Abstract
A photonic crystal (PC) surface is demonstrated as a high sensitivity platform for detection of a panel of twenty-one cancer biomarker antigens using a sandwich ELISA microarray format. A quartz-based PC structure fabricated by nanoimprint lithography, selected for its low autofluorescence, supports two independent optical resonances that simultaneously enable enhancement of fluorescence detection of biomarkers and label-free quantification of the density of antibody capture spots. A detection instrument is demonstrated that supports fluorescence and label-free imaging modalities, with the ability to optimize the fluorescence enhancement factor on a pixel-by-pixel basis throughout the microarray using an angle-scanning approach for the excitation laser that automatically compensates for variability in surface chemistry density and capture spot density. Measurements show that the angle-scanning illumination approach reduces the coefficient of variation of replicate assays by 20–99% compared to ordinary fluorescence microscopy, thus supporting reduction in limits of detectable biomarker concentration. Using the PC resonance, biomarkers in mixed samples were detectable at the lowest concentrations tested (2.1 – 41 pg/ml), resulting in a 3-log range of quantitative detection.
Introduction
Quantitation of cancer biomarkers in blood is expected to have substantial clinical impact for early detection, prognosis, and monitoring patients’ response to therapy.1–10 Among the techniques for detecting and measuring biomarkers, antibody microarrays have proven to be a powerful platform due to their capability for multiplexed detection, minimal reagent usage, and high sensitivity. Through the use of calibration standards, antibody microarrays provide highly quantitative measurements of analyte concentration.11, 12 Sandwich assays are used to increase the sensitivity and specificity of antibody microarrays through the use of a primary antibody to initially capture the analyte from serum, and a second primary antibody that is fluorophore-tagged that recognizes a separate epitope on the same analyte.13, 14 Several techniques have been successfully incorporated into assay protocols to amplify sandwich Enzyme-linked immunosorbent assay (ELISA) fluorescent output. These include chemical approaches (such as rolling circle amplification, tyramide amplification)14, 15 as well as electromagnetic amplification approaches that utilize special substrates to increase the electric field exposure of surface-bound fluorophores.16, 17
Recently, the optical resonances of photonic crystal (PC) surfaces have been demonstrated to provide substantial fluorescence excitation enhancement in addition to a 5–10x magnification of the emitted photon collection efficiency for surface-based fluorescent assays such as DNA microarrays and protein microarrays.18, 19 The optically active PC surface replaces the glass surface used as the assay solid support to reduce the limits of detection (LOD) and increase the signal-to-noise ratio (SNR), and thereby improve the ability to detect analytes that would otherwise fall below the detection threshold.17, 20–23 The PC is comprised of a periodically modulated sub-wavelength surface structure fabricated from a low refractive index material (such as plastic, glass, or quartz) coated with a high refractive index dielectric layer. For PC-enhanced fluorescence (PCEF), the surface is designed to perform as an optical resonator at the wavelengths of fluorescence excitation and fluorescence emission. At these two wavelengths, the PC supports optical standing waves that are confined in the high refractive index layer, but extend into the surrounding media as an evanescent electric field. The fluorescence amplification obtained through PCEF is attributed to two separate mechanisms that work independently of each other with multiplicative effects: enhanced excitation and enhanced extraction.17, 22, 23 Enhanced excitation is the result of the resonance at the wavelength of laser illumination, which results in amplified electric field intensity for surface-bound fluorophores, and thus greater photon output – an effect that is similar to providing illumination from a laser of greater power, but with the power confined only to the assay surface. Enhanced extraction uses the resonance at the fluorophore emission wavelength to efficiently direct photon output towards the detection instrument, resulting in increased collection efficiency. The simultaneous implementation of these two mechanisms has been demonstrated to enhance the fluorescence signal by three orders of magnitude20 as compared to the signal from an ordinary unpatterned glass substrate typically used as an assay surface using a detection instrument configured to optimize both effects simultaneously. In addition to providing a surface for fluorescence enhancement, detection of shifts in the resonant coupling conditions caused by attachment of biomolecules also allows a PC to serve as a platform for label-free detection.24, 25 A high resolution imaging system has been developed for PC surfaces to measure the density of DNA and protein microarray capture spots as a means for quality control,24 and the label-free image of microarray capture spots can be used as a template for identifying on-spot and off-spot regions for image processing of subsequent fluorescent images from PC surfaces.26
The initial demonstrations of PCEF utilized a PC surface that was produced from polymer materials using a molding process that can inexpensively produce PCs over large surface areas on flexible plastic substrates.21, 23 While this approach incorporated a SiO2 thin film buffer layer between the PC and the polymer to prevent resonant optical fields from extending into the polymer, generation of autofluorescence from the polymer materials provides a background intensity that is indistinguishable from fluorescent photons generated by the assay. The autofluorescence background intensity, if greater than the fluorescent intensity of labels in the assay, will serve to limit the detection of analytes at the lowest concentrations. Thus, we explored the design and fabrication of PC surfaces that could be fabricated from materials that exhibit little or no autofluorescence, such as quartz.20 The use of a quartz-based PC, in turn, enables us for the first time to use collimated (rather than focused) laser illumination to excite resonant modes of the PC. The use of collimated illumination is extremely important for PCEF, as the excitation wavelength/angle conditions can be tuned to precisely match the on-resonance coupling condition of the PC, and to resonantly couple nearly 100% of the incident light. In contrast, focused illumination, as typically provided by commercially available confocal microarray scanners, contains a wide range of incident angles and therefore only a fraction of all the excitation illumination can interact resonantly with the PC.27 Collimated illumination of the PC also allowed combination of label-free imaging and enhanced fluorescence imaging within the same detection instrument, enabling information from a label-free measurement of microarray capture spot density to be used directly to determine the optimal conditions for PCEF on a pixel-by-pixel basis over a large area.26
In this work, we demonstrate for the first time the combination of a quartz-based PC (for reduced autofluorescence) with a detection instrument designed to perform label-free (LF) and enhanced fluorescence (EF) imaging using collimated laser illumination. The EF/LF microscope is used to detect the output of a sandwich ELISA protein microarray with twenty-one breast cancer biomarker assays. The LF detection capability is used to quantify the density and variability of the immobilized capture antibodies, and to optimize the resonant coupling conditions throughout the microarray on a pixel-by-pixel basis. The incident angle scanning method reduces the coefficient of variation (CV) between replicate assays by 20–99% thus enabling more accurate quantification of analyte concentration and reduction of detection limits.
Materials and Methods
Reagents
(3-Glycidoxypropyl)trimethoxysilane (GPTS), phosphate buffered saline (PBS) powder, and Tween-20 were purchased from Sigma-Aldrich. Purified primary capture antibodies, antigens, and secondary detection antibodies were purchased from R&D Systems (Minneapolis, MN, USA), Lab Vision (Fremont, CA, USA), Fitzgerald (Concord, MA, USA), MBL International (Woburn, MA, USA), and BiosPacific (Emeryville, CA, USA).13 All detection antibodies were biotinylated by the supplier. Blocking solution containing 1% casein in PBS was purchased from Bio-Rad Laboratories. Streptavidin-conjugated Cy5 (SA-Cy5) was purchased from Amersham Bioscience.
Abbreviations of assays used in this work: amphiregulin (AMR); basic fibroblast growth factor (bFGF); cluster of differentiation 14 (CD14); epidermal growth factor (EGF); epidermal growth factor receptor (EGFR); E-selectin (Esel); heparin-binding epidermal growth factor (HBEGF); c-erbB-2 extracellular domain (Her2); hepatocyte growth factor (HGF); intracellular adhesion molecule 1 (ICAM); insulin-like growth factor 1 (IGF-1); matrix metalloprotease 1 (MMP1); matrix metalloprotease 2 (MMP2); matrix metalloprotease 9 (MMP9); platelet-derived growth factor AA (PDGF); prostate specific antigen (PSA); regulated on activation normal T cell expressed and secreted (RANTES); transforming growth factor alpha (TGFα); tumor necrosis factor alpha (TNFα); urokinase-type plasminogen activator receptor (uPAR); vascular endothelial growth factor (VEGF).
Quartz PC fabrication and characterization
The PC was fabricated by “step-and-flash” nanoimprint lithography (NIL)20 using a Molecular Imprints Imprio 50 system. In brief, the quartz substrate was spin coated with a layer of Transpin (Molecular Imprints Inc.) for planarizing the surface before the imprint. An imprint resist (MonoMat, Molecular Imprint Inc.) was dispensed onto the substrate and a template with a 8.75×8.75 mm2 of 1-D grating pattern was slowly pressed against the dispensed Monomat followed by a UV exposure. The template was then released from the MonoMat. The imprint process was repeated to create a 2×8 pattern on a quartz substrate as shown in Figure 1a. After the imprint, reactive ion etching was used to transfer the imprinted pattern into the quartz substrate. A dicing saw was used to cut the quartz substrate into 1×3 in2 standard microscope slides, in which the entire surface of the slide was covered with PC. Finally, a high refractive index layer of TiO2 (thickness = 130 nm, n = 2.35) was deposited on top of the imprinted grating structure by RF sputtering. The resulting grating pattern has a period of 400 nm with a depth of 40 nm and a duty cycle of ~50% as shown in Figure 1b.
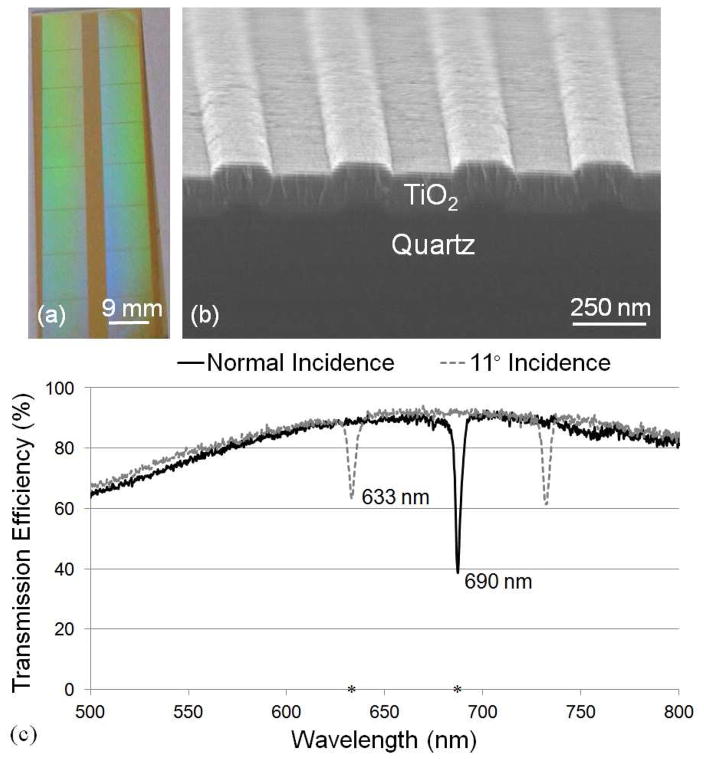
Figure 1.
(a) Photograph of a 1 × 3 in2 PC slide with an 8×2 pattern of 8.75 × 8.75 mm2 imprinted grating regions, (b) SEM image showing cross section of a PC and (c) transmission spectrum of both normal and 11° incidence. The resonance at λ=690 nm is used for label-free measurement and enhanced extraction of Cy5 emission. The excitation wavelength of λ=633 nm is used for enhance excitation of Cy5.
This device was designed to have two resonances at TM-polarization (electric field perpendicular to the grating structure) to support the EF and LF detection modalities through use of two laser illumination wavelengths. The resonance condition of the quartz PC can be observed by measuring the dip in the transmission spectrum when the PC is subjected to broadband illumination as shown in Figure 1c, where two transmission spectra are shown. When illuminated at normal incidence, the transmission dip is observed at λ~690 nm with a full width half maximum (FWHM) of Δλ = 3 nm. When light is incident at an angle of 11° there is a resonance at λ~633 nm with a FWHM of Δλ = 4 nm. The fiber-coupled broadband light source used in this experiment contains a moderate divergence; hence, the measured coupling efficiency is lower than that obtained via illumination by a collimated source, such as a laser.27 Laser coupling efficiency at the resonant angle/wavelength combination has been measured to be 90% (data not shown).
For the EF detection modality, the PC is designed specifically for the dye Cyanine-5 (Cy5) due to its strong quantum yield and high absorption efficiency at the λ=633 nm wavelength of HeNe lasers. The PC can be tuned to an on-resonance condition by illuminating its surface using a TM–polarized wavelength of λ=633 nm and corresponding incident angle of 11°. This illumination condition will result in an amplified near field electric field intensity on the PC surface for enhanced excitation. The large illumination angle is designed to prevent excitation light from coupling into the objective lens and subsequently reaching the detection system. Despite the use of an excitation filter (OD 7) to block the photons at the laser wavelength from reaching the imaging CCD, this phenomenon, if not accounted for, results in an elevated “background” signal that can be greater than the desired fluorescence signal. The PC exhibits a second resonance at λ = 690 nm, which spectrally overlaps with the emission spectra of Cy5. This resonance is used to more efficiently direct emitted photons towards the detector to obtain enhanced extraction.28 The resonance at λ = 690 nm at normal incidence angle is also used for the LF detection modality. Deposition of capture antibodies on the PC leads to a localized increase in the effective refractive index of the PC resulting in a shift in the resonance angle, where the shift in the angle of minimum transmission (AMT) efficiency is proportional to the density of the deposited antibody.
Microarray preparation and assay protocol
The microarray preparation and sandwich assay format has been described in detail previously.13, 19, 29 In brief, to bind the antibodies to the PC surface, a self-assembled monolayer is applied by a vapor-phase technique to form a monolayer of 3-glycidoxypropyl-trimethoxysilane (GPTS). Capture antibodies were diluted in PBS to a concentration of 0.8 mg/ml, and 4 replicate spots per assay were printed in each array on a quartz PC slide using a noncontact NanoPlotter NP2 printer (GeSiM, Germany). The resulting spot size is approximately 150 μm, which covers ~375 periods of the PC. Following the printing, the slide was incubated overnight at room temperature and 60% humidity. The slide was then blocked in 1% casein in phosphate buffered saline (PBS) solution. After washing in PBS with 0.05% Tween (PBS-T), the slide was incubated with a mixture of antigens and 0.1% casein in PBS with gentle agitation overnight. Dose-response curves were generated using three-fold dilution series of the antigen mixture for a total of seven concentrations, in addition to a blank (only dilution buffer, 0.1% casein in PBS). The blank spots are used as negative controls where the fluorescence intensities indicate the nonspecific binding. All the assays and the protocol used in this work have been previously verified and optimized to have minimum nonspecific binding between assays.13 The slide was then washed in PBS-T, followed by incubation with a mixture of biotinylated detection antibodies at 25 ng/ml in PBS-T with mild agitation. The slide was next washed with PBS-T and incubated in a solution of 1 μg/ml SA-Cy5 in PBS-T. Finally, the slides were washed and dried before fluorescence and LF measurements.
Fluorescence and label-free measurements
In order to measure both LF and EF intensities, the integrated EF/LF microscope system shown in Figure 2 was used. A detailed description of the instrument has been presented previously.25 In brief, unlike an imaging system that uses a focused beam to excite fluorophores, the instrument used in this work provides collimated laser illumination to provide efficient coupling to the PC. Rather than scanning the PC surface, the system captures an image of an entire field of view at once. The system is equipped with a 35 mW HeNe laser, (λ = 632.8 nm) for the EF modality, and a 50 mW AlGaAs semiconductor diode laser (λ = 690 nm) for the LF modality. Both excitation sources were TM polarized with respect to the PC grating lines. A beam expander produces an illuminated diameter of 4 mm with a divergence of 0.037°. A rotating diffuser is used to reduce speckle and fringes at the imaging plane by randomizing the spatial coherence of the laser. The incident angle was precisely controlled by a high precision angle-tuning gimbal-mounted mirror with a resolution of 0.005° incident angle increments. The sample was imaged by an electron multiplying charge coupled device (Hammamatsu, Japan) through a 4x microscope objective (N.A = 0.1). A bandpass filter (Semrock, λ = 690, Δλ = 20 nm) that overlaps with the emission bandwidth of the Cy5 was used to prevent photons at the excitation laser wavelength from reaching the CCD during fluorescence imaging.
Figure 2.
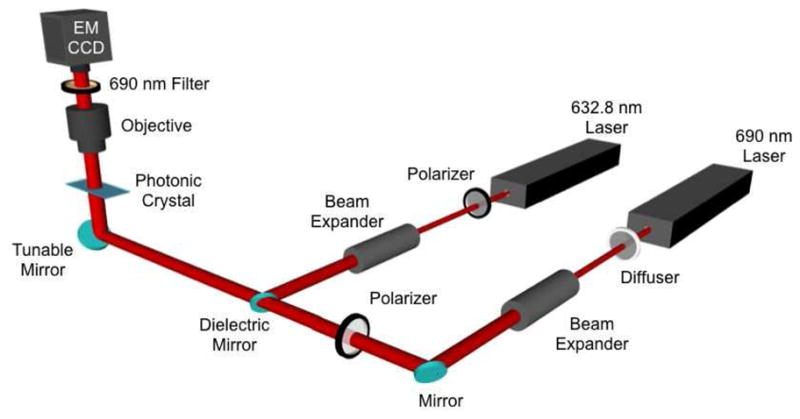
Schematic diagram of the instrument used for EF and LF measurement. The λ=632.8 laser is used for fluorescence excitation, while the λ=690 nm laser is used for label-free imaging. The system incorporates a computer-controlled tunable mirror that is capable of scanning the incident angle through a range of angles. A 4x objective images a 2×2 mm2 field of view of the PC. The CCD measures transmitted laser intensity as a function of incident angle at λ=690 nm for LF detection, and measures fluorescence emission intensity from the PC for EF detection.
For protein microarrays, the LOD is effected by the uniformity of spot intensities from replicate spots present across an array. For a fixed incident wavelength of λ = 633 nm, the PC resonant coupling condition has a full width half maximum in angle (FWHMθ) Δθ ~ 0.4°. While a narrow resonant coupling condition has been shown to provide the greatest fluorescence excitation enhancement factor, the stringent conditions for optimal laser-PC coupling means that a small deviation of Δθ = 0.4° from the true device resonant angle would result in a 50% reduction in fluorescence intensity.27 Therefore, a uniform enhancement effect over the entire array is critical. Variations in the optimal PC resonant coupling angle across an array can originate from the nonuniformity of the device during the fabrication process, nonuniformities in the surface chemistry layer, and nonuniformity in the density of antibody capture spots – both between spots and within an spot. In order to retain high signal amplification while achieving uniform signal enhancement across the whole microarray slide, an angle-scanning method that accounts for variations in the resonant angle across the slide was developed.27 Rather than gathering a single fluorescence image using only one incident angle, a sequence of fluorescence images are captured over a range of angles that includes the resonant angle. Image processing software compares the set of images taken at each angle, and selects the maximum intensity for each pixel. The maximum intensity for each pixel corresponds to the incident angle that matches the optimal resonant coupling condition. A composite fluorescence image is generated in which each pixel holds the maximum observed intensity value.
To obtain LF measurements of the printed antibodies, the λ=690 nm laser was used to illuminate the PC resonance over a range of incident angles near normal incidence. The angle of minimum transmission (AMT) is determined by software on a pixel-by-pixel basis, where shifts in the AMT correspond to locations on the PC with greater immobilized antibody density. Before immobilizing capture antibodies, the PC resonance is designed to occur at λ=690 nm (when illuminated at normal incidence), resulting in a minimum transmission as shown in Figure 1c. Using this technique, a high resolution spatial map of adsorbed captured antibody densities was generated as a function of position on the PC surface.
Data Analysis
Spot intensities of the LF images were analyzed using GenePix Pro 6.1 software. The locations of the spots from the LF image were overlaid with the EF image to quantify the spot fluorescence intensities. ProMAT, a software package developed by Pacific Northwest National Laboratory specifically for the analysis of microarray data, was used to generate concentration-response curves based on a four-parameter logistic model.11, 12 The LOD was also calculated by ProMAT, which is freely available at www.pnl.gov/statistics/ProMAT.
Results and Discussion
Label-free measurement of spot densities
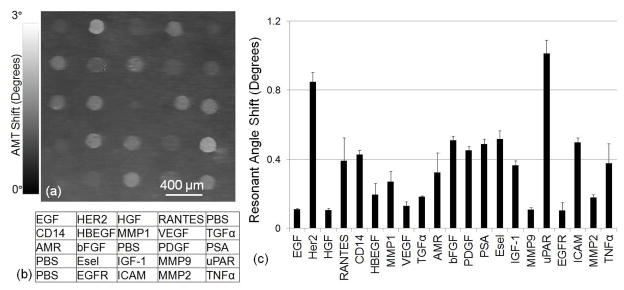
When antibodies are deposited onto a solid surface, they exhibit distinct substratum-binding characteristics due to antibody-specific diversity that results in binding density variability. The LF imaging modality of the detection instrument can be used to characterize the antibody capture spot density on the PC surface. As shown in Figure 1c, the PC exhibits a resonant reflection at λ = 690 nm near normal incidence. The dielectric permittivity of surface biomolecules results in an increase in the resonant coupling angle that is proportional to the adsorbed mass area density, and the coupling angle increase is spatially localized to the regions of the PC surface where the biomolecule attachment occurs. Using computer control, the angle scanning mirror of the detection instrument (Figure 2) can be rapidly scanned through a range of angles in small increments (0°< θ <3° with Δθ = 0.01°), and an image of the transmitted intensity can be gathered at each incident angle using the CCD imager. For each pixel, the AMT is determined by mathematically finding the minimum of the plot of transmitted intensity as a function of θ, to generate a spatial map of AMT versus position on the PC surface with pixel resolution of 16 μm. To account for any nonuniformity due to the PC fabrication or surface chemistry, two AMT images are gathered. An initial AMT image of the PC surface is obtained prior to deposition of antibody capture spots, and a second AMT is gathered after spot deposition. The two images are aligned and mathematically subtracted (after spots – before spots) to produce a spatial map of the AMT shift due only to the deposited capture antibodies. An AMT shift image of one set of immobilized capture antibody spots is shown in Figure 3a using the layout shown in Figure 3b. The average net AMT shift of four replicates for each antibody is plotted in Figure 3c, which depicts the variability of binding density obtained from the antibodies used in our array, despite the use of identical antibody concentrations, buffers, spotting conditions, and incubation conditions. After the deposition of antibodies, the AMT shifts roughly from 0.1° (for EGF and HGF) to 1° (for uPAR). This wide range of resonant coupling conditions suggests that, during the subsequent enhanced fluorescent imaging (that will be performed after exposing the array to analyte, secondary antibody, and Cy5/streptavidin labels) using a fixed incident angle to illuminate the entire array could not be expected to provide uniform fluorescence enhancement for every assay. In this work, the LF imaging of microarray capture spots was mainly used to quantify the resonance angle shifts caused by the printed antibody capture spots. The LF detection modality can also be used to identify the presence of missing capture spots, spots with nonuniform antibody density (either with heavy density in the spot center or around the spot periphery), or other spatial features that would render a capture spot useless for further analysis.24 Through identification of spot deposition errors, specific spots can be flagged for exclusion from consideration in assay replicate statistics, thus providing a valid means for reducing assay CV and LOD with information that is normally absent when using an ordinary glass surface.
Figure 3.
Label-free characterization of the density of antibody capture spots by measuring shifts in the λ=690 nm resonance. (a) Image of the shift in the Angle of Minimum Transmission (AMT) for a representative array of antibody capture spots, representing a label-free measurement of the capture spot density. (b) Layout of the capture antibodies within an array. (c) Average net AMT shift values for the capture antibodies with error bars representing one standard deviation of from four replicate arrays on the same chip. Measurements indicate a large variability in capture spot density that is dependent on the specific antibody, but excellent reproducibility for a particular antibody.
Angle scanning to achieve uniform enhancement
As discussed previously, the PC enhanced excitation effect is highly sensitive to the resonant angle for collimated illumination. For the PC used here, FWHMθ < 0.4°. Therefore, a small deviation in illumination angle from the actual resonant angle results in a significant drop in the signal enhancement. As indicated in the LF measurement, upon deposition of antibodies, the resonance angle can shift from 0.1°< Δθ <1°, depending on the density of the capture antibodies, thus it is necessary to adjust the incident angle of fluorescent illumination to achieve the maximum enhancement for each spot. Rather than using a single fixed angle to scan the entire array, a series of 300 fluorescent images were captured for incident angle range of 10.5°< θ < 12.5° at increment of Δθ = 0.01°. The 4× objective used in EF/LF microscopy system yields a 2×2 mm2 field of view which covers one of the replicates shown in Figure 4a. For the angle-scanning method, it takes 9 seconds to capture 300 images to generate a final composite image for one replicate. There are four replicates per concentration and eight concentrations for a slide, so it takes less than 5 min to capture all the images for a single slide. It takes less than 6 min to scan a whole slide including the time for stage movement.
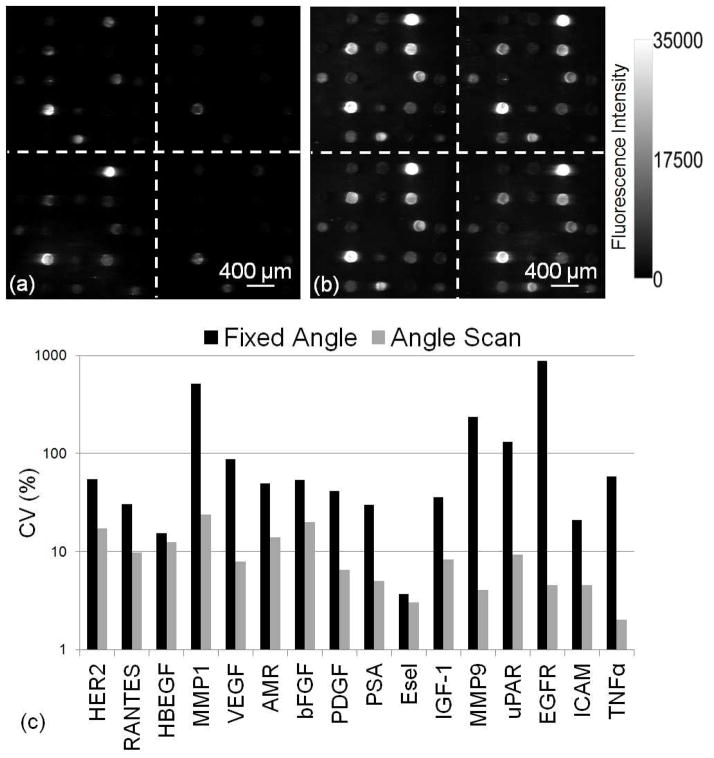
Figure 4.
(a) Fluorescence images from both (a) fixed angle method and (b) angle-scanning method from the third highest concentration. (c) Bar chart of CV comparison between two scanning methods shows that angle-scanning method greatly improves the uniformity of spot intensities.
The maximum intensity values for each pixel are used to generate a composite fluorescent image in which each pixel is represented at its optimal on-resonance coupling condition. For comparison, a single fixed angle intentionally selected at an off-resonance condition (θ = 12.2°) for an array is shown in Figure 4a, while the angle-scanning approach was used to generate the fluorescence image shown in Figure 4b. GenePix Pro 6.1 software was used to quantify the median spot intensity. The CV were calculated as the standard deviation divided by the average of these median spot intensities of four replicate spots for each assay from the third highest concentration well. The CV for each detection approach is compared in Figure 4c, in which the angle-scanning method improves the CV by up to 99%. We observe that assays with the greatest antibody capture spot density, as measured by the AMT shift image, do not correlate to higher fluorescence intensities for detection of biomarkers. We also observe that variability in the capture spot density does not correlate strongly with the CV of fluorescence signals (correlation coefficient ~0.5). Due to variability in capture molecule/analyte affinity and capture molecule activity, we do not observe a strong correlation between the fluorescent signal and the LF signal, therefore we do not apply a correction factor to the fluorescent measurement that compensates for capture antibody variability, so that, higher antibody density does not necessarily result in greater sensitivity for detection of the corresponding biomarker.
Standard curves and limit of detection
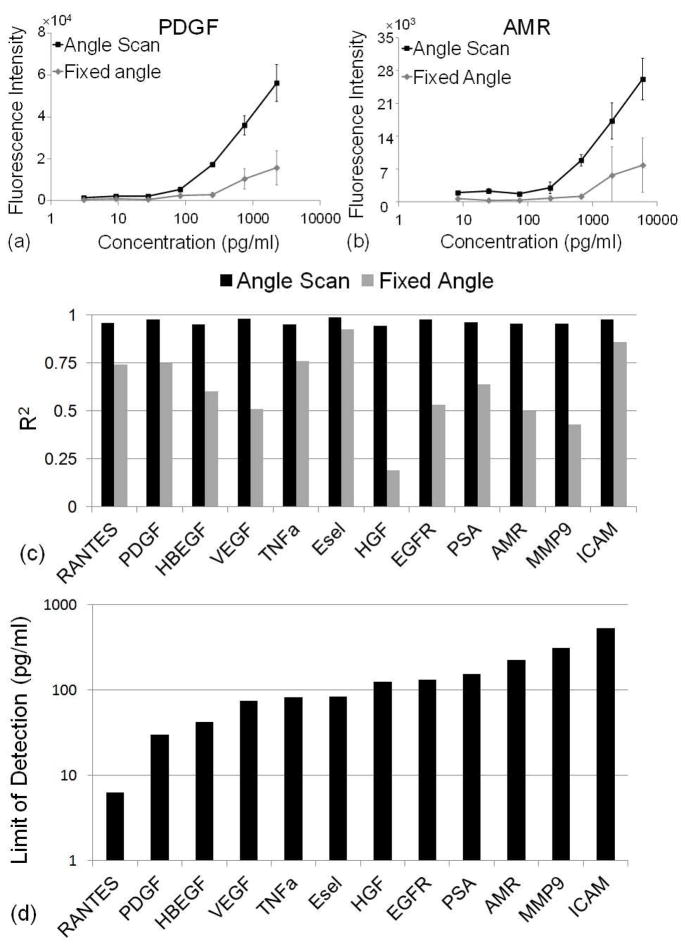
The fluorescence intensity as a function of biomarker concentration was used to generate standard curves for each assay. Standard curves for two exemplary assays (PDGF and AMR) are shown in Figures 5a-b, in which the angle-scanning and fixed angle (θ = 12.2°) methods are compared. These results highlight the effectiveness of the PC surface for enhancing the fluorescence output when illuminated in the on-resonance condition, when compared to off-resonance illumination for the fixed angle case. The plots also show that the angle-scanning approach provides a uniform enhancement effect, as evident by the low spot-to-spot CV for four replicate spots for each concentration. The coefficient of determination (R6) values of a 4-parameter logistic curve fitting algorithm for PDGF is R6 = 0.98 and R6 = 0.96 for AMR using the angle-scanning method. However, using the fixed angle method, the R6 values for these assays significantly drops to R6 = 0.75 for PDGF and R6 = 0.5 for AMR. Similar results are obtained for all the assays on the chip as shown in Figure 5c.
Figure 5.
Dose-response curves for (a) PDGF and (b) AMR where black and gray curves are from angle-scanning and fixed angle scanning method respectively. Error bars represent one standard deviation from four replicate spots. (c) Comparison of R6 for the 4-parameters logistic curve fitting algorithm for both angle scanning and fixed angle methods. (d) LODs based on the angle scanning method. Only assays with R6 > 0.95 from four parameter logistic curve fitting were listed here.
Detection of weak signals above background fluorescence (generated from the combination of autofluorescence from the substrate and nonspecific binding) is especially important for detection of low concentration biomolecules.20, 21, 30 For an enhanced fluorescence substrate such as a PC, any autofluorescence from the substrate can be enhanced in parallel with fluorophores tagged to analyte molecules. Due to the low autofluorescence of the quartz substrate and high spot fluorescence enhancement, all the spot signals are higher than the background signals. This indicates that by using a quartz substrate, the sensitivity of the microarray is limited by the nonspecific binding by the various protein reagents and not the substrate itself.
The limit of detection (LOD) is highly dependent on the affinity of the capture and detection antibodies for the biomarker, therefore a range of LODs is typically obtained for a multi-analyte array.31 As our array is being developed for early breast cancer screening, a wide range of potential biomarker proteins from a variety of protein families are included. These include assays for cytokines, proteases, growth factors, receptors, and adhesion proteins – each with different affinities between the capture antibodies and their corresponding antigens.
The LOD is calculated as the concentration corresponding to the mean of blank control spot intensities plus three standard deviations of the log-transformed fluorescence intensities from all concentration levels. Using the angle-scanning method for fluorescence detection, the LODs shown in Figure 5d were obtained, with values ranging between 6.3 and 500 pg/ml. Only assays with R6 > 0.95 from the four parameter logistic curve fitting were included here. Due to inter-spot variability, the fixed angle scanning method resulted in poor curve fitting, (R6 < 0.95).
Several assays are not included in Figure 5c–d. These either exhibit poor 4-parameter curve fitting (R6 < 0.95) or do not display sigmoidal dose-response curve behavior. For example, EGF generates fluorescence output that is not concentration dependent, indicating loss of function of either its capture antibody or detection antibody.
Conclusion
In this work, we demonstrate a novel optically active PC surface and detection method that can substantially augment protein biomarker microarray analysis in two distinct ways. For the first time, a quartz-based PC fabricated by nanoimprint lithography was demonstrated as a surface that can simultaneously support enhanced fluorescence and label-free measurement using a detection instrument that can provide collimated laser illumination that is scanned over a range of incident angles. The quartz-based PC structure was chosen specifically for its low autofluorescence properties relative to previously demonstrated PCEF surfaces that were fabricated from polymer. The device structure is designed to produce two distinct and narrow resonant modes that enable two complementary imaging modalities to be performed in the same detection instrument. The label-free detection modality is used to quantify and visualize binding density variability of immobilized capture antibodies as a means for providing quality control and spot-quality screening. We introduced an angle-scanning fluorescence imaging approach for PCEF that automatically corrects for variability in optimal resonant coupling conditions that originates from the observed variability in antibody capture spot density. The angle-scanning approach was demonstrated as an effective means for solving a critical problem, if PCEF is to be effectively applied to protein microarrays. The ability of the angle-scanning method to provide highly uniform fluorescent enhancement throughout an array was demonstrated through a substantial reduction in fluorescent intensity CV, as compared to measuring the same array with off-resonance illumination at a single angle. The combination of a quartz-based PC with a custom detection instrument that can optimally couple laser illumination into multiple PC resonances can be used as a platform for a wide variety of surface-based fluorescence assays that would benefit from reduction of detection limits and the ability to measure the density of capture molecules.
Acknowledgments
This work was supported by the National Institutes of Health (Grant No. GM086382A), and the National Science Foundation (Grant No. CBET 07-54122). Any opinions, findings, conclusions, or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Institutes of Health or the National Science Foundation.
References
- 1.Kumar S, Mohan A, Guleria R. Biomarkers. 2006;11:385–405. doi: 10.1080/13547500600775011. [DOI] [PubMed] [Google Scholar]
- 2.Levenson VV. Biochimica Et Biophysica Acta-General Subjects. 2007;1770(6):847–856. doi: 10.1016/j.bbagen.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Duffy MJ, Duggan C, Keane R, Hill ADK, McDermott E, Crown J, O’Higgins N. Clin Chem. 2004;50:559–563. doi: 10.1373/clinchem.2003.025288. [DOI] [PubMed] [Google Scholar]
- 4.Ebeling FG, Stieber P, Untch M, Nagel D, Konecny GE, Schmitt UM, Fateh-Moghadam A, Seidel D. Br J Cancer. 2002;86:1217–1222. doi: 10.1038/sj.bjc.6600248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumpulainen EJ, Keskikuru RJ, Johansson RT. Breast Cancer Res Treat. 2002;76:95–102. doi: 10.1023/a:1020514925143. [DOI] [PubMed] [Google Scholar]
- 6.Molina R, Filella X, Alicarte J, Zanon G, Pahisa J, Munoz M, Farrus B, Ballesta AM. Anticancer Res. 2003;23:1035–1041. [PubMed] [Google Scholar]
- 7.Cheung KL, Graves CRL, Robertson JFR. Cancer Treat Rev. 2000;26:2–102. doi: 10.1053/ctrv.1999.0151. [DOI] [PubMed] [Google Scholar]
- 8.Nicolini A, Carpi A. Tumor Biol. 2000;21:235–248. doi: 10.1159/000030129. [DOI] [PubMed] [Google Scholar]
- 9.Robertson JFR, Jaeger W, Syzmendera JJ, Selby C, Coleman R, Howell A, Winstanley J, Jonssen PE, Bombardieri E, Sainsbury JRC, Gronberg H, Kumpulainen E, Blamey RW Eropean Grp Serum Tumour Markers Breast C. Eur J Cancer. 1999;35:47–53. doi: 10.1016/s0959-8049(98)00297-4. [DOI] [PubMed] [Google Scholar]
- 10.VanDalen A, Heering KJ, Barak V, Peretz T, Cremaschi A, Geroni P, Gion M, Saracchini S, Molina R, Namer M, Stieber P, Sturgeon C, Leonard RCF, Einarsson R. Breast. 1996;5:82–88. [Google Scholar]
- 11.Daly DS, White AM, Varnum SM, Anderson KK, Zangar RC. Bmc Bioinf. 2005;6:Article No.: 17. doi: 10.1186/1471-2105-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White AM, Daly DS, Varnum SM, Anderson KK, Bollinger N, Zangar RC. Bioinformatics. 2006;22:1278–1279. doi: 10.1093/bioinformatics/btl093. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez RM, Seurynck-Servoss SL, Crowley SA, Brown M, Omenn GS, Hayes DF, Zangar RC. J Proteome Res. 2008;7:2406–2414. doi: 10.1021/pr700822t. [DOI] [PubMed] [Google Scholar]
- 14.Woodbury RL, Varnum SM, Zangar RC. J Proteome Res. 2002;1:233–237. doi: 10.1021/pr025506q. [DOI] [PubMed] [Google Scholar]
- 15.Schweitzer B, Roberts S, Grimwade B, Shao WP, Wang MJ, Fu Q, Shu QP, Laroche I, Zhou ZM, Tchernev VT, Christiansen J, Velleca M, Kingsmore SF. Nat Biotechnol. 2002;20:359–365. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaguchi T, Kaya T, Takei H. Anal Chem. 2007;364:171–179. doi: 10.1016/j.ab.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 17.Ganesh N, Block ID, Mathias PC, Zhang W, Chow E, Malyarchuk V, Cunningham BT. Opt Express. 2008;16:21626–21640. doi: 10.1364/oe.16.021626. [DOI] [PubMed] [Google Scholar]
- 18.Mathias PC, Jones SI, Wu HY, Yang F, Ganesh N, Gonzalez DO, Bollero G, Vodkin LO, Cunningham BT. Anal Chem. 2010;82:6854–6861. doi: 10.1021/ac100841d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang CS, George S, Lu M, Chaudhery V, Tan RM, Zangar RC, Cunningham BT. Anal Chem. 2011;83:1425–1430. doi: 10.1021/ac102989n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pokhriyal A, Lu M, Chaudhery V, Huang CS, Schulz S, Cunningham BT. Opt Express. 2010;18:24793–24808. doi: 10.1364/OE.18.024793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathias PC, Ganesh N, Cunningham BT. Anal Chem. 2008;80:9013–9020. doi: 10.1021/ac801377k. [DOI] [PubMed] [Google Scholar]
- 22.Ganesh N, Zhang W, Mathias PC, Chow E, Soares J, Malyarchuk V, Smith AD, Cunningham BT. Nat Nanotechnol. 2007;2:515–520. doi: 10.1038/nnano.2007.216. [DOI] [PubMed] [Google Scholar]
- 23.Mathias PC, Wu HY, Cunningham BT. Appl Phys Lett. 2009;95:021111–3. doi: 10.1063/1.3184573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George S, Block ID, Jones SI, Mathias PC, Chaudhery V, Vuttipittayamongkol P, Wu HY, Vodkin LO, Cunningham BT. Anal Chem. 2010;82:8551–8557. doi: 10.1021/ac101551c. [DOI] [PubMed] [Google Scholar]
- 25.Block ID, Mathias PC, Ganesh N, Jones SI, Dorvel BR, Chaudhery V, Vodkin LO, Bashir R, Cunningham BT. Opt Express. 2009;17:13222–13235. doi: 10.1364/oe.17.013222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaudhery V, Lu M, Huang C-S, Pokhriyal A, Polans J, Schulz SC, Cunningham BT. Opt Express. 2011 doi: 10.1364/OE.19.023327. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhery V, Lu M, Schulz SC, Cunningham BT. IEEE Sens J. 2011 in press. [Google Scholar]
- 28.Yang F, Cunningham BT. Opt Express. 2011;19:3908–3918. doi: 10.1364/OE.19.003908. [DOI] [PubMed] [Google Scholar]
- 29.Dorvel B, Reddy B, Block I, Mathias P, Clare SE, Cunningham B, Bergstrom DE, Bashir R. Adv Funct Mater. 2010;20:87–95. [Google Scholar]
- 30.Brown PO, Botstein D. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 31.Varnum SM, Woodbury RL, Zangar RC. Methods Mol Biol. 2004;264:161–172. doi: 10.1385/1-59259-759-9:161. [DOI] [PubMed] [Google Scholar]