter search queries. If one or more RadLex terms matched a specific reporting element, the user designated whether the match was exact or partial. The RadLex concept identifiers and match information were recorded in the XML-encoded template.
Using computed tomography (CT) and ultrasound (US) reporting templates as samples, the reporting templates in each category and the numbers of exactly matched, partially matched, and unmatched terms in each template were exported to an Excel spreadsheet to analyze the RadLex coverage of reporting terms in the templates and determine what kinds of terms are missing. The percentage of exactly matched, partially matched, and unmatched reporting terms were then determined.
The MDS visualization technique was used to analyze the radiology reporting terms that did not match with the RadLex terms to examine the term co-occurrence relationships and to visually reveal groups of radiological terms and semantic relationships among these terms. The statistical program SPSS was employed to conduct the MDS analysis in order to take advantage of the ease with which data input and interactive results manipulation is possible due to its interface, processing features, and powerful graphic display capabilities [9].
To conduct the term co-occurrence analysis, a frequently occurring unmatched term was manually identified as a starting point or seed term to retrieve related terms that co-occur with it in the designated template context. Reporting templates that contain the seed term were examined to define a group of the related terms. All the “element names” within the same section of the XML-encoded templates where the seed term is located were selected as related terms and formed an association term set. Then, the relationships among the terms in the association term set are described in a term relation matrix (TRM), shown in Eq. 1.
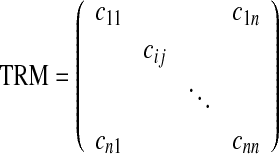 |
1 |
Both the number of the columns and the number of the rows of this matrix are equal to the size (n) of the association term set. A cell of the matrix defines the strength of two terms in the association term set. Given cij is a cell in the matrix, it indicates the strength between the term Ti and the term Tj in the association term set. Since the strength between Ti and Tj is the same as the strength between Tj and Ti, it means cij is always equal to cji. In other words, the matrix is a symmetric matrix.
As the element names in a XML-encoded reporting template are displayed like a hierarchy tree, those terms that share the same path in the hierarchy have close relationships, which usually means that they have the same subject characteristics in the reporting template. The farther away the two terms are located in the hierarchy, the less relevant the two terms are.
The strength between two terms (Ti and Tj) hinges on the distance between the two terms on the path. The farther the distance between the two terms, the weaker is the strength. The strength is defined in Eq. 2.
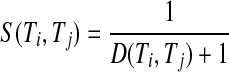 |
2 |
In Eq. 2, notice that the denominator is the distance between two terms plus 1. That is because if two terms are located in the same node, the distance between them is 0; adding 1 to the denominator would avoid the meaningless strength in the equation. More importantly, when the distance between two terms is 0, the corresponding strength is equal to 1, which reaches the maxium value of the strength. Since the defined distance between two terms is always larger than 0, it suggests that S(Ti, Tj) is always larger than 0 and smaller than 1. In other words, S(Ti, Tj) is normalized between 0 and 1.
If two terms Ti and Tj appear multiple times in a reporting template, the collective effects are considered in a proximity matrix (PM). Consequently, the final value of the cell cij is defined in Eq. 3. Here, h is the number of the times terms Ti and Tj appear in a reporting template.
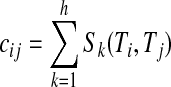 |
3 |
Each term in the related term set is examined in the reporting templates, the relationships between the term and other terms are determined, and the strength values between the term and other terms are calculated based on Eq. 3. As a result, the TRM is filled up and completed.
The finalized TRM is converted to a proximity matrix, shown in Eq. 4, which is the input of the later MDS analysis. The PM has the same structure as the TRM. In other words, the number of the columns and the number of the rows in the PM are the same as those in the TRM. The primary difference between the two matrices is the definition of the cells. Here, pij is a cell which defines the proximity value between Ti and Tj.
 |
4 |
There are various methods which can be used to convert a TRM to a PM. Each method relies on a similarity algorithm. There are many similarity algorithms available and each has its strengths and weaknesses [10]. Among them, the cosine similarity algorithm was selected in this study because it works best to identify the similarity between two objects which are proportionally similar [11]. Our pilot study demonstrated that the cosine algorithm outperformed other similarity algorithms such as the inner product similarity algorithm, Dice coefficient similarity algorithm, Jaccard coefficient similarity algorithm, and overlap coefficient similarity algorithm.
A cell in the PM is defined in Eq. 5 based on the cosine similarity algorithm.
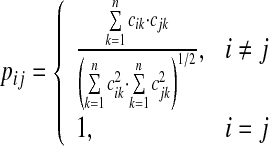 |
5 |
Equation 5 suggests that the proximity value (pij) between Ti and Tj is determined by the two corresponding rows of Ti and Tj in the TRM. That is, each row corresponds to a term vector. Like the TRM, the PM is a symmetric matrix. The cells located in the diagonal positions of the matrix are always equal to 1 because the proximity between a term and itself should reach the maximum value 1.
Results
From the 70 XML-encoded templates, RadMap extracted 6,489 element names, which represented 2,509 distinct terms. Some element names, such as “Report,” “Procedure,” “Clinical information,” “Findings,” and “Impression” appeared in every template. Duplicate terms were removed when we conducted the statistical analysis. Of the 2,509 unique terms, 1,017 terms (41%) matched exactly to RadLex terms, 660 (26%) were partial matches, and 832 reporting terms (33%) were unmatched to RadLex. Of 4,679 term occurrences, 623 (13%) were mapped to a post-coordinated term constructed from two or more RadLex terms.
Many of the exact matches related to anatomic terms and phrases. The reporting term “Liver,” for example, provided an exact match with the RadLex term “liver” (RID58). The reporting term “Right adrenal” mapped to a single RadLex term: “right adrenal” (RID30324), which is a direct subclass of the “adrenal gland” (RID88). The reporting term “central calcification” was mapped to two terms: “central” (RID5827) and “calcification” (RID5196). This new post-coordinated term was considered an exact match. The reporting term “Findings,” which was used to define the narrative section of the radiology report, was not found in RadLex, but was mapped to the synonymous RadLex term “observations section” as an exact match.
In the absence of an exact RadLex match, partial matches are considered. For example, the RadLex term “imaging technique” was selected as a partial match for the reporting term “Technique” because the reporting term had a broader meaning and included contrast administration as well as imaging technique. Additionally, there were instances where the report term was less granular than the information in RadLex. For example, the RadLex terms “anterior pararenal space” (RID432) and “posterior pararenal space” (RID433) are the component anatomic spaces that make up the the reporting term “pararenal space.” This represents a variation of a post-coordinated term and was considered an exact match.
A list of CT reporting templates is presented with their mapping results (Table 1). Table 2 shows a list of reporting terms in the US templates that are missing in RadLex. The mapping process revealed several general concepts such as “number,” “site,” and “location” that were not included in RadLex. These words themselves are not specific to radiology, but they can be parts of radiologic terms and thus useful for post-coordination.
Table 1.
Results of mapping CT exam templates to RadLex
| Template | No. of reporting elements | ||
|---|---|---|---|
| Exact match | Partial match | Unmatched | |
| CT Adrenal Mass | 37 | 11 | 8 |
| CT Brain | 37 | 15 | 14 |
| CT Brain Perfusion | 31 | 21 | 28 |
| CT Calcium Score | 37 | 20 | 17 |
| CT Cardiac | 119 | 47 | 65 |
| CT Cardiac Bypass Graft | 101 | 54 | 78 |
| CT Lung Nodule | 37 | 15 | 15 |
| CT Neck | 27 | 5 | 4 |
| CT Neck PostOp | 18 | 17 | 36 |
| CT Onco Followup | 37 | 17 | 16 |
| CT Onco Lung Mass | 37 | 17 | 12 |
| CT Onco Primary Liver Mass | 39 | 20 | 17 |
| CT Onco Primary Pancreas Mass | 47 | 15 | 25 |
| CT Onco Renal Mass | 36 | 14 | 9 |
| CT Pulmonary Veins | 57 | 8 | 35 |
| CT Renal Donor | 37 | 16 | 17 |
| CT Renal Mass | 41 | 12 | 20 |
| CT Renal Stone | 37 | 9 | 7 |
| CT Sinus | 24 | 5 | 4 |
| CT Spine | 130 | 18 | 10 |
| CT Temporal Bones | 21 | 3 | 9 |
| CT Urogram | 29 | 11 | 18 |
The numbers of exactly matched, partially matched, and unmatched terms in each template are indicated
Table 2.
List of terms that appeared in the Ultrasound reporting templates that were not included in RadLex
| Anteverted | Modifier |
| Elevated creatinine | Not seen |
| Empty | Postmenopausal |
| Fever | Recommendation |
| Focal parenchymal thinning | Resistance index |
| Hydronephrosis | Retroverted |
| LMP | Site |
| Location | Sonographic Murphy sign |
| Maximum | Waveform |
| Minimum |
Five ultrasound exam reporting templates (US Abdomen, US Pediatric Renal, US Pediatric Renal Doppler, US Pelvis, and US Retroperitoneum) were chosen as a sample to conduct the MDS visualization study. This sample was chosen because SPSS functions better with datasets that have fewer than 100 terms. The unmatched reporting element “Focal parenchymal thinning” was identified as a seed term because it is a term that is frequently used by radiologists and might be a possible addition to RadLex. Because “Focal parenchymal thinning” was selected from the “Findings” section of the XML-encoded reporting template, all the element names within the “Findings” section of the templates were selected as related terms and formed an association term set. As a result, 27 related terms were selected from the “Findings” secetion for the co-occurrence analysis (Table 3).
Table 3.
Seed term “focal parenchymal thinning” and its co-occurring terms in ultrasound reporting templates
| Abdominal aorta | Left kidney | Renal mass |
| Absent | Left ureter | Right kidney |
| AP diameter | Length | Right ureter |
| Contour | Location | Size |
| Diffuse | Mean | Standard deviation |
| Focal | Morphology | Stones |
| Focal parenchymal thinning | Normal | Transverse diameter |
| Hydronephrosis | Not seen | Urinary bladder |
| IVC | Renal length for age | Wall thickening |
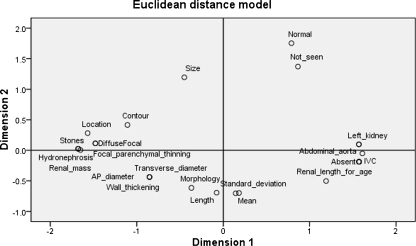
The MDS analysis results based on the seed term “focal parenchymal thinning” are shown in Fig. 1. The final stress value of the results is 0.01467. The RSQ value (r2 correlation value) is 0.99912. Both the stress value and the RSQ value are used to measure the quality of an MDS analysis. A stress value <0.10 and an RSQ >0.90 are considered sound and good results. In the 2D visual space shown in Fig. 1, one may determine the association strength between the seed term “focal parenchymal thinning” and its related co-occurring terms. It is clear that “hydronephrosis,” “renal mass,” and “stones” are clustered together with “focal parenchymal thinning,” while “length” and “morphology” are relatively far away from the seed term.
Fig. 1.
Graphical view of reporting term “Focal parenchymal thinning” and its co-occurring terms in a 2D visual space
Discussion
The use of standardized terminology in structured reports may reduce communication errors in radiology reporting. Mapping the elements of radiology reports to RadLex is a necessary step for indexing radiological information sources and verifying the applicability of the standard terminology in structured reporting. Our mapping results show that RadLex has covered most terms in the reporting templates. Much of RadLex has been built from a “top-down” view of which terms are needed in radiology. The unmatched and partially matched terms from the reporting templates provide a “bottom-up” inventory of entities used in radiology reports that can augment the RadLex vocabulary driven by use cases: real-world radiology problems in which terminology could enable solution. We view this effort as a constructive approach to improve the quality and coverage of both structured reporting templates and the RadLex vocabulary.
Information visualization was employed to reveal the connections and relationships among the investigated terms. Unlike traditional clustering analysis, the visualization method provides users with the proximity characteristics of terms that can be visually grouped into clusters. It presents the connections between a term and multiple relevant terms in a cluster and an overview of all involved terms and clusters [12].
Although there have been a number of studies using information visualization techniques, research to date has not addressed the visualization of radiological term usage. The MDS visualization technique, with its unique advantages over traditional data analysis approaches, offers promise for the visualization of term and topic relationships. It provides a semantic overview of the investigated reporting terms, vital visual contexts of the relationships among these terms, and interactive mechanisms for knowledge exploration [13]. The co-occurrences analysis gives a metric to determine the strength of an association between all of the terms returned from a seed term.
A weakness of the MDS analysis is its limited ability to effectively display a large dataset. When terms are gathered together onto the visual space and the clusters have blurred boundaries, it is difficult to identify hidden clusters in a given dataset and demonstrate sophisticated relationships among the identified clusters and connections among the terms in a cluster. To solve this problem, a traditional hierarchical clustering method was applied to the datasets for the purpose of comparison to confirm close proximities of the terms in the visual presentation. The clustering algorithms are used to find groups in the unlabeled data based on a similarity measure between the data elements. This means that similar patterns are placed together in the same cluster, which may clearly define and show the clusters for a given dataset regardless of the hidden cluster distribution. Therefore, comparing the MDS analysis results with the traditional clustering analysis results can verify clusters and the association strength among the co-occurring trems. Table 4 shows the clusters of reporting terms in the ultrasound reporting templates.
Table 4.
Clusters of reporting terms in the ultrasound reporting templates
| Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 |
|---|---|---|---|
| Transverse diameter | Diffuse | Right ureter | Normal |
| Wall thickening | Focal | Urinary bladder | Not seen |
| AP diameter | Hydronephrosis | Abdominal aorta | Size |
| Mean | Stones | Absent | |
| Standard deviation | Focal parenchymal thinning | Left ureter | |
| Length | Left kidney | ||
| Morphology | Renal mass | Right kidney | |
| Location | IVC | ||
| Contour | Renal length for age |
To preserve its utility as a standard for the radiology community, RadLex must continue to grow. Extracting terms from structured reporting templates may have the potential to increase the scope of the RadLex vocabulary more rapidly than manual addition by committees of experts. The terms used in radiology reporting templates should be terms in common clinical use in radiology and hence should be excellent candidates for inclusion into RadLex. RadLex lacks some terminology content such as definitions, relations, and categorical terms [14]. The MDS visualization study may help RadLex curators identify relationships between new and existing terms and may help them correctly place new terms into the RadLex hierarchy. Combined with a visual method of distributing and displaying lists of potential terms and relationships for domain expert validation, our techniques and results could support the ongoing development of RadLex.
Conclusions
Standard terminologies play an important role in radiology structured reporting. Sixty-seven percent of the terms used in 70 published structured reporting templates were mapped at least partially to terms in the RadLex vocabulary. The unmatched reporting terms will be submitted for addition to RadLex as new terms or synonyms. The semantic relationships among these terms revealed by the MDS visualization analysis may provide useful insights for ongoing development of RadLex. In this way, both the reporting templates and the RadLex vocabulary can evolve to become more complete and useful. The incorporation of RadLex terminology into structured reporting templates improves the consistency and quality of radiology reports. The use of uniform terminology for reporting across institutions enables aggregation and mining of radiology report data for research and quality improvement.
Acknowledgments
This research was supported in part by the National Institute of Biomedical Imaging and Bioengineering (NIBIB). We thank the RSNA Radiology Informatics Committee for leading and supporting the radiology reporting initiative, and we acknowledge the many RSNA volunteers who helped develop the reporting templates.
References
- 1.Langlotz CP. RadLex: a new method for indexing online educational materials. RadioGraphics. 2006;26:1595–1597. doi: 10.1148/rg.266065168. [DOI] [PubMed] [Google Scholar]
- 2.Rubin DL. Creating and curating a terminology for radiology: ontology modeling and analysis. J Digit Imaging. 2008;21:355–362. doi: 10.1007/s10278-007-9073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubin DL, Flanders A, Kim W, Siddiqui KM, Kahn CE., Jr Ontology-assisted analysis of web queries to determine the knowledge radiologists seek. J Digit Imaging. 2011;24:160–164. doi: 10.1007/s10278-010-9289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazen R, Esbroeck AP, Mongkolwat P, Channin DS. Automatic extraction of concepts to extend RadLex. J Digit Imaging. 2011;24:165–169. doi: 10.1007/s10278-010-9334-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kahn CE, Jr, Langlotz CP, Burnside ES, et al. Toward best practices in radiology reporting. Radiology. 2009;252:852–856. doi: 10.1148/radiol.2523081992. [DOI] [PubMed] [Google Scholar]
- 6.Robertson G, Card SK, Mackinlay JD: The cognitive coprocessor architecture for interactive user interfaces. Proceedings of the 2nd Annual ACM SIGGRAPH Symposium on User Interface Software and Technology, UIST '89. ACM Press, New York, NY, 1989, pp 10–18
- 7.Buja A, Swayne DF. Visualization methodology for multidimensional scaling. J Classification. 2002;19:7–43. doi: 10.1007/s00357-001-0031-0. [DOI] [Google Scholar]
- 8.Kahn CE Jr, Hong Y, Langlotz CP, Rubin DL: Encoding radiology report templates with RadLex terms. Radiological Society of North America (RSNA) 2010, exhibit SSG08-02
- 9.Zhang J, Wolfram D, Wang P, Hong Y, Gillis R. Visualization of health-subject analysis based on query term co-occurrences. J Am Soc Inform Sci Tech. 2008;59:1933–1947. doi: 10.1002/asi.20911. [DOI] [Google Scholar]
- 10.Zhang J, Rasmussen E. Developing a new similarity measure from two different perspectives. Inf Process Manag. 2001;37:279–294. doi: 10.1016/S0306-4573(00)00027-3. [DOI] [Google Scholar]
- 11.Korfhage RR. Information Storage and Retrieval. New York: Wiley; 1997. [Google Scholar]
- 12.Zhang J, Wolfram D, Wang P. Analysis of query keywords of sports-related queries using visualization and clustering. J Am Soc Inform Sci Tech. 2009;60(8):1550–1571. doi: 10.1002/asi.21098. [DOI] [Google Scholar]
- 13.Zhang J. Visualization for Information Retrieval. Berlin: Springer; 2008. [Google Scholar]
- 14.Marwede D, Schulz T, Kahn T. Indexing thoracic CT reports using a preliminary version of a standardized radiological lexicon (RadLex) J Digit Imaging. 2008;21:363–370. doi: 10.1007/s10278-007-9051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]