Abstract
The biogenesis of ribosomes is a fundamental cellular process, which provides the molecular machines that synthesize all cellular proteins. The assembly of eukaryotic ribosomes is a highly complex multi-step process that requires more than 200 ribosome biogenesis factors, which mediate a broad spectrum of maturation reactions. The participation of many energy-consuming enzymes (e.g. AAA-type ATPases, RNA helicases, and GTPases) in this process indicates that the expenditure of energy is required to drive ribosome assembly. While the precise function of many of these enzymes remains elusive, recent progress has revealed that the three AAA-type ATPases involved in 60S subunit biogenesis are specifically dedicated to the release and recycling of distinct biogenesis factors. In this review, we will highlight how the molecular power of yeast Drg1, Rix7, and Rea1 is harnessed to promote the release of their substrate proteins from evolving pre-60S particles and, where appropriate, discuss possible catalytic mechanisms. This article is part of a Special Issue entitled: AAA ATPases: structure and function.
Keywords: Ribosome assembly, AAA-ATPase, Drg1/Afg2, Rix7/NVL, Rea1/Mdn1/Midasin
Highlights
► Structural and functional properties of AAA-ATPases in ribosome biogenesis are summarized. ► The AAA-ATPases Rea1, Rix7 and Drg1 are essential for ribosome biogenesis in yeast. ► Rix7 and Drg1 are related to Cdc48, while Rea1 shares similarity to motor protein dynein. ► Rea1, Rix7 and Drg1 promote the release of biogenesis factors from nucleolar, nucleoplasmic and cytoplasmic pre-60S intermediates. ► The release of maturation factors by AAA-ATPases is critical for downstream maturation of pre-60S particles.
1. Introduction
Ribosomes are the molecular machines that translate the genetic information contained within messenger RNAs into proteins. While there is a detailed understanding of eukaryotic ribosomes at both a structural and functional level [1–3], much less is known about the intricate assembly process of the ribosomal subunits. The biogenesis of ribosomes is initiated in the nucleolus by the transcription of a common large precursor of mature ribosomal RNAs. The nascent precursor rRNA (pre-rRNA), which undergoes snoRNP-mediated modification of nucleotides, assembles with some ribosomal proteins and early biogenesis factors to form the first pre-ribosomal particles. Concomitant to or shortly after completion of transcription, the pre-rRNA undergoes endonucleolytic cleavages that separate the precursor particles to the mature 40S and 60S ribosomal subunits. These pre-40S and pre-60S particles mature further in the nucleolus and nucleoplasm before being exported to the cytoplasm, where final maturation events yield the translation-competent ribosomal subunits [4–10]. Proteomic analyses have identified many distinct pre-ribosomal particles that can be chronologically ordered along the maturation pathway from the nucleolus to the cytoplasm (Fig. 1). These landmark pre-ribosomal particles significantly differ in protein and (pre-)rRNA composition, thus highlighting the remarkable dynamics and complexity of shaping the rRNA and its associated ribosomal proteins into the correct structure.
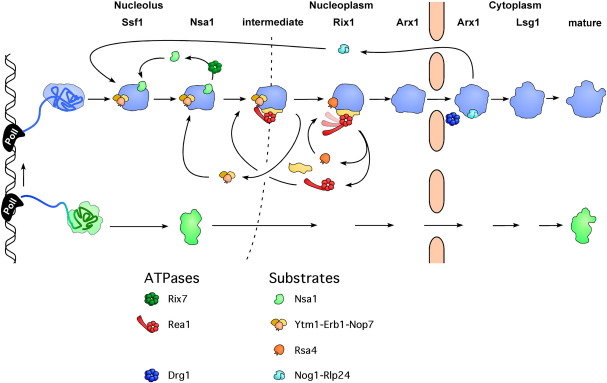
Fig. 1.
Biogenesis of the large (blue) and small (green) ribosomal subunit and the contribution of AAA-ATPases. Rix7, Rea1, and Drg1 are dedicated to the release and recycling of distinct biogenesis factors from different pre-60S particles. Major landmark of pre-60S ribosomes are depicted together with their corresponding bait proteins. These factors, namely Ssf1, Nsa1, Rix1, Arx1, and Lsg1, are only associated during a short time window with pre-60S particles and thus co-purify rather distinct pre-60S ribosomes. Ssf1 and Nsa1 purify nucleolar particles, whereas Rix1 purifies a nucleoplasmic intermediate. Arx1 represents an export-competent particle that carries export factors. Finally, Lsg1 is associated with an almost mature, cytoplasmic 60S particle. The three AAA-ATPases and their potential substrates are indicated.
The combination of genetic, cell biological, and proteomic methods has revealed that more than 200, mostly essential, non-ribosomal factors (also called biogenesis factors or protein trans-acting factors) contribute to eukaryotic ribosome biogenesis. While some of these factors are directly involved in the modification and processing of the pre-rRNA, others stabilize the pre-ribosomal particles, promote formation of productive RNA folding intermediates, or act as placeholders for selected ribosomal proteins that are recruited at a later time point in biogenesis. A further set of factors are essential for, or facilitate, the export of pre-ribosomes through the nuclear pore complex (NPC). A prominent number of biogenesis factors belong to different classes of energy-consuming enzymes, including ATP-dependent RNA helicases, GTPases, protein kinases, and three AAA-type ATPases [4,7]. Nucleotide binding or hydrolysis by these enzymes is believed to be instrumental for the promotion or regulation of key biogenesis steps, thus conferring directionality and accuracy to the assembly process. For recent reviews on non-ribosomal factors and their functions during ribosome biogenesis, see [4,5,7].
In this review, we focus on the molecular roles of the AAA-ATPases (ATPases associated with various cellular activities) involved in ribosome biogenesis. To date, a role in this process could be attributed to three essential AAA-ATPases, namely Rix7 (ribosome export), Rea1/Mdn1 (ribosome export associated/midasin), and Drg1/Afg2 (diazaborine resistance gene/ATPase family gene), which act at distinct steps during 60S subunit biogenesis in the yeast Saccharomyces cerevisiae. AAA-ATPases contain at least one structurally conserved ATPase module that assembles into functionally active ring structures. In addition to the conserved Walker A and Walker B motifs, they are characterized by class-specific elements that also contribute to ATP hydrolysis, such as the sensor-I, sensor-II and the arginine finger [11]. Within these molecular machines, cycles of nucleotide binding and hydrolysis induce conformational changes that affect a broad range of substrate proteins [11,12].
While the AAA-ATPases Rix7 and Drg1 are closely related to the well-characterized Cdc48 (p97 in mammals), Rea1, which is the largest yeast protein, shares similarity to the microtubule motor protein dynein heavy chain (Dyn1). Interestingly, all three AAA-ATPases promote the release of distinct biogenesis factors from nucleolar (Nsa1 by Rix7), nucleolar and nucleoplasmic (Ytm1-Erb1-Nop7 and Rsa4 by Rea1) and cytoplasmic (several shuttling factors by Drg1) pre-60S intermediates (Fig. 1) [13–16]. The release of these factors from pre-60S particles ensures their recycling and likely triggers conformational changes that are critical determinants for the progression of ribosome assembly, e.g. promoting export or subunit-joining competence.
2. The type II AAA-ATPases Drg1 and Rix7 are closely related to Cdc48/p97
Drg1 and Rix7 are essential, eukaryote-specific AAA-ATPases that belong to the classical AAA clade [12,17–19]. Proteins of the classical clade are structurally defined by the presence of a short additional α-helix between β-strand 2 and α-helix 2 within their AAA domains (see Fig. S1). The high sequence similarity of their AAA domains clearly separates this classical clade from the other AAA+ clades; i.e. (i) the generally conserved sensor-II arginine is replaced by an alanine, (ii) there is a short insertion within the arginine finger region leading to the occurrence of two conserved arginines (Fig. S1) [12,17]. Analysis of the improved structures of p97 suggests that the first of the two arginines, and not the conventional arginine, may be involved in catalysis [20].
Due to the strong sequence similarity of Drg1 and Rix7 to Cdc48/p97/VCP – they are in fact the closest relatives of Cdc48 – Drg1 and Rix7 can be further classified as members of the NSF/Cdc48/Pex family of AAA proteins [12]. These type II AAA-ATPases [21] notably contain two consecutive AAA domains (designated D1 and D2) that are preceded by a specific N-terminal domain, which is generally involved in substrate recognition (see below, Fig. 2). While the sequence conservation between Rix7 and Cdc48 is restricted to the two AAA domains, the similarity between Drg1 and Cdc48 extends to the N-terminal domain (see below, Fig. S2). Secondary and tertiary structure prediction suggests that the N-terminal domain of Drg1, like the N-terminal extensions of p97, NSF, PEX1, and the archaeal VCP-like AAA-ATPase VAT [22–28], folds into two sub-domains: a double-psi (ψ) beta (β) barrel Nn-domain and a four-stranded β-barrel Nc-domain (see Fig. S2).
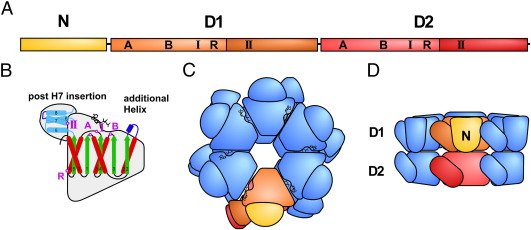
Fig. 2.
Schematic domain organization of Drg1 and Rix7. A, Linear representation of a protein monomer with the N-terminal, the D1 and the D2 domain depicted in yellow, orange and red, respectively. Note that the α-helical lid domains of D1 and D2 are shown in the respective dark orange and dark red color. The Walker A (A), Walker B (B) motifs, sensor-I (I) and II (II) and the arginine finger (R) of each domain are indicated. B, Representation of the secondary-structure element distribution within each ATPase domain with the α/β domain depicted in green (β-strands) and red (α-helices) and the α-helical elements of the α-helical lid domain depicted in light blue. The classic clade specific α-helix insertion is shown in dark blue. C and D, Two orthogonal views showing the hexameric organization of these AAA-ATPases. One protomer with its sub-domains is highlighted in the same colors as in A.
Intriguingly, the N-terminal domain of Rix7 differs structurally from the double-ψ β barrel/four-stranded β-barrel sub-domain organization that is prevalent among NSF/Cdc48/Pex family members. Secondary structure predictions suggest a primarily α-helical organization that is followed by a short linker region and a bipartite nuclear localization signal (NLS) (see Fig. S3A and B). A recently released NMR structure reveals that the first 74 amino acids of the Rix7 mouse orthologue NVL (nuclear VCP-like protein) fold into a three-helix bundle [29]. Even though the N-terminal domain of different Rix7 family members is only weakly conserved at the primary sequence level (Fig. S3), multiple sequence alignments together with secondary and tertiary structure prediction suggest that the first part of the N-terminal domain (N1) of Rix7/NVL proteins may generally fold into such a three-helix bundle. The second N-terminal region (N2) is predicted to contain one highly conserved α-helix, which indicates an overall conservation of the N1/N2 region among all eukaryotic species (see Fig. S3). In contrast to fungal Rix7, higher-eukaryotic orthologues, like NVL, contain an insertion (N3) between the end of the N2 region and the bipartite NLS (Fig. S3A).
Sequence alignments of the AAA domains of Cdc48, Drg1 and Rix7 reveal a few obvious differences between these proteins (Fig. S1): (i) Cdc48-family members notably contain a C-terminal extension to the D2 domain that mediates the interaction between Cdc48/p97 and certain substrate-processing cofactors [30–33]. (ii) Rix7-family members contain an insertion of around 40–50 amino acids after α-helix 7 within the D1 domain. Preliminary deletion analysis indicates that this insertion segment, despite its poor sequence conservation, is required for optimal Rix7 function (D. Kressler and E. Hurt, unpublished data). (iii) Finally, the D2 domain of Cdc48- and Rix7-family members harbors, again after α-helix 7, insertions of around 10–35 amino acids. This region seems to be rather flexible and devoid of strong secondary structure elements since it is not represented in the crystal structures of p97 [20].
2.1. Rix7 removes the biogenesis factor Nsa1 from nucleolar pre-60S particles
Rix7 was the first AAA-ATPase for which a role in 60S ribosome biogenesis could be established [19], and it is the earliest acting AAA-ATPase in 60S biogenesis (see Fig. 1). Rix7 was originally identified in an Rpl25-eGFP based screen for ribosomal export mutants (rix) of the large 60S subunit [19,34]. Strains defective in Rix7 accumulate the Rpl25-eGFP reporter predominantly in the nucleolus and they rapidly degrade the 27SB pre-rRNA, thus containing reduced amounts of mature 60S subunits [15,19]. These phenotypic observations suggest that Rix7 is involved in a structural rearrangement required for correct assembly, and therefore stability, of nucleolar pre-60S ribosomal particles. In agreement with a direct action of Rix7 on nuclear pre-60S ribosomes, Rix7-GFP localizes during exponential growth phase to the nucleoplasm and nucleolus [19]. Interestingly, Rix7-GFP exhibits a dynamic sub-nuclear localization. In stationary phase cells Rix7-GFP concentrates in the nucleolus, while, strikingly, upon transfer to fresh medium Rix7-GFP shows a transient perinuclear signal before regaining a uniform nuclear distribution after prolonged growth in fresh medium [19]. However, the functional basis of this nutrient-dependent localization of Rix7 remains so far elusive.
The predominantly nucleolar accumulation of the Rpl25-eGFP reporter upon mutational inactivation of Rix7 suggests that Rix7 may act on a nucleolar pre-60S particle. Indeed, Rix7 is specifically, albeit in sub-stoichiometric amounts, associated with a nucleolar, Nsa1-purified pre-60S intermediate that can be placed between the early nucleolar, Ssf1-defined and the nucleoplasmic, Rix1-defined pre-60S particle (see Fig. 1) [15]. Although direct in vitro evidence for a specific release of Nsa1 from pre-60S particles by Rix7 is currently lacking, cell biological and biochemical experiments revealed that, in vivo, Nsa1 cannot dissociate from pre-60S particles when Rix7 function is perturbed, as evidenced by the cytoplasmic accumulation of otherwise nucleolar Nsa1 on translation-competent 60S subunits [15]. In support of Rix7 acting directly on Nsa1, yeast two-hybrid interaction assays revealed a robust interaction between full-length Nsa1 and a Rix7 fragment (amino acids 166–557) lacking the N-terminal N1/N2 region and most of the D2 domain. Interestingly, Nsa1 appears to be very tightly bound to the pre-60S particle (D. Kressler and E. Hurt, unpublished data), which may explain the requirement for such a powerful molecular machine for its extraction. Altogether, these results strongly suggest that Nsa1 represents a potential pre-60S ribosomal substrate protein of the AAA-ATPase Rix7. The consequences of an inefficient release of Nsa1 upon mutational inactivation of Rix7 can be summarized as follows (see Fig. S4): (i) the majority of pre-60S particles do not evolve further and are degraded in the nucleus; (ii) a fraction of pre-60S particles are retained in the nucleolus and their composition is shifted towards one of earlier pre-60S particles; (iii) the pool of pre-60S particles that escape from turnover and nucleolar retention contains Nsa1, which accumulates over time on “aberrant” cytoplasmic 60S r-subunits [15]. Accordingly, Rix7 can be viewed as an energy-requiring trigger that powers progression of 60S r-subunit biogenesis by stripping Nsa1 from a late nucleolar pre-60S particle.
How does Rix7 recognize and dissociate its proposed substrate Nsa1 from pre-60S particles, and how exactly is the release of Nsa1 timed and triggered? Even though the N-terminal domain of Rix7 seems not to be required for the presumably direct interaction with Nsa1, its integrity is intimately, genetically and functionally, linked to Nsa1 [15]. Interestingly, Nsa1 is strongly poly-ubiquitinated in vivo (D. Kressler and E. Hurt unpublished data), which hints at a possible involvement of this modification in substrate recognition or processing. Maybe the analogy to Cdc48/p97 offers a hypothesis. The diverse cellular functions of Cdc48/p97 are all linked to the recognition of ubiquitinated substrates and their dissociation from unmodified binding partners [33]. The N-terminal domain of Cdc48/p97 represents a main interaction platform via which ubiquitinated substrate proteins are recognized, either directly or indirectly, through substrate-recruiting cofactors [33]. Moreover, the relative position of the N-terminal domain changes depending on the nucleotide bound to the D2 domain [20,35,36], suggesting that the movement of the substrate-bound N-terminal domain may lead to substrate release from its binding partner. Therefore, we speculate that the N-terminal domain of Rix7 may also contribute to the recognition and processing of ubiquitinated Nsa1. In analogy to the AAA-ATPases Cdc48 and Pex1/6 [37], the fate of Rix7-released Nsa1 – recycling or proteasomal degradation – may be determined by the length and nature of the attached ubiquitin chains. It may even be that ubiquitination of Nsa1 serves as a quality control during pre-60S biogenesis at the nucleolar–nucleoplasmic border.
Is the function of Rix7 restricted to the nucleolar release of Nsa1 from pre-60S particles? Given the fact that Rix7 localizes throughout the nucleus, it is tempting to speculate that Rix7 could participate, possibly through recognition of ubiquitinated substrates, in many different nuclear processes that await to be uncovered. In support of this view, studies on the human Rix7 orthologue suggest that NVL may directly interact with ribosomal protein L5 and the exosome-assisting RNA helicase hDOB1/hMTR4 [38,39]. However, the functional significance of these findings has not yet been explored in detail.
2.2. Drg1 promotes cytoplasmic maturation of pre-60S particles by stripping shuttling proteins
First indications for a function of the hexameric Drg1 in yeast came from the finding that the drug diazaborine causes a specific block in ribosome biogenesis, which can be relieved by diazaborine resistant alleles of DRG1 [40]. The involvement of Drg1 in ribosome biogenesis became obvious when the thermo-sensitive drg1-18 mutant was found to display nuclear pre-rRNA processing defects and reduced levels of 60S subunits. In apparent contradiction to a direct involvement of Drg1 in nuclear pre-60S maturation, Drg1 was found to be located exclusively in the cytoplasm [16].
This enigma was resolved by the finding that the shuttling biogenesis factors Rlp24, Arx1, Nog1 and Tif6 accumulate in the cytoplasm of drg1 mutant cells [16]. The fact that these factors remain associated to cytoplasmic pre-60S particles in the drg1-ts mutant shows that Drg1 is required for their release from pre-60S particles to allow their subsequent re-import. The pre-rRNA processing defects observed in the drg1-ts mutant are therefore likely a consequence of the nuclear depletion of the essential shuttling factors. Moreover, the persistence of these proteins on cytoplasmic pre-60S particles blocks downstream maturation steps [16]. Similar results were obtained upon overexpression of a dominant negative, D2 ATPase inactive Drg1, thus the catalytic activity is strictly required for the release of shuttling proteins and subsequent biogenesis events [16].
Very recently, it was shown that Mrt4 also accumulates in the cytoplasm of drg1-ts mutant cells [41]. Since the release of Mrt4 is required for loading of the ribosomal protein and stalk constituent Rpp0 [42–45], the formation of the characteristic 60S stalk structure is prevented in the drg1-ts mutant.
In summary, the function of Drg1 is required for the release of a whole set of shuttling proteins as well as for progression of 60S maturation [16,41]. The broad number of cytoplasmic maturation events affected by non-functional Drg1 makes it implausible that Drg1 is directly participating in all these maturation steps. More likely, Drg1 catalyzes one early reaction that is essential for the subsequent cascade of release and joining events. Consequently, the blockage in release of all these shuttling proteins, as well as the inability to assemble later factors in the drg1-ts mutant, suggests that Drg1 initiates cytoplasmic pre-60S maturation [16,41].
Which is (are) the direct substrate(s) of Drg1? Drg1 is physically associated with pre-60S particles purified via the shuttling proteins Arx1 or Alb1, but it is missing from late cytoplasmic particles purified by Rei1 or Lsg1 (Fig. 1). Therefore, factors that are still found on the Lsg1 particle can be most likely excluded from the list of potential substrates. A likely candidate is the shuttling protein Rlp24, which is released from the pre-60S particle shortly after export. Rlp24 is highly homologous to the ribosomal protein Rpl24 and is postulated to act as a placeholder until being replaced for Rpl24 in the cytoplasm [46]. Indeed, it was shown recently that Drg1 fails to associate with pre-60S particles isolated via a dominant negative, C-terminally truncated version of Rlp24 [41]. Moreover, Rlp24 interacts with Drg1 in vitro and stimulates its ATPase activity (L. Kappel and H. Bergler, unpublished results). Together, these results indicate that Rlp24 is required for recruitment of Drg1 to pre-60S particles and could be its direct substrate.
In line with Drg1 being required immediately after export of the pre-60S particle, Drg1 was recently found to interact biochemically and genetically with several FG-repeat containing nucleoporins localized at the cytoplasmic site of the NPC (L. Kappel and H. Bergler, unpublished results). One possible benefit of this interaction might be to increase the local concentration of the low abundance Drg1 protein to facilitate the recognition of the pre-60S substrate directly after export. Alternatively, this interaction may modulate nucleotide binding or ATP hydrolysis of the Drg1 ATPase.
2.3. Catalytic AAA domains within Drg1 and Rix7
Extensive mutational and biochemical analyses of Cdc48/p97 have revealed that the main ATPase activity resides in the D2 domain, whereas the D1 domain appears to be mainly important for hexamerisation ([35,47,48] and references therein). Genetic analysis of Drg1 and Rix7 variants harboring the classical Walker A (K > A, defective nucleotide binding) and Walker B mutations (E > Q, impaired ATP hydrolysis) (Fig. S1) [11] reveal a similar enzymatic organization as observed for Cdc48/p97 [16] (D. Kressler and E. Hurt, G. Zisser and H. Bergler unpublished data). The mutations in Walker A in D1 and D2, as well as Walker B in D2 do not support growth, whereas the Walker B mutation in D1 is viable. Moreover, overexpression of the Walker B D2 mutants exerts a dominant-lethal growth phenotype. These analyses indicate that nucleotide binding within D1 is essential, whereas ATP hydrolysis appears not to be required or does not strictly depend on the glutamate residue in the Walker B motif, suggesting a predominantly structural function. The D2 domain likely harbors the main catalytic activity since both ATP binding and hydrolysis is essential. Despite these functional parallels, there are differences between Drg1 and p97 with respect to nucleotide affinity, ATPase activity, oligomerisation, and hexamer stability [49–52]. Nevertheless, it is likely that each of these three AAA-ATPases utilizes a slight variation of a common mode of action. As proposed for p97, it is conceivable that in general the state of the bound nucleotide is transmitted to induce a relative movement between D1 and D2, which displaces the N-terminal substrate-binding domain and hence exerts the pulling force required for substrate extraction [20,36].
3. Rea1, a giant AAA-ATPase in 60S biogenesis
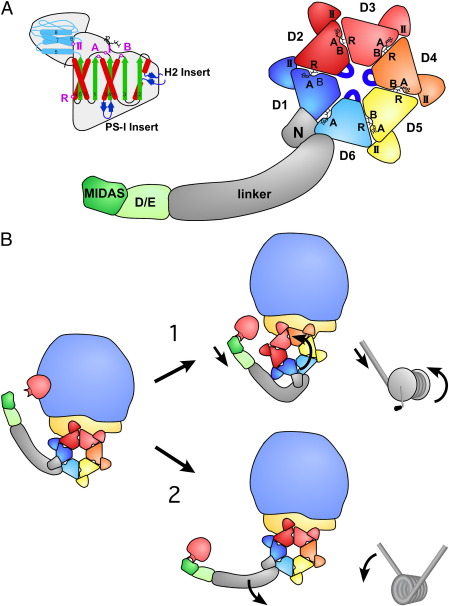
The best-studied ATPase in ribosome biogenesis is the huge Rea1 AAA-ATPase, which is conserved from yeast to human (Fig. S6) [53]. Rea1 is built up of several different domains: It has a weakly conserved N-terminal domain, followed by a dynein-like tandem array of six AAA-type ATPase domains [54], a large linker, a D/E rich region and a MIDAS (metal ion dependent adhesion site) domain (Fig. 4A). Since all six ATPase domains possess characteristic “pre-sensor-I insertion” (PSI insert) and “helix2 insertion” (H2 insert) elements (Fig. S5, [12,17]), Rea1 can be classified as a member of the H2-insert clade [17]. Whereas domains D1, D2, D3, D4, and D5 have a characteristic lid domain harboring a conserved sensor-II motif, the D6 domain lacks this region, but is connected directly to the large linker domain (Fig. 4A). Contrary to the classification of Rea1 into the “pre-sensor-II insert” (PS-II insert) clade by Erzberger and Berger [12], our sequence analysis did not reveal the presence of the characteristic α-helical PS-II insertion after α-helix 5 but rather suggests that α-helix 6 and 7 are connected by a linker of variable length (Fig. 4A, S5, S6). Clearly, structural information of the Rea1/Midasin ring domain is required to reveal the architecture of the lid domain.
Fig. 4.
Domain organization of Rea1. A, Rea1 consists of an N-terminal domain, six ATPase modules that form the ring domain, followed by a long α-helical linker, a D/E rich region and a MIDAS domain. The predicted structural organization of an ATPase module: the central α/β domain is shown in red (α-helices) and green (β-strands), including Walker A (A), and Walker B (B) motif, sensor-I (I) and the arginine finger (R) indicated by a purple dot. The lid domain including the sensor-II (II) region is depicted in light blue. The clade-specific helix2 insert and pre-sensor-I insert (PSI) are indicated in dark blue (colors are identical to secondary structure prediction of Figs. S5 and S6.) B, Possible mechanisms of Rea1-mediated release reactions. Mechanism 1 displays a ratchet-hoist model, where energy is utilized to move the Rea1 ATPase ring on the pre-ribosome, thus creating a tension that pulls off the substrate. In mechanism 2 (power-stroke model) the Rea1 ring domain stays firmly attached at the pre-ribosome. Conformational changes within the ATPase ring are causing a power stroke: i.e. an active movement of the linker domain with the attached D/E rich region and the MIDAS domain.
3.1. Rea1 promotes structural rearrangements of nucleolar and nucleoplasmic pre-60S ribosomes
Rea1 was first identified as a component of the Nug1-purified particle, a pre-60S ribosome at the transition from the nucleolus to the nucleoplasm [55]. Subsequently, it was discovered that Rea1 is enriched in the nucleoplasmic Rix1 particle (Fig. 1) [56–58]. Electron microscopy (EM) revealed that Rea1 consists of a ring-like domain, built up by the six ATPase modules, and an elongated tail structure carrying the MIDAS domain at its tip (Fig. 4A) [14]. Rea1 attaches, via the ATPase ring domain, to the Rix1 pre-ribosome in the vicinity of the Rix1-Ipi3-Ipi1 sub-complex with its tail protruding from the pre-ribosome [14,56]. However, whether the Rix1-Ipi3-Ipi1 sub-complex is a direct interaction partner of the ATPase ring remains elusive. Remarkably, the tail of Rea1 is flexible and the MIDAS domain is able to contact the pre-ribosome at a second distinct position where the pre-60S maturation factor Rsa4 is located. The MIDAS domain of Rea1 interacts in vivo and in vitro with the N-terminal MIDAS interacting domain (MIDO) of Rsa4 [14] and this interaction is essential for progression of 60S biogenesis [13,14].
In silico analysis of the Rsa4 MIDO domain revealed that Ytm1, a further 60S biogenesis factor, also harbors a MIDO domain at its N-terminus [13]. Subsequent genetic and biochemical analyses showed that the Ytm1 MIDO domain interacts with MIDAS with the same mechanistic properties underlying the Rea1–Rsa4 interaction [13]. However, Ytm1 associates with earlier, nucleolar pre-60S particles, whereas Rsa4 is associated with later, nucleoplasmic particles (Fig. 1) [13,14,59,60].
In vitro experiments showed that ATP treatment of the Rix1 particle results in the release of Rsa4 and Rea1 (Fig. 3) [14]. Moreover, Ytm1, together with its direct binding partners Erb1 and Nop7 [59,61,62] can be released from pre-ribosomal particles in a Rea1-dependent reaction (Fig. 3) [13]. Both release reactions require hydrolysable ATP and an intact MIDAS–MIDO interaction [13,14]. Thus, Rea1 releases at least two substrates from pre-ribosomal particles at different stages during 60S biogenesis. An active release of the Ytm1-Erb1-Nop7 sub-complex may weaken the interaction of neighboring biogenesis factors to the pre-ribosomal particle and thus facilitate their displacement or replacement by other factors. In this respect it should be considered that the transition of pre-60S particles from the nucleolus towards the nucleoplasm is marked by a substantial reduction in complexity with several nucleolar factors being released [15].
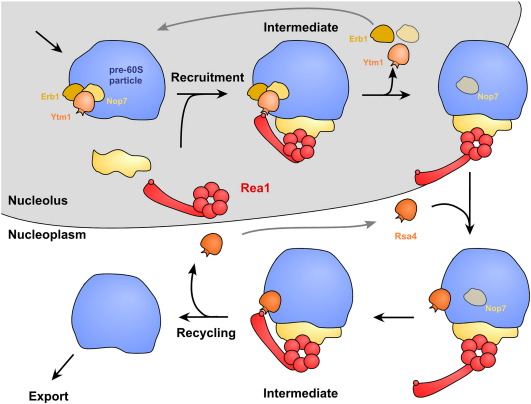
Fig. 3.
Model of a possible Rea1 cycle during 60S maturation. Rea1 associates with a nucleolar particle and releases the Ytm1-Erb1-Nop7 sub-complex. Then nucleoplasmic recruitment of Rsa4 to the pre-60S particle initiates the second Rea1-dependent reaction, which releases Rsa4 and Rea1. Note that several details of this multistep process are still elusive.
3.2. The MIDAS–MIDO interaction
MIDAS domains are best studied in integrin receptors and usually consist of six central β-strands sandwiched by amphipathic α-helices [63–65]. MIDAS domains contain the characteristic sequence motif DxSxS-x70-T-x30-DG (Fig. S6). These residues lie on three adjacent loops that coordinate a divalent metal ion, which is important for ligand binding. The sixth binding site of the metal ion is occupied by a conserved glutamate or aspartate residue of the MIDAS ligand. Mutational analysis of these residues within Rea1, Rsa4 and Ytm1 revealed their necessity for the MIDAS–MIDO interaction, the release reaction, and thus for 60S ribosome biogenesis [13,14]. In addition to the classical MIDAS domain, a highly conserved α-helical extension (amino acids 4622–4701 of S. cerevisiae Rea1, see Fig. S6) is essential for the MIDAS–MIDO complex formation [14] (C. Ulbrich and J. Baßler, unpublished data).
Rea1 has been proposed to be a mechanoenzyme that strips off biogenesis factors by physical force [13,14]. In order to allow an efficient detachment of the MIDO proteins, one would expect a strong interaction between MIDAS and MIDO domains. However, Rsa4 or Ytm1 do not stay associated with the Rea1 MIDAS domain after the release reaction [13,14]. Thus, there might be a molecular switch that allows a strong interaction during the release reaction but permits the subsequent release by a decreased affinity. Interestingly, it has been observed that the interaction of the integrin MIDAS with its ligand has to endure mechanical tension and that this interaction is further strengthened by tensile force [66,67]. A similar property may also exist for the Rea1 MIDAS–MIDO interaction.
MIDO mutants of Rsa4 and Ytm1 that cannot interact with Rea1 exhibit a dominant-lethal phenotype [13,14]. These mutant proteins are efficiently incorporated into nascent 60S pre-ribosomes, but completely block the release reaction and downstream pre-60S maturation [13,14]. In contrast, Rea1 MIDAS deletion mutants are lethal, but not dominant lethal (M. Thoms, J. Baßler, E. Hurt, unpublished data). This may reflect the fact that such mutant proteins are not efficiently incorporated into pre-60S particles. It remains elusive how Rea1 is recruited to pre-60S ribosomes and whether it first binds with the MIDAS or the ATPase ring domain. In addition, it is not clear whether Rea1, after releasing the Ytm1-Erb1-Nop7 sub-complex, remains bound to the particle via its ATPase ring domain or is released and re-joins the particle at a later stage to perform its nucleoplasmic function on Rsa4 (Fig. 3). Further mutational, biochemical, and structural investigations are required to answer these aspects of the Rea1 cycle.
3.3. Mechanism of the ATP-dependent release reaction
Structural and biochemical data suggest that, at an intermediate state, Rea1 is bound to two distinct sites. One contact is mediated via the motor domain and a second one via the MIDAS domain to Rsa4 or Ytm1, thereby creating a unique mechanochemical device. Analogous to other AAA-ATPases, the Rea1 AAA-modules may adopt different conformations in APO-, ATP- or ADP-bound states, which will affect associated sub-domains like the Rea1 tail or elements of the ring domain. Such conformational changes may ultimately promote the release of Rsa4 and Ytm1. Thus, ATP hydrolysis may create the mechanical force after Rea1 has docked at two sites on the pre-ribosome. Alternatively, ATP hydrolysis could be responsible for moving the tail towards Rsa4/Ytm1 at the pre-ribosome.
How are the conformational changes of the ATPase ring mechanically connected with the movement of the tail domain? Different mechanisms can be envisaged (Fig. 4B): the Rea1 ATPase ring might be fixed on the pre-60S particle and a conformational change inside the ATPase domains may generate a power stroke that moves the Rea1 tail. Such a power-stroke model has been proposed for the dynein-mediated movement along microtubules [68–70]. However, in contrast to dynein [70] the Rea1 tail emanates from a presumably inactive AAA-domain. Notably, the D6 subdomain shows a significantly lower degree of conservation in the Walker A motif and the Walker B motif deviates from the consensus sequence (DE > DN). Moreover the adjacent D1 domain lacks the arginine finger, suggesting that the D6 domain has a predominantly structural function (Figs. S5, S6) [53].
An alternative to the power stroke model might be described as a kind of ratchet-hoist model: conformational changes within the active ATPase modules that presumably interact directly with the pre-ribosome might twist the ATPase ring at the pre-ribosome, which would create a tension force pulling the MIDAS domain away from the pre-ribosome.
3.4. Regulation of the Rea1 ATPase activity
In general, it seems important to ensure that ATP hydrolysis is tightly coupled to ligand binding. For instance, it has been described that ligand binding to dynein causes conformational changes that are likely transmitted towards the ATPase ring, thereby modulating its ATPase activity [71–73]. Accordingly, Rea1 should only be activated after binding of the ATPase ring at the pre-ribosome and the binding of the MIDAS domain to Rsa4 or Ytm1. The direct binding partners of the ATPase ring are not yet known and it is unclear how they might influence ATPase activity. However, the integrin system offers a possible explanation of how ligand binding causes conformational changes within a MIDAS domain [64,74]. These conformational changes are then transmitted to the adjacent domains, and a model with bidirectional signal transmission has been proposed [63,64]. Also within Rea1, ligand binding has to be communicated along the linker domain to the ATPase ring. Sequence analysis indicates that the Rea1 linker domain is not simply a long connection, but also has sections with high sequence conservation, which might be important for such a proposed communication. However, there could be as yet unidentified co-factors that regulate Rea1 ATPase activity.
4. Concluding remarks
The common function of the AAA-ATPases involved in ribosome biogenesis appears to be the release of selected ribosome biogenesis factors. These release reactions likely mark major transitions on the pre-60S ribosome assembly pathway and seem to be critical for downstream maturation processes. Still, many questions remain to be answered: are the functions and targets of Drg1, Rix7, and Rea1 conserved among higher eukaryotes? Is there a relationship between ubiquitination and substrate processing for Drg1 and Rix7? Why are such big and powerful devices required for the release of selected ribosome biogenesis factors? Are these factors more tightly bound than others to pre-ribosomal particles, therefore requiring energy input for their release? Or are the release events coupled to energy-consuming structural rearrangements, which are necessary for downstream maturation reactions?
Moreover, a further fascinating question is how such huge molecular machines utilize ATP hydrolysis to perform mechanical work. Here, the substrates of these AAA-ATPases have to be pulled or pushed apart from pre-60S particles — “Give me a lever long enough and a fulcrum on which to place it, and I shall move the world.” [75]. In analogy, we expect these ATPases to interact with two distinct sites: an anchoring point and a working point. It is obvious from the structural investigations that Rea1 interacts with two different sites on the pre-ribosomal particle. Moreover, we propose that Drg1 and Rix7 also have to interact with two, maybe adjacent, sites which are shifted against each other during the ATP hydrolysis cycle, thereby generating the force to release one binding partner from the particle. Future structural and functional work should shed more light on the working mechanism of these highly interesting molecular machines.
Acknowledgments
We thank Dres. E. Thomson and B. Pertschy for fruitful discussion and helpful comments on the manuscript. J.B. and E.H. are recipients of grants from the Deutsche Forschungsgemeinschaft (Hu363/10 and Gottfried Wilhelm Leibniz Program) and Fonds der Chemischen Industrie. D.K. is supported by a grant from the Swiss National Science Foundation (PP00P3_123341) and H.B. by the Austrian Science Foundation FWF (P21991-B12).
Footnotes
This article is part of a Special Issue entitled: AAA ATPases: structure and function.
Supplementary data to this article can be found online at doi:10.1016/j.bbamcr.2011.06.017.
Contributor Information
Helmut Bergler, Email: helmut.bergler@uni-graz.at.
Jochen Baßler, Email: jochen.bassler@bzh.uni-heidelberg.de.
Appendix A. Supplementary data
Fig. S1: A, Multiple sequence alignment of Cdc48/p97, Drg1, and Rix7/NVL over the D1 and D2 AAA-ATPase domains. The alignment was generated with ClustalW [76] and displayed with Jalview [77]. Secondary structure elements (α-helices and β-strands) are according to the crystal structure of p97 (pdb 3CF2) [20]. In the D2 domain, the positions of α-helix 8 and of the last α-helix are according to secondary structure prediction (PSIPRED) [78]. Numbering of α-helices and β-strands as well as nomenclature of sequence elements is according to [17]. Elements of the ATPase core are colored in red (α-helices) and green (β-strands), the lid domain and its α-helices are depicted in light blue. The additional α-helix of classical clade AAA-ATPases and the post helix7 insertion are indicated in dark blue. Specific ATPase elements are labeled. The classical Walker A (K > A, defective nucleotide binding) and Walker B mutations (E > Q, impaired ATP hydrolysis) are also indicated [11]. Unique features of the classical clade are highlighted: (i) replacement of the generally conserved sensor-II arginine at the base of α-helix7 by an alanine; (ii) short insertion within the arginine finger region leading to the occurrence of two conserved arginines that are usually separated by proline and glycine. B, Representation of the secondary-structure distribution within each ATPase domain with the α/β domain depicted in green (β-strands) and red (α-helices) and the α-helical elements of the α-helical lid domain depicted in light blue. The classic-clade-specific additional α-helix and the post helix7 insertion are shown in dark blue.
Fig. S2: A, The N-terminal domain of Drg1 is composed of two sub-domains: a double-ψ β barrel Nn-domain and a four-stranded β-barrel Nc-domain. A, Multiple sequence alignment of Drg1 (S. cerevisiae, amino acids 31–249), Cdc48 (S. cerevisiae, amino acids 33–218) and p97 (M. musculus, amino acids 23–208). The alignment was generated with ClustalW [76] and displayed with Jalview [77]. Secondary structure elements (α-helices in red and β-strands in green) are derived from secondary structure prediction (PSIPRED) [78], a Drg1 structure model, or crystal structures of p97 (pdb 3CF2, 1R7R, and 1E32) [20,22,23]. Numbering of α-helices and β-strands is according to [22]. Note that the alignment was refined in order to properly position the predicted β-strand 10 of Drg1. B, Structure model of the N-terminal domain of Drg1. The structure model was calculated, based on HHpred alignments [79], by the MODELLER software from a p97 reference structure (pdb 3CF2) [20]. The PyMOL program was used to display the structure [80]. As in A, the structural elements of the Nc-domain are distinguished from the ones of the Nn-domain by the use of darker colors.
Fig. S3: Structural analysis of the N-terminal domain of Rix7. A, Schematic representation of the N-terminal domains of Rix7 and NVL. The distinct N-terminal regions are denoted by N1, N2 and N3 and they are followed by a bipartite nuclear localization signal (NLS). Relevant predicted α-helices are indicated. The two glycines (GG) indicate the start of the AAA-domain D1. B, Multiple sequence alignment of the N-terminal domains of fungal Rix7 proteins (S. cerevisiae, C. glabrata, A. gossypii and S. pombe). The alignment was generated with ClustalW [76] and displayed with Jalview [77]. Secondary structure elements (α-helices in red and β-strands in green) are derived from secondary structure prediction (PSIPRED) [78]. The conserved α-helix at the end of the N2 region is underlined in blue. C, Multiple sequence alignment of the N1 regions of fungal Rix7 (S. cerevisiae and S. pombe) and mammalian NVL (M. musculus). Secondary structure elements are derived from secondary structure prediction (PSIPRED) [78], a Rix7 (S. cerevisiae) structure model, or an NMR structure of mouse NVL (pdb 2RRE) [29]. D, Structure model of the N1 region of Rix7 (S. cerevisiae amino acids 1–113). The structure model was calculated, based on HHpred alignments [79], by the MODELLER software from a mouse NVL reference structure (pdb 2RRE) [29]. The PyMOL program was used to display the structure [80]. The color of the α-helices is as in A. E, Multiple sequence alignment of the conserved α-helix at the end of the N2 region of fungal Rix7 (S. cerevisiae and S. pombe) and mammalian NVL (H. sapiens and M. musculus). The position of this α-helix (red) is derived from secondary structure prediction (PSIPRED) [78].
Fig. S4: Effects of mutational inactivation of Rix7 on the fate of Nsa1-containing pre-60S particles. In wild-type cells (RIX7, upper green pathway), Rix7 releases Nsa1 from late nucleolar pre-60S particles. Upon mutational inactivation of Rix7 (rix7, lower red pathway), the majority of pre-60S particles do not further evolve and are degraded (1), while a fraction of pre-60S particles gain an earlier composition and are retained in the nucleolus (2). Nsa1 remains associated with pre-60S particles that have escaped from disassembly or nucleolar retention, and accumulates over time on ‘aberrant’ cytoplasmic 60S subunits (3). [15,19]. For simplicity, the Rea1-mediated release of the trimeric Ytm1-Erb1-Nop7 sub-complex has been omitted.
Fig. S5: Multiple sequence alignment of the Rea1 ATPase domains was done with ClustalW [76] and displayed with Jalview [77]. Secondary structure elements are depicted below (see also Fig. S6). Elements of the ATPase core are colored in red (α-helices) and green (β-strands), the lid domain is depicted in light blue and clade-specific insertions are indicated in dark blue. Specific ATPase elements are labeled.
Fig. S6: Multiple sequence alignment of Rea1 full-length was done with ClustalW [76] and displayed with Jalview [77]. Secondary structure prediction of S. cerevisiae Rea1, done with PSIPRED [78], is depicted below and ATPase specific elements are labeled (α-helices are shown in red, β-strands in green, the AAA-ATPase specific lid domain is indicated in light blue and the clade-specific helix2 insert and pre-sensor-I insert (PSI) are indicated in dark blue. A red line marks the MIDAS domain).
References
- 1.Armache J.P., Jarasch A., Anger A.M., Villa E., Becker T., Bhushan S., Jossinet F., Habeck M., Dindar G., Franckenberg S., Marquez V., Mielke T., Thomm M., Berninghausen O., Beatrix B., Soding J., Westhof E., Wilson D.N., Beckmann R. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc. Natl. Acad. Sci. U. S. A. 2010;107:19748–19753. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabl J., Leibundgut M., Ataide S.F., Haag A., Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Shem A., Jenner L., Yusupova G., Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 4.Kressler D., Hurt E., Baßler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Henras A.K., Soudet J., Gerus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 7.Strunk B.S., Karbstein K. Powering through ribosome assembly. RNA. 2009;15:2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tschochner H., Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 9.Panse V.G., Johnson A.W. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 2010;35:260–266. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granneman S., Baserga S.J. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 2004;296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 11.Hanson P.I., Whiteheart S.W. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 12.Erzberger J.P., Berger J.M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 13.Bassler J., Kallas M., Ulbrich C., Thoms M., Pertschy B., Hurt E. The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol. Cell. 2010;38:712–721. doi: 10.1016/j.molcel.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulbrich C., Diepholz M., Bassler J., Kressler D., Pertschy B., Pertschy B., G. K., Böttcher B., Hurt E. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunit. Cell. 2009;138:911–922. doi: 10.1016/j.cell.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 15.Kressler D., Roser D., Pertschy B., Hurt E. The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60S particles. J. Cell Biol. 2008;181:935–944. doi: 10.1083/jcb.200801181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pertschy B., Saveanu C., Zisser G., Lebreton A., Tengg M., Jacquier A., Liebminger E., Nobis B., Kappel L., van der Klei I., Hogenauer G., Fromont-Racine M., Bergler H. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol. Cell. Biol. 2007;27:6581–6592. doi: 10.1128/MCB.00668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer L.M., Leipe D.D., Koonin E.V., Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Thorsness P.E., White K.H., Ong W.C. AFG2, an essential gene in yeast, encodes a new member of the Sec18p, Pas1p, Cdc48p, TBP-1 family of putative ATPases. Yeast. 1993;9:1267–1271. doi: 10.1002/yea.320091114. [DOI] [PubMed] [Google Scholar]
- 19.Gadal O., Strauss D., Braspenning J., Hoepfner D., Petfalski E., Philippsen P., Tollervey D., Hurt E.C. A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J. 2001;20:3695–3704. doi: 10.1093/emboj/20.14.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies J.M., Brunger A.T., Weis W.I. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Patel S., Latterich M. The AAA team: related ATPases with diverse functions. Trends Cell Biol. 1998;8:65–71. [PubMed] [Google Scholar]
- 22.Zhang X., Shaw A., Bates P.A., Newman R.H., Gowen B., Orlova E., Gorman M.A., Kondo H., Dokurno P., Lally J., Leonard G., Meyer H., van Heel M., Freemont P.S. Structure of the AAA ATPase p97. Mol. Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 23.Huyton T., Pye V.E., Briggs L.C., Flynn T.C., Beuron F., Kondo H., Ma J., Zhang X., Freemont P.S. The crystal structure of murine p97/VCP at 3.6A. J. Struct. Biol. 2003;144:337–348. doi: 10.1016/j.jsb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 24.May A.P., Misura K.M., Whiteheart S.W., Weis W.I. Crystal structure of the amino-terminal domain of N-ethylmaleimide-sensitive fusion protein. Nat. Cell Biol. 1999;1:175–182. doi: 10.1038/11097. [DOI] [PubMed] [Google Scholar]
- 25.Yu R.C., Jahn R., Brunger A.T. NSF N-terminal domain crystal structure: models of NSF function. Mol, Cell. 1999;4:97–107. doi: 10.1016/s1097-2765(00)80191-4. [DOI] [PubMed] [Google Scholar]
- 26.Coles M., Diercks T., Liermann J., Groger A., Rockel B., Baumeister W., Koretke K.K., Lupas A., Peters J., Kessler H. The solution structure of VAT-N reveals a ‘missing link’ in the evolution of complex enzymes from a simple betaalphabetabeta element. Curr. Biol. 1999;9:1158–1168. doi: 10.1016/S0960-9822(00)80017-2. [DOI] [PubMed] [Google Scholar]
- 27.Shiozawa K., Maita N., Tomii K., Seto A., Goda N., Akiyama Y., Shimizu T., Shirakawa M., Hiroaki H. Structure of the N-terminal domain of PEX1 AAA-ATPase. Characterization of a putative adaptor-binding domain. J. Biol. Chem. 2004;279:50060–50068. doi: 10.1074/jbc.M407837200. [DOI] [PubMed] [Google Scholar]
- 28.DeLaBarre B., Brunger A.T. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- 29.Fujiwara Y., Fujiwara K., Goda N., Iwaya N., Tenno T., Shirakawa M., Hiroaki H. 2011. Structure and function of the N-terminal nucleolin binding domain of nuclear valocin containing protein like 2 (NVL2) harboring a nucleolar localization signal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu L., Pashkova N., Walker J.R., Winistorfer S., Allali-Hassani A., Akutsu M., Piper R., Dhe-Paganon S. Structure and function of the PLAA/Ufd3-p97/Cdc48 complex. J. Biol. Chem. 2010;285:365–372. doi: 10.1074/jbc.M109.044685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao G., Zhou X., Wang L., Li G., Schindelin H., Lennarz W.J. Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8785–8790. doi: 10.1073/pnas.0702966104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rumpf S., Jentsch S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol. Cell. 2006;21:261–269. doi: 10.1016/j.molcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 33.Jentsch S., Rumpf S. Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem. Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Hurt E., Hannus S., Schmelzl B., Lau D., Tollervey D., Simos G. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J. Cell Biol. 1999;144:389–401. doi: 10.1083/jcb.144.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pye V.E., Dreveny I., Briggs L.C., Sands C., Beuron F., Zhang X., Freemont P.S. Going through the motions: the ATPase cycle of p97. J. Struct. Biol. 2006;156:12–28. doi: 10.1016/j.jsb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 36.DeLaBarre B., Brunger A.T. Nucleotide dependent motion and mechanism of action of p97/VCP. J. Mol. Biol. 2005;347:437–452. doi: 10.1016/j.jmb.2005.01.060. [DOI] [PubMed] [Google Scholar]
- 37.Schliebs W., Girzalsky W., Erdmann R. Peroxisomal protein import and ERAD: variations on a common theme. Nat. Rev. Mol Cell Biol. 2010;11:885–890. doi: 10.1038/nrm3008. [DOI] [PubMed] [Google Scholar]
- 38.Nagahama M., Yamazoe T., Hara Y., Tani K., Tsuji A., Tagaya M. The AAA-ATPase NVL2 is a component of pre-ribosomal particles that interacts with the DExD/H-box RNA helicase DOB1. Biochem. Biophys. Res. Commun. 2006;346:1075–1082. doi: 10.1016/j.bbrc.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 39.Nagahama M., Hara Y., Seki A., Yamazoe T., Kawate Y., Shinohara T., Hatsuzawa K., Tani K., Tagaya M. NVL2 is a nucleolar AAA-ATPase that interacts with ribosomal protein L5 through its nucleolar localization sequence. Mol. Biol. Cell. 2004;15:5712–5723. doi: 10.1091/mbc.E04-08-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pertschy B., Zisser G., Schein H., Koffel R., Rauch G., Grillitsch K., Morgenstern C., Durchschlag M., Hogenauer G., Bergler H. Diazaborine treatment of yeast cells inhibits maturation of the 60S ribosomal subunit. Mol. Cell. Biol. 2004;24:6476–6487. doi: 10.1128/MCB.24.14.6476-6487.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo K.Y., Li Z., Bussiere C., Bresson S., Marcotte E.M., Johnson A.W. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell. 2010;39:196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kemmler S., Occhipinti L., Veisu M., Panse G.V. Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J. Cell Biol. 2009;186 doi: 10.1083/jcb.200904111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lo K.-Y., Li Z., Wang F., Marcotte E., Johnson A.W. Ribosome stalk assembly requires the dual specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J. Cell Biol. 2009;186 doi: 10.1083/jcb.200904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Mateos M., Garcia-Gomez J.J., Francisco-Velilla R., Remacha M., de la Cruz J., Ballesta J.P. Role and dynamics of the ribosomal protein P0 and its related trans-acting factor Mrt4 during ribosome assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:7519–7532. doi: 10.1093/nar/gkp806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez-Mateos M., Abia D., Garcia-Gomez J.J., Morreale A., de la Cruz J., Santos C., Remacha M., Ballesta J.P. The amino terminal domain from Mrt4 protein can functionally replace the RNA binding domain of the ribosomal P0 protein. Nucleic Acids Res. 2009;37:3514–3521. doi: 10.1093/nar/gkp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saveanu C., Namane A., Gleizes P.E., Lebreton A., Rousselle J.C., Noaillac-Depeyre J., Gas N., Jacquier A., Fromont-Racine M. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 2003;23:4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye Y., Meyer H.H., Rapoport T.A. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esaki M., Ogura T. ATP-bound form of the D1 AAA domain inhibits an essential function of Cdc48p/p97. Biochem. Cell Biol. 2010;88:109–117. doi: 10.1139/o09-116. [DOI] [PubMed] [Google Scholar]
- 49.Zakalskiy A., Hogenauer G., Ishikawa T., Wehrschutz-Sigl E., Wendler F., Teis D., Zisser G., Steven A.C., Bergler H. Structural and enzymatic properties of the AAA protein Drg1p from Saccharomyces cerevisiae. Decoupling of intracellular function from ATPase activity and hexamerization. J. Biol. Chem. 2002;277:26788–26795. doi: 10.1074/jbc.M201515200. [DOI] [PubMed] [Google Scholar]
- 50.Song C., Wang Q., Li C.C. ATPase activity of p97-valosin-containing protein (VCP). D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity. J. Biol. Chem. 2003;278:3648–3655. doi: 10.1074/jbc.M208422200. [DOI] [PubMed] [Google Scholar]
- 51.Wang Q., Song C., Li C.C. Hexamerization of p97-VCP is promoted by ATP binding to the D1 domain and required for ATPase and biological activities. Biochem. Biophys. Res. Commun. 2003;300:253–260. doi: 10.1016/s0006-291x(02)02840-1. [DOI] [PubMed] [Google Scholar]
- 52.Briggs L.C., Baldwin G.S., Miyata N., Kondo H., Zhang X., Freemont P.S. Analysis of nucleotide binding to P97 reveals the properties of a tandem AAA hexameric ATPase. J. Biol. Chem. 2008;283:13745–13752. doi: 10.1074/jbc.M709632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garbarino J.E., Gibbons I.R. Expression and genomic analysis of midasin, a novel and highly conserved AAA protein distantly related to dynein. BMC Genomics. 2002;3:18–28. doi: 10.1186/1471-2164-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neuwald A.F., Aravind L., Spouge J.L., Koonin E.V. AAA+: a class of chaperone-like ATPases associated with the assembly, operation,and dissassembly of proteins complexes. Genome Res. 1999:27–43. [PubMed] [Google Scholar]
- 55.Bassler J., Grandi P., Gadal O., Leßmann T., Tollervey D., Lechner J., Hurt E.C. Identification of a 60S pre-ribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 56.Nissan T.A., Galani K., Maco B., Tollervey D., Aebi U., Hurt E. A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Mol Cell. 2004;15:295–301. doi: 10.1016/j.molcel.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 57.Galani K., Nissan T.A., Petfalski E., Tollervey D., Hurt E. Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60S subunits. J. Biol. Chem. 2004;279:55411–55418. doi: 10.1074/jbc.M406876200. [DOI] [PubMed] [Google Scholar]
- 58.Nissan T.A., Bassler J., Petfalski E., Tollervey D., Hurt E.C. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miles T.D., Jakovljevic J., Horsey E.W., Harnpicharnchai P., Tang L., Woolford J.L., Jr. Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell. Biol. 2005;25:10419–10432. doi: 10.1128/MCB.25.23.10419-10432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De la Cruz J., Sanz-Martinez E., Remacha M. The essential WD-repeat protein Rsa4p is required for rRNA processing and intra-nuclear transport of 60S ribosomal subunits. Nucleic Acids Res. 2005;33:5728–5739. doi: 10.1093/nar/gki887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang L., Sahasranaman A., Jakovljevic J., Schleifman E., Woolford J.L., Jr. Interactions among Ytm1, Erb1, and Nop7 required for assembly of the Nop7-subcomplex in yeast preribosomes. Mol. Biol. Cell. 2008;19:2844–2856. doi: 10.1091/mbc.E07-12-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hölzel M., Rohrmoser M., Schlee M., Grimm T., Harasim T., Malamoussi A., Gruber-Eber A., Kremmer E., Hiddemann W., Bornkamm G.W., Eick D. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J. Cell Biol. 2005;170:367–378. doi: 10.1083/jcb.200501141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arnaout M.A., Mahalingam B., Xiong J.P. Integrin structure, allostery, and bidirectional signaling. Annu. Rev. Cell Dev. Biol. 2005;21:381–410. doi: 10.1146/annurev.cellbio.21.090704.151217. [DOI] [PubMed] [Google Scholar]
- 64.Luo B.H., Carman C.V., Springer T.A. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takagi J. Structural basis for ligand recognition by integrins. Curr. Opin. Cell Biol. 2007;19:557–564. doi: 10.1016/j.ceb.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 66.Craig D., Gao M., Schulten K., Vogel V. Structural insights into how the MIDAS ion stabilizes integrin binding to an RGD peptide under force. Structure. 2004;12:2049–2058. doi: 10.1016/j.str.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 67.Astrof N.S., Salas A., Shimaoka M., Chen J., Springer T.A. Importance of force linkage in mechanochemistry of adhesion receptors. Biochemistry. 2006;45:15020–15028. doi: 10.1021/bi061566o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roberts A.J., Numata N., Walker M.L., Kato Y.S., Malkova B., Kon T., Ohkura R., Arisaka F., Knight P.J., Sutoh K., Burgess S.A. AAA+ ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burgess S.A., Walker M.L., Sakakibara H., Knight P.J., Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 70.Numata N., Kon T., Shima T., Imamula K., Mogami T., Ohkura R., Sutoh K. Molecular mechanism of force generation by dynein, a molecular motor belonging to the AAA+ family. Biochem. Soc. Trans. 2008;36:131–135. doi: 10.1042/BST0360131. [DOI] [PubMed] [Google Scholar]
- 71.Carter A.P., Vale R.D. Communication between the AAA+ ring and microtubule-binding domain of dynein. Biochem. Cell Biol. 2010;88:15–21. doi: 10.1139/o09-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kon T., Imamula K., Roberts A.J., Ohkura R., Knight P.J., Gibbons I.R., Burgess S.A., Sutoh K. Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nat. Struct. Mol. Biol. 2009;16:325–333. doi: 10.1038/nsmb.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carter A.P., Garbarino J.E., Wilson-Kubalek E.M., Shipley W.E., Cho C., Milligan R.A., Vale R.D., Gibbons I.R. Structure and functional role of dynein's microtubule-binding domain. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shimaoka M., Xiao T., Liu J.H., Yang Y., Dong Y., Jun C.D., McCormack A., Zhang R., Joachimiak A., Takagi J., Wang J.H., Springer T.A. Structures of the alpha L I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation. Cell. 2003;112:99–111. doi: 10.1016/s0092-8674(02)01257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Archimedes, of, Syracuse, Greek mathematician, physicist, engineer, inventor, and astronomer, (287 BC–212 BC).
- 76.Myers E.W., Miller W. Optimal alignments in linear space. Comput. Appl. Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. http://www.ebi.ac.uk/Tools/msa/clustalw12/ [DOI] [PubMed] [Google Scholar]
- 77.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. http://www.jalview.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bryson K., McGuffin L.J., Marsden R.L., Ward J.J., Sodhi J.S., Jones D.T. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–W38. doi: 10.1093/nar/gki410. http://bioinf.cs.ucl.ac.uk/psipred/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Söding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. http://toolkit.tuebingen.mpg.de/hhpred [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schrödinger L.L.C. 2010. The PyMOL Molecular Graphics System, Version 1.3r1. in. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: A, Multiple sequence alignment of Cdc48/p97, Drg1, and Rix7/NVL over the D1 and D2 AAA-ATPase domains. The alignment was generated with ClustalW [76] and displayed with Jalview [77]. Secondary structure elements (α-helices and β-strands) are according to the crystal structure of p97 (pdb 3CF2) [20]. In the D2 domain, the positions of α-helix 8 and of the last α-helix are according to secondary structure prediction (PSIPRED) [78]. Numbering of α-helices and β-strands as well as nomenclature of sequence elements is according to [17]. Elements of the ATPase core are colored in red (α-helices) and green (β-strands), the lid domain and its α-helices are depicted in light blue. The additional α-helix of classical clade AAA-ATPases and the post helix7 insertion are indicated in dark blue. Specific ATPase elements are labeled. The classical Walker A (K > A, defective nucleotide binding) and Walker B mutations (E > Q, impaired ATP hydrolysis) are also indicated [11]. Unique features of the classical clade are highlighted: (i) replacement of the generally conserved sensor-II arginine at the base of α-helix7 by an alanine; (ii) short insertion within the arginine finger region leading to the occurrence of two conserved arginines that are usually separated by proline and glycine. B, Representation of the secondary-structure distribution within each ATPase domain with the α/β domain depicted in green (β-strands) and red (α-helices) and the α-helical elements of the α-helical lid domain depicted in light blue. The classic-clade-specific additional α-helix and the post helix7 insertion are shown in dark blue.
Fig. S2: A, The N-terminal domain of Drg1 is composed of two sub-domains: a double-ψ β barrel Nn-domain and a four-stranded β-barrel Nc-domain. A, Multiple sequence alignment of Drg1 (S. cerevisiae, amino acids 31–249), Cdc48 (S. cerevisiae, amino acids 33–218) and p97 (M. musculus, amino acids 23–208). The alignment was generated with ClustalW [76] and displayed with Jalview [77]. Secondary structure elements (α-helices in red and β-strands in green) are derived from secondary structure prediction (PSIPRED) [78], a Drg1 structure model, or crystal structures of p97 (pdb 3CF2, 1R7R, and 1E32) [20,22,23]. Numbering of α-helices and β-strands is according to [22]. Note that the alignment was refined in order to properly position the predicted β-strand 10 of Drg1. B, Structure model of the N-terminal domain of Drg1. The structure model was calculated, based on HHpred alignments [79], by the MODELLER software from a p97 reference structure (pdb 3CF2) [20]. The PyMOL program was used to display the structure [80]. As in A, the structural elements of the Nc-domain are distinguished from the ones of the Nn-domain by the use of darker colors.
Fig. S3: Structural analysis of the N-terminal domain of Rix7. A, Schematic representation of the N-terminal domains of Rix7 and NVL. The distinct N-terminal regions are denoted by N1, N2 and N3 and they are followed by a bipartite nuclear localization signal (NLS). Relevant predicted α-helices are indicated. The two glycines (GG) indicate the start of the AAA-domain D1. B, Multiple sequence alignment of the N-terminal domains of fungal Rix7 proteins (S. cerevisiae, C. glabrata, A. gossypii and S. pombe). The alignment was generated with ClustalW [76] and displayed with Jalview [77]. Secondary structure elements (α-helices in red and β-strands in green) are derived from secondary structure prediction (PSIPRED) [78]. The conserved α-helix at the end of the N2 region is underlined in blue. C, Multiple sequence alignment of the N1 regions of fungal Rix7 (S. cerevisiae and S. pombe) and mammalian NVL (M. musculus). Secondary structure elements are derived from secondary structure prediction (PSIPRED) [78], a Rix7 (S. cerevisiae) structure model, or an NMR structure of mouse NVL (pdb 2RRE) [29]. D, Structure model of the N1 region of Rix7 (S. cerevisiae amino acids 1–113). The structure model was calculated, based on HHpred alignments [79], by the MODELLER software from a mouse NVL reference structure (pdb 2RRE) [29]. The PyMOL program was used to display the structure [80]. The color of the α-helices is as in A. E, Multiple sequence alignment of the conserved α-helix at the end of the N2 region of fungal Rix7 (S. cerevisiae and S. pombe) and mammalian NVL (H. sapiens and M. musculus). The position of this α-helix (red) is derived from secondary structure prediction (PSIPRED) [78].
Fig. S4: Effects of mutational inactivation of Rix7 on the fate of Nsa1-containing pre-60S particles. In wild-type cells (RIX7, upper green pathway), Rix7 releases Nsa1 from late nucleolar pre-60S particles. Upon mutational inactivation of Rix7 (rix7, lower red pathway), the majority of pre-60S particles do not further evolve and are degraded (1), while a fraction of pre-60S particles gain an earlier composition and are retained in the nucleolus (2). Nsa1 remains associated with pre-60S particles that have escaped from disassembly or nucleolar retention, and accumulates over time on ‘aberrant’ cytoplasmic 60S subunits (3). [15,19]. For simplicity, the Rea1-mediated release of the trimeric Ytm1-Erb1-Nop7 sub-complex has been omitted.
Fig. S5: Multiple sequence alignment of the Rea1 ATPase domains was done with ClustalW [76] and displayed with Jalview [77]. Secondary structure elements are depicted below (see also Fig. S6). Elements of the ATPase core are colored in red (α-helices) and green (β-strands), the lid domain is depicted in light blue and clade-specific insertions are indicated in dark blue. Specific ATPase elements are labeled.
Fig. S6: Multiple sequence alignment of Rea1 full-length was done with ClustalW [76] and displayed with Jalview [77]. Secondary structure prediction of S. cerevisiae Rea1, done with PSIPRED [78], is depicted below and ATPase specific elements are labeled (α-helices are shown in red, β-strands in green, the AAA-ATPase specific lid domain is indicated in light blue and the clade-specific helix2 insert and pre-sensor-I insert (PSI) are indicated in dark blue. A red line marks the MIDAS domain).