Abstract
Although acute myocardial infarction (MI) is consistently among the top causes of death in the United States, the spatial distribution of lipids and metabolites following MI remains to be elucidated. This work presents the investigation of an in vivo rat model of MI using mass spectrometric imaging (MSI) and multivariate data analysis. MSI was conducted on cardiac tissue following a 24-hour left anterior descending coronary artery ligation in order to analyze multiple compound classes. First, the spatial distribution of a small metabolite, creatine, was used to identify areas of infarcted myocardium. Second, multivariate data analysis and tandem mass spectrometry were used to identify phospholipid (PL) markers of MI. A number of lysophospholipids demonstrated increased ion signal in areas of infarction. In contrast, select intact PLs demonstrated decreased ion signal in the area of infarction. The complementary nature of these two lipid classes suggest increased activity of phospholipase A2, an enzyme that has been implicated in coronary heart disease and inflammation.
Keywords: mass spectrometric imaging, myocardial infarction, multivariate data analysis, lipids, tandem mass spectrometry, coronary artery disease
Introduction
Coronary heart disease (CHD) has remained the number one cause of death in the United States over the past four decades.1, 2 Many deaths occurring in patients with CHD arise from acute myocardial infarction (MI), more commonly known as a heart attack. The onset of MI is most often the result of atheromatous occlusion of coronary arteries restricting blood supply to cardiac tissue.3 The lack of oxygenated blood to cardiac tissue results in severe and often irreversible damage that may ultimately lead to heart failure. Although MI affects millions of people each year, the blood-borne and tissue-specific biochemical changes that occur following MI are not fully characterized, and a comprehensive understanding of these events may provide additional biochemical markers for use in clinical applications. To date, increases in two sets of blood-borne protein markers, creatine kinase (CK-MB) and troponins, are commonly used for the detection of myocardial necrosis (tissue death) following MI; however, these biomarker concentrations return to normal levels within days.4 The discovery of additional biomarkers may improve both clinical diagnosis capabilities and our understanding of this condition. Furthermore, identifying biochemical changes in infarcted myocardium may provide mechanistic insights for new therapeutic intervention targets designed to limit myocardial tissue loss. Thus, there is a growing need to study the biochemical changes resulting from both CHD and MI.
Mass spectrometry (MS) is an analytical tool with the potential to provide mechanistic detail for CHD characterization, biomarker discovery, and clinical diagnosis.5 MS was utilized as early as 1996 to characterize protein epitopes for MI6 as well as alterations in fatty acid metabolism in ischemic myocardium.7 Since these early studies, the focus of CHD MS research has shifted to proteomic and metabolomic analyses of plasma, serum, and urine, revealing a number of proteins8-10 and metabolites11, 12 as potential biomarkers and/or predictors for MI resulting from CHD. In addition, an extensive list of potential protein markers in serum was recently compiled, with the most abundant 25% projected to be amenable to tandem MS platforms.13 Although there has been extensive study on biological fluids, MS analysis of intact cardiac tissue from CHD and MI positive specimens has not been spatially detailed. Thus, an analytical method that characterizes small molecule biochemical changes locally in cardiac tissue following MI may yield valuable insight.
Mass spectrometric imaging (MSI) is a microprobe technique that generates chemically selective images from thin tissue sections,14, 15 and matrix-assisted laser desorption/ionization (MALDI) is a soft ionization technique well-suited for the analysis of small and large biomolecules.16, 17 The coupling of MALDI and MSI allows for tissue imaging by rastering a sample with respect to a stationary laser beam, collecting mass spectra at discrete positions. An image is generated by plotting the intensity of a selected mass-to-charge (m/z) versus the X,Y position, thereby preserving both chemical and spatial information. To date, MALDI MSI analyses have identified the spatial distribution of endogenous and exogenous compounds including proteins,14, 18 peptides,19, 20 lipids,21-23 and drugs.24-26 One advantage of this technique is the ability to elucidate relative intensity changes and spatial distributions resulting from external stimuli such as administration of an exogenous drug24 or injury models.27 Thus, MALDI MSI should be an ideal tool to delineate local alterations in lipids and metabolites in infarcted myocardium following a model such as a left anterior descending (LAD) coronary artery ligation. This model permits comparison of perfused (non-affected) and affected zones of tissue within a single coronary tissue section, eliminating much of the variability inherent in tissue-to-tissue comparisons.
Although MALDI MSI tissue experiments can generate a wealth of data, interpretation of multi-dimensional data sets can be time-consuming, tedious, and/or inconclusive. Multivariate data analysis has long been a staple of the ‘omics’ workflows, and has recently come of interest in the MSI community.28-30 Common multivariate techniques such as principal components analysis (PCA) and partial least squares discriminant analysis (PLS-DA) can be utilized for data reduction in multi-dimensional data sets. Briefly, these techniques separate sample groupings, or regions of tissue for MSI, based on the axes of maximum variation within the multi-dimensional data set, known as components. Although PCA and PLS-DA both determine the direction of maximum variance within a data set, the techniques’ definitions of maximum variance differs. PCA is an unsupervised technique that finds directions maximizing the total variance within a data set, whereas PLS-DA is a supervised technique that separates samples based on the covariance determined by both the data set and membership groupings.31 For visualization, samples are represented on two-dimensional or three-dimensional scores plots, and the weighting factors for each variable are displayed on loadings plots. Variables with the largest weighting factors contribute significantly to the grouping separation; thus, data sets containing large numbers of variables are reduced to a specified number of variables, simplifying data interpretation. This work reports the combined use of MALDI-MSI and multivariate data analysis to study lipids and the small metabolite creatine in cardiac tissue following LAD coronary artery ligation.
Experimental
Chemicals and Reagents
2,5-dihydroxybenzoic acid (DHB) was purchased from Sigma-Aldrich (St. Louis, MO). Sodium acetate (NaOAc), potassium acetate (KOAc), anhydrous creatine, HPLC-grade water (H2O), and HPLC-grade methanol (MeOH) were purchased from Fisher Scientific (San Jose, CA). 100% ethanol (EtOH) was purchased from Decon Labs (King of Prussia, PA). 1-heptadecanoyl-2-hydroxy-sn-glycero-3-phosphocholine (LPC 17:0) was purchased from Avanti Polar Lipids. A creatine standard was prepared in H2O to a final concentration of 100 ppm. LPC 17:0 was dissolved and diluted in EtOH to a concentration of 100 ppm. A MALDI matrix solution consisting of 40 mg/mL DHB in 70:30 MeOH:H2O (v/v) was prepared for the analysis of creatine (Matrix 1). Two other matrix solutions were prepared as above, except NaOAc or KOAc was added to a final concentration of 10 mM for Matrix 2 or Matrix 3, respectively.
Biological Sample Preparation
All animal procedures were conducted in accordance with guidelines published in the Guide for the Care and Use of Laboratory Animals (National Research Council, National Academy Press, Washington, DC, 2010) and were approved by the Animal Care Committee of Saint Louis University. Ligation of the LAD was performed as previously described.32 In brief, male Sprague-Dawley rats (250–300 g body weight) were injected with ketamine/xylazine (55 mg/ml, 7 mg/ml; 0.1 ml/100 g, i.p.). Rats were subsequently intubated and injected with Buprenex (0.05 mg/ml; 0.1 ml/100 g, i.p.). Animals were then ventilated with air at a tidal volume of 3–4 ml and a rate of 50–60 breaths/min (Harvard Apparatus). A left lateral thoracotomy was then performed. The thoracic cage was exposed and the intercostal space between ribs 4 and 5 was separated with a retractor. The left atrial appendage was retracted and a 6-0 suture was placed around the proximal LAD. This LAD suture was tied tightly or loosely (controls) to produce an infarction or sham surgery, respectively. Then, the thoracic incision was closed with a 5-0 suture. After recovery from surgery, rats were weighed and individually housed. Twenty-four hours following LAD occlusion (or control sham surgery), rats were euthanized with pentobarbital (~800 mg/kg, i.p.), and subsequently hearts were removed and flash-frozen in liquid nitrogen. Organs were stored at −80 °C until further preparation.
Heart tissue was bisected along a transverse plane that passed through the left and right ventricles. The upper half of the heart was subjected to 2,3,5-triphenyltetrazolium chloride (TTC) staining to distinguish perfused and damaged tissue.33 The remaining lower half of the heart was utilized for MALDI MSI analysis. Cardiac tissue was sectioned using a Microm HM 505E cryostat (Waldorf, Germany). Optimal cutting temperature polymer (OCT) was not used for tissue mounting as OCT is reported to produce abundant ion signals in MALDI analysis of thin tissue sections, resulting in analyte ion suppression.34 Instead, tissue was held atop a drop of HPLC-grade H2O on the cutting stage and placed into the cryostat chamber held at −25 °C, thereby freezing the drop of water and fusing the organ to the cutting stage. Subsequently, 10-μm thick sections were thaw-mounted atop glass microscope slides and stored at −80 °C. Prior to matrix coating, tissue sections were placed in a vacuum desiccator for approximately 45 minutes to remove excess H2O.
Serial cardiac tissue sections were coated with either Matrix 1 or Matrix 2. MALDI matrix was spray coated atop tissue sections using a glass Type A Meinhard Nebulizer (Golden, CO). Nitrogen was used as the nebulizing gas at a pressure of 30 PSI and the matrix solution was delivered at a flow rate of 3 mL/min. In this method, approximately six passes were conducted over each slide followed by 15 seconds of waiting time in order to avoid excessive wetting of the tissue sections. The process was repeated until a homogenous layer of matrix crystals was obtained over the entire tissue. Approximately 2 mL of MALDI matrix solution were used per microscope slide to obtain a suitable matrix layer on the tissue sections.
Mass Spectrometry and Imaging
All experiments were performed using a Thermo Scientific LTQ XL linear ion trap mass spectrometer (San Jose, CA). The instrument was equipped with a MALDI ionization source, consisting of a Lasertechnik Berlin MNL 106-LD N2 laser (λ=337nm) (Berlin, Germany). The laser has a repetition rate of 60 Hz and produces a laser spot diameter of approximately 100 μm. Creatine and LPC standards were deposited onto a 96-well stainless steel MALDI plate using a modified dried-droplet method.16 In this method, 1 μL of the prepared standard followed by 1 μL of matrix were pipetted onto the well plate. Solvent was evaporated from the mixture using gentle heat, leaving behind MALDI matrix and analyte co-crystals. MS, MS2, and MS3 spectra were acquired using a laser energy of 5 μJ and 3 laser shots per laser stop, and 100 spectra were averaged to produce one standard spectrum.
Following analysis of standards, MSI in MS and MS2 modes was performed on prepared cardiac tissue sections. The tissue sections were rastered with respect to the laser at a horizontal and vertical step size of 100 μm. MS spectra (m/z 100–250) were collected in profile mode over tissue sections coated with Matrix 1. Additionally, MS spectra (m/z 200–2000) were collected in centroid mode (to minimize the size of the data files) over tissue coated with Matrix 2. A laser energy of 4 μJ and 3 laser shots per laser stop were utilized for all MS and MS2 imaging experiments. Automatic gain control (AGC) was toggled off for all experiments in order to maintain a constant number of laser shots at each position. Images were generated using Thermo ImageQuest v1.0.1 software (San Jose, Ca). All MS images were generated by plotting ion intensity divided by the total ion current (TIC) at each pixel. MS2 images were generated by plotting fragment ion intensity at each pixel; however, these images were not normalized to the TIC.
Compound identification and MS2 imaging acquisition for lipid species from cardiac tissue was performed using tandem MS with collision-induced dissociation (CID) or pulsed Q collision-induced dissociation (PQD). For MS2 experiments, an isolation width of 1.2 amu and a collision energy of 35 AU (arbitrary units normalized to m/z 400) was utilized. In instances where MS3 was necessary to identify ions, the same settings were utilized for the second stage of MS, and the third stage of MS was performed with an isolation width of 1.5 amu and a collision energy of 35 AU. In instances where the linear ion trap’s low-mass cut-off (LMCO) hindered ion identification (fragmenting intact PLs), PQD was utilized with an isolation width of 1.2 amu and a collision energy of 25 AU.35
Statistical Analysis
Multivariate data analysis and significance testing were performed to determine significant m/z values (features) differing between perfused and infarcted tissue within the same tissue section from hearts subjected to LAD ligation. PCA and PLS-DA were performed using Metaboanalyst web server.31, 36 Five samples, each consisting of an average of 25 mass spectra, were selected from each tissue zone. Mass-to-charge (from centroid data) and intensity lists were exported from QualBrowser into Microsoft Excel and saved as .csv files. Since Metaboanalyst does not support mean-centering without further normalization (e.g., normalization to the range or standard deviation of the mean), the data were processed in two different methods: the first method mean-centered the intensity at each m/z value before it was imported into Metaboanalyst and the second method left the data unprocessed before it was imported. After the processed m/z and intensity lists were imported, a mass tolerance of 0.5 m/z was utilized to counteract possible mass shifts due to space charge effects. Next, m/z values known to arise from MALDI matrix ions were removed from all samples. In this instance, the intense matrix ions observed in the mass spectra were m/z 273 and 274, representing the [2M-2H2O+H]+ and its 13C isotope, respectively. Following MALDI matrix peak removal, signal intensities were normalized to the TIC within each sample to account for signal variability inherent in MALDI MS tissue analysis. Additionally, the utility of scaling techniques (e.g., autoscaling) was investigated. After multivariate data analysis, a Student’s t-test was performed on selected features using Microsoft Excel.
Results
TTC Staining
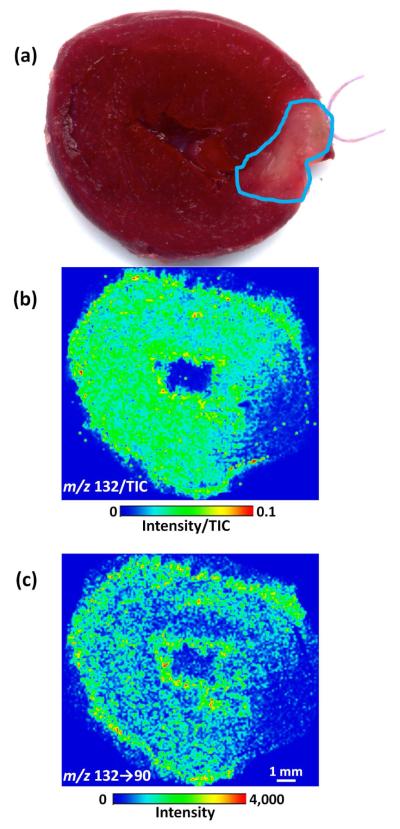
Prior to preparation for MSI analysis, the myocardial tissue sections were submitted to TTC staining, which is utilized to identify regions of tissue necrosis.37 In its original state, TTC is colorless; however, in the presence of cellular dehydrogenases of healthy tissue, enzymatic reduction alters TTC to a formazan, producing a brick red color.33 Infarcted tissues lack the dehydrogenases necessary for enzymatic reduction of TTC; thus, the tissue remains unstained.38 TTC staining of the heart illustrated infarcted myocardium in the affected area of the left ventricle, which was previously supplied arterial blood by the LAD (Figure 1a).
Figure 1.
Photograph of the upper half of a heart stained with 2,3,5-triphenyltetrazolium chloride (TTC) following LAD coronary artery ligation (a). Unstained tissue, outlined in blue, indicates tissue damage from myocardial necrosis. MS image of m/z 132 intensity divided by the TIC (b) and MS2 image of m/z 132 → 90 intensity (c) from the lower half of the same heart depicted in (a). Lower ion signal was observed in the infarcted region near the right side of the tissue section.
Identification of Infarcted Myocardium by MSI via Creatine
Biochemically, creatine kinase (CK) consumes one molecule of adenosine triphosphate (ATP) to catalyze the conversion of one molecule of creatine to one molecule of phosphocreatine, which serves as a high energy phosphate buffer for ATP in muscle tissue. Three to six hours following a heart attack, CK-MB, a creatine kinase selective to cardiac tissue, is released into the interstitial fluid, and consequently the bloodstream, resulting in an elevated plasma level of CK-MB.4 Although CK-MB can be detected using biological assays, the molecular weight of CK-MB (~86 kDa) and the limited mass range of a linear ion trap hinder direct detection of this enzyme. Conversely, the enzyme’s substrate, creatine, is well within the mass range of the linear ion trap. One might expect decreased levels of creatine within infarcted or damaged tissues if the damaged tissue also leaks water-soluble metabolites.
Prior to tissue analysis, a creatine standard was characterized in MS and MS2 modes. MS analysis yields analyte ions at m/z 132 and m/z 154 corresponding to the [M+H]+ and [M+Na]+ ions, respectively. The ion at m/z 154 is isobaric with the [M]•+ for DHB; thus, the addition of sodium acetate to the MALDI matrix would not be beneficial for MS studies of creatine, as it may drive creatine ion signal to the [M+Na]+. The MS2 spectrum of m/z 132 demonstrates a single abundant fragment ion at m/z 90 (protonated n-methylglycine, more commonly known as sarcosine) resulting from the loss of CH2N2.
Following authentic creatine characterization, MS and MS2 were applied to assess the spatial localization of creatine in cross-sectional tissue sections from hearts sliced at the mid-ventricle level. Both control rats and rats subjected to 24h of regional ischemia as a result of LAD coronary artery occlusion were analyzed. The MS image of m/z 132 normalized to the TIC (Figure 1b) demonstrates a lower signal in the infarcted region of tissue, positively correlating with the TTC image (Figure 1a). Similarly, the MS2 image mapping the transition from m/z 132 → 90 also demonstrates a lower signal in the infarcted region of tissue (Figure 1c). In contrast, control tissues demonstrated a constant signal throughout cardiac tissue sections (Figure S-1). The lower creatine signal may reflect the state of the plasma membrane of cells within infarcted myocardium. Upon rupture of these plasma membranes, water-soluble enzymes (e.g., CK) and possibly small metabolite substrates such as creatine may leach out from infarcted myocardium.
Phospholipase A2
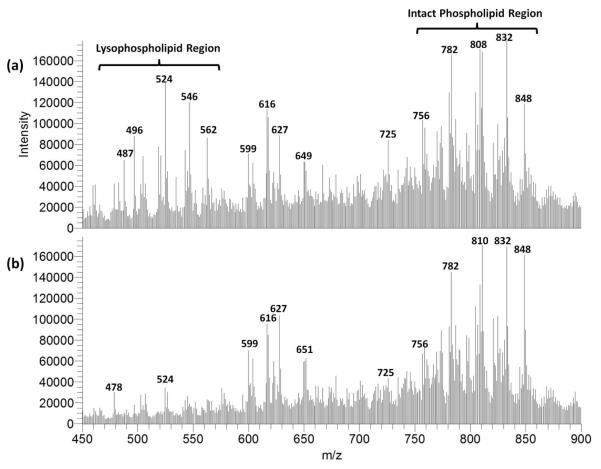
Phospholipase A2 (PLA2) hydrolyzes the sn-2 acyl bond of intact phospholipids (PLs) including phosphatidylcholines (PCs) and phosphatidylethanolamines (PEs), yielding free fatty acids and lysophospholipids (lysoPLs) including lysophosphatidylcholines (LPCs) and lysophosphatidylethanolamines (LPEs). Multiple investigations have suggested an important role of accelerated PLA2 activity as an enzymic mediator of the pathophysiological sequelae of myocardial ischemia.39-41 Although LPCs and LPEs have been considered important arrhythmogenic lipids generated by PLA2, their spatial localization in ischemic myocardium has not been delineated. Accordingly, MSI experiments were performed on tissue sections of hearts subjected to LAD coronary artery ligation. For these experiments, NaOAc was added to the matrix to enhance alkali metal adduction, as previous investigations have indicated that the analysis of sodium adducts enhance MSn identification of lipid species.22 Representative mass spectra (an average of 50 scans) from the lipid region (m/z 450–900) for infarcted and perfused tissue are shown in Figure 2. Previous research utilizing MALDI time-of-flight MS to monitor the in-solution digestion of PC by PLA2 has identified a number of characteristic LPC ions in the lysoPL region (m/z 450–600).42 An increase in relative intensity is observed for various ions in this lysoPL region within infarcted zones of these sections; however, few other major differences were elucidated from the averaged mass spectra from the two zones.
Figure 2.
Mass spectra recorded in the lipid region for infarcted (a) and perfused zones of cardiac tissue (b).
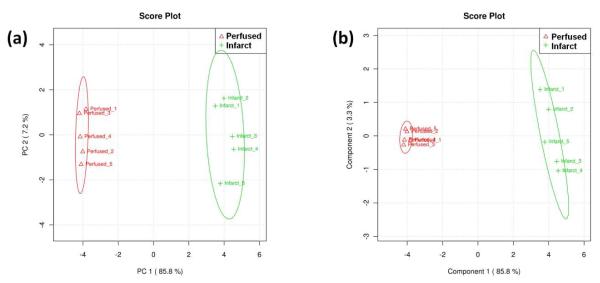
PCA and PLS-DA were conducted on both tissue zones of a single infarcted section. A single tissue section was chosen to minimize any variance that might occur from section to section or specimen to specimen. Multiple samples were generated from a single cardiac section by choosing five horizontal lines, each line consisting of 25 mass spectra, within a selected tissue region. Scores plots from PCA and PLS-DA, shown in Figure 3, were generated using Metaboanalyst. Separation between perfused and infarcted zones was observed in both multivariate data analysis techniques. In PCA of the mean-centered data, principal component 1 (PC1) and principal component 2 (PC2) carried 85.8% and 7.2% of the total variation, respectively (Figure 3a). The separation between infarcted and perfused tissue was largely dictated by PC 1, whereas PC 2 accounted for the variation within the sample groupings. The autoscaled PCA plots demonstrated a similar separation (data not shown).
Figure 3.
PCA scores plot (a) and PLS-DA scores plot (b) for mean-centered data. Each sample consists of 25 spectra taken along a horizontal line within either infarcted (green crosses) or perfused tissue (red triangles) from a single tissue section following an LAD coronary artery ligation. The ovals indicate the 95% confidence interval for the sample groupings.
In contrast to PCA, PLS-DA is a supervised technique that separates samples based on the largest covariance in the data set (i.e., the sample groupings are known). The PLS-DA scores plot for the TIC-normalized peak list is shown in Figure 3b. Similar to PCA, component 1 carries 85.8% of the covariance, and sufficiently separates the two tissue zones. Although the PCA and PLS-DA scores plots show similar separation along the first component, PLS-DA demonstrated a tighter cluster within each zone along the second component. PLS-DA was also conducted on the autoscaled data (data not shown). The generated scores plot also shows sufficient separation between the perfused and infarcted zones along component 1.
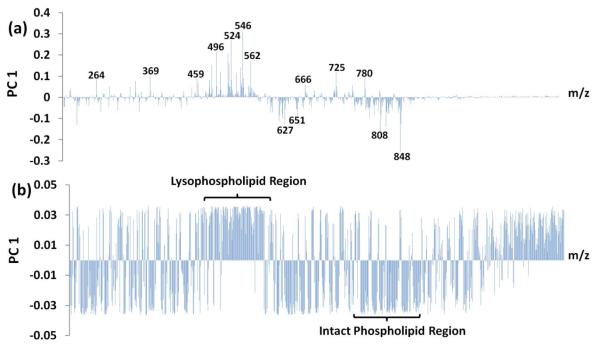
The loadings plots for these four separation methods were then analyzed for significant m/z values that influenced separation (Figure 4). In all four methods, the separation was dictated by PC 1 (for PCA) or Component 1 (for PLS-DA). Thus, analysis of the loadings plots can be simplified to the first PC or component for identifying significant m/z values. When analyzing the loadings for the mean-centered data without autoscaling (Figure 4a), m/z values in the lysoPL region were positively correlated with infarcted tissue (e.g., m/z 496, 524, 546, and 562). In contrast, some m/z values present in the intact PL region were positively correlated with the perfused tissue (e.g., m/z 810 and 848). Analysis of the loadings for the autoscaled data proved to be more difficult, as many m/z values proved to have similar loadings values (Figure 4b). In principle, any variable that differs significantly between sample sets should influence the separation; however, a major drawback for autoscaling is that low intensity signals, which may have large relative variances, are weighted equally with high intensity variables. Upon analysis of the autoscaled loadings values, the majority of the m/z values with high loadings values appeared to be of low intensity; therefore, autoscaling was not utilized for feature identification. Loadings plots from PLS-DA produced many of the same m/z values as PCA.
Figure 4.
PCA loadings plots from mean-centered data (a) and from autoscaled data (b).
Significance testing was then performed on selected m/z values identified by PCA and PLS-DA from the mean-centered data sets (without autoscaling). Additionally, significance testing was performed on a MALDI matrix ion (m/z 273) for comparison. A one-tailed Student’s t-test was performed on the signal intensities at three significance levels (95%, 98%, and 99%). The results (Table 1) demonstrate a significant difference for many of the features identified by PCA and PLS-DA in the lysoPL region, including m/z 496, 518. 524, 544, and 546, each positively correlated with infarcted tissue. Additionally, two m/z values in the intact PL region, m/z 832 and 848, correlated negatively with infarcted tissue and passed the t-test at a significance level of 95% or greater. The opposite trends for these two lipid classes may lend evidence towards increased PLA2 activity.
Table 1.
Significance testing and putative MSn identifications for lipids in perfused and infarcted zones of cardiac tissue.
| m/z | Infarcted Signal/TIC |
Perfused Signal/TIC |
Pooled Standard Deviation |
Direction | t | MSn Identification |
Ion |
|---|---|---|---|---|---|---|---|
| 273 | 0.01490 | 0.01330 | 0.00176 | ↓ | 1.12 | DHB | [2M-2H2O+H]+ |
| 459 | 0.00088 | 0.00037 | 0.00019 | ↑ | 3.26† | LPC 16:0 | [M-C3H9N+Na]+ |
| 487 | 0.00163 | 0.00051 | 0.00049 | ↑ | 2.80† | LPC 18:0 | [M-C3H9N+Na]+ |
| 496 | 0.00236 | 0.00085 | 0.00077 | ↑ | 3.84‡ | LPC 16:0 | [M+H]+ |
| 518 | 0.00297 | 0.00083 | 0.00053 | ↑ | 4.97‡ | LPC 16:0 | [M+Na]+ |
| 520 | 0.00261 | 0.00069 | 0.00080 | ↑ | 2.96† | LPE 18:0 LPC 18:2 |
[M+K]+ [M+H]+ |
| 524a | 0.00748 | 0.00196 | 0.00208 | ↑ | 3.26† | LPC 18:0 | [M+H]+ |
| 534 | 0.00130 | 0.00072 | 0.00037 | ↑ | 1.91 | LPC 16:0 | [M+K]+ |
| 544 | 0.00317 | 0.00241 | 0.00028 | ↑ | 3.29† | LPC 18:1 LPC 20:4 |
[M+Na]+ [M+H]+ |
| 546 | 0.00619 | 0.00150 | 0.00134 | ↑ | 4.29‡ | LPC 18:0 | [M+Na]+ |
| 782 | 0.00571 | 0.00600 | 0.00085 | ↓ | 0.41 | PC (16:0/18:1) | [M+Na]+ |
| 810 | 0.00651 | 0.00612 | 0.00056 | ↑ | 0.85 | PC (18:0/18:1) | [M+Na]+ |
| 832 | 0.00767 | 0.01190 | 0.00196 | ↓ | 2.64* | PC (18:0/20:4) | [M+Na]+ |
| 848 | 0.00350 | 0.00818 | 0.00029 | ↓ | 19.50‡ | PC (18:0/20:4) | [M+K]+ |
Significant at 95% confidence interval
Significant at 98% confidence interval
Significant at 99% confidence interval
Unidentified isobaric LPE also present
Targeted MSn studies for ion identification were conducted on significant features within the region of interest utilizing CID and PQD. MS2 experiments revealed the major contribution of all significant features to be PC, LPC, or LPE species. Previous work has demonstrated that MS2 differentiates PEs, alkali metal adducts of PCs, and protonated ions of PCs; a neutral loss (NL) of 43 or 59 indicates the presence of a PE or PC, respectively,43, 44 whereas a fragment ion at m/z 184 indicates a protonated PC.22 Protonated PCs and LPCs yield relatively uninformative fragmentation; however, the mass of the fatty acid tail can be inferred for LPCs, as only one fatty acid tail is present. Fragmentation of alkali metal adducts of PCs and LPCs produces more relevant structural information. MS3, fragmenting the daughter ion resulting from a NL of 59, was used to determine the presence of sodium or potassium in adduction. In this instance, a NL of 146 indicates the presence of a sodium adduct, whereas a NL of 162 indicates the presence of a potassium adduct.44 For LPCs, the remaining mass can be accounted for by the glycerol backbone, a hydroxyl group in the sn-2 position, and the lone fatty acid tail in the sn-1 position. Low abundance neutral losses corresponding to the mass of the fatty acid tails, which are crucial for structural elucidation of intact PCs containing two fatty acid tails, may also be observed in alkali metal adduct PC fragmentation.
An LPC 17:0 standard, spotted with either Matrix 2 or Matrix 3, was characterized in MS, MS2, and MS3 modes to verify these findings. MS spectra of the LPC standard demonstrate an abundant [M+H]+ ion and an abundant alkali metal adduct of either [M+Na]+ or [M+K]+ for Matrix 2 or Matrix 3, respectively (Figure S-2). Additionally, source fragments are present at 59 Da lower than the alkali metal adduct in each respective spectrum. The fragmentation pathways described above for the sodium and potassium adducts were confirmed following MSn characterization (Figures S-3 and S-4). Knowledge of these fragmentation pathways was crucial for identifying lipid species from tissue. The tentative identification for selected features is listed in Table 1. MSn revealed isobaric lipids at both m/z 520 and m/z 544, specifically an LPC and LPE at m/z 520 and two LPCs at m/z 544. Representative spectra for m/z 546 and 848 are shown in Figures S-5 and S-6, respectively.
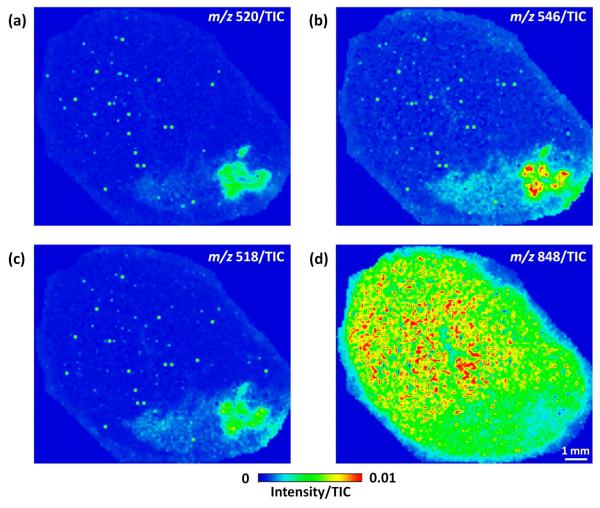
Mass spectrometric images were generated from infarcted cardiac sections for each of the identified LPCs and PCs. Representative images for three lysoPLs (Figure 5a–c) and one intact PC (Figure 5d) are shown. For each LPC or LPE, an intense ion signal is observed in the core of infarcted myocardium, and an area of less intense ion signal is observed moving outward from this area. In contrast to lysoPLs, pertinent intact PCs, such as PC (18:0/20:4), demonstrate the reverse trend: an area of relatively low signal is present near the core of infarcted myocardium, and an area of more intense signal is observed moving outward from this area. These trends suggest that intact PCs are being converted to LPCs via the action of PLA2 within the infarct zone.
Figure 5.
MS images for the [M+K]+ of LPE 18:0 at m/z 520 (a), the [M+Na]+ of LPC 18:0 at m/z 546 (b), the [M+Na]+ of LPC 16:0 at m/z 518 (c), and the [M+K]+ of PC (18:0/20:4) at m/z 848 (d) from a heart following LAD coronary artery ligation. All images were generated by plotting the ion intensity divided by the TIC.
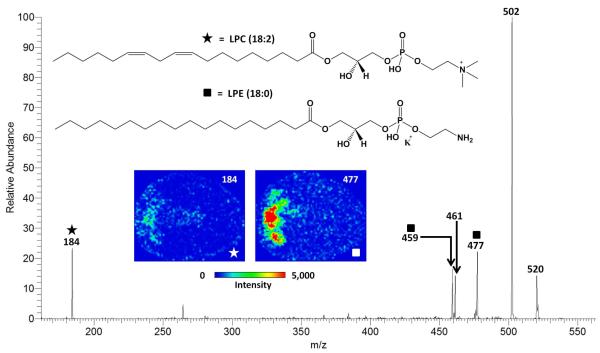
MS2 imaging experiments were conducted on features found to contain multiple lysoPL species at a nominal m/z to determine the major contributor to signal in infarcted myocardium. The MS2 spectrum of m/z 520 and images of fragment ions from m/z 520 are shown in Figure 6. Targeted MS2 analysis previously identified two lipids present at m/z 520: the [M+K]+ of LPE 18:0 and the [M+H]+ of LPC (18:2). MS2 imaging revealed that the LPE fragment ion (NL of 43) contributed more ion signal relative to LPC fragment ion (m/z 184) within infarcted myocardium. The mechanism for the formation of LPC 18:2 is not certain, as unsaturated fatty acids typically occur on the sn-2 position of the glycerol backbone, making this lysoPL an unlikely candidate for enzymatic formation by PLA2. In contrast, LPE 18:0 contains a saturated fatty acid tail, which is hypothesized to occur on the sn-1 position of the glycerol backbone. The localization of this fragment ion coupled with the hypothesized location of the fatty acid tail indicates that LPE 18:0 may have arisen via the action of PLA2 within infarcted myocardium.
Figure 6.
MS2 spectrum using CID of m/z 520 from infarcted cardiac tissue indicating two isobaric lipid ions. Fragment ions from [M+K]+ of LPE 18:0 and the [M+H]+ of LPC 18:2 are indicated with squares and stars, respectively. The structures of each lipid and MS2 images of each species’ characteristic fragment (i.e., m/z 520→477 for LPE 18:0 and m/z 520→184 for LPC 18:2) from cardiac tissue following LAD coronary artery ligation are shown as an inset. The LPE contributes more ion signal within the infarcted zone than the LPC.
Conclusions
This research is the first presentation of MSI displaying potential markers for MI within intact cardiac tissue. First, creatine, a metabolite involved in the creatine kinase metabolic pathway, demonstrated decreased ion signal within areas of infarcted tissue, agreeing well with TTC-stained hearts following LAD coronary artery ligation. Furthermore, a combination of multivariate data analysis and tandem mass spectrometry conducted on an MSI data set was used to identify PC, LPC, and LPE markers of MI. These studies indicated that many LPCs and select LPEs demonstrated increased ion signal within the infarction zone and select PCs demonstrated decreased ion signal within the infarction zone. It is also interesting that two intact PCs found to decrease in infarcted tissue presumably contain arachidonic acid in the sn-2 position. It has been reported that PLA2 and arachidonic acid (AA) may play a role in the cellular response to myocardial infarction, specifically involving protection against ischemic cell death.45 The complementary localization of LPC 18:0 at m/z 546 and its precursor, PC (18:0/20:4), at m/z 848, support this hypothesis. The PL findings of this study suggest that PLA2 activity increased in infarcted myocardium following LAD ligation.
It should be noted that tissue-specific markers do not immediately result in blood or plasma markers of MI. Nevertheless, it may be beneficial to investigate these tissue-specific markers in biological fluids using targeted, sensitive methods (e.g., liquid chromatography/tandem mass spectrometry (LC/MS/MS) using selected reaction monitoring). In particular, small metabolites such as creatine that may leak into the interstitial fluid following infarction may demonstrate an increase in blood-borne concentration. The discovery of one such small molecule marker would offer a valuable alternative to the traditional protein markers utilized in a clinical setting.
MALDI MSI offers advantages over traditional MI characterization techniques such as histological staining and biological assays. In particular, the ability to detect, identify, and localize small molecular species allows for identification of putative markers for MI with a single analytical technique. Furthermore, a wealth of data was collected relatively quickly (e.g., approximately one hour for a single cardiac section), making these experiments prime for multivariate data analysis techniques commonly used in the ‘omics’ platforms. For these reasons, MALDI MSI, coupled with multivariate data analysis has the potential to be applied to a wide variety of tissue applications, making it a powerful tool for diseased-state characterization and biomarker discovery.
Supplementary Material
Acknowledgements
This work was partially supported by the NSF MS-PIRE grant (OISE-0730072) and NIH grants HL074214 (DAF) and HL111906 (DAF).
Footnotes
Supporting Information This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Xu J, Kochanek KD, Murphy SL, Tejada-Vera B. Natl. Vital Stat. Rep. 2010;58:1–135. [PubMed] [Google Scholar]
- (2).Jemal A, Ward E, Hao YP, Thun M. JAMA, J. Am. Med. Assoc. 2005;294:1255–1259. doi: 10.1001/jama.294.10.1255. [DOI] [PubMed] [Google Scholar]
- (3).Hansson GK. N. Engl. J. Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- (4).Morrow DA, Cannon CP, Jesse RL, Newby LK, Ravkilde J, Storrow AB, Wu AHB, Christenson RH, Apple FS, Francis G, Tang W, Members NWG, Members NC. Clin. Chem. (Washington, DC, U. S.) 2007;53:552–574. doi: 10.1373/clinchem.2006.084194. [DOI] [PubMed] [Google Scholar]
- (5).Gerszten RE, Wang TJ. Nature. 2008;451:949–952. doi: 10.1038/nature06802. [DOI] [PubMed] [Google Scholar]
- (6).Macht M, Fiedler W, Kurzinger K, Przybylski M. Biochemistry. 1996;35:15633–15639. doi: 10.1021/bi961727w. [DOI] [PubMed] [Google Scholar]
- (7).Ford DA, Han X, Horner CC, Gross RW. Biochemistry. 1996;35:7903–7909. doi: 10.1021/bi960552n. [DOI] [PubMed] [Google Scholar]
- (8).Marshall J, Kupchak P, Zhu WM, Yantha J, Vrees T, Furesz S, Jacks K, Smith C, Kireeva I, Zhang R, Takahashi M, Stanton E, Jackowski G. J. Proteome Res. 2003;2:361–372. doi: 10.1021/pr030003l. [DOI] [PubMed] [Google Scholar]
- (9).Delles C, Schiffer E, von zur Muhlen C, Peter K, Rossing P, Parving HH, Dymott JA, Neisius U, Zimmerli LU, Snell-Bergeon JK, Maahs DM, Schmieder RE, Mischak H, Dominiczak AF. J. Hypertens. 2010;28:2316–2322. doi: 10.1097/HJH.0b013e32833d81b7. [DOI] [PubMed] [Google Scholar]
- (10).von zur Muhlen C, Schiffer E, Zuerbig P, Kellmann M, Brasse M, Meert N, Vanholder RC, Dominiczak AF, Chen YC, Mischak H, Bode C, Peter K. J. Proteome Res. 2009;8:335–345. doi: 10.1021/pr800615t. [DOI] [PubMed] [Google Scholar]
- (11).Zhang HY, Chen X, Hu P, Liang QL, Liang XP, Wang YM, Luo GA. Talanta. 2009;79:254–259. doi: 10.1016/j.talanta.2009.03.045. [DOI] [PubMed] [Google Scholar]
- (12).Sabatine MS, Liu E, Morrow DA, Heller E, McCarroll R, Wiegand R, Berriz GF, Roth FP, Gerszten RE. Circulation. 2005;112:3868–3875. doi: 10.1161/CIRCULATIONAHA.105.569137. [DOI] [PubMed] [Google Scholar]
- (13).Anderson L. J. Physiol.-London. 2005;563:23–60. doi: 10.1113/jphysiol.2004.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Caprioli RM, Farmer TB, Gile J. Anal. Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- (15).Garrett TJ, Yost RA. Practical Aspects of Trapped Ion Mass Spectrometry. Volume V. CRC Press; 2009. pp. 417–438. [Google Scholar]
- (16).Karas M, Hillenkamp F. Anal. Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- (17).Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T, Matsuo T. Rapid Commun. Mass Spectrom. 1988;2:151–153. [Google Scholar]
- (18).Chaurand P, Schwartz SA, Caprioli RM. Anal. Chem. 2004;76:86A–93A. [PubMed] [Google Scholar]
- (19).Taban IM, Altelaar AFM, van der Burgt YEM, McDonnell LA, Heeren RMA, Fuchser J, Baykut G. J. Am. Soc. Mass Spectrom. 2007;18:145–151. doi: 10.1016/j.jasms.2006.09.017. [DOI] [PubMed] [Google Scholar]
- (20).Guenther S, Römpp A, Kummer W, Spengler B. Int. J. Mass Spectrom. 2011;305:228–237. [Google Scholar]
- (21).Landgraf RR, Conaway MCP, Garrett TJ, Stacpoole PW, Yost RA. Anal. Chem. 2009;81:8488–8495. doi: 10.1021/ac901387u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Garrett TJ, Prieto-Conaway MC, Kovtoun V, Bui H, Izgarian N, Stafford G, Yost RA. Int. J. Mass Spectrom. 2007;260:166–176. [Google Scholar]
- (23).Garrett T, Menger R, Dawson W, Yost R. Anal. Bioanal. Chem. 2011:1–11. doi: 10.1007/s00216-011-5044-x. [DOI] [PubMed] [Google Scholar]
- (24).Hsieh Y, Casale R, Fukuda E, Chen JW, Knemeyer I, Wingate J, Morrison R, Korfmacher W. Rapid Commun. Mass Spectrom. 2006;20:965–972. doi: 10.1002/rcm.2397. [DOI] [PubMed] [Google Scholar]
- (25).Hopfgartner G, Varesio E, Stoeckli M. Rapid Commun. Mass Spectrom. 2009;23:733–736. doi: 10.1002/rcm.3934. [DOI] [PubMed] [Google Scholar]
- (26).Reich RF, Magparangalan DP, Garrett TJ, Yost RA. Mass Spectrometry in Drug Metabolism and Disposition. John Wiley & Sons, Inc.; 2011. pp. 449–482. [Google Scholar]
- (27).Hankin JA, Farias SE, Barkley R, Heidenreich K, Frey LC, Hamazaki K, Kim HY, Murphy RC. J. Am. Soc. Mass Spectrom. 2011;22:1014–1021. doi: 10.1007/s13361-011-0122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Trim PJ, Atkinson SJ, Princivalle AP, Marshall PS, West A, Clench MR. Rapid Commun. Mass Spectrom. 2008;22:1503–1509. doi: 10.1002/rcm.3498. [DOI] [PubMed] [Google Scholar]
- (29).McCombie G, Staab D, Stoeckli M, Knochenmuss R. Anal. Chem. 2005;77:6118–6124. doi: 10.1021/ac051081q. [DOI] [PubMed] [Google Scholar]
- (30).Bonnel D, Longuespee R, Franck J, Roudbaraki M, Gosset P, Day R, Salzet M, Fournier I. Anal. Bioanal. Chem. 2011;401:149–165. doi: 10.1007/s00216-011-5020-5. [DOI] [PubMed] [Google Scholar]
- (31).Xia J, Psychogios N, Young N, Wishart DS. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Fishbein MC, Maclean D, Maroko PR. Am. J. Pathol. 1978;90:57–70. [PMC free article] [PubMed] [Google Scholar]
- (33).Benedek A, Moricz K, Juranyi Z, Gigler G, Levay G, Harsing LG, Matyus P, Szenasi G, Albert M. Brain Res. 2006;1116:159–165. doi: 10.1016/j.brainres.2006.07.123. [DOI] [PubMed] [Google Scholar]
- (34).Schwartz SA, Reyzer ML, Caprioli RM. J. Mass Spectrom. 2003;38:699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- (35).Armenta JM, Hoeschele I, Lazar IM. J. Am. Soc. Mass Spectrom. 2009;20:1287–1302. doi: 10.1016/j.jasms.2009.02.029. [DOI] [PubMed] [Google Scholar]
- (36).Xia J, Wishart DS. Nat. Protoc. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- (37).Ytrehus K, Liu Y, Tsuchida A, Miura T, Liu GS, Yang XM, Herbert D, Cohen MV, Downey JM. Am. J. Physiol. Heart Circ. Physiol. 1994;267:H2383–H2390. doi: 10.1152/ajpheart.1994.267.6.H2383. [DOI] [PubMed] [Google Scholar]
- (38).Goldlust EJ, Paczynski RP, He YY, Hsu CY, Goldberg MP. Stroke. 1996;27:1657–1662. doi: 10.1161/01.str.27.9.1657. [DOI] [PubMed] [Google Scholar]
- (39).Ford DA. Prog. Lipid Res. 2002;41:6–26. doi: 10.1016/s0163-7827(01)00014-5. [DOI] [PubMed] [Google Scholar]
- (40).Ford DA, Hazen SL, Saffitz JE, Gross RW. J. Clin. Invest. 1991;88:331–335. doi: 10.1172/JCI115296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Mancuso DJ, Abendschein DR, Jenkins CM, Han XL, Saffitz JE, Schuessler RB, Gross RW. J. Biol. Chem. 2003;278:22231–22236. doi: 10.1074/jbc.C300033200. [DOI] [PubMed] [Google Scholar]
- (42).Petkovic M, Muller J, Muller M, Schiller J, Arnold K, Arnhold J. Anal. Biochem. 2002;308:61–70. doi: 10.1016/s0003-2697(02)00236-1. [DOI] [PubMed] [Google Scholar]
- (43).Han X, Gross RW. J. Am. Soc. Mass Spectrom. 1995;6:1202–1210. doi: 10.1016/1044-0305(95)00568-4. [DOI] [PubMed] [Google Scholar]
- (44).Garrett TJ, Yost RA. Anal. Chem. 2006;78:2465–2469. doi: 10.1021/ac0522761. [DOI] [PubMed] [Google Scholar]
- (45).Starkopf J, Andreasen TV, Bugge E, Ytrehus K. Cardiovasc. Res. 1998;37:66–75. doi: 10.1016/s0008-6363(97)00240-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.